Abstract
Antimicrobials are sometimes given to food animals at low doses in order to promote faster growth. However, the mechanisms by which those drugs improve performance are not fully understood. This study aimed to investigate the impact of zinc bacitracin (55g/ton), enramycin (10g/ton); halquinol® (30g/ton); virginiamycin (16,5g/ton) and avilamycin (10g/ton) on the cecal microbiota of broiler chicken, compared to a control group. Six hundred and twenty four chicks (Cobb 500) arriving to an experimental unit were randomly assigned into each treatment with four repetitions per treatment. The cecal content of 16 animals per treatment (n = 96) was used for DNA extraction and sequencing of the V4 region of the 16S rRNA gene using Illumina technology. The use of antimicrobials induced significant changes in membership but not in structure of the cecal microbiota compared to the control group, suggesting a greater impact on the less abundant species of bacteria present in that environment. Halquinol was the only drug that did not affect microbial membership. Firmicutes comprised the major bacterial phylum present in the cecum of all groups. There was no statistical difference in relative abundances of the main phyla between treated animals and the control group (all P>0.05). Treatment with enramycin was associated with decreased richness and with lower relative abundance of unclassified Firmicutes, Clostridium XI, unclassified Peptostreptococcaceae (all P<0.001) and greater abundance of Clostridium XIVb (P = 0.004) and Anaerosporobacter spp. (P = 0.015), and treatment with bacitracin with greater relative abundance of Bilophila spp. (P = 0.004). Several bacterial genera were identified as representative of usage of each drug. This study used high throughput sequencing to characterize the impact of several antimicrobials in broiler chicken under controlled conditions and add new insights to the current knowledge on how AGPs affect the cecal microbiota of chicken.
Introduction
Antibiotic growth promoters (AGPs) have been widely used to improve performance of food animals. Antimicrobials are given to broiler chicken in order to control diseases such as necrotic enteritis caused by Clostridium perfringens, and also to promote faster growth and improve conversion rates [1–3]. The effects of those drugs are not fully understood, but the potential of the intestinal microbiota in increasing feed efficiency has been shown [4–7].
This practice has recently raised concerns regarding emergence of antibiotic resistant strains of bacteria that could potentially spread to humans [8]. Indeed, AGPs usage has been banned in the European Union and there is increasing pressure for stricter regulations in North America [9].
The intestinal microbiota has been shown to have a tremendous influence on host health and disturbances in its balance (dysbiosis) have been associated with various diseases [10]. While several factors, such as diet, environment and genetics can induce changes in the intestinal microbiota, the use of antimicrobials is one of the most important [11]. The different spectrum of selection depending on the active ingredients present in each compound should induce predictable changes on the intestinal microbiota [12]. However, the in order to adequately address those changes, controlled environmental conditions should be used for the characterizations of changes induced by those drugs.
Changes in the cecal environment are of importance since cecal bacteria are responsible for food fermentation and short chain fatty acids (SCFA) production in chickens [13–15]. Therefore, a better characterization of how AGPs impact the cecal microbiota of chickens could be the keystone for the development of alternative methods to improve growth efficacy in this species [16].
The effects of AGPs on the chicken microbiota have recently been investigated [3,17,18]. However, since several factors can impact the intestinal microbiota, comparison between studies is difficult and in order to adequately compare the effect of different drugs molecules, it is essential to secure that experimental conditions are rigorously controlled. To date, most studies using high throughput sequencing have compared a limited number of drugs, with special emphasis given to virginiamycin and bacitracin [3,19–21].
This study was designed to test the hypothesis that different antimicrobial drugs would differentially affect the cecal microbiota of broiler chicken.
Materials and methods
Study design
This study was approved by the University of Londrina’s Animal Care and Use Committee (process number: 10482014.57).
Six hundred and twenty four 2 day-old chicks (Cobb 500) arriving to the poultry farm of the experimental unit of the Londrina University (Londrina, Paraná State, Brazil) were allocated into cages of 1.45 x 1.45 m (26 animals per cage). Cages were randomly assigned into six treatment groups with four repetitions per treatment: control (no antibiotic), zinc bacitracin (55g/ton), enramycin (10g/ton); halquinol (30g/ton); virginiamycin (16,5g/ton) and avilamycin (10g/ton). All animals received a standard diet recommended for this breed from 1 to 21 days of life and another from 22 to 42 days of life, constituted mainly of corn (63%), grinded soy (28%) and soy oil (5%) (S1 Table). Antimicrobials were given to animals during the whole period of the trial.
Chicks were vaccinated at the hatchery against Marek’s disease and at the experimental unit when were 10 day-old against gumboro (Bursine 2, Zoetis, New Jersey USA). Litter consisted of wood shavings was added to the cages to a height of 6 cm. Approximately 300 grams of litter from a commercial farm were mixed to the clean litter in order to increase challenge with disease strains present in the field.
Weight gain and feed intake were recorded and feed conversion and viability calculated within each group. Four animals per cage (n = 16/group; 96 total) were randomly selected at the time of slaughter (43 days) and had their cecal contents collected into sterile plastic tubes that were promptly refrigerated (for a maximum of 1.5 hour) and frozen at -80°C until DNA extraction. Chickens were rendered unconscious by electrical stunning just before slaughtering by exsanguination.
DNA extraction and sequencing
DNA was extracted with a commercial kit (E.Z.N.A. Stool DNA Kit, Omega Bio-Tek) according to the manufacturer’s instructions. The V4 region of the gene 16S rRNA was amplified by PCR using the forward S-D-Bact-0564-a-S-15 and the reverse primers S-D-Bact-0785-b-A-18 [22] containing an overlapping region of the Illumina sequencing primers. PCR was carried in two steps: first, 2.5μL of DNA were added to a mixture containing 9μL of water, 12.5μL of Kapa 2X ReadMix (Kapa Biosystems. MA) and 0.5μL of each 16S primer (10 pmol/μL). The reaction was carried according to the PCR conditions: 3 min at 94°C for denaturing, and 26 cycles of 45s at 94°C for denaturing, 60s at 53°C for annealing and 90s at 72°C for elongation followed by a final period of 10 min at 72°C and kept at 4°C. PCR products were purified with 20 μL of Agencourt AMPure XP (Beckman Coulter) magnetic beads and eluted in 52.5 μl Tris buffer (10mM, pH 8.5). The second PCR was carried by adding 4μL of the purified product to a mixture with 9.6μL of water, 20μL of 2X Ready Mix and 3.2μL of each Illumina index primer (2.5pmol/μL), which was submitted to the following PCR conditions: 3 min at 94°C, and 7 cycles of 45s at 94°C, 60s at 50°C and 90s at 72°C and a final period of 10 min at 72°C and kept at 4°C. A second purification was performed by using 40μL of AMPure beads and eluting samples with 32μL of Tris buffer (10mM, pH 8.5). Sequencing was performed with an Illumina MiSeq platform with the V2 reagents kit for 250 cycles from each end at the Genomics Facility of the University of Guelph.
Data were made publicly available at the NCBI Sequence Read Archive under accession number SRP096720.
Statistical analysis
Bioinformatic analysis was performed using the software mothur (v.1.36.1) following the standard operational protocol recommended by Kozich et al. [23]. In short, after data clean up sequences were assigned into phylotypes at the genus level (94% similarity) with taxonomic classification obtained from the Ribosomal Database Classifier (March 2012) [24].
Relative abundances of the main phyla and genera (abundance >1%) and the Firmicutes:Bacteroidetes (F:B) and Firmicutes:Proteobacteria ratios (F:P) found in each treatment were represented by column charts. The effect of treatment on each variable was investigated using the ANOVA on ranks with the Kruskal-Wallis non-parametric test and the Tukey test for multiple comparison correction considering a P<0.010 as statistically significant to decrease false discovery rate. The ANOVA with Tukey’s test (considering a P<0.05 as significant) was used to investigate interactions between treatments and performance indexes.
In order to decrease bias caused by non-uniform sequence numbers, a subsample from the main dataset was used for alpha and beta diversity analyses and the Good´s coverage bootstrap was calculated in order to ensure that the cutoff adopted was representative of original samples. Richness was estimated by the Chao index and by the number of observed genera. The Shannon and the Simpson’s indices were used to estimate diversity. The effect of treatment on those variables was investigated using the ANOVA on ranks with the Kruskal-Wallis non-parametric test.
Community membership (that considers the different genera present in each sample) and structure (that considers the different genera and their evenness in each sample) were addressed respectively by the classic Jaccard and by the Yue and Clayton index [25]. The similarity between community membership and structure present in each sample was represented by dendrograms visualized with FigTree (v1.4.2) (http://tree.bio.ed.ac.uk/software/figtree/), and by the Principal Coordinate Analysis (PCoA) with two dimensions. The Parsimony test and the analysis of molecular variance (AMOVA) were used for statistical comparison of communities’ membership and structure between the groups, and the Benjamini-Hochberg for multiple comparisons adjustment using a false discovery rate of 0.20.
The “indicator analysis” implemented in mothur [26] was used with a cutoff of 0.01 in order to identify the most representative bacterial taxa present within each group. In addition, the linear discriminant analysis (LDA) Effective Size (LefSe) was used to detect meaningful biological differences between treatments [27]. P values <0.05 and logarithmic LDA scores higher than 2.0 were considered as significant.
Results
A total of 5,695,309 good quality reads were used for the analysis, from which 15,510 reads per sample were randomly subsampled to normalize sequence numbers. The subsampling yielded coverage of 99.9%, indicating that it was representative of the total population.
Alpha diversity
Average and standard deviation of alpha diversity indices found in each group are presented in Table 1. The statistical analysis revealed that only enramycin was associated with decreased richness estimated by the number of observed genera (P = 0.007) and by the Chao index (P = 0.031) compared to controls. There was no statistical difference between the other treatment groups regarding the number of observed genera, estimated richness (Chao), or diversity (Simpson and Shannon indices).
Table 1. Average and standard deviation (in brackets) of the number of different genera and results of Chao, Simpson and Shannon indexes present in the cecum of broiler chicken after treatment with different antibiotic growth promoters.
| # genera | Chao | Simpson | Shannon | |
|---|---|---|---|---|
| Avilamycin | 92.94 (5.32) | 108.84 (6.88) | 10.34 (1.55) | 2.88 (0.11) |
| Bacitracin | 93.67 (2.91) | 109.67 (4.92) | 10.05 (2.17) | 2.84 (0.16) |
| Halquinol | 94.04 (8.78) | 108.31 (11.86) | 9.92 (2.28) | 2.80 (0.19) |
| Enramycin | 87.48 (4.32) | 102.11 (7.85) | 10.25 (2.11) | 2.83 (0.16) |
| Virginiamycin | 91.73 (4.09) | 107.62 (5.32) | 9.25 (1.80) | 2.79 (0.16) |
| Control | 93.13 (7.04) | 110.69 (8.77) | 9.89 (2.21) | 2.79 (0.19) |
Beta diversity
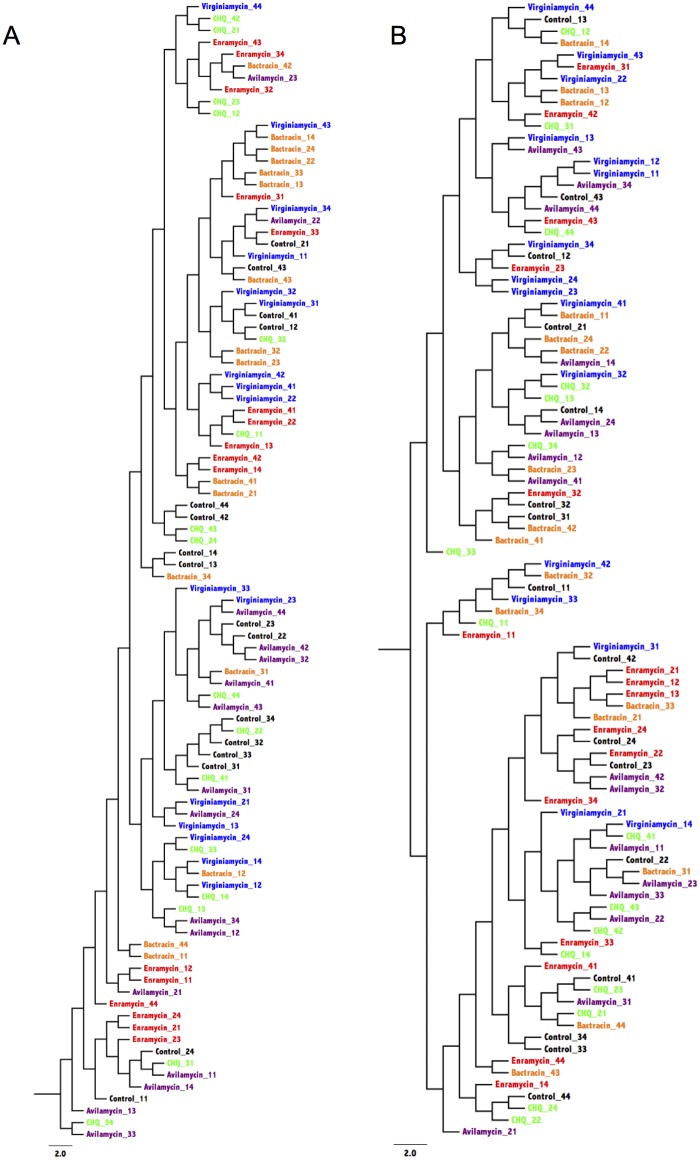
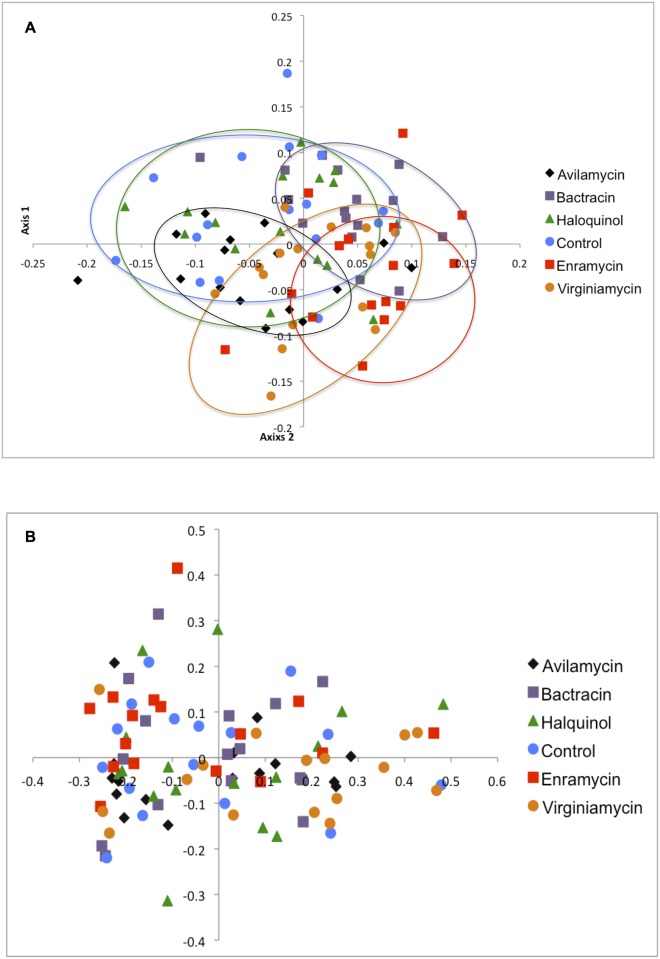
The similarity between bacterial communities present in each sample is represented by dendrograms in Fig 1 and Principal Coordinate Analysis (PCoA) in Fig 2. Fig 2A represents microbial membership present in each sample and is indicative that each antimicrobial drug tended to have a specific effect on the selection of cecal species, evidenced by the formation of clusters according to the different treatments. Noteworthy is the overlapping between controls and animals treated with halquinol, indicating a limited apparent impact of this drug on the microbiota. The lack of action on microbial community structure is evident from Fig 2B, with overlapping of samples from the different treatments.
Fig 1. Dendrograms representing the similarity between membership (A) and structure (B) of bacterial communities found in cecum of broiler chicken treated with zinc bacitracin (orange), enramycin (red); halquinol (green); virginiamycin (blue), avilamycin (purple) and in a control group (black).
CQH: Halquinol.
Fig 2. Principal Coordinate Analysis (PCoA) representing the similarity between membership (A) and structure (B) of bacterial communities found in cecum of broiler chicken treated with several antibiotic growth promoters.
The effect of antimicrobials on microbial membership is further evidenced by Parsimony and AMOVA results (Table 2). Halquinol was the only drug that did not affect microbial membership compared to the control group. Interestingly, treatment with antimicrobials did not affect microbial structure in the cecum of any of the studied groups. This, along with the changes caused in membership, suggests that those drugs had a greater impact on rare species present in that environment.
Table 2. P values obtained from the comparison of microbial membership (gray background) and structure (white background) using the Parsimony and AMOVA tests.
| Parsimony | ||||||
| Avilamycin | Bacitracin | Halq | Enramycin | Virginiamycin | Control | |
| Avilamycin | 0.380 | 0.625 | 0.012 | 0.356 | 0.365 | |
| Bacitracin | 0.004 | 0.340 | 0.358 | 0.357 | 0.967 | |
| Halquinol | 0.356 | 0.056 | 0.362 | 0.366 | 0.852 | |
| Enramycin | 0.003 | 0.014 | 0.010 | 0.151 | 0.964 | |
| Virginiamycin | 0.045 | 0.050 | 0.351 | 0.006 | 0.628 | |
| Control | 0.041 | 0.046 | 0.134 | 0.007 | 0.050 | |
| AMOVA | ||||||
| Avilamycin | Bacitracin | Halq | Enramycin | Virginiamycin | Control | |
| Avilamycin | 0.231 | 0.551 | 0.098 | 0.058 | 0.724 | |
| Bacitracin | <0.001 | 0.573 | 0.388 | 0.042 | 0.739 | |
| Halquinol | <0.001 | <0.001 | 0.147 | 0.142 | 0.809 | |
| Enramycin | <0.001 | <0.001 | <0.001 | 0.018 | 0.212 | |
| Virginiamycin | <0.001 | <0.001 | 0.002 | <0.001 | 0.065 | |
| Control | <0.001 | <0.001 | 0.142 | <0.001 | <0.001 | |
Statistically significant results after the Benjamini-Hochberg adjustment are in bold.
Halq: Halquinol
Relative abundances
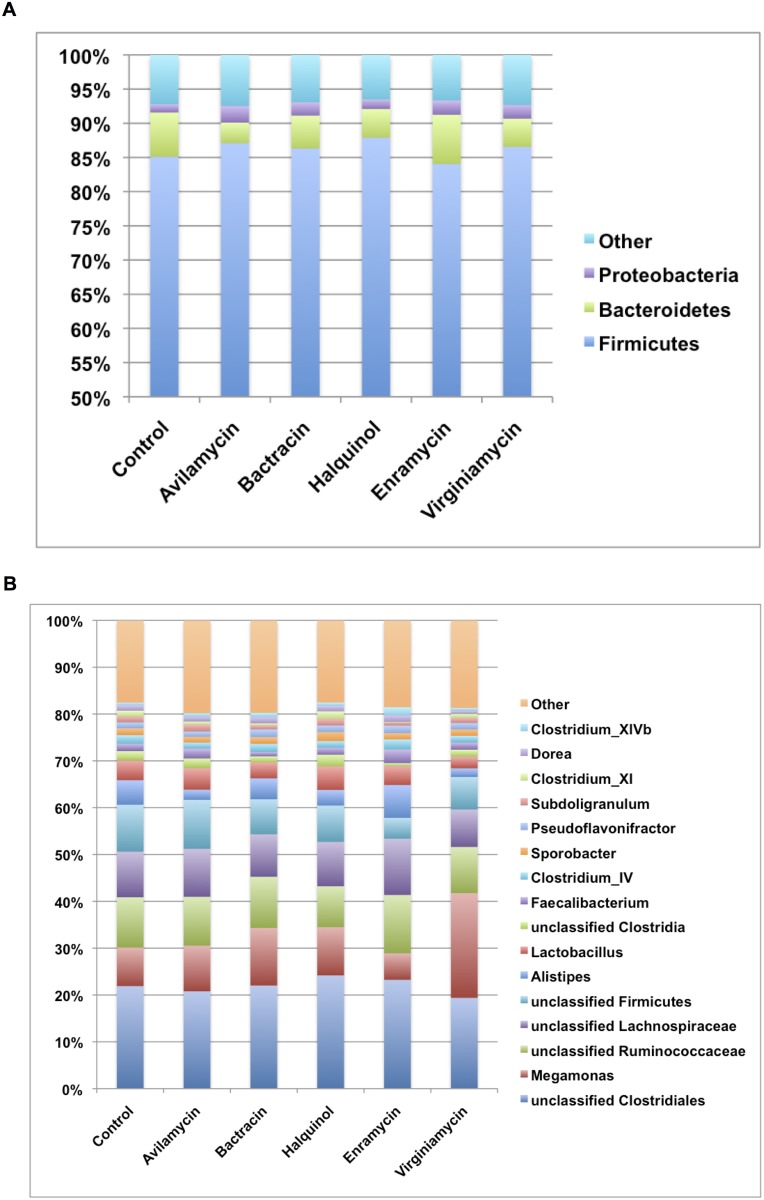
Relative abundances of the main phyla and genera are presented in Fig 3 and S1 Fig. Firmicutes comprised the major bacterial phylum present in the cecum of all groups. Along with Bacteroidetes and Proteobacteria, these three phyla accounted for more than 90% of all sequences. There was no statistical difference in the relative abundances between treated animals and the control group (all P>0.05).
Fig 3. Relative abundances at the phylum (A) and genus (B) level of the main bacteria found in the cecum of broiler chicken treated with zinc bacitracin, enramycin; halquinol; virginamycin, avilamycin and in a control group.
Compared to the control group, animals treated with enramycin had statistically lower relative abundance of unclassified Firmicutes, Clostridium XI, unclassified Peptostreptococcaceae (all P<0.001) and greater abundance of Clostridium XIVb (P = 0.004) and Anaerosporobacter (P = 0.015). In addition, animals treated with bacitracin had greater abundance of Bilophila (P = 0.004) compared to controls.
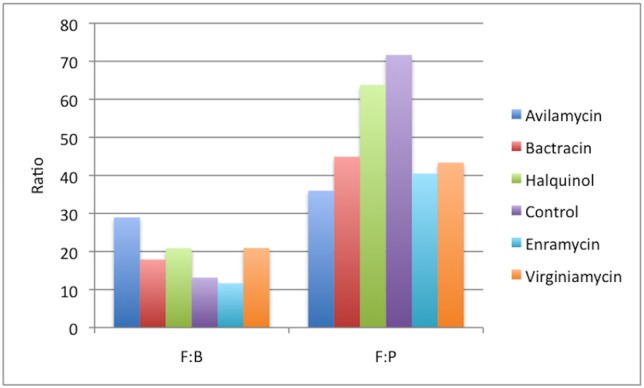
The Firmicutes:Bacteroidetes and Firmicutes:Proteobacteria ratios were calculated within each treatment group and are presented in Fig 4. Firmicutes:Bacteroidetes ratio did not differ between groups (all P>0.05), but all groups treated with AGPs had numerically lower Firmicutes:Proteobacteria ratios compared to controls.
Fig 4. Firmicutes to Bacteroidetes (F:B) and Firmicutes to Proteobacteria (F:P) ratios present in the cecum of broiler chicken treated with different antibiotic growth promoters.
Specific taxa associated with each treatment
The taxa considered more likely to be representative of each group (P>0.001) as per the indicator analysis are presented in Table 3. No significantly discriminative features could be identified using the LefSe analysis.
Table 3. Bacterial taxa found to be significantly associated with as representative of each treatment as per the indicator analysis.
| Treatment | Taxa |
|---|---|
| Avilamycin | Unclassified Sutterellaceae |
| Bacitracin | Faecalibacterium, Anaerofilum, Hydrogenoanaerobacterium |
| Halquinol | Turicibacte, Clostridium_XI, unclassified Firmicutes |
| Enramycin | Clostridium_XlVb, Anaerosporobacter, Planococcaceae, Holdemania, Eubacterium |
| Virginiamycin | Meniscus, Lachnospira, Megamonas |
| Control | Cronobacter, unclassified Hyphomonadaceae |
Table 4 contains data from the performance indexes calculated for each treatment. avilamycin was the only drug associated with higher weight gain and feed conversion.
Table 4. Weight gain (in grams), feed conversion and viability (in percentage) observed in chickens treated with different antibiotic growth promoters.
| Treatment | Weight gain (g)* | Feed conversion* | Viability (%) |
|---|---|---|---|
| Control | 2915 ab | 1,70 a | 98,07 |
| Avilamycin | 3066 a | 1,61 b | 97,11 |
| Bacitracin | 2841 ab | 1,70 a | 97,11 |
| Halquinol | 2668 b | 1,75 a | 93,27 |
| Enramycin | 2707 b | 1,71 a | 89,42 |
| Virginiamycin | 2723 b | 1,74 a | 95,19 |
* different letters in the same column represent significant difference between treatments.
Discussion
None of the antibiotics selected for this study caused significant changes in cecal community structure (related to the genera comprising a community and how they are distributed) compared to the control group, but did affect microbial membership (the different genera present in the cecum). Thus, while there were significant changes in the specific bacterial members that were present (Jaccard index), the lack of an overall impact on the microbial structure (Yue and Clayton index) would suggest that changes affected rare members of the microbiota. This could be related to the fact that most evident changes would be expected to happen at earlier ages and in the most proximal compartments of the intestinal tract [18]. Nevertheless, these findings are particularly important for a better understanding of how AGPs affect cecal bacterial populations and might be used in the future for microbiota manipulation.
Although microbial membership was significantly different from controls regardless the statistical test applied (Table 2), clustering of samples according to the different treatments was not as strong as it has been shown with the use of therapeutic doses [28]. Yet, some clustering can still be noticed in the principal coordinate analysis (Fig 2A).
Halquinol was the only drug that did not alter significantly the membership of the cecal microbiota in any detectable way. The similarity between animals treated with the drug and controls is evidenced by the overlapping of samples observed in Fig 2A. However, at this point, it is not clear whether this “lack of action” is desirable or not. Guasti [26] reported lower weight gain in chicken receiving halquinol compared to groups that also received avilamycin and a prebiotic, which could indicate that this drug may not adequately select species associated with better efficiency for harvesting feed energy. In agreement with those results, weight gain from animals treated with halquinol in the present study did not differ from the control group, but further studies are necessary before solid conclusions can be made.
The group treated with avilamycin was the only to have statistically greater weight gain and better feed conversion compared to controls. Although no genera of biological significance could be identified using the LefSe analysis, but an unclassified Sutterellaceae was associated with the use of this drug pointed by the indicator analysis and may be of importance for the development of new alternative approaches to decrease the use of antimicrobials for growth promotion, such as probiotics [3,29].
Enramycin was associated with decreased diversity and greatly affected the relative abundance of several genera compared to controls. Pedroso et al. [30] also reported changes in composition of the intestinal microbiota of chicken treated with this drug, but the characterization of those changes was not possible since molecular fingerprinting was used in that study.
Fasina et al. [3] showed that treatment with bacitracin suppressed experimental infection with C. perfirngens, since treated animals presented lower abundance of the Clostridiaceae family. This drug has also been reported to deplete members of the Lactobacillaceae family [3,20,21], but the same trend could not be observed in the present study.
Virginiamycin was associated with enrichment of Faecalibacterium and Lactobacillus spp. and with decreased diversity in the cecal microbiota [21]. Also, the use of this drug along with monensin was reported to cause depletion of Firmicutes compared to a control group [31]. However, those findings could not be confirmed by our results and that might be related to differences in diet, geographical location and methodological analyses.
Greater Firmicutes:Bacteroidetes ratios have been associated with bacterial profiles with higher capacity of energy harvesting [7,32,33]. Firmicutes are also reported to be the main phylum in commercial broilers, while free range chicken seems to present with increased Bacteroidetes and Proteobacteria [34]. Interestingly, the group with the highest Firmicutes:Bacteroidetes ratio in this study was the only one to gain statistically more weight. However, differences in ratios between groups were not significant, and further investigations are required before any assumption can be made.
Firmicutes, along with Bacteroidetes and Proteobacteria comprised the major bacterial phyla present in the cecum of all groups. Despite selection bias towards Verrucomicrobia detection usually observed with sequencing of the V4 region of the 16S gene, this phylum was not amongst the most abundant in this study, which further supports the findings of other researchers [35,36]. While results of this study is in agreement with other reports that showed Clostridium, Lactobacillus and Ruminococcus spp. as the most abundant genera in the chicken cecum [31,37], the high abundance of Megamonas spp. was not expected. This genus has been recently classified and is considered a commensal organism present in the intestinal tract of mammals and birds [38].
Since the 1940’s low doses of antimicrobials have been given to food animals to increase weight gain, but the mechanisms of how this drugs favor the increase in productivity are still under investigation. The role that the intestinal microbiota plays in this process has been demonstrated [4] and further studies should focus on gut microbiota manipulation in order to improve productivity and animal health.
The results of this study add new insights to the current knowledge on how AGPs affect the cecal microbiota of chicken.
Conclusions
The use of several antimicrobials at growth promotion doses caused significant changes in the cecal microbial membership of broiler chicken, but not in microbial structure, suggesting that those drugs have a stronger impact on the rare species of bacteria present in that environment.
Supporting information
(DOCX)
(TIFF)
Data Availability
Data were made publicly available at the NCBI Sequence Read Archive under accession number SUB1906187.
Funding Statement
This work was funded by Coordenacao de Aperfeicoamento de Pessoal de Nivel Superior (CAPES), Brasil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Castanon JIR. History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci. 2007;86: 2466–2471. 10.3382/ps.2007-00249 [DOI] [PubMed] [Google Scholar]
- 2.Stutz MW, Lawton GC. Effects of diet and antimicrobials on growth, feed efficiency, intestinal Clostridium perfringens, and ileal weight of broiler chicks. Poult Sci. 1984;63: 2036–2042. [DOI] [PubMed] [Google Scholar]
- 3.Fasina YO, Newman MM, Stough JM, Liles MR. Effect of Clostridium perfringens infection and antibiotic administration on microbiota in the small intestine of broiler chickens. Poult Sci. 2015. [DOI] [PubMed] [Google Scholar]
- 4.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158: 705–721. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han GG, Kim EB, Lee J, Lee J-Y, Jin G, Park J, et al. Relationship between the microbiota in different sections of the gastrointestinal tract, and the body weight of broiler chickens. Springerplus. 2016;5: 911 10.1186/s40064-016-2604-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latorre JD, Hernandez-Velasco X, Wolfenden RE, Vicente JL, Wolfenden AD, Menconi A, et al. Evaluation and Selection of Bacillus Species Based on Enzyme Production, Antimicrobial Activity, and Biofilm Synthesis as Direct-Fed Microbial Candidates for Poultry. Front Vet Sci. 2016;3: 95 10.3389/fvets.2016.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh KM, Shah T, Deshpande S, Jakhesara SJ, Koringa PG, Rank DN, et al. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep. 2012;39: 10595–10602. 10.1007/s11033-012-1947-7 [DOI] [PubMed] [Google Scholar]
- 8.Maron DF, Smith TJS, Nachman KE. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Global Health. 2013;9: 48 10.1186/1744-8603-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller M. The White House Forum on Antibiotic Stewardship impacts labs across the U.S. MLO Med Lab Obs. 2015;47: 28. [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444: 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 11.Yegani M, Korver DR. Factors affecting intestinal health in poultry. Poult Sci. 2008;87: 2052–2063. 10.3382/ps.2008-00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa MC, Stampfli HR, Arroyo LG, Allen-Vercoe E, Gomes RG, Weese JS. Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Veterinary Research. 2015;11: 19 10.1186/s12917-015-0335-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson JA, Burkholder KM. Application of prebiotics and probiotics in poultry production. Poult Sci. 2003;82: 627–631. [DOI] [PubMed] [Google Scholar]
- 14.Rehman HU, Vahjen W, Awad WA, Zentek J. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch Anim Nutr. 2007;61: 319–335. 10.1080/17450390701556817 [DOI] [PubMed] [Google Scholar]
- 15.Sergeant MJ, Constantinidou C, Cogan TA, Bedford MR, Penn CW, Pallen MJ. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE. 2014;9: e91941 10.1371/journal.pone.0091941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley D, Denman SE, Hughes RJ, Geier MS, Crowley TM, Chen H, et al. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl Microbiol Biotechnol. 2012;96: 1361–1369. 10.1007/s00253-011-3847-5 [DOI] [PubMed] [Google Scholar]
- 17.Pourabedin M, Guan L, Zhao X. Xylo-oligosaccharides and virginiamycin differentially modulate gut microbial composition in chickens. Microbiome. 2015;3: 15 10.1186/s40168-015-0079-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumonceaux TJ, Hill JE, Hemmingsen SM, Van Kessel AG. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Applied and Environmental Microbiology. 2006;72: 2815–2823. 10.1128/AEM.72.4.2815-2823.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Hofacre C, Smith F, Lee MD. Effects of feed additives on the development on the ileal bacterial community of the broiler chicken. Animal. 2008;2: 669–676. 10.1017/S1751731108001894 [DOI] [PubMed] [Google Scholar]
- 20.Wise MG, Siragusa GR. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. Journal of Applied Microbiology. 2007;102: 1138–1149. 10.1111/j.1365-2672.2006.03153.x [DOI] [PubMed] [Google Scholar]
- 21.Neumann AP, Suen G. Differences in major bacterial populations in the intestines of mature broilers after feeding virginiamycin or bacitracin methylene disalicylate. Journal of Applied Microbiology. 2015. [DOI] [PubMed] [Google Scholar]
- 22.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41: e1–e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Applied and Environmental Microbiology. 2013;79: 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Research. 2014;42: D633–42. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa MC, Arroyo LG, Allen-Vercoe E, Stampfli HR, Kim PT, Sturgeon A, et al. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS ONE. 2012;7: e41484 10.1371/journal.pone.0041484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dufrene M, Legendre P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr 1997;67: 345–66. [Google Scholar]
- 27.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12: R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Videnska P, Faldynova M, Juricova H, Babak V, Sisak F, Havlickova H, et al. Chicken faecal microbiota and disturbances induced by single or repeated therapy with tetracycline and streptomycin. BMC Veterinary Research. 2013;9: 30 10.1186/1746-6148-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, et al. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Applied and Environmental Microbiology. 2011;77: 5868–5878. 10.1128/AEM.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedroso AA, Menten JFM, Lambais MR, Racanicci AMC, Longo FA, Sorbara JOB. Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult Sci. 2006;85: 747–752. [DOI] [PubMed] [Google Scholar]
- 31.Danzeisen JL, Kim HB, Isaacson RE, Tu ZJ, Johnson TJ. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE. 2011;6: e27949 10.1371/journal.pone.0027949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102: 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE. 2014;9: e85423 10.1371/journal.pone.0085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancabelli L, Ferrario C, Milani C, Mangifesta M, Turroni F, Duranti S, et al. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ Microbiol. 2016. [DOI] [PubMed] [Google Scholar]
- 35.Wei S, Morrison M, Yu Z. Bacterial census of poultry intestinal microbiome. Poult Sci. 2013;92: 671–683. 10.3382/ps.2012-02822 [DOI] [PubMed] [Google Scholar]
- 36.Reed S, Neuman H, Moscovich S, Glahn RP, Koren O, Tako E. Chronic Zinc Deficiency Alters Chick Gut Microbiota Composition and Function. Nutrients. 2015;7: 9768–9784. 10.3390/nu7125497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong J, Si W, Forster RJ, Huang R, Yu H, Yin Y, et al. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol Ecol. 2007;59: 147–157. 10.1111/j.1574-6941.2006.00193.x [DOI] [PubMed] [Google Scholar]
- 38.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I. Important Metabolic Pathways and Biological Processes Expressed by Chicken Cecal Microbiota. Applied and Environmental Microbiology. 2015;82: 1569–1576. 10.1128/AEM.03473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIFF)
Data Availability Statement
Data were made publicly available at the NCBI Sequence Read Archive under accession number SUB1906187.