Abstract
The hypoxia adaptation to high altitudes is of considerable interest in the biological sciences. As a breed with adaptability to highland environments, the Tibetan chicken (Gallus gallus domestics), provides a biological model to search for genetic differences between high and lowland chickens. To address mechanisms of hypoxia adaptability at high altitudes for the Tibetan chicken, we focused on the Endothelial PAS domain protein 1 (EPAS1), a key regulatory factor in hypoxia responses. Detected were polymorphisms of EPAS1 exons in 157 Tibetan chickens from 8 populations and 139 lowland chickens from 7 breeds. We then designed 15 pairs of primers to amplify exon sequences by Sanger sequencing methods. Six single nucleotide polymorphisms (SNPs) were detected, including 2 missense mutations (SNP3 rs316126786 and SNP5 rs740389732) and 4 synonymous mutations (SNP1 rs315040213, SNP4 rs739281102, SNP6 rs739010166, and SNP2 rs14330062). There were negative correlations between altitude and mutant allele frequencies for both SNP6 (rs739010166, r = 0.758, p<0.001) and SNP3 (rs316126786, r = 0.844, P<0.001). We also aligned the EPAS1 protein with ortholog proteins from diverse vertebrates and focused that SNP3 (Y333C) was a conserved site among species. Also, SNP3 (Y333C) occurred in a well-defined protein domain Per-AhR-Arnt-Sim (PAS domain). These results imply that SNP3 (Y333C) is the most likely casual mutation for the high-altitude adaption in Tibetan chicken. These variations of EPAS1 provide new insights into the gene’s function.
Introduction
Low oxygen is one of the more severe environmental challenges for animals living in high-altitude regions such as the Tibetan Plateau, where the average altitude is above 4000 m. The partial pressure of oxygen at 4000 m is approximately 50% of that at sea-level, and the hypoxia imposes severe constraints on aerobic metabolism and leading to high-altitude illness[1, 2][3] and the mechanisms of hypoxic adaptation [4].
The Tibetan chicken (Gallus gallus; TC) is an aboriginal breed found in the Qinghai-Tibetan Plateau (QTP), where the altitude range from 2200 to 4100 m. They are smaller, mature at older ages and lay smaller eggs than breed at lower altitude (Table A in S1 File). Also, TC has higher red blood cells numbers, blood oxygen affinity, and hemoglobin concentrations [5], and loser mean corpuscular volume. Hatchability at the high altitude is greater for TC [6] than that of lowland chickens [7] and they are is less sensitive to pulmonary hypertension syndrome (PHS) and hypocapnia [8, 9]. It is obviously that TC process unique physiological properties and there provide a model for studying genetic mechanisms of hypoxia adaptation.
Endothelial PAS domain protein 1 (EPAS1) (also known as hypoxia inducible factor-2a (HIF-2a)), belongs to a member of the transcription factor family characterized by a basic helix-loop-helix (bHLH) and by a (Per-AhR-Arnt-Sim) PAS domain. Thus, it plays a prominent role in the process of adaptation to hypoxia and in the promotion of livability during hypoxia stress [10, 11]. EPAS1 is a prime regulator during chronic hypoxia and directly regulates key genes such as Erythropoietin (EPO), and vascular endothelial growth factor (VEGF) [12]. Also, EPAS1 has been implicated processes such as erythropoiesis, iron homeostasis, pulmonary hypertension and vascular permeability [13, 14].
Genome-wide scans and whole genome re-sequencing analyses to identify selection signatures of high-altitude adaptation in vertebrates have been performed across a range of species, including humans[15], dogs [16, 17], grey wolves [18], yaks [19], Tibetan antelopes [20], and cattle [21]. In the humans, dogs, and grey wolf convergent evolution for EPAS1 gene has been noted [15–18]. Because of an association between the allele frequency of the SNPs near or in EPAS1 and the altitude. Other studies have focused on the effects of mitochondrial DNA polymorphisms in adaptation of TC to high-altitude. MT-CO3 [22] and ATP-6 [23] are candidate genes of hypoxia adaptation. Genes involved in the Ca2+ signaling pathway might be related to the adaptation of TC to hypoxic conditions at high altitude[24], and genome re-sequencing, also identified unique hypoxia adaptation of TC, especially genes related to development of the cardiovascular and respiratory systems, DNA repairs, responses to radiation, inflammation, and immune responses [24].
In the present study, we hypothesis that the sequence variations in the EPAS1 gene may contribute to the high altitude adaptation in TC. Here, we compared on the polymorphisms of EPAS1 coding sequence in populations that included 157 Tibetan chickens and 139 lowland chickens to determine whether there were genetic differences between high and low altitude chickens in EPAS1 gene.
Materials and methods
Sampling
In total, 157 Tibetan chickens (TC) were collected from 8 different locations in Qinghai, Tibet, Yunnan, and Sichuan provinces. They included 4 Tibet areas (Shigatse (RKZ), Lhoka (SN), Lhasa (LS), and Nyingchi (LZ)), 2 Sichuan areas (Garze (GZ) and Aba (AB)), 1 Yunnan area (Diqing (DQ)), and 1 Qinghai area (Haibei (QH)). Also, 139 lowland chickens (LC) from 7 different Chinese native chicken breeds were sampled (below 1800 m). Details of sample number, breed, altitude, latitude, and longitude are provided in Table 1. A sample of 1 mL blood from each individual was obtained from the brachial vein. No animal was injured in during capture and blood collection. The protocol was approved by the Committee of the Care and Use of Laboratory Animals of the State-level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University (Approval ID: DKY-S20143136).
Table 1. Altitude, longitude and latitude of the sampling locations for the 8 Tibetan and 7 Lowland chicken populations.
| Breed | Sampling location | Population | Sample Size | Altitude (m) | Longitude (E) | Latitude (N) |
|---|---|---|---|---|---|---|
| Tibetan | ||||||
| Shigatse,Tibet | Shigatse (RKZ) | 30 | 3900 | 89°60' | 28°92' | |
| Lhoka,Tibet | Lhoka (SN) | 14 | 3700 | 90°03' | 28°27' | |
| Lhasa,Tibet | Lhasa (LS) | 24 | 3650 | 91°01' | 29°26' | |
| Garze,Sichuan | Garze (GZ) | 16 | 3390 | 99°22' | 28°34' | |
| Aba,Sichuan | Aba (AB) | 25 | 3300 | 102°33' | 31°27' | |
| Diqing,Yunnan | Diqing (DQ) | 13 | 3280 | 99°53' | 28°08' | |
| Haibei,Qinghai | Haibei (QH) | 7 | 2700 | 100°86' | 36°85' | |
| Nyingchi,Tibet | Nyingchi (LZ) | 20 | 2400 | 94°26' | 29°25' | |
| Lowland | ||||||
| Emei black | Emei,Sichuan | Emei (EM) | 23 | 1800 | 103°41' | 29°49' |
| Shimian | Yaan,Sichuan | Shimian (SM) | 14 | 1120 | 102°13' | 29°40' |
| Jiuyuan black | Wanyuan,Sichuan | Jiuyuan (JY) | 25 | 900 | 108°21' | 31°84' |
| Pengxian yellow | Yaan,Sichuan | Pengxian (PX) | 12 | 600 | 102°98' | 29°98' |
| Blue-shelled Dongxiang | Yaan,Sichuan | Dongxiang(LK) | 14 | 600 | 102°98' | 29°98' |
| Muchuan black-bone | Muchuan,Sichuan | Muchuan (MC) | 20 | 540 | 103°90' | 29°02' |
| Wenchang | Wenchang,Hainan | Wenchang(WC) | 31 | 10 | 110°87' | 19°72' |
DNA extraction, PCR amplification, and genotyping
DNA was extracted by the phenol-chloroform method [25]. The high altitude DNA pool was constructed from 50 samples, each containing 100 ng DNA from randomly selected TC. The LC pool was constructed in the same way. These two gene pools were used to detect SNPs.
After blasting chicken EPAS1 mRNA sequence (Ensembl accession ENSGALT00000016253) in the chicken genome, we designed 15 primer pairs (Table 2) to amplify the 15 exons using Primer V6.0 software [26]. PCR was performed in 25 μL of reaction volume. The ingredient contain 50 ng DNA template, 1×buffer (including 1500 μmol L-1 Mg2Cl2, 200 μmol L-1 dNTPs, and 1.5 U of Taq DNA polymerase) and 1 μmol L-1 of each primer. Besides, the PCR procedure was as following: initial denaturation for 5 min at 9500, followed by 35 cycles of 95°C for 30 s; annealing at prescribed annealing temperature for 30 s; and primer extension at 72°C for 45 s. The final extension was performed at 72°C for 7 min. PCR products were sequenced on an ABI 3730 DNA sequencer. Then, the SNPs were determined and pairs of 4 primers (P1, P7, P12, and P14) were used for individual genotyping. PCR procedures and systems were the same as above.
Table 2. Primer information for detecting SNPs in coding regions.
| Names | Sequences (5'-3') | Sizes of product (bp) | Annealing temperatures (°C) | Target region | Purpose |
|---|---|---|---|---|---|
| P1 | F1:CAAGGGGTCTGAAGGCTGAT | 534 | 55 | Exon1 | Identification of SNP and individual genotyping |
| R2:TGGGAAAACAAAACAAAAACTG | |||||
| P2 | F:GTTGCCTTGTTATTGGATGTGAT | 473 | 56 | Exon2 | Identification of SNP |
| R:GGGGGAGTTGTCTTTTCTTGAT | |||||
| P3 | F:CCCCCACTAATTGTCTCCTG | 395 | 55 | Exon3 | Identification of SNP |
| R:TCCACCTGTTTTAACTTGATGAA | |||||
| P4 | F:GATGGTCTCTGCAGCTATGTTAC | 422 | 55 | Exon4 | Identification of SNP |
| R:AAGTGTGAGGAGGGCAAGTGT | |||||
| P5 | F:GAGTCGGTTTTTCGTTCACAA | 443 | 55 | Exon5 | Identification of SNP |
| R:AGCAGGGGTACCATTTTCTCTAA | |||||
| P6 | F:ACTGTGGTTTTAATCCCTTTTGTT | 263 | 55 | Exon6 | Identification of SNP |
| R:CTACTGGTTGTGGCTTATTATTTG | |||||
| P7 | F:ATTGGCCACCCTGTTGATG | 579 | 55 | Exon7 | Identification of SNP and individual genotyping |
| R:AGAAGAAAAAGGCTATGGTGTAAT | |||||
| P8 | F:GGAACTGTGCCTTGCCTGTCA | 530 | 57.8 | Exon8 | Identification of SNP |
| R:GCCTGGTTCTGCCCTCATTCA | |||||
| P9 | F:AGGGCAGAACCAGGCTTTTT | 420 | 55 | Exon9 | Identification of SNP |
| R:TGGGATTTGCATAGAGAACATAAC | |||||
| P10 | F:ACTGTAGTTGTCTTGGCTTCTTAT | 438 | 55 | Exon10 | Identification of SNP |
| R:AGGCTGCTGCTTCTATTTGT | |||||
| P11 | F:AGCTGGCCACTTTCCTTTAC | 740 | 55 | Exon11 | Identification of SNP |
| R:AGCCATGCCTGTCTCCTT | |||||
| P12 | F:GTGGGATGACTGACAAAAGGAA | 397 | 55 | Exon12 | Identification of SNP and individual genotyping |
| R:TGAGGTGGGTTAATGACAGATGG | |||||
| P13 | F:GAGTGGGGTAATCATAAGAA | 434 | 55 | Exon13 | Identification of SNP |
| R:ATTTGCCTCCCACATAAG | |||||
| P14 | F:TGTGCAGGCATTTCAGAG | 443 | 55 | Exon14 | Identification of SNP and individual genotyping |
| R:TGCCAACATTTTTCAGC | |||||
| P15 | F:GGTATAAATAGAAGAGGGAAGAGA | 503 | 55 | Exon15 | Identification of SNP |
| R:ATAAAGGGAGGGTAATCAAACT |
1.2 F and R are the abbreviation of forward and reverse primer, respectively.
Data analysis
The sequences were edited and aligned by DNAstar Software (DNAstar Inc. Madison, WI, USA). Sequence variations were identified using MEAG5 [27]. The complete EPAS1 sequence of Red Jungle Fowl (G.g gallus, Ensemble accession number: ENSGALG00000010005) was used as reference for determining the variable sites of chicken. Hardy-Weinberg equilibrium, pairwise linkage disequilibrium (LD), and association analysis were conducted by Haploview software (version 4.2, http://www.broad.mit.edu/mpg/haploview/). We calculated the nucleotide diversity (Pi) and average number of nucleotide differences (K) for the TC and LC populations by DNAspV5 [28]. The Fixation index (Fst) between the high (≥3300m) and low altitude groups (≤600m) were calculated by Arlequin 3.5 [29].
Differences of allele frequencies between TC and LC groups were analyzed using Pearson’ Chi-square test, odds ratio (OR), 95% confidence intervals (95% CLs), and MAF (Minor allele frequencies) were calculated. Correlation coefficients between the altitude and frequencies of mutant alleles were calculated. All these parameters were calculated by SPSS software, Version 22 [30] and P < 0.05 was considered as statistical significance.
Protein secondary structure prediction and variable sites analysis
The protein domains of EPAS1 were predicted by the Pfam web service [31]. The EPAS1 ortholog proteins of 16 vertebrates were retrieved from Ensembl [32]. Multiple sequence alignments were performed by Clustal X (V2.1) [33] and displayed by Jalview (v2.8) [34]. To find the best model for a Maximum Likelihood (ML) based tree, we use Prottest V3.4 to compare all models and chose the ML tree as starting topology. Then we used BEAST V1.8.3 to generate the trees by the best model. To find the best fit model, we use Prottest V3.4 [35] to exam all the models and chose the Maximum likelihood tree as starting topology. Then BEAST 2 [36] were used to generate the trees by the best model. The protein sequence of chicken EPAS1 was searched in Ensembl database (ENSGALP00000016234).
Results
Sequence variations in EPAS1 gene
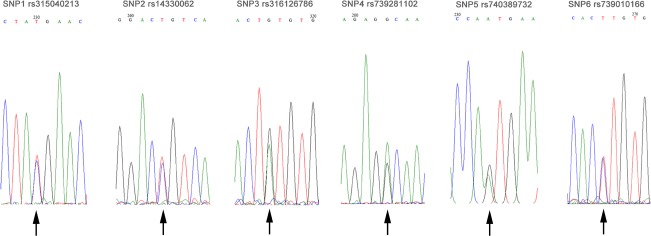
The length of EPAS1 exon sequences was 2730 bp (Ensembl accession numbers ENSGALT00000016253; no insertion/deletions were detected). A total of 6 single-nucleotide polymorphisms (SNPs) were found in the exon of EPAS1 gene from the 15 chicken populations (Table 3 and Fig 1). There were four synonymous mutations (SNP1: rs315040213, SNP2: rs14330062, SNP4: rs739281102, and SNP6: rs739010166) and two non-synonymous substitutions (SNP3: rs316126786 and SNP5: rs740389732).
Table 3. Four synonymous mutations and two non-synonymous mutations in EPAS1.
| SNP locus1 | Genomic location2 | Nucleotide mutations | Amino acid mutations | ||
|---|---|---|---|---|---|
| Reference allele | Mutation allele | Reference | Mutation | ||
| SNP1 rs315040213 | Chr3:25981719 (+) | C | T | Tyr | Tyr |
| SNP2 rs14330062 | Chr3:25976150 (+) | T | C | Thr | Thr |
| SNP3 rs316126786 | Chr3:25976203 (+) | A | G | Tyr | Cys |
| SNP4 rs739281102 | Chr3:25980904 (+) | A | G | Arg | Arg |
| SNP5 rs740389732 | Chr3:25981687 (+) | A | G | Met | Val |
| SNP6 rs739010166 | Chr3:25981719 (+) | C | T | Thr | Thr |
1 SNP loci are exported from dbSNP.
2The genomic location of EPAS1 mutation is based on Ensembl Gallus gallus version 84.4 (galgal4) Chromosome 3.
Fig 1. The sequencing images of 6 SNPs in the coding region of chicken EPAS1 gene.
The position and base substitutions in coding regions refer to the sequence of ensemble ENSGALT00000016253.
SNP genotypes
Genotype frequencies are presented in Table 4 and Fig 2. To detect SNP sites which had putative associations with a high-altitude environment, we focused on genotypes which difference (P < 0.05) between TC and LC. Excluding SNP2 (P = 0.062), SNP4 (P = 0.108, OR = 0.623, 0.408 ≤ 95% CI ≤ 0.952), and SNP5 (P = 0.119, OR = 0.675, 0.437 ≤ 95% CI ≤ 1.045), the other SNPs, which were the SNP1 (P = 0.001, OR = 1.826, 1.246 ≤ 95% CI ≤ 2.676), SNP3 (P = 2.16E-06, OR = 0.257, 0.142 ≤ 95% CI ≤ 0.464), and SNP6 (P = 3.76E-07, OR = 0.212, 95% CI = 0.120–0.374) were different between TC and LC groups. Results of odds ratios shared that the probability of the ability to adapt hypoxia for chickens with a C allele at SNP1 polymorphic site was more than 1.826 times that of chickens with T allele. The odds ratios of allele A at SNP3 and allele C at SNP6 were less 0.257 and 0.212 fold than chickens with G and T alleles, respectively.
Table 4. Genotype of the EPAS1 gene SNPs.
| SNP locus | Genotypes | p value in Pearson Chi-square test | ||
|---|---|---|---|---|
| SNP1 rs315040213 | CC | CT | TT | 0.001 |
| TC1 | 74 (47.10%)3 | 70 (44.50%) | 13 (8.20%) | |
| LC2 | 94 (67.60%) | 36 (25.80%) | 9 (6.40%) | |
| SNP2 rs14330062 | TT | TC | CC | 0.062 |
| TC | 152 (96.80%) | 5 (3.10%) | 0 (0%) | |
| LC | 139 (100%) | 0 (0%) | 0 (0%) | |
| SNP3 rs316126786 | AA | AG | GG | 2.16E-06 |
| TC | 142 (90.40%) | 14 (8.90%) | 1 (0.60%) | |
| LC | 94 (67.60%) | 42 (30.20%) | 3 (2.10%) | |
| SNP4 rs739281102 | AA | AG | GG | 0.108 |
| TC | 119 (75.70%) | 30 (19.10%) | 8 (5%) | |
| LC | 90 (64.70%) | 38 (27.30%) | 11 (7.90%) | |
| SNP5 rs740389732 | AA | AG | GG | 0.119 |
| TC | 122 (77.70%) | 26 (16.50%) | 9 (5.70%) | |
| LC | 94 (67.60%) | 36 (25.80%) | 9 (6.40%) | |
| SNP6 rs739010166 | CC | CT | TT | 3.76E-07 |
| TC | 141 (89.80%) | 15 (9.50%) | 1 (0.60%) | |
| LC | 91 (65.40%) | 37 (26.60%) | 11 (7.90%) | |
1TC and
2LC abbreviations for Tibetan and lowland chickens, respectively.
3Values represent the genotype number and percentage of the population.
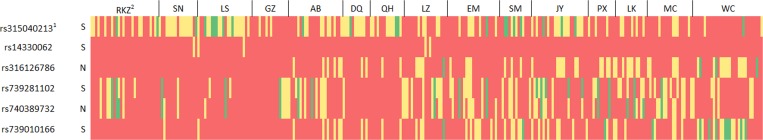
Fig 2. All samples genotypes.
These genotypes were derived from Sanger sequencing of 157 Tibetan and 139 Lowland chickens and were marked as red block (homozygous reference, abbreviated as ref), yellow block (heterozygote, abbreviated as het), and green block (homozygote alternative, abbreviated as alt). N: non-synonymous; S: synonymous.1The mutation sites of EPAS1. 2The abbreviations of sample locations, shown are in Table 1.
Correlation coefficients between altitudes and allele frequencies of the EPAS1 gene
Table 5 contains the observed allelic frequencies of each polymorphic site for each sample from the 15 populations (Allele and genotype frequencies of all SNPs were shown in Tables B, C, and D in S1 File.). We found that SNP2 was unique to TC with the allele frequency of 1.5%. The other five SNPs (SNP1, SNP4 and SNP6, SNP3, and SNP5) were shared by TC and LC. Using Pearson chi-square test and Fisher exact test except the SNP5 (P = 0.077), the other four SNPs whose allele frequencies were significantly different between TC and LC, implying that these four SNPs were associated with high-altitude adaptation. Notably, The non-synonymous substitutions SNP3 (P = 1.94E-06) was significantly different between TC and LC. There were significant negative linear correlations between altitude and the frequency of mutant allele at SNP6 (rs739010166, r = 0.758, p<0.001) and SNP3 (rs316126786, r = 0.844, P<0.001). There was no correlation for the other SNPs.
Table 5. Allele frequency, correlation and odds ratio of mutation loci in the EPAS1 gene.
| SNP sites | Allele distribution | p value in Pearson Chi-square test | OR1 | 95% CI2 | MAF3 | R | Pearson r | P-value8 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele | TC5 | LC6 | ||||||||
| SNP1 rs315040213 | C | 218 (69.4%)7 | 224 (80.5%) | 0.001 | 1.826 | 1.246-~2.676 | 0.25337 | 0.231 | 0.48 | 0.07 |
| T | 96 (30.6%) | 54 (19.4%) | ||||||||
| SNP2 rs14330062 | T | 5 (1.5%) | 0 (0%) | 0.034 | NA4 | NA-NA | 0.0084 | 0.132 | 0.363 | 0.184 |
| C | 309 (98.4%) | 278 (100%) | ||||||||
| SNP3 rs316126786 | A | 298 (94.9%) | 230 (82.7%) | 1.94E-06 | 0.257 | 0.142–0.464 | 0.1081 | 0.712 | 0.844 | 0.000* |
| G | 16 (5%) | 48 (17.2%) | ||||||||
| SNP4 rs739281102 | A | 268 (85.3%) | 218 (78.4%) | 0.028 | 0.623 | 0.408–0.952 | 0.179 | 0.223 | 0.472 | 0.076 |
| G | 46 (14.6%) | 60 (21.5%) | ||||||||
| SNP5 rs740389732 | A | 270 (85.9%) | 224 (80.5%) | 0.077 | 0.675 | 0.437–1.045 | 0.1655 | 0.17 | 0.413 | 0.126 |
| G | 44 (14%) | 54 (19.4%) | ||||||||
| SNP6 rs739010166 | C | 297 (94.5%) | 219 (78.7%) | 9.53E-09 | 0.212 | 0.120–0.374 | 0.1283 | 0.575 | 0.758 | 0.001* |
| T | 17 (5.4%) | 59 (21.2%) | ||||||||
1OR represents odds ratio
2Cl represents confidence interval
3MAF represents the minor allele frequency.
4 NA means cannot be calculated.
5 TC abbreviations for Tibetan chicken.
6 LC abbreviations for Lowland chicken.
7 Values represent the allele frequency and percentage in the population.
8P-value was calculated by Pearson r.
*represent significant level P<0.05.
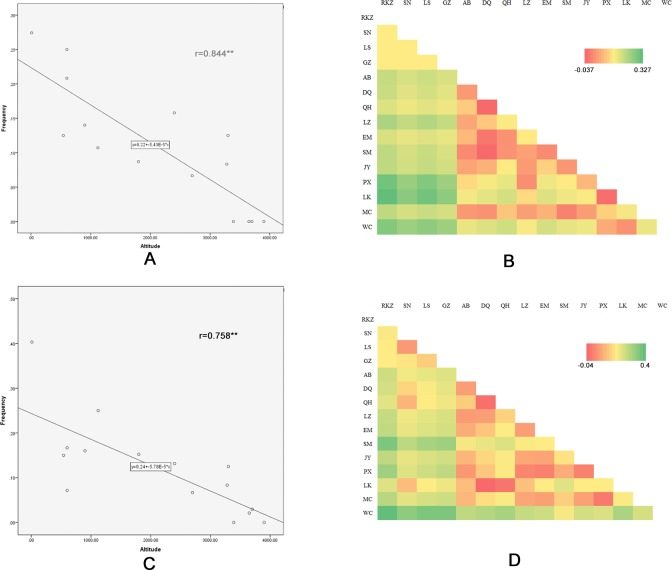
There was a striking change of allele frequency along with the altitude elevation in SNP3. As shown in Fig 3A, the high mutant allele frequencies of SNP3 (Y333C) in the populations WC (27.4%, n = 31, 10 m), LK (25%, n = 14, 600 m), and PX (20.8%, n = 12, 600 m) were found. Then, the frequencies declined in the populations JY (14%, n = 25, 900 m), SM (10.7%, n = 14, 1120 m), and EM (8.7%, n = 23, 1800 m). And they dropped the populations QH (6.7%, n = 15, 2700 m) and DQ (8.3%, n = 12, 3280 m). Moreover, when the altitude was more than 3,400 m, the mutation essentially disappeared, as shown in the populations GZ (0%, n = 16, 3390 m), LS (0%, n = 24, 3650 m), SN (0%, n = 14, 3700 m), and RKZ (0%, n = 30, 3900 m). The same tendency was found in SNP6 (Fig 3C).
Fig 3. The correlation analysis between frequencies and the altitude for SNP3 and SNP6.
(A) Plot of the correlation analysis between frequencies of the SNP3 and the altitudes among fifteen population, “r” represents the correlation coefficient (r = 0.844, P<0.001). (B) The Fst of 15 different populations from TC and LC were calculated based on SNP3 of EPAS1 gene. (C) Plot of the correlation analysis between frequencies of the SNP6 and the altitudes among fifteen chicken population, “r” represents the correlation coefficient (r = 0.578, P = 0.001). (D) The Fst of 15 different populations from TC and LC were calculated based on SNP6 of EPAS1 gene.
These results suggested that the frequencies of SNP3 and SNP6 in EPAS1 decreased with the elevation of altitude.
The Hardy-Weinberg equilibrium, nucleotide diversity, and population differentiation of the EPAS1
To test whether all of the SNPs at EPAS1 loci were in equilibrium in each population, the Hardy-Weinberg equilibrium (HWE) tests was performed on each SNP locus (Table 6). Except for SNP4 and SNP5, all alleles in TC were in accordance with Hardy-Weinberg equilibrium (P>0.05). In contrast, there was a departure from Hardy-Weinberg equilibrium (P < 0.05) for LC in SNP1, SNP4, SNP5, and SNP6. Notably, SNP3 in both populations was in accordance with Hardy-Weinberg equilibrium. Besides, there were no significant diferences between TC and LC for nucleotide diversity (Pi) and the average number of nucleotide differences (K) (Table E in S1 File). Then we calculated the LD block of Tibetan chicken and Lowland chicken and we found close linkage between SNP3 and SNP6 in TC (Figure A in S1 File). Besides, the observed heterozygosity of all SNPs was from 0.03 to 0.44 in TC and from 0 to 0.30 in LC (Table 6). The minor allele frequencies (MAF) of all mutations were more than 0.005 (Table 5).
Table 6. The Hardy-Weinberg equilibrium (HWE) tests of the candidate SNPs in EPAS1 in TC and the LC.
| SNP locus | TC4 | LC5 | Fst | P-value3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | Pearson's p | Ho1 | He2 | χ2 | Pearson's p | Ho | He | |||
| SNP1 rs315040213 | 0.396 | 0.528 | 0.44586 | 0.42588 | 4.141 | 0.0418* | 0.25899 | 0.31416 | 0.18057 | <0.000** |
| SNP2 rs14330062 | 0.001 | 0.839 | 0.03185 | 0.03144 | 0 | 1 | 0 | 0 | 0.01252 | 0.16031 |
| SNP3 rs316126786 | 0.955 | 0.328 | 0.08917 | 0.09703 | 0.461 | 0.497 | 0.30216 | 0.28673 | 0.22956 | <0.000** |
| SNP4 rs739281102 | 8.735 | 0.0031** | 0.19108 | 0.25087 | 5.143 | 0.023* | 0.27338 | 0.33971 | 0.0224 | 0.07038 |
| SNP5 rs740389732 | 15.36 | 0.00090** | 0.16561 | 0.24175 | 4.141 | 0.041* | 0.25899 | 0.31416 | 0.02805 | 0.02933 |
| SNP6 rs739010166 | 0.707 | 0.400 | 0.09554 | 0.10275 | 5.78 | 0.016* | 0.26619 | 0.33558 | 0.27826 | <0.000** |
*Represent significant P-value of less than 0.05.
** represent significant P-value of less than 0.001.
1.2 Ho and He represent the observed heterozygosity and expected heterozygosity.
3 The P-value is calculated by Fst value.
4 TC abbreviations for Tibetan chickens.
5 LC abbreviations for Lowland chickens.
To fully understand the population differentiation that occurred between TC and LC, we analyzed the distribution of alleles at the EPAS1 for the high altitude (≥3300m) and low altitude groups (≤600m). The Fixation index (Fst) was calculated to evaluate the population differentiation degree. As shown in Table 6, the Fst values for SNP1 (0.181, P < 0.001), SNP3 (0.229, P < 0.001), and SNP6 (0.278, P < 0.001) for two groups reflected remarkable population differentiation.
Protein secondary structure prediction and non-synonymous sites analysis
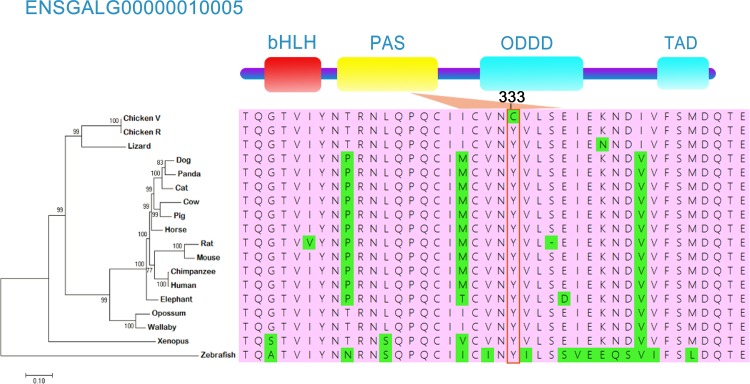
SNP3 (Y333C) was located in a well-defined protein domain (PAS domain, Fig 4). Using the crystal structure model of PAS domain for human EPAS1, we found that mutation SNP3 in chicken (Y333C) occurred in a beta sheet which may affect the thermodynamic stability of the domain. To evaluate the its functional, we aligned the mutant EPAS1 protein with its ortholog proteins of diverse vertebrates (Fig 4) and found that the amino acid mutation (Y333C) were conserved among species. By contrast, M765V was not found in other species. Because mutant functional effects showed that Y333C was deleterious, while M765V were tolerated implied that Y333C was the most likely casual mutation for the EPAS1 in LC but not TC.
Fig 4. EPAS1 mutation in coding regions.
Structural and evolutionary analysis of the 4 amino acid variants. The protein coordinate is based on Ensembl ID ENSGALP00000016234. The upper layer displayed the Pfam domains of the protein. The orthologous proteins from 17 different vertebrates were aligned with the residues shown as green. Cow, ENSBTAP00000004836; pig, ENSSSCP00000009011; human, ENSP00000406137; dog, ENSCAFP00000003819; elephant, ENSLAFP00000010336; mouse, ENSMUSP00000024954; opossum, ENSMODP00000001136; lizard, ENSACAP00000004025; xenopus, ENSXETP00000031612; zebrafish, ENSDARP00000074832; wallaby, ENSMEUT00000004049; horse, ENSECAT00000015683; cat, ENSFCAT00000013633; rat, ENSRNOT00000034991; panda, ENSAMET00000006660; chimpanzee, ENSPTRT00000022124. The ML tree is shown in the left area. (PAS) Per-Arnt-Sim; (HIF) hypoxia-inducible factor; (ODDD) O2-dependent degradation domain; (TAD) C-terminal transactivation domain.
Discussion
Long-term exposure to hypoxia reduced metabolic activity, retarded development, and increased embryonic mortality in animal fetes [37] and chronic hypoxia decreased growth of developing chick embryos [38, 39]. The Tibetan chicken, a Chinese native chicken breed, adapts itself well to the low pressure and high altitude plateau environment [7, 40]. Here, we investigated specific SNPs in EPAS1 gene and discuss their possible contributions to the high-altitude adaptation of the Tibetan chicken.
Amino acid replacement at key sites can difference protein function [41]. EPAS1 protein has an important role in regulating transcriptional responses to hypoxia in physiological and pathologic conditions including up-regulating the expression of erythropoietin (EPO) [42], Thus, it is not surprising that Mutations at EPAS1 are associated with hematologic phenotypes [43].
Here, we found 4 synonymous mutations and 2 non-synonymous substitutions which resulted in amino acid mutations of Tyr to Cys at SNP3 and Met to Val at SNP5, respectively. Except for SNP5, the other 5 SNPs were significantly different between TC and LC for genotypic frequencies. Both genotypic and allelic results for SNP1, SNP3, and SNP6 revealed significant differences between two chicken groups. Except for SNP4 and SNP5, all the alleles in the TC were in accordance with Hardy-Weinberg equilibrium. While for LC there was a significant departure from Hardy-Weinberg equilibrium (P < 0.05) in SNP1, SNP4, SNP5, and SNP6. These results implied an association of SNP1 and SNP6 might be correlate with high-altitude adaptation. Also there is linkage between SNP3 and SNP6 in TC (Figure A in S1 File). The Fst values for SNP1, SNP3, and SNP6 of EPAS1 between the high altitude group (HIC≥3300m) and low altitude group (LOC ≤ 600m) reflected remarkable population differentiation. Overall, both the Fst value and the LD block demonstrate a remarkable population differentiation between the TC and LC. We deduce reasons why the previous study[24] did not find SNP3 are that compared to other SNPs, SNP3 may have low frequency in the TC population and sample size, only 40 Tibetan chickens from 3 different areas were involved.
A correlation coefficient is a number that quantifies some type of association between random variables and observed data [44]. Pearson correlation between altitude and allele frequency showed that in TC there was a significant relationship in synonymous mutation SNP6 and non-synonymous mutation SNP3. The frequencies of SNP3 (Y333C) and SNP6 were high in LC but decreased along with the elevation of altitude. The same relationship also found for the EPAS1 gene in cattle [45]. There was an association of double variants in the oxygen degradation domain (ODDD) of EPAS1 in Angus cattle with High-altitude pulmonary hypertension (HAPH). These mutations were prevalent in lowland cattle, but they disappeared for the animals located at high altitudes[45]. Like Angus cattle, the mutation we found was located in a well-defined domain and quite conserved.
Human EPAS1 is a basic helix-loop-helix transcription factor that contains a Per-AhR-Arnt-Sim (PAS) domain and shares 48% homology [46, 47]. The transcriptional response to hypoxia appears in several physiological and pathologic conditions, including up-regulation in the expression of erythropoietin (EPO) [48]. Mutations at EPAS1 in humans are associated with hematologic phenotypes [43]. HIF proteins are constantly degraded with sufficient oxygen availability, but one released under hypoxia condition. HIFs are protected by inhibition of oxygen-dependent hydroxylation of specific residues in the oxygen-dependent degradation domain under hypoxic condition. This prevents interactions with the von Hippel-Lindau ubiquitin ligase complex and proteasome destruction. HIF2a is found in all human tissues and its effects on downstream cascades include regulation of angioenic factors VEGF and TGFa, and cell permeability and stimulation of erythropoietic and glycolytic protein [46, 49, 50]. Clinical studies found that several gain-of-function mutations that occurred in the primary hydroxylation site (Pro531) at EPAS1 caused erythroctosis and pulmonary hypertension [51].While, the unique EPAS1 mutation in the Tibetan people were associated with lower hemoglobin concentrations [52, 53], suggesting that it played a loss-of-function role to high blood viscosity and cardiovascular disorders. As erythrocytosis is a common symptom of chronic mountain sickness which can lead to high blood viscosity and cardiovascular disorders, a decrease of hemoglobin level may provide a protective mechanism for Tibetan people [43]. Nonetheless, it is worth noting that all EPAS1 variations detected preciously in the Tibetan population are in introns [52, 53].
Transgenic mice carrying a G536W variant in EPAS1 developed high right ventricular systolic pressure and medial hypertrophy of pulmonary arteries in addition to erythrocytosis under normoxic condition [54]. The key amino acid mutation G305S was identified at EAPS1 PAS domain in dogs was predicted to be damaging, which is also likely to cause the loss of function of EPAS1 and it is also associated with blood flow resistance [16]. Recently, the missense mutation Q597L was identified in EPAS1 in the Tibetan Cashmere goat, which occurred next to the hypoxia-inducible factor-1 (HIF-1) domain, and was exclusively enriched in the high-altitude population [55]. Thus, HIF2a dysregulation is well recognized in association with pulmonary hypertension, frequency in context of erythrocytosis, with or without severe hypoxemia. Here, we found a non-synonymous mutation SNP3 (Y333C) in PAS domain which is similar to the mutation in dogs and Angus cattle. In order to adapt to the same selection pressure, mutations that produce an adaptive change in one species may preclude possibilities in other species because of differences in genetic background and experiments involving resurrected ancestral proteins have revealed continuous effects of historical substitutions [56]. Although the function of mutation (SNP3 Y333C) in chickens is unknown, based on the previously studies, we inferred that SNP3 (Y333C) might also be harmful to chickens at high altitudes. The lower oxygen and extreme environment selectively swept out mutation SNP3 (Y333C). Taking together, these results indicate that EAPS1 gene may play a key role in the adaptation of the Tibetan chicken to a high-altitude environment at the molecular level. The adaptation patterns however, may differ in human, dog, grey wolf, and Tibetan Cashmere goat.
In conclusion, genetic analysis in the present study for EPAS1 gene in TC and LC populations indicated that the non-synonymous SNP3 (Y333C) may have critical role in the high-altitude adaptation of TC. Although the adaptation patterns may not be the same in humans and dogs, these novel variations of EPAS1 could put new insights into the gene function.
Supporting information
Figure A. The LD block of Tibetan chicken. TC means Tibetan chicken.
Table A. The partial production performance of Tibetan and 7 lowland chicken breeds. The production performance data of 8 breeds of chicken is based on poultry genetic resources in China. Table B. Allele and genotype frequencies of the SNP1 rs315040213 and SNP2 rs14330062 in the EPAS1. “C” represents the reference allele and “T” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table C. Allele and genotype frequencies of the SNP3 rs316126786 and SNP4 rs739281102 in the EPAS1. “A” represents the reference allele and “G” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table D. Allele and genotype frequencies of the SNP5 rs740389732 and SNP6 rs739010166 in the EPAS1. “A” represents the reference allele and “G” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. “C” represents the reference allele and “T” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table E. Nucleotide diversities of EPAS1 gene in Tibetan chicken and Lowland chicken. 1Pi is the abbreviation of nucleotide diversity. 2K is the abbreviation of average number of nucleotide differences.3TC means Tibetan chicken.4LC means Lowland chicken.
(DOCX)
Acknowledgments
We would like to express our gratitude to Prof Paul Siegel from Virginia Tech, USA for reading our manuscript, providing useful comments and revising the language patiently.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by National Natural Science Foundation of China (Grant No: 31402070) and the China Agriculture Research System (CARS-41).
References
- 1.Quirouette R, Arch B. Air pressure and the building envelope: Citeseer; 2004. [Google Scholar]
- 2.Storz JF, Sabatino SJ, Hoffmann FG, Gering EJ, Moriyama H, Ferrand N, et al. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3(3):e45 10.1371/journal.pgen.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao M, Kong QP, Wang HW, Peng MS, Xie XD, Wang WZ, et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci U S A. 2009;106(50):21230–5. 10.1073/pnas.0907844106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6(9):e1001116 10.1371/journal.pgen.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Wu CX, Chamba Y, Ling Y. Blood characteristics for high altitude adaptation in Tibetan chickens. Poult Sci. 2007;86(7):1384–9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Wu C, Qiangba Y, Ma X, Tang X, Pu B. Curve analysis of embryonic mortality in chickens incubation at high altitude. Journal of China Agricultural University. 2004;10(4):109–14. [Google Scholar]
- 7.Hao Z, Changxin W, Yangzom C, Yao L, Zhang L. Adaptability to high altitude and NOS activity of lung in Tibetan chicken. Journal of China Agricultural University. 2006;11(1):35. [Google Scholar]
- 8.Wideman R. Pathophysiology of heart or lung disorders: pulmonary hypertension syndrome in broiler chickens. World’s Poult Sci J. 2001;57(3):289–307. [Google Scholar]
- 9.Wideman RF Jr., Erf GF, Chapman ME, Wang W, Anthony NB, Xiaofang L. Intravenous micro-particle injections and pulmonary hypertension in broiler chickens: acute post-injection mortality and ascites susceptibility. Poult Sci. 2002;81(8):1203–17. [DOI] [PubMed] [Google Scholar]
- 10.Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Pathol. 2011;6:165–92. 10.1146/annurev-pathol-011110-130321 [DOI] [PubMed] [Google Scholar]
- 11.Qing G, Simon MC. Hypoxia inducible factor-2alpha: a critical mediator of aggressive tumor phenotypes. Curr Opin Genet Dev. 2009;19(1):60–6. 10.1016/j.gde.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92(3):967–1003. 10.1152/physrev.00030.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Patot MC, Gassmann M. Hypoxia: adapting to high altitude by mutating EPAS-1, the gene encoding HIF-2alpha. High Alt Med Biol. 2011;12(2):157–67. 10.1089/ham.2010.1099 [DOI] [PubMed] [Google Scholar]
- 14.Maxwell PH. Hypoxia-inducible factor as a physiological regulator. Exp Physiol. 2005;90(6):791–7. 10.1113/expphysiol.2005.030924 [DOI] [PubMed] [Google Scholar]
- 15.Lorenzo FR, Huff C, Myllymaki M, Olenchock B, Swierczek S, Tashi T, et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 2014;46(9):951–6. 10.1038/ng.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D, et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24(8):1308–15. 10.1101/gr.171876.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang GD, Fan RX, Zhai W, Liu F, Wang L, Zhong L, et al. Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the tibetan plateau. Genome Biol Evol. 2014;6(8):2122–8. 10.1093/gbe/evu162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Fan Z, Han E, Hou R, Zhang L, Galaverni M, et al. Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 2014;10(7):e1004466 10.1371/journal.pgen.1004466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44(8):946–9. 10.1038/ng.2343 [DOI] [PubMed] [Google Scholar]
- 20.Ge RL, Cai Q, Shen YY, San A, Ma L, Zhang Y, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4:1858 10.1038/ncomms2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman JH, Holt TN, Cogan JD, Womack B, Phillips JA 3rd, Li C, et al. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat Commun. 2015;6:6863 10.1038/ncomms7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Zhong H, Chen SY, Yao YG, Liu YP. Association between MT-CO3 haplotypes and high-altitude adaptation in Tibetan chicken. Gene. 2013;529(1):131–7. 10.1016/j.gene.2013.06.075 [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Wu N, Zhu Q, Gaur U, Gu T, Li D. High-altitude adaptation of Tibetan chicken from MT-COI and ATP-6 perspective. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(5):3280–8. 10.3109/19401736.2015.1015006 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Gou W, Wang X, Zhang Y, Ma J, Zhang H, et al. Genome resequencing identifies unique adaptations of tibetan chickens to hypoxia and High-Dose ultraviolet radiation in high-altitude environments. Genome Biol Evol. 2016;8(3):765–76. 10.1093/gbe/evw032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullenbach R, Lagoda PJ, Welter C. An efficient salt-chloroform extraction of DNA from blood and tissues. Trends Genet. 1989;5(12):391 [PubMed] [Google Scholar]
- 26.Singh VK, Mangalam AK, Dwivedi S, Naik S. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques. 1998;24(2):318–9. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 29.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–7. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 30.Priyatno D. SPSS 22 Pengolah Data Terpraktis. Penerbit ANDI, Yogyakarta. 2014.
- 31.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–6. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 34.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–91. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–5. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamitomo M, Alonso JG, Okai T, Longo LD, Gilbert RD. Effects of long-term, high-altitude hypoxemia on ovine fetal cardiac output and blood flow distribution. Am J Obstet Gynecol. 1993;169(3):701–7. [DOI] [PubMed] [Google Scholar]
- 38.Baumann R, Padeken S, Haller EA, Brilmayer T. Effects of hypoxia on oxygen affinity, hemoglobin pattern, and blood volume of early chicken embryos. Am J Physiol. 1983;244(5):R733–41. [DOI] [PubMed] [Google Scholar]
- 39.Dzialowski EM, von Plettenberg D, Elmonoufy NA, Burggren WW. Chronic hypoxia alters the physiological and morphological trajectories of developing chicken embryos. Comp Biochem Physiol A Mol Integr Physiol. 2002;131(4):713–24. [DOI] [PubMed] [Google Scholar]
- 40.Bao HG, Zhao CJ, Li JY, Zhang H, Wu C. A comparison of mitochondrial respiratory function of Tibet chicken and Silky chicken embryonic brain. Poult Sci. 2007;86(10):2210–5. [DOI] [PubMed] [Google Scholar]
- 41.Golding GB, Dean AM. The structural basis of molecular adaptation. Mol Biol Evol. 1998;15(4):355–69. [DOI] [PubMed] [Google Scholar]
- 42.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2α in development and disease. Cell Death & Differentiation. 2008;15(4):628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tissot van Patot MC, Gassmann M. Hypoxia: adapting to high altitude by mutating EPAS-1, the gene encoding HIF-2α. High altitude medicine & biology. 2011;12(2):157–67. [DOI] [PubMed] [Google Scholar]
- 44.Lee Rodgers J, & Nicewander W. A. Thirteen ways to look at the correlation coefficient. The American Statistician. 1988;1(42):59–66. [Google Scholar]
- 45.Newman JH, Holt TN, Cogan JD, Womack B, Phillips JA III, Li C, et al. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nature communications. 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimoda LA, Semenza GL. HIF and the lung: role of hypoxia-inducible factors in pulmonary development and disease. Am J Respir Crit Care Med. 2011;183(2):152–6. 10.1164/rccm.201009-1393PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2alpha in development and disease. Cell Death Differ. 2008;15(4):628–34. 10.1038/cdd.2008.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimoda LA, Semenza GL. HIF and the lung: role of hypoxia-inducible factors in pulmonary development and disease. American journal of respiratory and critical care medicine. 2011;183(2):152–6. 10.1164/rccm.201009-1393PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickey MM, Simon MC. Regulation of angiogenesis by hypoxia and hypoxia-inducible factors. Curr Top Dev Biol. 2006;76:217–57. 10.1016/S0070-2153(06)76007-0 [DOI] [PubMed] [Google Scholar]
- 51.Percy MJ, Furlow PW, Lucas GS, Li X, Lappin TR, McMullin MF, et al. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med. 2008;358(2):162–8. 10.1056/NEJMoa073123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–64. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–8. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan Q, Kerestes H, Percy MJ, Pietrofesa R, Chen L, Khurana TS, et al. Erythrocytosis and pulmonary hypertension in a mouse model of human HIF2A gain of function mutation. J Biol Chem. 2013;288(24):17134–44. 10.1074/jbc.M112.444059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song S, Yao N, Yang M, Liu X, Dong K, Zhao Q, et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genomics. 2016;17:122 10.1186/s12864-016-2449-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natarajan C, Hoffmann FG, Weber RE, Fago A, Witt CC, Storz JF. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science. 2016;354(6310):336–9. 10.1126/science.aaf9070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A. The LD block of Tibetan chicken. TC means Tibetan chicken.
Table A. The partial production performance of Tibetan and 7 lowland chicken breeds. The production performance data of 8 breeds of chicken is based on poultry genetic resources in China. Table B. Allele and genotype frequencies of the SNP1 rs315040213 and SNP2 rs14330062 in the EPAS1. “C” represents the reference allele and “T” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table C. Allele and genotype frequencies of the SNP3 rs316126786 and SNP4 rs739281102 in the EPAS1. “A” represents the reference allele and “G” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table D. Allele and genotype frequencies of the SNP5 rs740389732 and SNP6 rs739010166 in the EPAS1. “A” represents the reference allele and “G” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. “C” represents the reference allele and “T” represents the mutant allele. Numbers represent allele/genotype frequency, with the figures in brackets representing the number of individuals with each genotype. Table E. Nucleotide diversities of EPAS1 gene in Tibetan chicken and Lowland chicken. 1Pi is the abbreviation of nucleotide diversity. 2K is the abbreviation of average number of nucleotide differences.3TC means Tibetan chicken.4LC means Lowland chicken.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.