Abstract
Inflammatory bowel disease (IBD) is characterized by flares of inflammation with periodic need for increased medication and sometimes even surgery. IBD etiology is partly attributed to a deregulated immune response to gut microbiome dysbiosis. Cross-sectional studies have revealed microbial signatures for different IBD diseases, including ulcerative colitis (UC), colonic Crohn’s Disease (CCD), and ileal CD (ICD). Although IBD is dynamic, microbiome studies have primarily focused on single timepoints or few individuals. Here we dissect the long-term dynamic behavior of the gut microbiome in IBD and differentiate this from normal variation. Microbiomes of IBD subjects fluctuate more than healthy individuals, based on deviation from a newly-defined healthy plane (HP). ICD subjects deviated most from the HP, especially subjects with surgical resection. Intriguingly, the microbiomes of some IBD subjects periodically visited the HP then deviated away from it. Inflammation was not directly correlated with distance to the healthy plane, but there was some correlation between observed dramatic fluctuations in the gut microbiome and intensified medication due to a flare of the disease. These results help guide therapies that will re-direct the gut microbiome towards a healthy state and maintain remission in IBD.
Both the state and the dynamics of the human gut microbiome in healthy individuals are highly personalized1–7. Although cross sectional studies have revealed dysbiosis of the gut microbiome in IBD8–12, little is known about the individual nature of microbiome dynamics in IBD, beyond a study of 3 UC patients before and after ileostomy, and two small studies of IBD patients in remission or during changes in disease activity13–15. Here we studied the long-term dynamics of the gut microbiome from an IBD cohort of 128 individuals (49 CD, 60 UC, 4 lymphocytic colitis (LC), 15 collagenous colitis (CC)) and 9 healthy controls (HC). We sampled at three-month intervals, collecting 1–10 samples per individual for a total of 683 samples (Supplementary Table 1). The microbiome composition in each sample was determined by sequencing the V4 region of the 16S rRNA gene for a total of 248 million 16S rRNA gene amplicons. To determine links between the gut microbiome and clinical factors, we collected clinical data, including fecal calprotectin (f-calprotectin) concentration and surgical resection status. To control sampling bias, we restricted our statistical analyses of volatility to a subset of the cohort that had sequence data from the first four time points and that had matching f-calprotectin concentrations; yielding 276 samples from 69 patients (Supplementary Table 1, Supplementary Information 1; results were similar when all subjects were considered). We also included patient genetic load scores (GLS) based on 163 known IBD risk loci for 29 patients to assess potential links between the host genetics, IBD and the microbiome16.
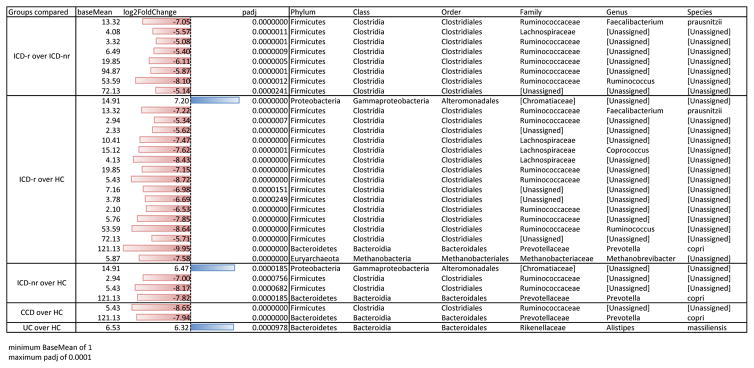
As expected from previous work10,11, we found that HC and IBD subtypes formed distinct clusters by Principal coordinates analysis (PCoA) of unweighted UniFrac distances, with ICD patients least similar to healthy controls (Supplementary Fig. 1, ADONIS stratified by time point, p < 0.001). As in previous studies10,12,13, we found differences in alpha and beta diversity of the microbiome according to IBD subtype and between subtypes and healthy controls and identified several families that correlated with health or disease state, e.g. Enterobacteriaceae with ICD and Ruminococcaceae with HC (see Supplementary Figs. 1 and 2). Individual taxa that are differentially abundant between IBD subtypes compared to HC are listed in Table 1 (DESeq2, log2 fold change). The IBD microbiomes contained significantly lower abundances of putative beneficial OTUs present in HC, as previously reported8–13, including Prevotella copri and the butyrate-producing bacterium Faecalibacterium prauznitzii (Table 1; Supplementary Fig. 3).
Table 1.
Differential abundance in specific taxa according to disease phenotype comparisons (DESeq2)
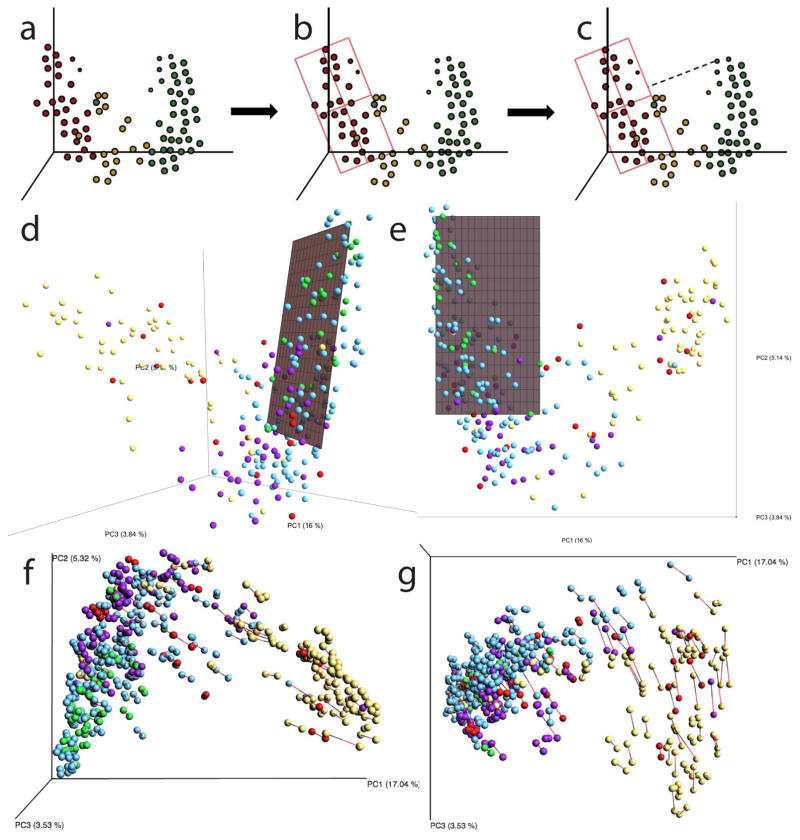
From the animated ordination of the samples (Supplementary Video 1), we observed that although microbiome samples from the healthy individuals varied over time, they were restricted to a small volume of the ordination space. In contrast, IBD subtypes traversed far more of the total volume, sporadically visiting the area where healthy samples resided. To summarize these dynamics, we identified a “healthy plane” (hereafter referred to as HP). Briefly, this plane is calculated in a space derived from Principal Coordinate Analysis (PCoA) of unweighted UniFrac distances of healthy subjects (Supplementary Video 1). We constructed a model using the samples from the HC patients, and fit them to a two-dimensional plane embedded in a three-dimensional space using the least squares method (Fig. 1). The plane is then restricted to only span the three-dimensional ranges of the HC samples. This plane was used as a proxy to represent the normal microbial variation within healthy subjects and to summarize the abnormal, intermittent dysbiosis associated with IBD.
Figure 1. Defining a healthy plane.
Diagram summarizing the procedure for creating a representative plane for a group of samples S: a, sample selection, b, model fitting and c, distance calculations for all samples. The healthy plane is then located in UniFrac space by d, fitting a line to the major axis of the points, and e, defining a least-squares fit to identify a plane that minimizes the sum of squares of distances to the nearest point on the plane. f, Verification that the position of the healthy plane is not driven by proteobacteria-dominated outliers: Procrustes Analysis comparing original samples and those with Proteobacteria removed. A vector connects each original sample (red) with the same samples after Proteobacteria have been omitted (black). p < 0.001, M2 = 0.018, 999 permutations. g, The short length of most vectors indicates that the relative composition of most samples does not change when proteobacteria are filtered out.
The procedure was as follows: let S be a set of n samples s1, s2, … sn corresponding to a group of trajectories, each trajectory pertaining to a subject with at least four samples collected at distinct points in time. Each sample is represented as a three-dimensional vector corresponding to sample coordinates in ordinated space i.e. s1 = (x1, y1, z1); s2 = (x2, y2, z2); … sn = (xn, yn, zn). We fit a linear model to S by the least squares method to obtain coefficients for the equation of a three dimensional surface T , next we restricted a segment of this surface to the ranges given by [minx(S), maxx(S)] and [miny(S), maxy(S)] , we defined this to be a plane representative of S or Ps. When S is the set of samples from healthy subjects in an ordination space we, refer to Ps as the healthy plane (HP). Finally, we defined dk to be the Euclidean distance from a sample k to the nearest point lying on Ps. After measuring dk for all samples in our study, we grouped samples according to their diagnosis, and compared the distributions of distances. Fig. 1 a–c demonstrates this procedure, and Fig. 1d–e demonstrates the placement of the HP in the context of our full IBD dataset. Samples from the HP are co-located with one another, while many samples from IBD patients are further away. To investigate whether this effect is due to ‘outlier’ groups of samples that are dominated by taxa that are typically rare in healthy individuals, we excluded all Proteobacteria from the dataset, and compared the location of samples in unweighted UniFrac space with and without Proteobacteria using Procrustes analysis. As shown in Figure 1f–g, the omission of Proteobacteria is significant (p < 0.01), but this effect is largest in the already dysbiotic ICD-r patients and aligns with PC3. The effect on healthy controls is minimal, and healthy samples are still located only near the HP (Fig. 1f–g).
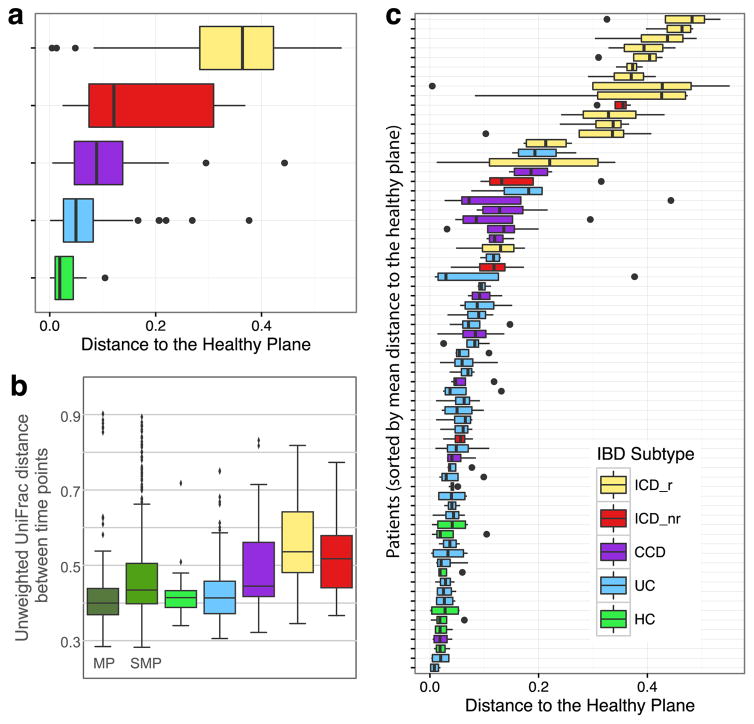
Calculation of the mean Euclidean distance from each sample to the HP revealed that all subtypes of IBD significantly deviated from the HP (Generalized linear mixed effects model (GLM), all p < 0.00268, Fig. 2a). Samples from patients with CCD and UC were closer to the HP, and some samples did not differ significantly from healthy controls, whereas ICD samples were the most distant from the HP (Supplementary Video 1; Fig. 2). The highest volatility was observed for ICD patients that had previously undergone ileocecal resection (ICD-r), followed by ICD patients without surgery (ICD-nr) based on UniFrac distances between successive samples (Fig. 2b). ICD-r patients also had low gut microbial richness (Supplementary Fig. 2). Ileocecal resection is a major modifier of intestinal physiology, and the observed pronounced volatility in ICD-r patients might be partly explained by removal of the ileocecal valve per se. Ileal inflammation may also influence bile salt uptake and, consequently, colonic transit time and microbiome volatility. Interestingly, several IBD patients had complex trajectories that sporadically moved to and from the HP (Fig. 2c, Supplementary Video 1).
Figure 2. The gut microbiomes of different IBD subtypes display different distributions relative to a healthy plane (HP).
a, Median distances from HP for each IBD subtype. All IBD subtypes were significantly different from healthy controls (GLM, all p < 0.00261). b, UniFrac distances between subsequent samples. c, Distance to HP for each individual patient. HP was defined using data shown in Supplemental Video 1. See Supplementary Table 1 for composition of downstream analysis cohort. Boxes show interquartile range (IQR). Whiskers denote the lowest and highest values within 2.5 × IQR of the median. Circles represent outliers.
Although our study represents the largest longitudinal analysis of the IBD microbiome to date, the small number of patients in specific subgroups limited some statistical comparisons. Furthermore, the number of healthy controls was lower than the number of IBD patients. To address this limitation of unequal sampling of healthy individuals and IBD patients, we compared the volatility of our healthy controls to healthy participants in two published studies, the Student Microbiome Project (SMP) and the Moving Pictures (MP) datasets6,17. There was less variability over time in healthy individuals across all three cohorts compared to those with IBD (Figure 2b). This result emphasizes that IBD is characterized by volatile dysbiosis not found in healthy people, and confirms earlier preliminary results and meta-analysis of much smaller studies13–15,18.
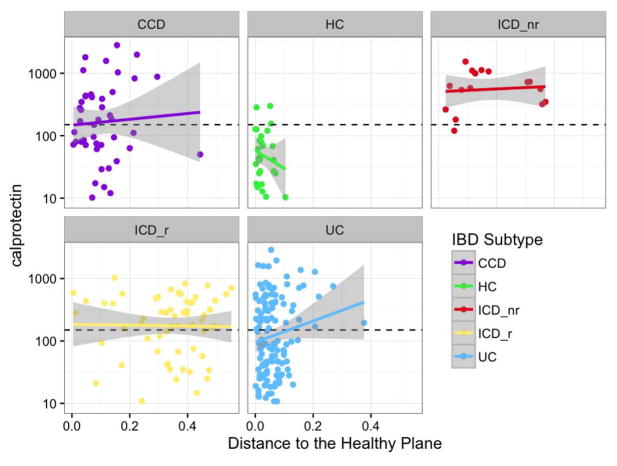
To extend our understanding of the mechanisms underlying the microbiome dynamics, we explored the correlation between the dynamics and inflammatory activity in each sample using f-calprotectin > 150 μg/g as a surrogate for inflammatory activity. The concentration of f-calprotectin in stool samples has previously been correlated with endoscopic and histopathologic activity, and is used in daily clinical practice because it is non-invasive19. We observed that concentrations of f-calprotectin were higher in all IBD subtypes than in healthy controls (Fig. 3). However, we did not observe a significant correlation between f-calprotectin and distance from the HP (Fig. 3, GLM p = 0.275). Although recent microarray analyses of the gut microbiome in a cohort of anti-TNF treated pediatric IBD patients20 and experiments with gnotobiotic fecal transplants suggest that microbial composition and function are causally associated with inflammatory activity21, our differential abundance testing revealed only weak trends and no specific OTUs that varied significantly with active inflammation, using f-calprotectin > 150 μg/g as cut-off. However, the use of f-calprotectin as a proxy for inflammatory activity might have introduced bias, because f-calprotectin is a less accurate marker of ileal than colonic inflammation22 and ICD patients displayed the greatest distance from the HP.
Figure 3. Correlation between fecal calprotectin concentrations and distance to a defined healthy plane (HP) in 3D ordination space.
Data represent a correlation of f-calprotectin levels and distance to the here defined healthy plane in 3D ordination space (see Supplementary Video 1) for each individual and time point for different inflammatory bowel disease (IBD) subtypes. To compare the relationship between f-calprotectin and the healthy plane, a generalized linear mixed effects model was fit, with a conditional Gamma distribution, using f-calprotectin and disease type as fixed effects and including a random subject effect; f-calprotectin was not significant (p = 0.27501).
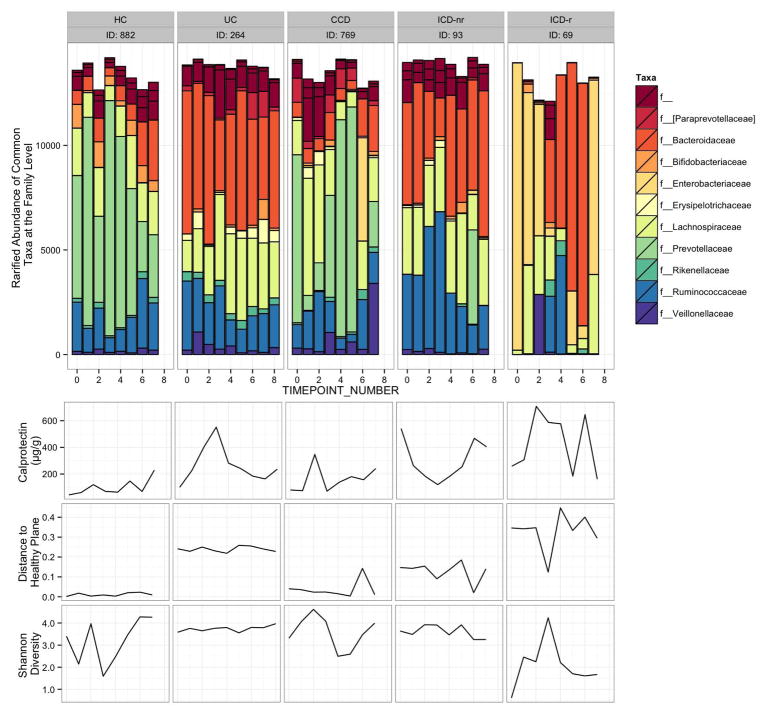
Examples of the microbiome dynamics for one representative HC and one from each IBD subgroup are shown in addition to changes in f-calprotectin concentrations, distance to HP, and Shannon diversity in Figure 4 (all individual profiles are shown in Supplementary Fig. 3), illustrating the more stable dynamics over time for HC and UC compared to the other clinical phenotypes of IBD, with the most fluctuations occurring for patient 69 that had undergone surgical resection. The f-calprotectin levels were low and relatively stable for the HC compared to the IBD patient (Fig. 4). In these examples, there were also substantial fluctuations in diversity for the HC, ICD-r, and CCD by contrast to UC and ICD-nr patients.
Figure 4. Microbiome dynamics of selected individuals from each IBD subtype and a healthy control.
From each IBD subtype and healthy control group, representative individuals sampled over the most time points and having complete clinical and sequence data were selected. Data represent f-calprotectin values, distance to the healthy plane, and Shannon diversity and rarified abundances of most common taxa at the family level. Note that taxa unclassified at the family level are represented in the ‘f__’ category.
We further explored the individual dynamics of the gut microbiome in IBD patients who experienced increased clinical disease activity, according to the physician’s global assessment (Supplementary Fig. 4). Recently, the short-term (6 weeks) dynamics of the microbiome in pediatric patients with active IBD treated with anti-TNF indicated that initiation of medical treatment changes the microbial composition at the genus level20. Because we had few anti-TNF exposed patients, and our patients received a course of corticosteroids at flare as first line therapy, we explored how corticosteroid administration influenced microbiome dynamics. Our data demonstrate that change in medication influenced the volatility of the microbiome (Supplementary Data Fig. 4). Patients receiving a course of oral corticosteroids (n=7) had more microbiome fluctuations than patients on stable medication (n=49), based on calculations of unweighted UniFrac distance between time points (Wilcoxon Signed-rank test; p=0.04). Our dynamic model suggests that beyond the association with IBD subtype and the weak correlation with inflammation, the dynamics of the microbiome composition are influenced by changes in medication. The extent to which other factors, such as dietary changes and smoking, may have influenced the observed volatility remains speculative, because the collected information was insufficiently detailed to include these factors as covariates.
To evaluate the microbiome as a predictive tool, we combined the microbial and clinical data and used a supervised learning Random Forests model to predict IBD subtypes23,24. To avoid overfitting, our models were built using OTU abundances from the first time points only, along with clinical metadata (BMI, f-calprotectin concentrations, sex, and Distance to the HP). Accuracy was evaluated using the remaining three time points, which were not used to train the model. Using this model, the IBD subtypes were discriminated from healthy controls and correctly predicted for 66.6% of samples (Supplementary Table 2), consistent with the findings reported in Gevers et al. (2014). Feature importance scores from this model revealed several potential microbial indicators of IBD subtypes, including OTUs matching to Lachnospira, Clostridium, Oscillospira, and many unidentified Ruminococcaceae (Supplementary Table 3). Intriguingly, the accuracy of the model increased slightly if f-calprotectin concentrations were omitted, but decreased by at least 10% if the distance to the HP was removed (Supplementary Tables 2 and 3), suggesting that the HP is a more important factor in the model. Comparable levels of accuracy were previously achieved using rectal samples11, but here we show that the same can be achieved with fecal samples, which are easier to collect. When immunochip data were included for a subset of 29 IBD individuals and the Random Forests model was repeated, the samples were still classified into the four IBD subtypes (UC, CCD, ICD-r, ICD-nr) (Supplementary Tables 2 and 3). While distance to the HP remained as the single most important feature for classification, genetic load scores (GLS) were more predictive than sex, f-calprotectin, or BMI when included in the model (Supplementary Table 2). However, including GLS only increased the overall accuracy of the model by about 2%, demonstrating the predictive potential of the microbiome. Interestingly, in a recent study of obesity covering 339,224 individuals, the 97 risk loci for obesity accounted for only 2.7% of BMI variation25, whereas the microbiome classified lean from obese individuals with 90% accuracy26, providing precedent for the predictive value of the gut microbiome over human genetics in chronic disease.
In summary, by analyzing fecal samples collected every 3 months from a large IBD cohort, we determined the long-term volatility of the gut microbiome in IBD. Our data revealed that although the microbiome of healthy individuals varied it was only within a newly defined HP, whereas there was considerable volatility away from the HP for several of the IBD cohorts. Devising improved methods to detect the healthy state with non-invasive sampling, to predict when the healthy state will be departed, and to sustain the microbiome in this healthy state by erecting barriers that prevent the slip back into dysbiosis, will be an important focus of future work.
METHODS
Cohort Demographics
Patients with CD or UC, the two major forms of IBD, attending the outpatient clinic were consecutively invited to take part. After obtaining written consent, BMI was recorded and patients were asked to provide a fecal sample and to fill in a questionnaire with clinical disease activity, present medication, dietary habits, use of antibiotics and use of NSAIDs. Disease phenotype was classified according to the Montreal classification27. Individuals were then followed prospectively, asked to provide fecal samples and to fill in the questionnaire every third month for a two-year period. If a patient did not provide a fecal sample at any of the three months periods, a reminder letter was sent. In total 109 patients with IBD (CD; n=49 and UC; n=60) took part. Nine additional individuals with no IBD or any other gastrointestinal conditions were recruited as HC as well as 19 patients with other chronic inflammatory gastrointestinal diseases (4 lymphocytic colitis (LC) and 15 collagenous colitis (CC)). All 137 individuals were Caucasians and together they provided 683 fecal samples during the two-year period (Supplementary Table 4). The study was approved by the Ethical Committee of the Medical Faculty, Uppsala University (2007/291).
Sample Collection
Fecal samples were self-collected in sterile plastic containers and stored at −80 °C until shipping on dry ice and processing.
Fecal calprotectin
To assess the degree of inflammatory activity at the collection of each fecal sample, the concentration of f-calprotectin was assessed by commercially available ELISA, Calprotectin Elisa Buhlmann Laboratories AG, Basel, Switzerland, according to the manufacturer’s protocol.
DNA Extraction and Amplification
Genomic DNA was extracted from 0.25 g of fecal material from each sample using the Earth Microbiome DNA extraction protocol28. Briefly, DNA was extracted using the 96-well format MoBio Powersoil DNA kit on an EpMotion 5075 robot with vacuum (Eppendorf, Hamburg, Germany). DNA was quantified with the Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
PCR amplification and library preparation were performed similarly to the protocol described by Caporaso et al.29. 515F/806R Illumina primers with unique reverse primer barcodes were used to target the V4 region of the 16S rRNA gene. Samples were amplified in triplicate and cleaned using the MO BIO 69 htp PCR cleanup kit. Each PCR reaction included 1X PCR buffer, 10 μM each forward and reverse primer, 200 μM dNTPs, 1 U/ml Taq polymerase, 15 ng template DNA, and PCR grade water, with a total reaction volume of 25 μL. Reactions were kept at 94°C for 3 minutes for denaturation to occur. Amplification was performed by 25 cycles of 94 °C for 45s, 58 °C for 60s, and 72 °C for 90s. The V4 amplicons were sequenced on the Illumina HiSeq 2000 platform, yielding single end, 100 base pair reads. Sequencing and quality assessment were performed at the Yale Center for Genome Analysis.
Phylogenetic Analysis
Sequence data were processed using QIIME 1.9.0-dev through the online platform QIITA (https://qiita.ucsd.edu)30. Four HiSeq lanes of data were demultiplexed with default quality filtering settings and subsequently combined, resulting in 248,547,926 total sequences. These sequences were clustered using SortMeRNA at 97% identity against the Greengenes rRNA reference database May 2013 release31,32. Sequences that failed to match the database were discarded. 237,653,256, or approximately 95.6% of the sequences, clustered against the Greengenes reference dataset. Even sampling was performed at 14,553 sequences per sample for beta diversity and supervised learning analyses. The beta diversity principal coordinates plot of unweighted UniFrac distances was constructed using the same rarefied OTU table, and visualized in Emperor33,34. Clinical matched metadata, including f-calprotectin concentrations, were included when available. From each patient, the first four microbiome samples with matched f-calprotectin concentrations were sub-selected for use in downstream analysis.
Statistical Analysis
The R package phyloseq was used to import and graph data, while the packages DESeq2, randomForest, and vegan were used to perform differential abundance testing and supervised learning24,35–38. Statistical significance of unweighted UniFrac distance matrices comparing healthy controls and IBD subtypes was assessed using the ADONIS test. The variation of the microbial community over time was calculated with vector lengths produced by summing the total distance between each subject’s time points over the first three PCoA axes of unweighted UniFrac space. The Random Forests model was constructed using the first time point from each patient in the downstream analysis cohort and prediction accuracy was measured using the subsequent three time points (CCD = 11 patients, ICD without resection (ICD-nr) = 4 patients, ICD with resection (ICD-r) = 15 patients, UC = 30 patients, and HC = 7 patients). The following metadata categories used as features for supervised learning: BMI, f-calprotectin (continuous), sex, and Distance to the Healthy Plane. For classification on the subset of samples with immunochip data, the above process was used while adding genetic load scores (GLS). We used a dataset of 29 IBD samples (25 CD, 4 UC) with available immunochip data to estimate a Genetic Load Score (GLS) for each sample. In particular, GLS was calculated summing the number (counts) of risk alleles (0,1,2) at lead SNP from each IBD risk locus (n = 163) according to Jostins et al (2012)16.
After performing principal coordinates analysis of the unweighted UniFrac distance matrix, the samples of HC were used to fit using the least squares method on the first three principal coordinates. The distance from each sample to this plane was measured and added to the sample metadata both for weighted and unweighted UniFrac. To compare distribution of samples relative to this healthy plane, a generalized linear mixed effects model was fit, with a conditional Gamma distribution, using disease type as a fixed effect and including a random subject effect
To assess the variation between samples in an orderly manner (as described by the time point), we measured the UniFrac (weighted and unweighted) distance between samples that occur sequentially for any given subject; samples representing the final collection point do not have a value in these columns, as there is no subsequent sample to compare to.
We examined the power of differential abundance tests for the 16S data. DESeq224 assumes that counts can be modeled as a negative binomial distribution with a mean parameter, allowing for size factors, and a dispersion parameter. The test for differential abundances fits a generalized linear model with a negative binomial family and a log link function.
A power analysis was conducted by using samples from the downstream analysis cohort (Supplementary Table 4). The power of the differential abundance test is dependent on the sample size of groups, difference in mean counts, type 1 error rate, and the dispersion value. Here we used a conventional type 1 error rate of 0.05 and assumed that we have two groups with sample sizes of n1 = 25 and n2 = 80. A total of 1000 OTUs were randomly selected from the data and dispersion parameters were estimated for each OTU. Data was then simulated from a negative binomial distribution, as specified by DESeq2, for each estimated dispersion value with means giving 2 fold, 1.5 fold, and 1.25 fold differences. For each mean fold difference value and dispersion value, a total of 5000 data simulations were done and a Wald test for difference in means using a generalized linear model was conducted. Based on these simulations, where data is generated with true differences, the fraction of times that the null hypothesis is correctly rejected (the power), was calculated and our comparisons were well powered given our subset sample sizes (Supplementary Table 4).
Data Availability
Microbiome data from this study is available on Qiita under study ID 1629 (https://qiita.ucsd.edu/study/description/1629) and using the EBI accession number ERP020401. Patient clinical information is available on Qiita and in Supplemental Information 1.
Code Availability
Our analysis methods make use of standard, open source software. R software packages are available on CRAN (cran.r-project.org), bioconductor (bioconductor.org), and GitHub (github.com/joey711/phyloseq). Python software is available in the bioconda and biocore Conda channels, and is maintain on GitHub (github.com/biocore, github.com/ElDeveloper/reference-plane).
Supplementary Material
Supplementary Fig. 1. The gut microbiomes of different IBD subtypes differ in community composition
Supplementary Fig. 2, Alpha diversity of IBD-subtypes and healthy controls.
Supplementary Fig. 3, Microbiome dynamics of all individuals used in downstream analysis.
Supplementary Fig. 4, Shifts in microbiome structure in IBD patients with unstable medications.
Supplementary Table 1. Summary of the patient cohort
Supplementary Table 2. Clinical features of the Random Forests model
Supplementary Table 3. Top features of the Random Forests model
Supplementary Table 4. Power Analysis for differential abundance tests
Supplementary Video 1. Dynamics of healthy controls and IBD subtypes within ordinated UniFrac space. This video illustrates the dynamics along a “healthy plane” for healthy controls. By contrast, the IBD subjects move in a 3rd dimension away from the healthy plane. The extent of this deviation from the healthy plane is quantified and significant for the different IBD subtypes (data shown in Figure 2).
Supplementary Information 1. Clinical metadata for patients and samples. This QIIME compatible mapping file lists the genetic and clinical context of each sample used in this study.
Acknowledgments
This research was partially supported by the United States National Institute of Health (grant NIH IU54DE023798-01) and by the Pacific Northwest National Laboratory under Contract DE-AC05-76RLO1830, by the Crohns and Colitis Foundation of America. Financial support was also provided by the Örebro University Hospital Research Foundation (grant OLL-507001), and the Swedish Research Council (521-2011-2764). Additional support was provided by a grant to Juniata College from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program and the National Science Foundation (NSF Award # DBI-1248096). R.K. was a Howard Hughes Institute Early Career Scientist for part of the duration of this project.
Footnotes
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Genetic data from this study is available on QIITA under study ID 1629.
Author contributions: JH and JKJ designed the study. JH carried out the clinical study, collected samples and clinical data. CJB, YV-B, WAW, DM, MDA, FB, AG, RL, EEM, LB and MFD performed bioinformatics and statistical analyses of the data. CJB, YV-B, WAW, and AG made the figures. JH, CJB, YV-B, JKJ and RK wrote the majority of the text with input from all coauthors.
References
- 1.Eckburg PB, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, et al. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Consortium. Structure Function and Diversity of the Healthy Human Microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez I, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores GE, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaura E, et al. Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces. mBio. 2015;6:e01693–1615. doi: 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willing B, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 10.Willing BP, et al. A Pyrosequencing Study in Twins Shows That Gastrointestinal Microbial Profiles Vary With Inflammatory Bowel Disease Phenotypes. Gastroenterology. 2010;139:1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 11.Gevers D, et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajca S, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis. 2014;20:978–986. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 13.Wills ES, et al. Fecal Microbial Composition of Ulcerative Colitis and Crohn’s Disease Patients in Remission and Subsequent Exacerbation. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0090981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young VB, et al. Multiphasic analysis of the temporal development of the distal gut microbiota in patients following ileal pouch anal anastomosis. Microbiome. 2013;1:9. doi: 10.1186/2049-2618-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez C, et al. Unstable Composition of the Fecal Microbiota in Ulcerative Colitis During Clinical Remission. Am J Gastroenterol. 2008;103:643–648. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 16.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 19.Lewis JD. The Utility of Biomarkers in the Diagnosis and Therapy of Inflammatory Bowel Disease. Gastroenterology. 2011;140:1817–1826.e2. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolho KL, et al. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am J Gastroenterol. 2015;110:921–930. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 21.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sipponen T, et al. Correlation of faecal calprotectin and lactoferrin with an endoscopic score for Crohn’s disease and histological findings. Aliment Pharmacol Ther. 2008;28:1221–1229. doi: 10.1111/j.1365-2036.2008.03835.x. [DOI] [PubMed] [Google Scholar]
- 23.Breiman L. Random Forests. Mach Learn. 45:5–32. [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe. 2011;10 doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silverberg MS, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol J Can Gastroenterol. 2005;19(Suppl A):5A–36A. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 28.EMP DNA Extraction Protocol. 2011 [Google Scholar]
- 29.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 32.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299. [Google Scholar]
- 36.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 38.Oksanen J, et al. vegan: Community Ecology Package. 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. The gut microbiomes of different IBD subtypes differ in community composition
Supplementary Fig. 2, Alpha diversity of IBD-subtypes and healthy controls.
Supplementary Fig. 3, Microbiome dynamics of all individuals used in downstream analysis.
Supplementary Fig. 4, Shifts in microbiome structure in IBD patients with unstable medications.
Supplementary Table 1. Summary of the patient cohort
Supplementary Table 2. Clinical features of the Random Forests model
Supplementary Table 3. Top features of the Random Forests model
Supplementary Table 4. Power Analysis for differential abundance tests
Supplementary Video 1. Dynamics of healthy controls and IBD subtypes within ordinated UniFrac space. This video illustrates the dynamics along a “healthy plane” for healthy controls. By contrast, the IBD subjects move in a 3rd dimension away from the healthy plane. The extent of this deviation from the healthy plane is quantified and significant for the different IBD subtypes (data shown in Figure 2).
Supplementary Information 1. Clinical metadata for patients and samples. This QIIME compatible mapping file lists the genetic and clinical context of each sample used in this study.
Data Availability Statement
Microbiome data from this study is available on Qiita under study ID 1629 (https://qiita.ucsd.edu/study/description/1629) and using the EBI accession number ERP020401. Patient clinical information is available on Qiita and in Supplemental Information 1.