Abstract
The tubby protein (Tub), a putative transcription factor, plays important roles in the maintenance and function of neuronal cells. A splicing defect-causing mutation in the 3′-end of the tubby gene, which is predicted to disrupt the carboxy-terminal region of the Tub protein, causes maturity-onset obesity, blindness, and deafness in mice. Although this pathological Tub mutation leads to a loss of function, the precise mechanism has not yet been investigated. Here, we found that the mutant Tub proteins were mostly localized to puncta found in the perinuclear region and that the C-terminus was important for its solubility. Immunocytochemical analysis revealed that puncta of mutant Tub co-localized with the aggresome. Moreover, whereas wild-type Tub was translocated to the nucleus by extracellular signaling, the mutant forms failed to undergo such translocation. Taken together, our results suggest that the malfunctions of the Tub mutant are caused by its misfolding and subsequent localization to aggresomes.
Keywords: Aggresome, Misfolding, Mutation, Obesity, Tubby
INTRODUCTION
The tubby gene (TUB) was first identified from a spontaneous adult-onset obese mouse strain (called tubby mice), and other members of the TUB family were subsequently cloned in humans and mice (1–4). The members of the tubby-like protein (TULP) family share highly conserved signature carboxy-terminal (C-terminal) tubby domains, but vary in the sequences and functions of their amino (N) termini. TULPs (including Tub, TULP1, TULP2 and TULP3) are evolutionally conserved in various multicellular organisms from plants to humans. Their levels of gene expression in vertebrates are varied and highly regulated (5, 6), and mutations of TULP family members have been strongly associated with various diseases in mammals. A mutation of TULP1, which is selectively expressed in retina, causes retinitis pigmentosa in humans and retinal degeneration in mice (7–9), whereas loss of TULP3 which is expressed ubiquitously in the mouse embryo, yields embryonic lethality with defects in the dorsoventral patterning of the spinal cord (10, 11). TUB is highly expressed in the brain, particularly in the hypothalamus, and tubby mice show deafness and blindness in addition to an obese phenotype. Tubby mice have been shown to carry a splice defect mutation in the 3′ coding region of TUB, leading to the generation of a longer transcript containing the unspliced introns, and the replacement of 44 C-terminal amino acids with an intron-encoded sequence (2, 3).
The C-terminal domain of Tub, which comprises the signature tubby domain, binds to phosphatidylinositol 4,5-bisphosphate (PIP2), thereby localizing the protein to the plasma membrane. Upon G-protein activation or insulin signaling, Tub translocates to the nucleus (12, 13). Boggon et al. have suggested that the C-terminus of Tub contains a DNA binding domain (DBD) (14), but we do not yet know the specific DNA sequence recognized by Tub or its transcriptionally activated targets. Moreover, although Stubdal et al. showed TUB knockout mice showed a phenotype identical to that of naturally arising tubby mice (15), it is unclear how the latter mutation leads to loss of function.
In this study, we manipulated mouse TUB-encoding constructs to produce wild-type, C-terminal-domain mutant, and C- and N-terminal-domain Tub proteins, and examined their expressions in the Neuro-2A mouse neuroblastoma cell line. Our results revealed that mutant Tub proteins localized to the perinucleus as puncta exhibited a structure different from that of wild-type, and was associated with aggresomes.
RESULTS AND DISCUSSION
C-terminal mutation of tubby induces its aggregation as Tx-100 insoluble puncta
The C-terminal core domain of the mouse Tub protein (about 260 amino acids) forms a unique structure in which a central hydrophobic helix at the C-terminus transverses the interior of a closed 12-stranded β-barrel (14). In tubby mice, a single nucleotide substitution (G → T) in the 3′ coding region of the TUB causes a splicing defect that replaces the 44 C-terminal amino acids of the wild-type protein with a new sequence of 20 amino acids (3). This change might abrogate the hydrophobicity of the C-terminal central helix (data not shown), leading to a loss of function. However, no previous study has examined this mutant Tub protein with respect to its processing for protein maturation, its localization, or how its function might be interrupted.
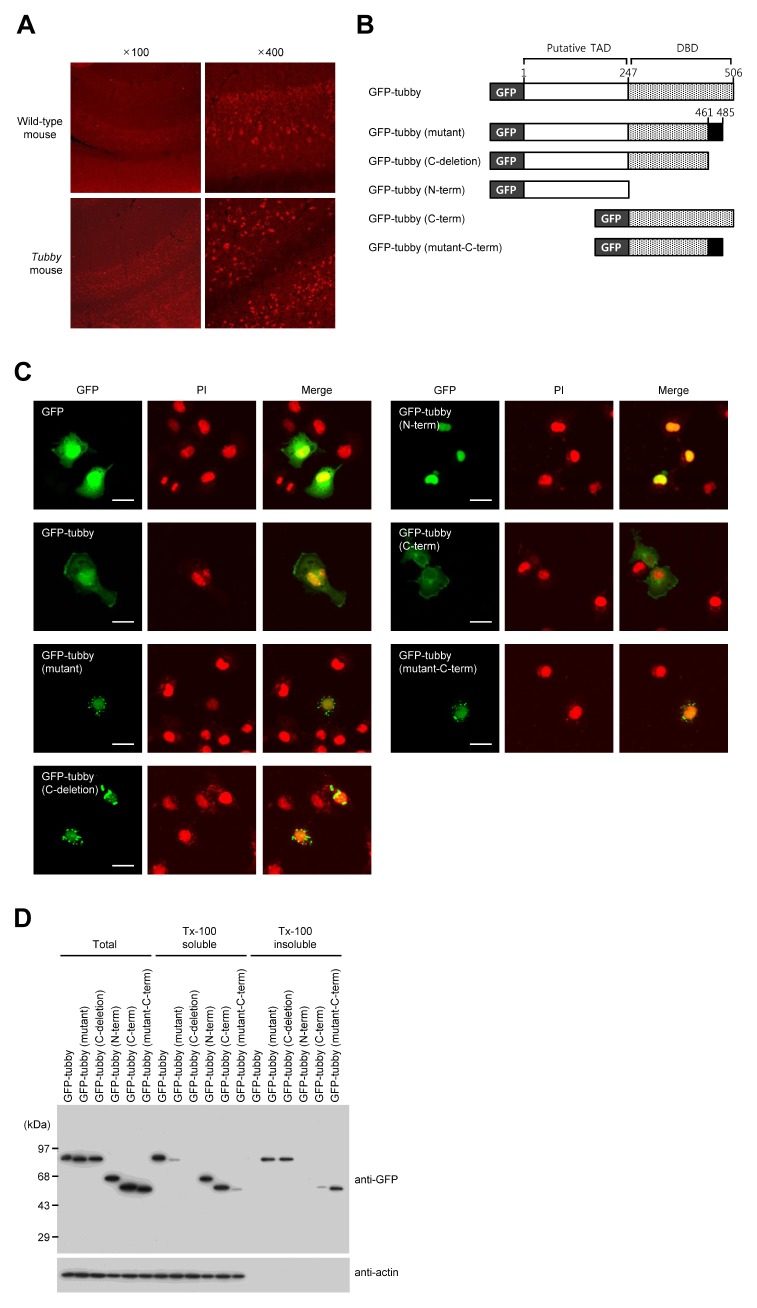
We first used immunohistochemical analysis with an antibody against tubby to examine the distribution pattern of wild-type or mutant Tub proteins in the mouse brain. Mutant Tub proteins showed punctate expression in the hippocampal regions of tubby mouse brains, whereas wild-type Tub protein was evenly distributed in the hippocampal regions of normal mouse brains (Fig. 1A). To evaluate the cellular distribution patterns of the wild-type, mutant, N-terminal domain, and C-terminal domain Tub proteins, we obtained cDNA of Tub or mutant Tub from a wild-type or tubby mouse respectively, generated constructs in which they were fused with green fluorescent protein (GFP), expressed the recombinant proteins in COS-7 cells, and observed their localizations (Fig. 1B). Consistent with previous reports, wild-type Tub localized to both the plasma membrane and the nucleus, the N-terminal domain (the putative transcriptional-activation domain; TAD) localized to the nucleus, and the C-terminal domain (containing the DBD) localized to the plasma membrane (12). In contrast, mutant Tub exhibited a punctate distribution in the perinuclear region, as did fusion proteins with deletion of the C-terminal 44 amino acids and a mutant C-terminal domain (Fig. 1C). To further characterize the biochemical properties of the various Tub proteins, we investigated their solubilities in detergent-containing solution. After transfection for 48 h, the COS-7 cell extract was separated into Tx-100-soluble and -insoluble fractions. We found that the wild-type, N-terminal domain, and C-terminal domain-mutant Tub proteins localized to the Tx-100 soluble fraction, whereas the majority of the mutant, C-terminal-deleted, and C-terminal-domain-mutant proteins localized to the insoluble fraction. Similar patterns were observed using Flag- or His-tagged tubby proteins (Fig. S1). Taken together, these observations suggest that C-terminal region of the Tub protein is important for its proper folding and subcellular localization.
Fig. 1.
The C-terminal region of tubby is essential for its subcellular localization. (A) Frozen sections of mouse hippocampus were subjected to immunohistochemical analysis using an anti-tubby antibody. (B) A schematic representation of the utilized GFP-fused wild-type tubby, mutant tubby, and truncated tubby proteins. Numbers indicate positions with respect to the amino acid sequence. (C) Representative fluorescence micrographs of COS-7 cells are expressing GFP-fused proteins. COS-7 cells were transfected with constructs encoding GFP-tagged tubby proteins. Twenty-four hours later, the cells were starved overnight, fixed, and visualized by confocal microscopy. Nuclei were visualized with propidium iodide (PI). Scale bars: 20 μm. (D) At 48 h post-transfection, the cells were lysed and separated into Tx-100 soluble (supernatant) and insoluble (pellet) fractions. The fractions and total lysates were analyzed by Western blotting using anti-GFP and anti-actin antibodies.
Mutant tubby forms aggresome-like structures
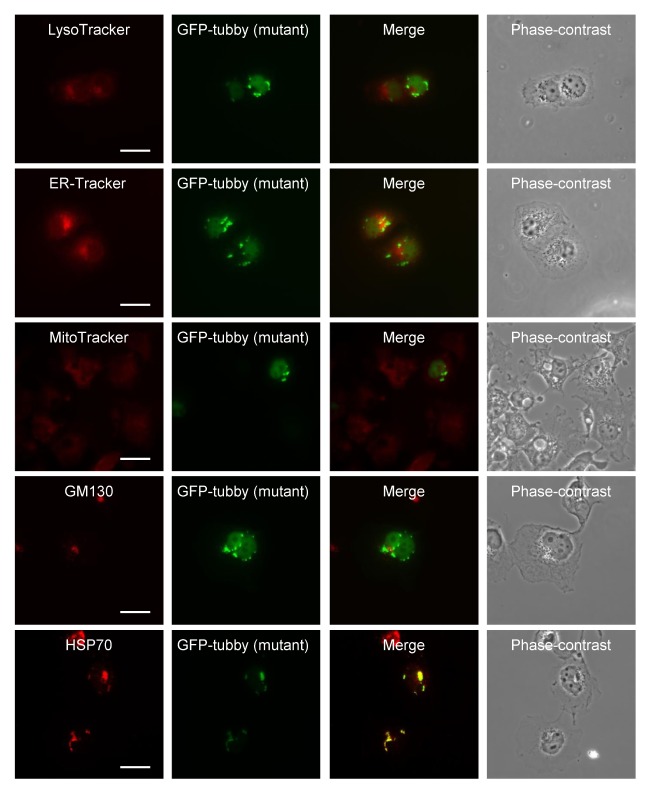
We investigated the subcellular localization of the mutant Tub protein with fusing fluorescent organelle markers. We found that the ER-Tracker signal partly overlapped with that of mutant Tub protein, while those of LysoTracker, MitoTracker, and GM130 (a Golgi marker) did not overlap (Fig. 2).
Fig. 2.
Mutant tubby aggregates co-localize with Hsp70 (an aggresome marker), but not with markers for lysosomes (LysoTracker), the ER (ER-Tracker), mitochondria (MitoTracker), or the Golgi apparatus (GM130). COS-7 cells expressing GFP-mutant tubby were fixed and stained. Scale bars: 20 μm.
Misfolded proteins are typically degraded by the sophis ticated proteasomal system. An imbalance between the generation and clearance of misfolded proteins and the subsequent accumulation of insoluble protein aggregates has been strongly associated with neurodegenerative diseases, including Parkinson’s disease and Huntington’s disease (16). Numerous reports have shown that misfolded proteins produced under stress conditions or genetic mutation form aggregates, move to the perinuclear region along microtubules, and accumulate as a type of microtubule-dependent inclusion body that is called an “aggresome” (17, 18). Various proteins, such as histone deacetylase 6, parkin, ataxin-3, dynein complex motor, and ubiquilin-1 are involved in aggresome formation, and all major chaperones (e.g., Hsc70, Hsp40, Hsp70 and small Hsp family members) are ubiquitously associated with aggresomes. Immunofluorescence analyses indicated that aggresomes colocalize with γ-tubulin, vimentin, and chaperones, including Hsp70 (19, 20). Here, we assessed co-immunostaining with Hsp70 to examine whether mutant Tub proteins observed in the perinuclear region were localized in aggresomes. As shown in Fig. 2, Hsp70 did indeed colocalize with GFP-mutant tubby in the perinuclear region, indicating that the protein was associated with aggresomes.
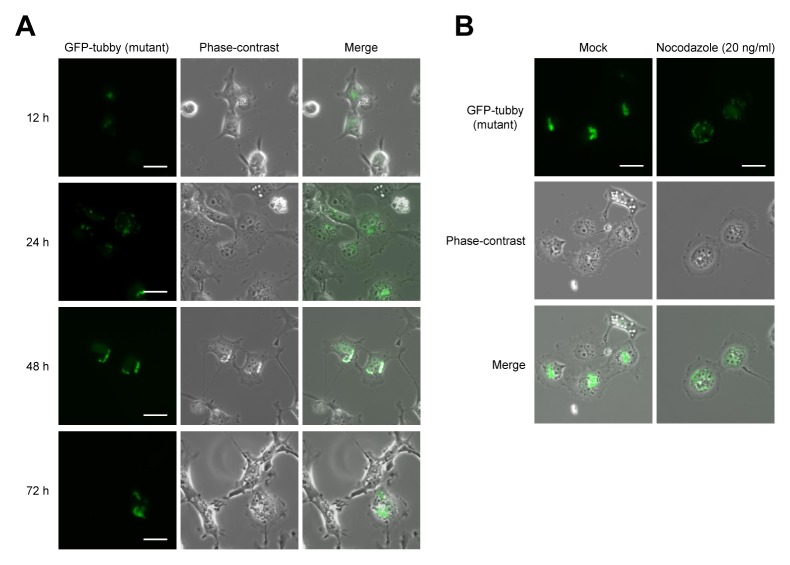
To form an aggresome, small aggregations of misfolded proteins located throughout the cytoplasm are recruited to the dynein motor complex and transported along microtubules toward the perinucleus near the microtubule organizing center (MTOC) (17, 18). Therefore, we analyzed the cellular distribution of mutant Tub proteins over time. After COS-7 cells had been transfected with GFP-mutant tubby, we observed diffuse fluorescence signals at 12 h post-transfection, many small aggregates in the cytoplasm at 24 h, and the appearance of larger perinuclear accumulations (and the corresponding loss of diffuse fluorescence) at 48 h and 72 h (Fig. 3A).
Fig. 3.
Overexpression of mutant tubby leads to aggresome formation. (A) COS-7 cells were transfected with GFP-mutant tubby. At 12, 24, 48, and 72 h post-transfection, the cells were fixed and imaged for GFP. (B) At 12 h post-transfection, the cells were incubated with or without 20 ng/ml nocodazole for 24 h and then imaged for GFP. Scale bars: 20 μm.
Since the movement of protein aggregates to the perinuclear region is dependent on microtubules (20), microtubule inhibitors (e.g., nocodazole) can block aggresome formation. To determine whether microtubule disruption could prevent the formation of mutant Tub aggresomes, COS-7 cells expressing GFP-mutant tubby were treated with 20 ng/ml nocodazole at 24 h post-transfection and then viewed at 48 h using confocal microscopy. While non-inhibitor-treated cells exhibited large perinuclear aggresome structures, nocodazole-treated cells displayed diffuse proteins and small cytoplasmic aggregates (Fig. 3B). These observations suggested that the aggregation of mutant Tub is microtubule-dependent.
Mutant tubby is not translocated to the nucleus in response to extracellular signals
The Tub protein localizes to the plasma membrane via the binding of its C-terminus to PIP2 and its association with the G protein, αq. The activation of αq by a G-protein coupled receptor (GPCR)-ligand releases Tub protein from the plasma membrane through the action of phospholipase C-beta (PLC-β), resulting in the nuclear translocation of the Tub protein (12). Moreover, insulin and leptin can phosphorylate the Tub protein through their receptors, thereby also inducing the PLC-β mediated nuclear translocation of Tub (13, 21).
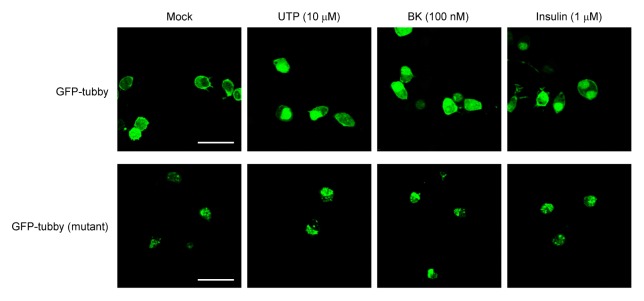
To investigate the translocation of Tub or mutant Tub in response to extracellular signaling, we transfected Neuro-2A cells with vectors encoding GFP-tubby or GFP-mutant tubby and treated the cells with UTP, bradykinin (BK), or insulin (Fig. 4). We found that GFP-tubby was localized predominantly to the plasma membrane under basal conditions, whereas treatment with 10 μM UTP, 100 nM BK, or 1 μM insulin strongly triggered the translocation of GFP-tubby to the nucleus. These results confirm that PLC is activated and PIP2 in Neuro-2A cells stimulated with these agents. In contrast, GFP-mutant tubby remained in the perinuclear region upon extracellular stimulation with these agents; it was not translocated to the nucleus, and thus would fail to function as a transcription factor.
Fig. 4.
The C-terminal region of tubby is important for its nuclear translocation. Neuro-2A cells were transfected with vectors encoding GFP-tubby or GFP-mutant tubby, and grown for 24 h. After being starved for overnight, the cells were treated with 10 μM UTP, 100 nM BK, or 1 μM insulin for 2 h, and the cellular localizations of wild-type and mutant tubby were observed under confocal microscopy. Scale bars: 20 μm.
The C-terminal region of the Tub protein is essential for the function of this protein, as identical phenotypes are associated with the C-terminal mutation of tubby mice and knockout of the Tub protein. However, to our knowledge, no previous study had examined Tub proteins carrying C-terminal mutations. Here, we reveal that mutant Tub proteins colocalized with aggresomes in the perinuclear region and failed to undergo nuclear translocation in response to extracellular signals. This might offer a mechanistic basis for the loss of function in the C-terminally mutated protein.
MATERIALS AND METHODS
Ethics statement
All experiments of the current study were performed following an animal protocol approved by the Institutional Animal Care and Utilization Committee (IACUC) of Ulsan National Institute of Science and Technology (UNIST).
Antibodies
The antibodies were obtained as follows: anti-tubby, anti-GFP, anti-actin, and the anti-goat IgG-TRITC secondary antibody from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-GM130 from BD Biosciences (Palo Alto, CA, USA); anti-Hsp70 from Cell Signaling (Danvers, MA, USA); anti-Flag from Sigma (St. Louis, MO, USA); horseradish peroxidase-conjugated goat anti-rabbit IgG and goat anti-mouse IgA, IgM, and IgG from Kirkegaard & Perry Laboratories (Gaithersburg, MD, USA).
Animals and immunohistochemistry
Homozygous mutant tubby mice (tub/tub) were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and bred with wild-type C57BL/6J (wt/wt) mice (SLC Inc., Seoul, Korea). The obtained heterozygous tubby mice (tub/wt) were used as breeding parents. Mouse strains were bred and housed in the Animal Research Facility at UNIST under specific pathogen-free conditions in accordance with the AAALAC International Animal Care Policy. Animals were maintained in a controlled 12 h light-dark cycle at a temperature of 22°C ± 1°C and were given unrestricted access to food and water. All surgery was performed under chloral hydrate anesthesia, and all efforts were made to minimize suffering. All animals at the end of the experiment were sacrificed by cervical decapitation.
For immunohistochemistry, 10-week-old wild-type or tubby mice were anesthetized and perfused with PBS followed by 4% paraformaldehyde (PFA)/PBS. The mice were sacrificed by cervical decapitation. The brains were removed and placed in a standard Cryomold (Sakura Finetek, Tokyo, Japan), which was then filled with optimum cutting temperature (OCT) compound (Tissue-Tek; Sakura Finetek) and placed in dry ice. Once preserved, the frozen tissue samples were stored at −80°C until analysis. For immunostaining, cryostat sections (10-μm) were obtained from the frozen tumor blocks, fixed with 4% paraformaldehyde for 10 min, and washed three times with PBS for 3 min each. The tissue sections were blocked with 20% FBS, incubated for 2 h with the anti-tubby antibody, and then incubated for 1 h with the anti-goat IgG-TRITC secondary antibody. The tissue-bearing slides were mounted with mounting medium and examined under a confocal microscope (LSM 510 Meta; Zeiss, Oberkochen, Germany).
Construction of plasmids
The full-length coding sequences for tubby or mutant tubby were generated by reverse transcription-PCR (RT-PCR) of total RNAs from wild-type or tubby mouse brains. Fragments of tubby (N-term, C-term, C-deletion) and mutant tubby (mutant-C-term) were produced by PCR. To generate GFP-, FLAG-, and His×6 proteins, appropriate PCR products were cloned into the pEGFP-C1 (BD Biosciences), pCMV2-Flag (Sigma), and pRSET C (Invitrogen) vectors, respectively.
PCR was performed using the following specific primers: wild type tubby, (5′-CCCGAATTCTATGACTTCCAAGCCGCA TTCCGAC-3′) and Tub-R (5′-CCCGAATTCCTACTCGCAGGC CAGCTTGC-3′); for mutant tubby, Tub-F and Mut-R (5′-CCCGAATTCTCAGGGGATTGGGGGTGGGGTG-3′); for Nterminal-domain tubby, Tub-F and Ndom-R (5′-CCCGAATTCTCAGACCTCAATATCCACTGGTGGC-3′); for C-terminal-domain tubby, Cdom-F (5′-CCCGAATTCTCAGGATCTAGAGGAGTTTGCAC-3′) and Tub-R; and for C-deletion mutant tubby, Tub-F and Cdel-R (5′-CCCGAATTCGTCATTGCCGTGGATCTGG-3′).
Cell culture and transfection
COS-7 and Neuro-2a cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. The cells were grown in a humidified atmosphere containing 5% CO2. For transient expression of the various constructs, the cells were plated on 6-well plates or coverslips coated with 10 μg/ml of poly-L-lysine and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Analysis of Triton X-100 soluble and insoluble fractions
COS-7 cells were transfected with GFP-tubby, GFP-tubby (mutant), GFP-tubby (C-deletion), GFP-tubby (N-term), or GFP-tubby (mutant-C-term). At 48 h post-transfection, the cells were washed with ice-cold PBS and lysed for 30 min on ice with 200 μl of Triton X-100 (Tx-100)-containing lysis buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1% Tx-100) supplemented with a protease-inhibitor cocktail. Insoluble and soluble fractions were separated by centrifugation at 13,000 g for 15 min at 4°C. Once the supernatant was removed to a fresh tube, the insoluble pellet was resuspended in 200 μl of 1% SDS in lysis buffer and sonicated for 20 s with a microtip-sonicator. For comparison, total cell lysates were prepared by sonicating cells with 200 μl of 1% SDS in lysis buffer. Equal volumes of each fraction were boiled for 5 min in SDS-PAGE sample buffer and analyzed by SDS-PAGE as previously described (22).
Immunofluorescence
Cells were grown on coverslips, transfected as described, rinsed four times with PBS, and then fixed for 20 min with 4% (w/v) paraformaldehyde. For direct fluorescence, the cells were washed, subjected to nuclear staining with propidium iodide (PI), and mounted with mounting medium. For indirect immunofluorescence, cells were permeabilized with 0.1% Tx-100 for 30 min, washed with PBS, blocked with 1% goat serum, and incubated with anti-GM130 or -Hsp70. The cells were then washed and incubated with a secondary antibody (anti-mouse TRITC for GM130 or anti-rabbit TRITC for HSP70), washed, and mounted as described above. Confocal images were acquired using a Zeiss LSM 510 Meta confocal microscope.
Staining of lysosomes, the ER, and mitochondria
COS-7 cells were seeded on glass coverslips, transfected with vectors encoding GFP-mutant tubby, and stained with LysoTracker Red DND-99 (100 nM; Invitrogen), ER-Tracker (1 mM; Molecular Probes, Eugene, OR, USA), or MitoTracker Red CMXRos diluted in HBSS (500 mM; Invitrogen) at 37°C to detect lysosomes, ER, and mitochondria, respectively. After 30 min, the cells were washed with PBS, fixed with 4% formaldehyde, washed with PBS, and mounted as described above.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by research grant from the National Cancer Center Grant (NCC1410270) and the National Research Foundation of Korea (NRF) Grant funded by the Korea Government (MISP) (No.2010-0028684).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Coleman D, Eicher E. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered. 1990;81:424–427. doi: 10.1093/oxfordjournals.jhered.a111019. [DOI] [PubMed] [Google Scholar]
- 2.Kleyn PW, Fan W, Kovats SG, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/S0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 3.Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 4.North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci U S A. 1997;94:3128–3133. doi: 10.1073/pnas.94.7.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll K, Gomez C, Shapiro L. Tubby proteins: the plot thickens. Nat Rev Mol Cell Biol. 2004;5:55–64. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay S, Jackson PK. The tubby family proteins. Genome Biol. 2011;12:225. doi: 10.1186/gb-2011-12-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagstrom SA, North MA, Nishina PM, Berson EL, Dryja TP. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 8.Banerjeel P, Kleyn PW, Knowles JA, et al. TULP1 mutation in two extended Dominican kindreds with autosomal recessive Retinitis pigmentosa. Nat Genet. 1998;18:177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda S, Shiva N, Ikeda A, et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet. 2000;9:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum Mol Genet. 2001;10:1325–1334. doi: 10.1093/hmg/10.12.1325. [DOI] [PubMed] [Google Scholar]
- 11.Patterson VL, Damrau C, Paudyal A, et al. Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects, and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum Mol Genet. 2009;18:1719–1739. doi: 10.1093/hmg/ddp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santagata S, Boggon TJ, Baird CL, et al. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 13.Prada PO, Quaresma PG, Caricilli AM, et al. Tub has a key role in insulin and leptin signaling and action in vivo in hypothalamic nuclei. Diabetes. 2013;62:137–148. doi: 10.2337/db11-1388. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Boggon TJ, Shan W-S, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 15.Stubdal H, Lynch CA, Moriarty A, et al. Targeted deletion of the tub mouse obesity gene reveals that tubby is a loss-of-function mutation. Mol Cell Biol. 2000;20:878–882. doi: 10.1128/MCB.20.3.878-882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia B, Wu Y, Zhou Y. 14-3-3 and aggresome formation: implications in neurodegenerative diseases. Prion. 2014;8:173–177. doi: 10.4161/pri.28123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/S0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 19.García-Mata R, Bebök Z, Sorscher EJ, Sztul ES. Characterization and Dynamics of Aggresome Formation by a Cytosolic GFP-Chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debure L, Vayssière J-l, Rincheval V, Loison F, Le Dréan Y, Michel D. Intracellular clusterin causes juxta-nuclear aggregate formation and mitochondrial alteration. J Cell Sci. 2003;116:3109–3121. doi: 10.1242/jcs.00619. [DOI] [PubMed] [Google Scholar]
- 21.Kapeller R, Moriarty A, Strauss A, et al. Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J Biol Chem. 1999;274:24980–24986. doi: 10.1074/jbc.274.35.24980. [DOI] [PubMed] [Google Scholar]
- 22.Heo K, Park KA, Kim YH, et al. Sphingosine 1-phosphate induces vascular endothelial growth factor expression in endothelial cells. BMB Rep. 2009;42:685–690. doi: 10.5483/BMBRep.2009.42.10.685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.