Summary
We propose that the essential function of the most highly-conserved protein in bacterial cytokinesis, FtsZ, is not to generate a mechanical force to drive cell division. Rather, we suggest that FtsZ acts as a signal-processing hub to coordinate cell wall synthesis at the division septum with a diverse array of cellular processes, ensuring that the cell divides smoothly at the correct time and place, and with the correct septum morphology. Here, we explore how the polymerization properties of FtsZ, which have been widely attributed to force generation, can also be advantageous in this signal processing role. We suggest mechanisms by which FtsZ senses and integrates both mechanical and biochemical signals, and conclude by proposing experiments to investigate how FtsZ contributes to the remarkable spatial and temporal precision of bacterial cytokinesis.
Keywords: cell division, cell wall synthesis, cytokinesis, cytoskeleton, force generation, FtsZ, dynamics, signaling
Introduction
Bacterial cytokinesis is carried out by a ring-like assembly termed the ‘divisome’. The major structural components of the divisome are polymeric filaments formed by FtsZ [1], an essential GTPase with structural homology to eukaryotic tubulin [2-4]. FtsZ filament polymerization is mediated by an N-terminal globular domain that binds and hydrolyzes GTP (Fig. 1A). The C-terminus of FtsZ is capped by a peptide ‘landing pad’ that interacts with a multitude of other divisome constituents and regulatory proteins [5]. This landing pad peptide is separated from the globular domain by a long, intrinsically-disordered linker that varies widely in length and sequence among different bacterial species [6].
Figure 1.
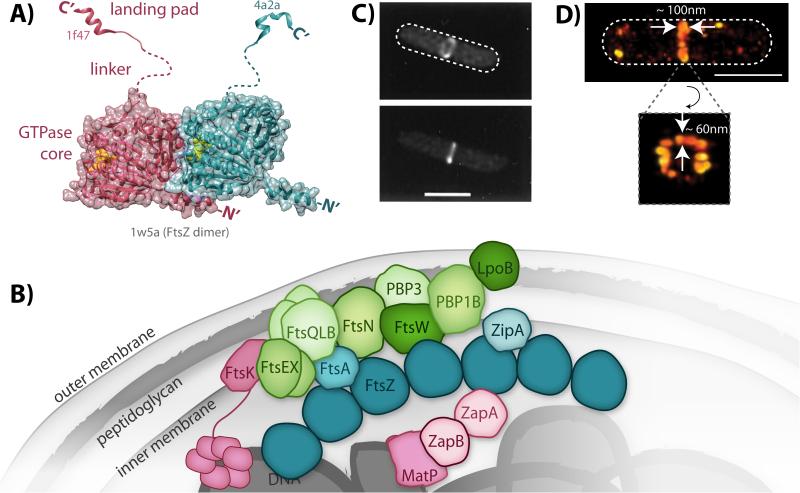
Structure and organization of FtsZ and the divisome. A: Model of a FtsZ dimer based on solved crystal structures. The two FtsZ subunits within the dimer are colored individually in either magenta or teal. Bound GTP (yellow spheres) is observed at the polymerization interface in the dimer structure of the globular GTPase core domains (PDB ID: 1w5a). The two different landing pad structures were solved in complex with either ZipA (magenta, PDB ID: 1f47) or FtsA (teal, PDB ID: 4a2a). The unstructured linker domains are represented with dashed lines. B: Schematic of the network of interacting divisome proteins involved in cytokinesis. For simplicity, only FtsZ (dark teal) is shown as an oligomer although other divisome constituents may also oligomerize. Divisome constituents are broadly grouped as the Z-ring (FtsZ and its membrane tethers, teal), nucleoid-associated (magenta), and peptidoglycan-associated (green), although some proteins span multiple categories. C: Early images of the Z-ring obtained by conventional fluorescence microscopy of E. coli cells expressing FtsZ-GFP [108]. (© 1996 National Academy of Sciences) D: Recent superresolution images of Z-ring heterogeneity in E. coli obtained by photoactivated localization microscopy [21]. White dashed lines depict approximate cell outlines. Scale bars, 1 μm.
One essential role of FtsZ in cytokinesis is the recruitment of all other divisome proteins once FtsZ coalesces into the ‘Z-ring’ at midcell [7, 8]. Cytoplasmic FtsZ first polymerizes by associating with membrane proteins (such as FtsA and ZipA in Escherichia coli [9, 10]) via FtsZ's C-terminal landing pad peptide. These FtsZ polymers accumulate into the midcell Z-ring, whose assembly and location are regulated by a wide variety of proteins that interact with FtsZ in response to various cellular signals [5, 11, 12]. Z-ring assembly triggers a series of downstream interactions that recruit dozens of other divisome proteins to midcell, forming a multi-layered protein network (Fig. 1B) that includes enzymes involved in cell wall synthesis (e.g. PBPs, or penicillin binding proteins), proteins that regulate PBP activity, and at least in E. coli cells, proteins in the cytoplasm that link the divisome to the nucleoid DNA to coordinate cytokinesis with nucleoid segregation [13, 14]. As such, the fully assembled divisome spans the entire cell envelope and can extend onto the nucleoid DNA.
Understanding the spatial organization of the divisome, and in particular the arrangement of FtsZ filaments within the Z-ring, is crucial to unraveling the mechanisms of cytokinesis. However, ultrastructural characterization of assembled FtsZ filaments in vivo has been difficult due to the small size of bacterial cells. By conventional fluorescence microscopy, the Z-ring appears to be a continuous, homogenous structure (Fig. 1C), but higher resolution studies using electron cryotomography and superresolution fluorescence microscopy have revealed that the ring most likely comprises discontinuous, short filaments that are bundled together into clusters within a toroidal zone of 80 - 100 nm axial width and 40 - 60 nm radial width (Fig. 1D) [15-23].
FtsZ filaments within the discontinuous Z-ring are dynamic on the seconds timescale. Fluorescence recovery after photobleaching (FRAP) experiments have shown that FtsZ subunits within the Z-ring turn over in ~ 10 s in both E. coli and Bacillus subtilis [20, 24, 25]. Additionally, recent time-lapse imaging experiments have revealed another mode of FtsZ dynamics on the ~ 100 s timescale in vitro [26] and in live E. coli [27] and B. subtilis [17, 28] cells. This second mode of dynamics describes the rearrangement of FtsZ clusters within the Z-ring via treadmilling, or continuous polymerization at one end and depolymerization at the other end without directional movement of individual FtsZ molecules.
The coalescence of dynamic FtsZ filaments into a ring at midcell is conceptually similar to the assembly of dynamic actomyosin filaments within the eukaryotic cytokinetic ring [29]. Thus it is natural to ask whether the Z-ring shares functional similarity with the actomyosin ring, which actively generates a contractile force as myosin motors pull actin filaments together, constricting the eukaryotic cell. The question is – does the Z-ring also generate a constrictive force that drives bacterial cytokinesis?
The force-generating role of FtsZ has garnered much attention in the past two decades. However recent evidence, as we will discuss below, suggests that the critical, conserved role of FtsZ may not be its ability to generate a driving force for cytokinesis, but to regulate the spatiotemporal dynamics of other divisome components, in particular proteins involved in septal cell wall synthesis [30]. In this role, FtsZ may significantly impact the composition and morphology of septal cell wall synthesis.
In addition to this proposed role in guiding septal cell wall synthesis, FtsZ also participates in many molecular interactions that regulate Z-ring structure in response to various cellular cues. Thus, FtsZ receives and integrates signals that modulate its influence on cell wall synthesis, making the Z-ring a complex signal-processing hub. Here, we discuss how the properties of FtsZ polymerization, and the resulting Z-ring organization and dynamics that have been widely attributed to FtsZ's ability to generate force, can provide considerable advantages in this signal processing role. We suggest that if the Z-ring does generate a force, the force does not drive constriction progress, but rather helps FtsZ guide cell wall synthesis to promote a smooth, well-defined septum shape, thereby promoting the mechanical stability and integrity of the division septum. These signaling and regulatory functions point to a designation of FtsZ's role in cytokinesis as ‘master orchestrator’ [31] rather than force generator.
Does the Z-ring generate a constrictive driving force?
Possible Z-ring force generation mechanisms
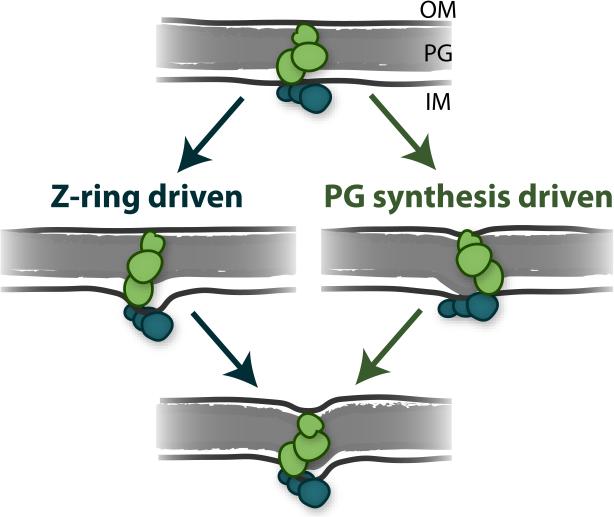
Bacteria lack a cognate motor protein that would perform the analogous function of myosin in a contractile Z-ring. Therefore, FtsZ itself has been widely proposed to be the source of a cytokinetic contractile force [8]. This proposal is bolstered by several experiments demonstrating that purified FtsZ can indeed deform and constrict membrane liposomes [32-35]. This FtsZ-generated constrictive force could drive cytokinesis by pulling on the cytoplasmic face of the inner membrane, allowing subsequent inward progression of cell wall synthesis (Fig. 2, on the left).
Figure 2.
Two proposed mechanisms for driving bacterial cytokinesis. The midcell divisome contains FtsZ filaments (teal ovals) tethered to the inner membrane (IM) and cell wall synthesis enzymes (green ovals) tethered to the cell wall peptidoglycan (PG) and the outer membrane (OM). Inward septum growth may be driven by FtsZ-centric pulling on the inner membrane (left) and/or by inward pushing by peptidoglycan synthesis (right).
The source of such an ‘FtsZ-centric’ force may be the chemical energy released by GTP hydrolysis, which has been shown to increase the curvature of FtsZ polymers [15, 36-40]. Additionally, the intrinsic affinity that FtsZ filaments have to bundle with each other could also promote the condensation of the Z-ring into smaller and denser structures, thereby generating a constrictive force on the attached membrane [41]. These two mechanisms (‘hydrolyze and bend’ and condensation) have been incorporated into many computational models suggesting that the Z-ring can generate forces up to ~ 100 pN [37, 38, 41-43]. These predictions are consistent with the estimated 20 - 90 pN of bending force that purified FtsZ filaments can impose onto membrane liposomes [43, 44], but are considerably smaller than what would be required to balance against the cell's inner turgor pressure and cell wall tension [45].
Evidence that cell wall synthesis, not FtsZ, drives cytokinesis
We recently looked for evidence that a variety of proposed FtsZ force-generation mechanisms indeed drive cytokinesis in E. coli cells. We assessed the effects of altered FtsZ GTPase activity, Z-ring density, and Z-ring assembly dynamics on the rate of cell envelope constriction during cytokinesis [21]. However, we found that these alterations did not substantially alter the rate or progress of cytokinesis in E. coli, indicating that an FtsZ-derived force does not limit cell envelope constriction. Instead, we observed that the rate of cytokinesis linearly scaled with cell growth rate, which is proportional to cell wall synthesis rate, and that impairing cell wall synthesis via mutation to the enzyme PBP3 (aka FtsI) substantially reduced the rate of constriction. These results suggest that the chemical energy involved in cell wall synthesis and remodeling [46] provides the driving force (Fig. 2, on the right).
These findings are consistent with recent computational studies indicating that most, if not all, of the > 400 pN force required to significantly deform the inner membrane can be attributed to cell wall synthesis and remodeling [45, 47, 48]. Recent microscopy studies also showed that FtsZ departs the division site prior to complete septum closure in E. coli, indicating that any FtsZ-derived force is dispensable at least at the final stages of cytokinesis [49, 50].
Evolutionarily, a prominent role for cell wall synthesis in driving bacterial cytokinesis is consistent with mechanisms of FtsZ-mediated constriction in chloroplasts and mitochondria. These organelles have lost their cell walls since endosymbiosis, and thus require only membrane constriction to divide. Yet in species where FtsZ is utilized for chloroplast or mitochondrial division, eukaryotic dynamins have been recruited to facilitate division from the external surface of the organelles [51]. The recruitment of external dynamins suggests that FtsZ filaments alone do not provide sufficient constrictive force to divide the organelle membranes, and is thus consistent with a limited role for FtsZ-derived force in constricting bacterial membranes as well.
Similarly, FtsZ is not essential in ‘L-form’ strains of B. subtilis, E. coli, Staphylococcus aureus, and Corynebacterium glutamicum that have lost their cell walls via laboratory selection [52]. Furthermore, deletion of ftsZ from L-forms followed by additional selection has recently led to E. coli strains that divide in the presence of cell wall without FtsZ, albeit in an altered, non-binary manner [53]. Together with results indicating that bacteria such as Chlamydiae and Planctomycetes have evolved to grow and divide without FtsZ while synthesizing low levels of cell wall PG [54,55], these findings support a role for FtsZ in modulating cell wall synthesis rather than in driving membrane constriction during cytokinesis.
Interestingly, a dominant role for cell wall synthesis in driving cytokinesis has also been demonstrated for eukaryotic species such as the fission yeast Schizosaccharomyces pombe [56-58]. The actomyosin ring of S. pombe can exert a contractile force [59], but is not required for all stages of cytokinesis [56]. Furthermore, recent studies indicate that the amount of tension produced by the actomyosin ring is not large enough to influence the overall rate of cytokinesis in S. pombe [58], and that the rate of cytokinesis is instead determined by cell wall synthesis [56-58]. The mechanistic similarity between bacterial and eukaryotic species, which differ in the composition of both their cell wall and cytokinetic protein network, suggests that the progress of cytokinesis may be highly influenced by more general physicochemical properties of a stress-bearing cell wall.
The time scales of FtsZ dynamics and cell wall remodeling
The ability of FtsZ to drive membrane constriction in vitro [32-35], but not cell envelope constriction in E. coli cells [21], is likely related to the physical properties of the cell wall. Studies of E. coli cells deformed by a hydrodynamic flow force have indicated that the cell wall is elastic (i.e. it bounces back to its original shape) on the 10 s timescale, and that permanent changes to cell wall shape require hydrodynamic forces maintained over many minutes [60]. This slow remodeling of the cell wall in response to applied forces is consistent with the slow, ~ 25 min turnover time of the cytoske letal protein crescentin [61], which produces the curvature in the crescent shape of Caulobacter crescentus cells.
Furthermore, these studies suggest that the cell wall has properties of visco-elastic materials [62], which behave as solids on timescales faster than their turnover dynamics, but as fluids on longer timescales that allow the materials to remodel. This view is consistent with the slow turnover rate of cell wall materials governed by the action of cell wall synthases and hydrolases [14, 63], and the observation that permanent cell wall deformation requires forces applied on the minutes scale [60]. Thus the fast dynamics of FtsZ subunit turnover [20, 24, 25] and treadmilling [27, 28] on the seconds-scale suggest that the Z-ring is unlikely to produce sustained cell wall deformation through direct mechanical bending.
Instead, recent discoveries suggest that the Z-ring influences cell wall geometry more indirectly by guiding new enzymatic synthesis or modification of cell wall. Some cell wall synthesis enzymes (PBP3/FtsI in E. coli and PBP2b in B. subtilis) were shown to exhibit FtsZ treadmilling-dependent directional movement along division septa [27, 28], indicating that FtsZ may influence cell wall synthesis activity through its dynamics. Interestingly, in E. coli the speed of FtsZ treadmilling only influenced the spatiotemporal distribution of new synthesis but not the overall synthesis rate [27], whereas in B. subtilis both were influenced [28]. These different behaviors may reflect differences between Gram negative and Gram positive cell wall organizations and/or mechanisms of communication between FtsZ and cell wall synthesis machineries.
The Z-ring as a dynamic signaling hub
If the widely-conserved Z-ring does not drive invagination of the cell envelope, then what is FtsZ's essential role? We suggest that FtsZ's abilities to polymerize [23,64] and interact with a large protein network [5, 11-13] reflect its key function as the central signaling hub for cytokinesis. Below, we explore the inputs and outputs of this signaling system and their relationships to Z-ring structure and dynamics.
Z-ring inputs: Protein regulators of FtsZ polymerization
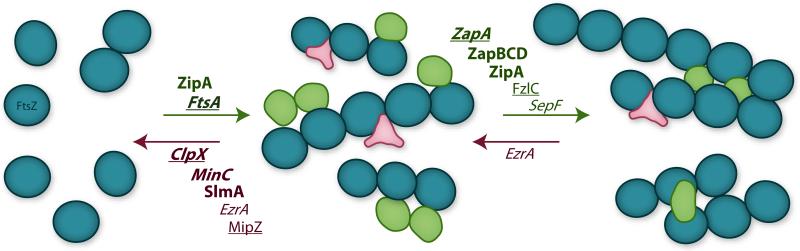
Bacterial cytokinesis is carried out with remarkable precision in both time and space [65]. This precision is regulated by many proteins that promote or inhibit Z-ring assembly to influence the timing and location of cell division (Fig. 3) [5, 11-13]. For example, proteins such as ZapA, ZapC, and ZipA are known to promote bundling of E. coli FtsZ filaments [66-68], and proteins such as MinC and SlmA are known to promote filament breakage [69, 70]. Interactions between FtsZ and these regulatory inputs occur simultaneously in the cell such that an individual FtsZ polymer could interact with both positive and negative regulators at different subunit positions. The polymer structure thus allows FtsZ to integrate the combined input signals from both positive and negative regulators to define both Z-ring dynamics (rates of FtsZ subunit turn over, polymerization and de-polymerization) and Z-ring structure (FtsZ filament length, curvature, orientation, and density).
Figure 3.
Modulation of FtsZ filament structure. FtsZ can reversibly assemble into a wide variety of filament structures depending on the local environment. Monomers (left) can polymerize into single-stranded filaments (middle), which can further associate into bundled and cross-linked structures (right). Regulatory proteins can either promote (green) or inhibit (magenta) assembly of these higher-order structures. Examples of positive and negative regulators from three model species are listed above each arrow. Regulators found in E. coli are listed in bold, those found in C. crescentus are underlined, and those found in B. subtilis are italicized.
The identity and mechanism of FtsZ's positive and negative regulators vary widely across different species [5, 12], perhaps reflecting the variation in signaling pathways necessitated by differences in species environments and lifestyles. Due to the cytoplasmic nature of FtsZ, all known regulators of FtsZ polymerization reside in the cytoplasm or the cytoplasmic face of the inner membrane such that the direction of signaling is from the inside out.
It is important to point out that while these regulators modulate FtsZ filament organization as described above, they do not appear to, or at least are not strong enough to, define the overall structural dimensions or dynamics of the Z-ring. The axial width of the Z-ring (along the cell's long axis) was shown to be unaltered by removal of the regulators MinC or MatP in E. coli [21]. Removal of either ZapA or ZapB in E. coli often led to dispersion of some Z-rings into separate, parallel FtsZ clusters, but these clusters retained the same intrinsic width [18]. Moreover, removing any known individual regulator (SlmA, SulA, MinC, ClpX, ClpA) or stabilizer (ZapA, ZapB, ZapC, ZapD, MatP) of FtsZ did not alter FtsZ's dynamics significantly [27]. Only an FtsZ variant with lowered GTPase activity, D212A, was shown to lead to wider Z-rings under highly over-expressed conditions [23], and perturbations to FtsZ's GTPase activity led to slowed subunit turnover and treadmilling dynamics [27]. Thus, intrinsic properties of FtsZ filaments modulated by its GTP binding or hydrolysis activity most likely serve regulatory roles in defining Z-ring dimensions and dynamics, consistent with the observed similarity of Z-ring width [16, 71-73] and dynamics [24, 27, 28] across species with varying regulators and cell geometries.
Z-ring outputs: Septal cell wall synthesis
Z-ring structure and dynamics established by regulatory inputs likely influence divisome organization on a nanometer spatial scale and seconds to minutes time scale. Thus, the Z-ring may output a spatiotemporal signal to other divisome proteins, including those involved in septal cell wall synthesis (Fig. 1B). The ability of FtsZ to communicate with cell wall synthesis complexes in this way is supported by recent observations that the motion and local activity of essential cell wall synthesis enzymes are coupled to FtsZ treadmilling dynamics [27, 28]. Previous evidence of FtsZ-guided cell wall synthesis includes the observation that division septa always occur at sites of FtsZ polymerization, even when mutations to FtsZ cause aberrant Z-ring structures such as arcs and spirals [74, 75]. Furthermore, FtsZ-dependent cell wall synthesis has been observed at midcell prior to any observed septum invagination [76, 77], and even along the lateral wall outside the midcell [78]. Interestingly, the axial septum width appears to be similar to axial Z-ring width: the septum appears to be ~ 80 nm wide in AFM images of Gram negative E. coli sacculi [79] and 60 - 80 nm in electron micrographs of Gram positive B. subtilis [80, 81], while Z-ring width has been shown to be 80 – 100 nm in Gram negative E. coli and C. crescentus, and in Gram positive Streptococcus pneumoniae [16, 18, 21-23, 71-73, 82]. Thus, the ultimate output of Z-ring signaling may be to define the composition and morphology of septal cell wall synthesis, mediated by signaling interactions between FtsZ and other divisome constituents.
Influence of Z-ring structure on signal transduction
FtsZ filaments integrate many low-affinity interactions
One key advantage of the polymeric structure of FtsZ filaments in a signaling role is that polymerization increases the avidity of intrinsically weak input and output interactions by increasing the local concentration of FtsZ binding partners. For example, ZipA exhibits a low affinity interaction to two sites on E. coli FtsZ (Kd = 30 and 140 μM), but when FtsZ polymerizes, the apparent affinity increases to Kd = 0.4 μM [83]. Similarly, FtsA exhibits a low affinity for individual monomers of E. coli FtsZ (Kd > 40 μM [84]), such that FtsA can only recruit polymerized, but not monomeric, FtsZ to the membrane [26].
Intrinsically weak interactions reduce the risk of spurious signal propagation when only a few binding events occur, and allow the state of FtsZ polymerization to control when and where these weak interactions are favored. Furthermore, by increasing the local concentration of all its binding partners, FtsZ could also promote weak interactions between other divisome proteins, such as members of cell wall synthesis complexes, ensuring that functional output complexes only form in the vicinity of the Z-ring. This type of polymer-organized assembly of low affinity interaction complexes has been described as a ‘signalosome’ in some eukaryotic signaling pathways [85], and is likely a widespread mode of regulating signal transduction.
The three-dimensional toroidal organization of the Z-ring facilitates signaling
The Z-ring has been shown to be a patchy, discontinuous structure with FtsZ clusters loosely occupying a three-dimensional toroidal zone with cross-sectional dimensions of ~ 100 × 60 nm (Fig. 1D) [15-23]. As described above, divisome proteins can be concentrated near this zone through direct or indirect interactions with FtsZ, promoting productive interactions between them. Concentration of a Z-ring-proximal zone of cytoplasmic divisome proteins is supported by in vivo measurements of Z-ring dimensions, and the dimensions of rings formed by regulatory input proteins such as ZapA and ZapB [20]. Both proteins predominantly localize to midcell zones that overlap with that of FtsZ [20], even though ZapB does not bind FtsZ itself, but only indirectly through ZapA [86].
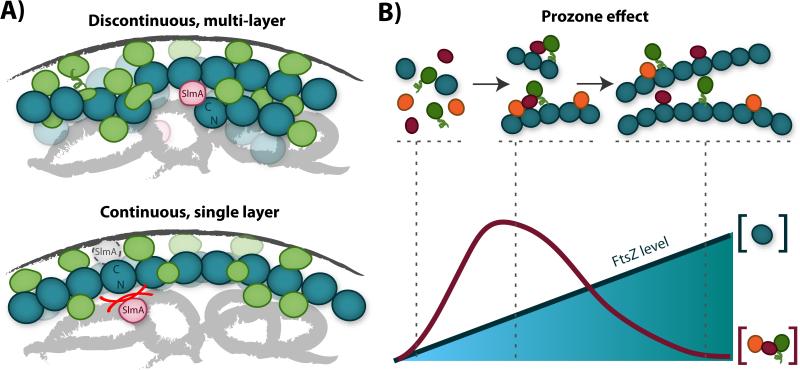
The three-dimensional nature of the Z-ring may offer a further advantage over a single-layer polymer organization by allowing a higher accumulation of cytoplasmic signaling proteins per unit surface area (Fig. 4A). Given that proteins receiving signals from FtsZ reside at the inner membrane, then the Z-ring's output capacity is likely coupled to its membrane-facing surface area rather than to its total cytoplasmic volume. Thus, a three-dimensional Z-ring structure allows FtsZ to consolidate the effects of regulatory input signals from multiple layers of FtsZ filaments onto a smaller membrane-proximal interface. This organization provides flexibility in the amounts and types of regulatory inputs, and may promote more effective downstream interactions at the Z-ring/membrane interface than would a single-layer structure.
Figure 4.
Influence of FtsZ filament structure on signaling. A: Signaling advantages of the discontinuous, multi-layer Z-ring structure (upper) compared to a model structure with a single, continuous layer (lower). The discontinuous, multi-layer structure can accommodate larger numbers of binding partners (green and pink) within the same region of membrane surface area (dark gray curve). Furthermore, the discontinuous structure allows ample room for proteins such as SlmA (pink) to bind both the nucleoid DNA (light gray swirls) and the C-terminal face of FtsZ (teal), while the single-layer structure sterically hinders this interaction (lower, red lines). B: Formation of signaling complexes on FtsZ filaments is described by the prozone effect [89]. As overall FtsZ levels rise (lower, teal line and shading), levels of polymerized FtsZ also rise (upper, teal circles). At a fixed level of interacting signaling partners (orange, magenta, green), the likelihood of forming a complete complex with all three partners first increases, then decreases, with increasing FtsZ levels (lower, magenta line). As such, FtsZ filament organization can control complex formation, and random fluctuations in FtsZ levels may prevent robust signaling. If FtsZ primarily guides the formation of signaling complexes at the membrane, then the multi-layer Z-ring structure provides a mechanism to buffer FtsZ levels at the membrane while overall FtsZ levels vary.
The discontinuous nature of the Z-ring also provides an advantage in FtsZ's role as a dynamic signaling scaffold. By maintaining ample empty space in the toroidal zone, the Z-ring can retain sufficient room to accommodate all the simultaneous input and output interactions required for its function (Fig. 4A). A tightly-packed Z-ring, on the other hand, may have difficulty associating with all its binding partners at once. For example, Z-ring discontinuity can alleviate some mechanistic questions about how regulatory input proteins such as SlmA can simultaneously bind both DNA and FtsZ [70]. Discontinuities in the ring allow ample space for nucleoid-bound SlmA to contact FtsZ polymers at cluster peripheries (Fig. 4A). Furthermore, a loose structure may also facilitate the conformational dynamics or protein associations required for FtsZ exchange dynamics [39, 40, 87].
Finally, the three-dimensional organization of the Z-ring may also serve as a buffering system against intrinsic fluctuations in FtsZ levels to maintain a proper signaling surface area. By maintaining a three-dimensional zone that can accommodate multiple FtsZ layers, FtsZ clusters can maintain the same output-facing surface area under increased FtsZ levels. The existence of such a buffering system for increased FtsZ levels is supported by the continued punctate nature of the Z-ring even at 8-fold overexpression [23], and by the higher tolerance of E. coli for FtsZ overexpression (viable at levels up to 8-fold [16]) compared to FtsZ depletion (not viable below ~ 50% FtsZ levels [88]). On the contrary, if FtsZ filaments could only form a single tightly-packed layer, an 8-fold increase in FtsZ levels would in turn lead to an 8-fold increase in the membrane-exposed Z-ring surface area, likely diluting the concentrations of other divisome proteins per unit surface area.
This three-dimensional buffering system can be viewed as a mechanism to regulate the so-called ‘prozone’ effect (Fig. 4B), which describes the influence of increasing the concentration of one core protein in a multi-protein complex or network [89]. When the core protein (FtsZ) is scarce, complete complexes do not form efficiently because many bridging connections cannot be made. However, when the core protein is too abundant, it promotes the formation of smaller, partial complexes rather than the larger, complete complex by diluting out its binding partners (Fig. 4B). A membrane-facing Z-ring surface area that is robust to fluctuations in FtsZ levels thus allows the system to maintain the same signaling capacity across a large range of FtsZ levels. Note that although FtsZ does not participate in all protein-protein interactions in the divisome, it could still deliver the prozone effect by interacting with other hub proteins such as FtsA [90].
Influence of Z-ring dynamics on signal transduction
Z-ring dynamics influence Z-ring structure
Z-ring dynamics likely play an important role in the signaling function of FtsZ. One intuitive way is through the coupling between Z-ring dynamics and the Z-ring structural organization described above. For example, we recently showed that increases in turnover rates of ZapA/ZapB (~ 50% faster) and FtsZ (~ 15 % faster) caused by deletion of matP were associated with decreases in Z-ring density in E. coli [20, 21], supporting a relationship between Z-ring density and FtsZ turnover dynamics. Moreover, Z-ring organization appears more continuous and homogenous under conditions of reduced GTPase activity and increased FtsZ expression levels [23, 35], which are also associated with slowed dynamics [24]. These studies suggest that FtsZ turnover and polymerization dynamics may establish the size of clusters within the ring. In E. coli, these clusters span < 100 nm on each dimension [21], which is consistent with the filament length expected for diffusion-limited polymerization of FtsZ subunits during the ~ 10 s Z-ring turnover half-time (~ 200 nm maximum) [24]. Thus, FtsZ turnover dynamics may govern the maximal size of the Z-ring, and in turn the precision with which the size of the toroidal signaling zone is established.
Z-ring dynamics allow communication across membrane compartments
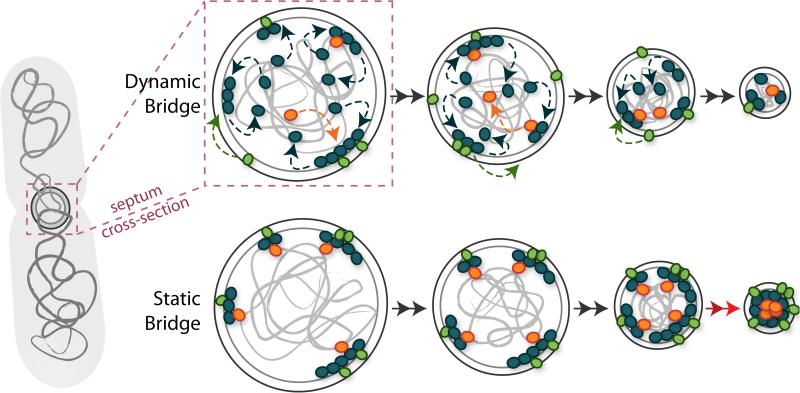
Other clear benefits of Z-ring dynamics arise if they are able to influence the dynamics of other divisome constituents. The dynamics of other divisome proteins, including ZapA, ZapB, FtsA, and ZipA in E. coli [20, 25, 91] and EzrA and PBP2 in S. aureus [17], have been shown to be similar to that of FtsZ, despite the fact that these proteins also interact with other entities such as the nucleoid DNA or cell wall peptidoglycan. Furthermore, the speed with which cell wall synthesis enzymes (PBP3/FtsI in E. coli and PBP2b in B. subtilis) move along the division septum has been shown to depend on the speed of FtsZ treadmilling [27, 28]. Such a dynamic divisome that constantly turns over would have the benefit of accommodating the gradually shrinking septal diameter while maintaining a time-averaged signaling bridge from the cytoplasmic nucleoid (input side) to the outer membrane (output side), but without creating a static link that might hinder the progress of either septum invagination or nucleoid segregation (Fig. 5).
Figure 5.
A dynamic divisome accommodates the changing septum shape. Cross-sectional views of the division septum as it closes during cytokinesis. At the septum, divisome proteins (small colored ovals) bridge the cell envelope (large gray circles) to the nucleoid DNA (light gray swirls) via cell-wall-associated (green ovals) and DNA-associated (orange ovals) proteins. Dynamic turnover of individual divisome proteins (dashed arrows) driven by treadmilling of FtsZ (blue dashed arrows) allows the cell to maintain a time-averaged bridge throughout the course of cytokinesis (left to right), and also to adjust to the shrinking septal volume. If the divisome were static (lower), the bridges could generate steric blockages that may hinder the progress of cytokinesis (lower right, red arrows).
Dynamic exchange can promote efficient formation of cell wall synthesis complexes
Dynamic redistribution of divisome proteins may be particularly important for those that are expressed at only hundreds of molecules per cell, such as FtsA, FtsI, FtsK, PBP1A, PBP1B, and ZipA in E. coli [92-95]. Production of these divisome proteins at low levels may prevent accidental activation of septal cell wall synthesis by limiting the random diffusion-limited association of intact synthesis complexes. This mechanism is analogous to the ability of weak binding interactions to prevent spurious complex formation. However, low copy numbers are associated with larger fluctuations in both protein levels and spatial distributions, as described for E. coli PBP2, which is involved in cell elongation (100s per cell) [94]. PBP2's fluctuations are thought to be buffered by its intrinsic fast turnover dynamics, which allow frequent disruption of non-productive complexes and thus more opportunities for the formation of productive complexes [96]. By analogy, FtsZ's intrinsic dynamics may provide buffering against low copy number fluctuations of other divisome constituents, allowing more opportunities for productive complexes to form.
FtsZ dynamics correlate with PG synthesis dynamics
If FtsZ's output signal indeed influences cell wall synthesis, then the speed, dwell time and/or dimensions of signaling clusters within the Z-ring might be expected to correlate with those of cell wall synthesis. Indeed, at least in E. coli the speed of PBP3/FtsI movement is linearly correlated with FtsZ treadmilling speed [27]. The dwell time of FtsZ clusters, likely reflected in the ~ 100 s time-scale dynamics observed in B. subtilis [17,28] and E. coli [27], is also on par with the ~ 40 s incorporation time for newly inserted PG strands in E. coli cells growing with a doubling time of 30 min [97]. These inserted PG strands contain 50 - 60 disaccharides on average [97], corresponding to 50 - 60 nm based on a ~ 1 nm disaccharide length [98], which is remarkably similar to the 30 – 50 nm FtsZ cluster length we observed in E. coli [21]. As such, the length and duration of processive PG synthesis, and the movement speed of PG synthesis enzymes, correlate well with the size and dynamics of FtsZ clusters, likely supporting a signaling connection between FtsZ and cell wall synthesis.
A role for FtsZ filament curvature in signal transduction
Our discussion so far has focused on a biochemical role for FtsZ polymerization and dynamics in promoting productive protein interactions. Below, we discuss the possibility that FtsZ's ability to detect and/or impose membrane curvature may also influence its signal propagation to the cell wall, thereby promoting a well-formed septum shape.
FtsZ curvature may be essential for a septum homogenizing function
Recently, mutations to FtsZ's flexible linker region (Fig. 1A) have been shown to result in altered cell wall composition and septum morphology in C. crescentus and B. subtilis [99, 100]. Interestingly, some of these linker variants, and others in B. subtilis FtsZ, form aberrantly straight super-structures in vivorather than the curved wild-type Z-ring structure [99, 100]. These studies suggest a possible link between FtsZ filament curvature and cell wall synthesis, which may be mediated by the linker between FtsZ and its landing pad peptide that binds membrane anchor proteins. Furthermore, an important role for FtsZ curvature is supported by the discovery of a Z-ring regulator that modulates FtsZ curvature [101].
If wild-type FtsZ filaments do prefer curved orientations over straight orientations in vivo, then they may preferentially bind to or treadmill in regions along the septum that have higher local curvature than the average septum curvature. These regions of higher FtsZ density may be more likely to recruit complete cell wall synthesis complexes than regions of lower FtsZ density, resulting in higher PG synthesis at regions of higher local curvature. As such, FtsZ could guide cell wall synthesis such that both concave and convex imperfections are smoothed out (Fig. 6). This role is conceptually similar to that proposed for MreB. During cell elongation, MreB filaments preferentially bind to regions of local negative curvature and promote cell wall synthesis in these regions [102].
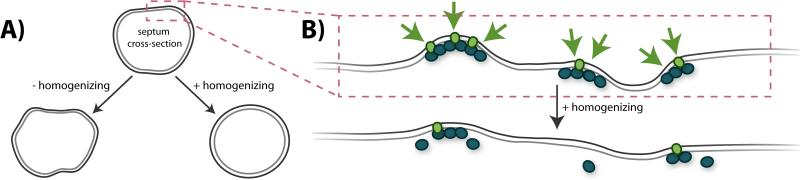
Figure 6.
FtsZ's homogenizing influence on septum morphology. A: A division septum (shown in cross-section) with minor imperfections (upper) can develop into a deformed and possibly lethal shape (lower left) without the influence of a homogenizing force (perhaps from FtsZ). In the presence of a balancing, homogenizing force, the minor imperfections can be corrected before they are amplified (lower right). B: Schematic of a proposed mechanism by which FtsZ could help to homogenize the septum shape. Upper: FtsZ (teal) may preferentially localize to regions with higher negative curvature along the septum surface (gray lines). Higher density regions of FtsZ would likely recruit higher levels of cell wall synthesis enzymes (green ovals), and thus generate more cell wall synthesis activity (green arrows). Lower: After the FtsZ-directed cell wall synthesis has smoothed out the local membrane imperfections, turnover dynamics allows redistribution of the cell wall synthesis enzymes to other regions of the septum.
A homogenizing influence on cell wall synthesis, by MreB and perhaps FtsZ, is crucial to cell wall integrity. Small heterogeneities in the bacterial cell wall can amplify to large imperfections if not balanced by a homogenizing force [103], eventually leading to lethal cracks and bulges in the cell envelope [104]. This homogenization may become even more important during cytokinesis than during cell elongation because the rate of cell wall synthesis increases during cell division [105].
Alternatively, FtsZ filaments can perform a homogenizing function without a curvature-mediated bias in their localization. As we discussed above, turnover of FtsZ subunits via treadmilling may dynamically redistribute cell wall synthesis complexes. Such dynamic redistribution may ensure that cell wall synthesis complexes do not dwell too long in one location, which may cause large amounts of cell wall to be deposited at these sites while other sites experience negligible cell wall synthesis, leading to increased septal heterogeneity. Thus, FtsZ dynamics are essential for this homogenizing role, which may also be enhanced by a curvature-mediated bias in FtsZ localization.
A dynamic homogenizing function of the Z-ring is analogous to the emerging view of cytokinesis in walled eukaryotes such as S. pombe, in which the rate of cytokinesis is determined by cell wall synthesis, and actomyosin ring tension is thought to organize the spatial distribution of cell wall synthesis within the septum to repair local rough patches, ensuring a symmetric shape [56-58]. In this way, the S. pombe actomyosin ring, and perhaps the bacterial Z-ring, homogenizes the leading septum edge, acting as a proof-reading mechanism to preserve septum shape fidelity.
Conclusions and prospects
We have proposed that the Z-ring does not serve as a major force generator to drive bacterial cytokinesis but likely integrates and transduces signals to the cell wall. While we have presented evidence that cytokinesis is instead driven by cell wall synthesis and remodeling, direct evidence of force-generation by these processes has yet to be reported. Application of biophysical tools such as atomic force microscopy or fluorescence-based molecular force sensors [106] may provide fruitful avenues in this endeavor. Furthermore, the molecular details of signal transmission from FtsZ to cell wall components remain obscure, and will likely be worked out through a combination of in vivo genetic and in vitro biochemical assays.
We have borrowed analogies from eukaryotic biology to propose mechanisms by which FtsZ may regulate the spatiotemporal dynamics and organization of cell wall synthesis complexes. In light of these proposals, it will be interesting to investigate whether there is indeed a spatial correlation between FtsZ clusters and cell wall synthesis, and whether these are both preferentially localized to regions of higher membrane curvature. High-resolution studies using fluorescent markers for cell wall synthesis [107] and precise analytical methods for membrane curvature assessment [102] will likely be useful tools for these studies.
Similarly, we would anticipate that alterations to FtsZ cluster dynamics or dimensions may result in corresponding alterations to cell wall synthesis dynamics or processivity lengths. These alterations are likely to be on length scales only accessible by super-resolution fluorescence microscopy or electron microscopy. Further application of these tools to characterize the localization and dynamics of other divisome constituents, as well as their effects on Z-ring organization and cell wall synthesis will greatly improve our understanding of the mechanisms of bacterial cytokinesis and the role of FtsZ.
Abbreviations
- PBP
penicillin binding protein
- PG
peptidoglycan
References
- 1.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–6. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 3.RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP- binding protein. Nature. 1992;359:251–4. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 4.Nogales E, Downing KH, Amos LA, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–8. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 5.Huang KH, Durand-Heredia J, Janakiraman A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195:1859–68. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 7.Goehring NW, Beckwith J. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–R26. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 8.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–28. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–34. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 10.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–85. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 11.Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: Poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–54. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monahan LG, Liew ATF, Bottomley AL, Harry EJ. Division site positioning in bacteria: one size does not fit all. Front Microbiol. 2014;5:19. doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–53. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 14.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–36. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu G, Huang T, Buss J, Coltharp C, et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, et al. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: Implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buss J, Coltharp C, Huang T, Pohlmeyer C, et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowlett Veronica W, Margolin W. 3D-SIM Super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J. 2014;107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buss J, Coltharp C, Shtengel G, Yang X, et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 2015;11:e1005128. doi: 10.1371/journal.pgen.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci USA. 2016;113:E1044–53. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vedyaykin AD, Vishnyakov IE, Polinovskaya VS, Khodorkovskii MA, et al. New insights into FtsZ rearrangements during the cell division of Escherichia coli from single-molecule localization microscopy of fixed cells. MicrobiologyOpen. 2016;5:378–86. doi: 10.1002/mbo3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu Z, Coltharp C, Yang X, Xiao J. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers. 2016;105:725–34. doi: 10.1002/bip.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–81. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geissler B, Shiomi D, Margolin W. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology. 2007;153:814–25. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Lyu Z, Miguel A, McQuillen R, et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell-wall synthesis. bioRxiv. 2016 doi: 10.1126/science.aak9995. doi: 10.1101/077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisson Filho AW, Hsu Y-P, Squyres G, Kuru E, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. bioRxiv. 2016 doi: 10.1126/science.aak9973. doi: 10.1101/077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramanian MK, Srinivasan R, Huang Y, Ng KH. Comparing contractile apparatus- driven cytokinesis mechanisms across kingdoms. Cytoskeleton. 2012;69:942–56. doi: 10.1002/cm.21082. [DOI] [PubMed] [Google Scholar]
- 30.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton. 2012;69:778–90. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monahan LG, Robinson A, Harry EJ. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol Microbiol. 2009;74:1004–17. doi: 10.1111/j.1365-2958.2009.06914.x. [DOI] [PubMed] [Google Scholar]
- 32.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–4. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–84. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–4. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szwedziak P, Wang Q, Bharat TA, Tsim M, et al. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–70. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh B, Sain A. Origin of contractile force during cell division of bacteria. Phys Rev Lett. 2008;101:178101. doi: 10.1103/PhysRevLett.101.178101. [DOI] [PubMed] [Google Scholar]
- 38.Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc Natl Acad Sci USA. 2009;106:145–50. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Erickson HP. Conformational changes of FtsZ reported by tryptophan mutants. Biochemistry. 2011;50:4675–84. doi: 10.1021/bi200106d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Hsin J, Zhao L, Cheng Y, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–5. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, et al. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci USA. 2009;106:121–6. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surovtsev IV, Morgan JJ, Lindahl PA. Kinetic modeling of the assembly, dynamic steady state, and contraction of the FtsZ ring in prokaryotic cytokinesis. PLoS Comput Biol. 2008;4:e1000102. doi: 10.1371/journal.pcbi.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paez A, Mateos-Gil P, Horger I, Mingorance J, et al. Simple modeling of FtsZ polymers on flat and curved surfaces: correlation with experimental in vitro observations. PMC Biophys. 2009;2:8. doi: 10.1186/1757-5036-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horger I, Campelo F, Hernandez-Machado A, Tarazona P. Constricting force of filamentary protein rings evaluated from experimental results. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81:031922. doi: 10.1103/PhysRevE.81.031922. [DOI] [PubMed] [Google Scholar]
- 45.Lan G, Wolgemuth CW, Sun SX. Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci USA. 2007;104:16110–5. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch AL. The Bacterium's Way for Safe Enlargement and Division. Appl Environ Microbiol. 2000;66:3657–63. doi: 10.1128/aem.66.9.3657-3663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Si F, Margolin W, Sun SX. Mechanical control of bacterial cell shape. Biophys J. 2011;101:327–35. doi: 10.1016/j.bpj.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee S, Scherer NF, Dinner AR. Shape dynamics of growing cell walls. Soft Matter. 2016;12:3442–50. doi: 10.1039/c5sm02991k. [DOI] [PubMed] [Google Scholar]
- 49.Soderstrom B, Skoog K, Blom H, Weiss DS, et al. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol Microbiol. 2014;92:1–9. doi: 10.1111/mmi.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soderstrom B, Mirzadeh K, Toddo S, von Heijne G, et al. Coordinated disassembly of the divisome complex in Escherichia coli. Mol Microbiol. 2016;101:425–38. doi: 10.1111/mmi.13400. [DOI] [PubMed] [Google Scholar]
- 51.Miyagishima S-y, Nishida K, Kuroiwa T. An evolutionary puzzle: chloroplast and mitochondrial division rings. Trends Plant Sci. 2003;8:432–8. doi: 10.1016/S1360-1385(03)00193-6. [DOI] [PubMed] [Google Scholar]
- 52.Mercier R, Kawai Y, Errington J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife. 2014;3:e04629. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercier R, Kawai Y, Errington J. Wall proficient E. coli capable of sustained growth in the absence of the Z-ring division machine. Nat Microbiol. 2016;1:16091. doi: 10.1038/nmicrobiol.2016.91. [DOI] [PubMed] [Google Scholar]
- 54.Pilhofer M, Aistleitner K, Biboy J, Gray J, et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeske O, Schuler M, Schumann P, Schneider A, et al. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 2015;6:7116. doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Proctor Stephen A, Minc N, Boudaoud A, Chang F. Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr Biol. 2012;22:1601–8. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Z, Munteanu EL, He J, Ursell T, et al. The contractile ring coordinates curvature dependent septum assembly during fission yeast cytokinesis. Mol Biol Cell. 2015;26:78–90. doi: 10.1091/mbc.E14-10-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiyagarajan S, Munteanu EL, Arasada R, Pollard TD, et al. The fission yeast cytokinetic contractile ring regulates septum shape and closure. J Cell Sci. 2015;128:3672–81. doi: 10.1242/jcs.166926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stachowiak Matthew R, Laplante C, Chin Harvey F, Guirao B, et al. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 29:547–61. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amir A, Babaeipour F, McIntosh DB, Nelson DR, et al. Bending forces plastically deform growing bacterial cell walls. Proc Natl Acad Sci USA. 2014;111:5778–83. doi: 10.1073/pnas.1317497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esue O, Rupprecht L, Sun SX, Wirtz D. Dynamics of the bacterial intermediate filament crescentin in vitro and in vivo. PLoS One. 2010;5:e8855. doi: 10.1371/journal.pone.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasza KE, Rowat AC, Liu J, Angelini TE, et al. The cell as a material. Curr Opin Cell Biol. 2007;19:101–7. doi: 10.1016/j.ceb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popp D, Iwasa M, Narita A, Erickson HP, et al. FtsZ condensates: An in vitro electron microscopy study. Biopolymers. 2009;91:340–50. doi: 10.1002/bip.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan KR, Shapiro L. Temporal and spatial regulation in prokaryotic cell cycle progression and Development. Annu Rev Biochem. 2003;72:367–94. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- 66.Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–66. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, et al. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry. 2009;48:11056–66. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hale CA, Shiomi D, Liu B, Bernhardt TG, et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dajkovic A, Lan G, Sun SX, Wirtz D, et al. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18:235–44. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 70.Du S, Lutkenhaus J. SlmA Antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet. 2014;10:e1004460. doi: 10.1371/journal.pgen.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmon G, Fishov I, Feingold M. Oriented imaging of 3D subcellular structures in bacterial cells using optical tweezers. Opt Lett. 2012;37:440–2. doi: 10.1364/OL.37.000440. [DOI] [PubMed] [Google Scholar]
- 72.Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. 2014;111:4566–71. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacq M, Adam V, Bourgeois D, Moriscot C, et al. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. MBio. 2015;6:e01108–15. doi: 10.1128/mBio.01108-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–7. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 75.Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol. 1992;174:5414–23. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–34. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Potluri LP, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol. 2012;194:5334–42. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 2007;189:5692–704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turner DJ, Portman I, Dafforn TR, Rodger A, et al. The mechanics of FtsZ fibers. Biophys J. 2012;102:731–8. doi: 10.1016/j.bpj.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koch AL, Burdett ID. Normal pole formation during total inhibition of wall synthesis of Bacillus subtilis. J Gen Microbiol. 1986;132:3441–9. doi: 10.1099/00221287-132-12-3441. [DOI] [PubMed] [Google Scholar]
- 81.Tocheva EI, López-Garrido J, Hughes HV, Fredlund J, et al. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–86. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biteen JS, Goley ED, Shapiro L, Moerner WE. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem. 2012;13:1007–12. doi: 10.1002/cphc.201100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du S, Park K-T, Lutkenhaus J. Oligomerization of FtsZ converts the FtsZ tail motif (conserved carboxy-terminal peptide) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol Microbiol. 2015;95:173–88. doi: 10.1111/mmi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–60. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci. 39:487–95. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol. 2010;76:1514–26. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- 87.Hsin J, Gopinathan A, Huang KC. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109:9432–7. doi: 10.1073/pnas.1120761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palacios P, Vicente M, Sanchez M. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–8. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 89.Bray D, Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA. 1997;94:13493–8. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Egan AJF, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 91.Mosyak L, Zhang Y, Glasfeld E, Haney S, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–91. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mukherjee A, Donachie WD. Differential translation of cell division proteins. J Bacteriol. 1990;172:6106–11. doi: 10.1128/jb.172.10.6106-6111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bisicchia P, Steel B, Mariam Debela MH, Lowe J, et al. The N-terminal membrane-spanning domain of the Escherichia coli DNA translocase FtsK hexamerizes at midcell. MBio. 2013;4:e00800–13. doi: 10.1128/mBio.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dougherty TJ, Kennedy K, Kessler RE, Pucci MJ. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J Bacteriol. 1996;178:6110–5. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vischer NOE, Verheul J, Postma M, van den Berg van Saparoea B, et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front Microbiol. 2015;6:586. doi: 10.3389/fmicb.2015.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee TK, Tropini C, Hsin J, Desmarais SM, et al. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proc Natl Acad Sci USA. 2014;111:4554–9. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glauner B, Höltje JV. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990;265:18988–96. [PubMed] [Google Scholar]
- 98.Labischinski H, Barnickel G, Bradaczek H, Giesbrecht P. On the secondary and tertiary structure of murein. Eur J Biochem. 1979;95:147–55. doi: 10.1111/j.1432-1033.1979.tb12949.x. [DOI] [PubMed] [Google Scholar]
- 99.Sundararajan K, Miguel A, Desmarais SM, Meier EL, et al. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat Commun. 2015;6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buske PJ, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol. 2013;89:249–63. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goley ED, Dye NA, Werner JN, Gitai Z, et al. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol Cell. 2010;39:975–87. doi: 10.1016/j.molcel.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ursell TS, Nguyen J, Monds RD, Colavin A, et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA. 2014;111:E1025–E34. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun SX, Jiang H. Physics of bacterial morphogenesis. Microbiol Mol Biol Rev. 2011;75:543–65. doi: 10.1128/MMBR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, et al. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105:19282–7. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wientjes FB, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989;171:3412–9. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Freikamp A, Cost AL, Grashoff C. The piconewton force awakens: Quantifying mechanics in cells. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.07.005. in press, doi: 10.1016/j.tcb.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 107.Kuru E, Hughes HV, Brown PJ, Hall E, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed. 2012;51:12519–23. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–3003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]