Abstract
We report a molecular dynamics investigation of the structure, function and inhibition of geranylgeranyl diphosphate synthase (GGPPS), a potential drug target, from the malaria parasite Plasmodium vivax. We discovered several GGPPS inhibitors, benzoic acids, and solved their structures crystallographically. We then used molecular dynamics simulations to investigate the dynamics of three such inhibitors and two bisphosphonate inhibitors—zoledronate and a lipophilic analog of zoledronate—as well as the enzyme's product, GGPP. We were able to identify the main motions that govern substrate binding and product release as well as the molecular features required for GGPPS inhibition by both classes of inhibitor. The results are of broad general interest since they represent the first detailed investigation of the mechanism of action, and inhibition, of an important anti-malarial drug target, geranylgeranyl diphosphate synthase, and may help guide the development of other, novel inhibitors as new drug leads.
Graphical Abstract
Introduction
Malaria is a leading cause of morbidity and mortality.[1] The most dangerous and widespread form of malaria is caused by the apicomplexan parasite Plasmodium falciparum and it is typically treated with an artemisinin combination therapy. Malaria caused by Plasmodium vivax is also common and is increasing[2–3] and, regardless of species, the emergence of drug resistance is a common threat. This is evidenced by e.g. the sequential occurrence of resistance by malaria parasites to quinine, then chloroquine and now, artemisinin, leading to the need for new anti-malarials, acting on new targets.[4–5] One potential target is geranylgeranyl diphosphate synthase (GGPPS), an essential enzyme involved in (C20) isoprenoid biosynthesis.[6–9] Isoprenoid biosynthesis in general is the target for a very diverse range of drugs. For example, one of the most widely prescribed drugs, the statin Lipitor, blocks cholesterol biosynthesis at the level of HMGCoA reductase; bisphosphonate drugs such as risedronate (Actonel) and zoledronate (Zometa), used to treat bone resorption diseases, inhibit farnesyl diphosphate (FPP; C15) synthase, and azoles such as miconazole and posaconazole inhibit lanosterol 14α-demethylase, inhibiting ergosterol biosynthesis in yeasts and fungi.[10] There is, therefore, considerable interest in the discovery and development of novel isoprenoid biosynthesis inhibitors as new drug leads for malaria and in earlier work we reported that several lipophilic bisphosphonates targeted Plasmodium GGPPS and had in vivo activity in a mouse model of infection.[7, 9] However, the bisphosphonate class of drugs bind very tightly to bone mineral—a desirable feature for a bone drug, but not in general an anti-infective, since these drugs are rapidly removed from the circulation. In recent work, Jahnke et al. reported a series of non-bisphosphonate inhibitors of farnesyl diphosphate synthase (FPPS) that do not bind to bone mineral leading us here to seek new, non-bisphosphonate inhibitors of GGPPS.[11]
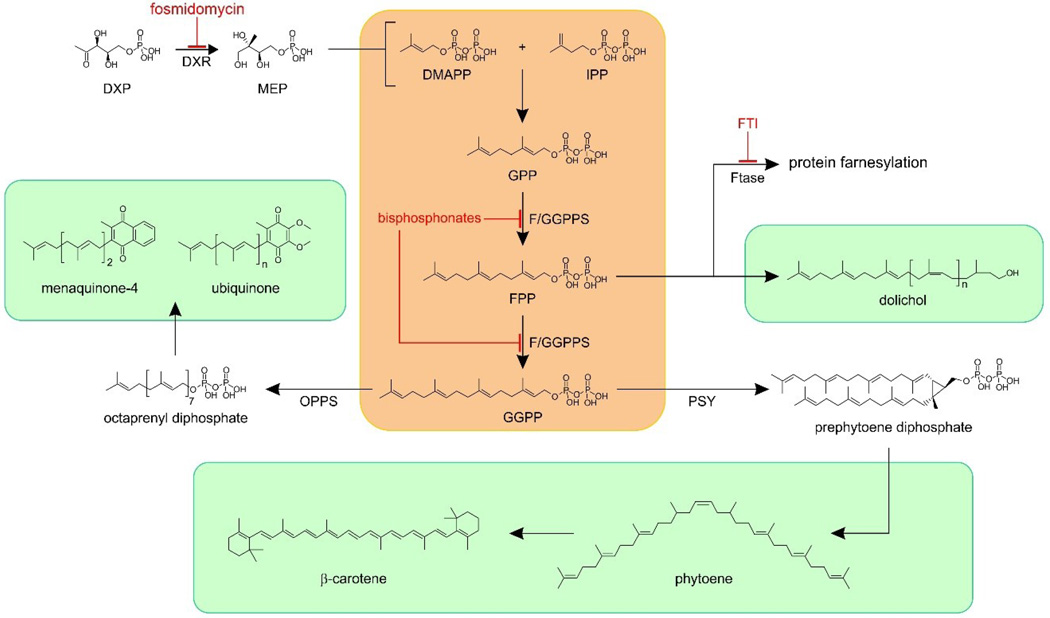
Isoprenoids contain C5 (isoprene) subunits and are made by the condensation of two compounds: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (Figure 1, in pink). These molecules are biosynthesized in the mevalonate pathway in humans, or in the methylerythritol phosphate (MEP) pathway in malaria parasites, and in most bacteria. In Plasmodium species, DMAPP and IPP then condense to form geranyl (C10) and farnesyl (C15) diphosphates in addition to (C20) geranylgeranyl diphosphate in reactions catalyzed by a bi-functional farnesyl/geranylgeranyl diphosphate synthase, typically abbreviated as GGPPS (since GGPP appears to be the major product).[8, 12–13] FPP and GGPP are then used in a wide variety of reactions including protein prenylation, quinone, dolichol and, apparently, carotenoid biosynthesis, as shown in Figure 1. Inhibitors of these pathways include fosmidomycin, bisphosphonates, as well as protein farnesyl transferase inhibitors (FTIs) with fosmidomycin having been used clinically (in combination with clindamycin) against malaria.[14]
Figure 1.
Isoprenoid biosynthesis in malaria parasites. The central stages (in pink) are performed by a single multi-functional farnesyl/geranylgeranyl diphosphate synthase (F/GGPPS). FPP and GGPP are precursors for the biosynthesis of many important isoprenoid products (in light green). DXP = deoxyxylulose-5-phosphate; DXR = deoxyxylulose-5-phosphate reductoisomerase; FTase = protein farnesyl transferase; OPPS = octaprenyl diphosphate synthase; PSY = phytoene synthase.
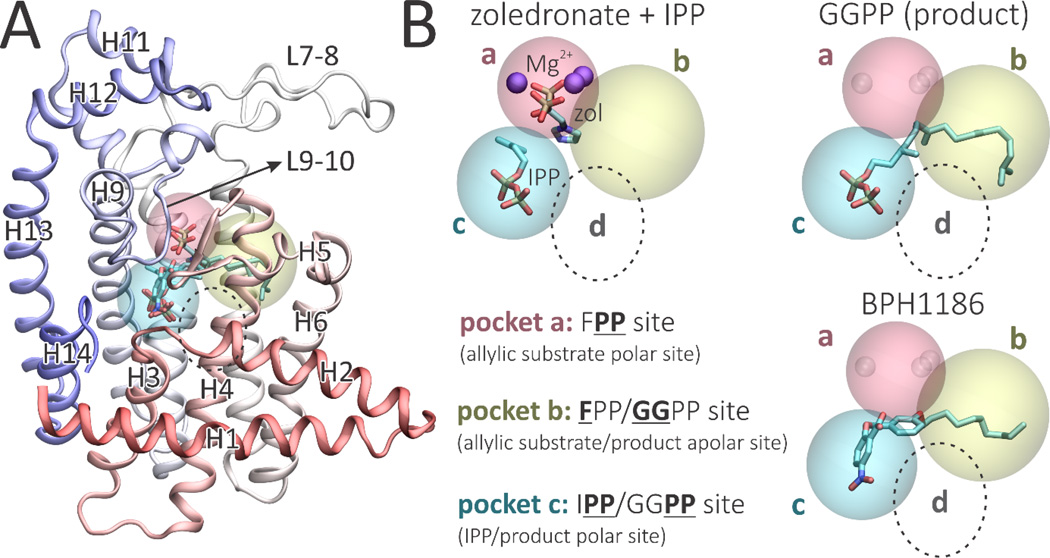
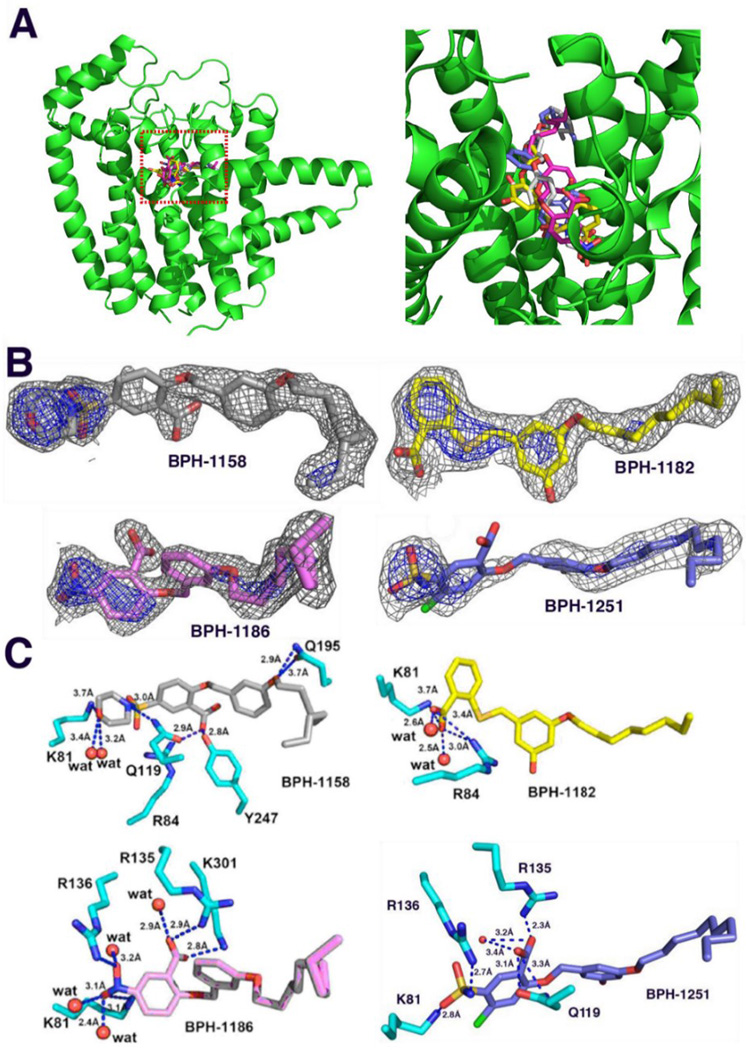
Although Plasmodium vivax GGPPS (PvGGPPS) primarily synthesizes GGPP in vitro,[8] paradoxically, its tertiary structure is more similar to that of human FPPS as compared to human GGPPS.[15–17] In addition, PvGGPPS is potently inhibited by bisphosphonates such as zoledronate, while human GGPPS is only weakly inhibited.[6] Structurally, PvGGPPS is a homodimer with each monomer formed by 14 α-helices connected by loops of varying length (Figure 2A). In PvGGPPS, the ligand binding cavity consists of 3 pockets that can accommodate either the substrates, intermediates or the final product, as shown in Figure 2B (and in Figure S1, in more detail).
Figure 2.
Structure and binding sites of Plasmodium vivax geranylgeranyl diphosphate synthase. A, Structure of PvGGPPS monomer showing helix and loop designations used in the Text. Ligand orientations correspond to PBD ID codes 3LDW, 3PH7 and 5HN9.8 B, The a, b and c ligand binding pockets found in PvGGPPS. The d pocket, found only in yeast and human GGPPS, is depicted by the dotted circle.
The diphosphate groups of the DMAPP substrate or GPP/FPP intermediates bind to site a via 3 Mg2+, essential for diphosphate ionization. The hydrophobic pocket b accommodates the hydrophobic tail of the allylic substrates or products.[8] Pocket c can accommodate the IPP substrate, or the diphosphate groups of the allylic products/intermediates. In this way, instead of being released, an intermediate product need only to have its diphosphate group move from pocket c to pocket a to be ready for a second round of catalysis. In human GGPPS, a fourth hydrophobic pocket that can bind the GGPP sidechain as well as several inhibitors is present,[15] indicated as d in Figure 2B. However, this pocket is either absent or not occupied in the more FPPS-like PvGGPPS.
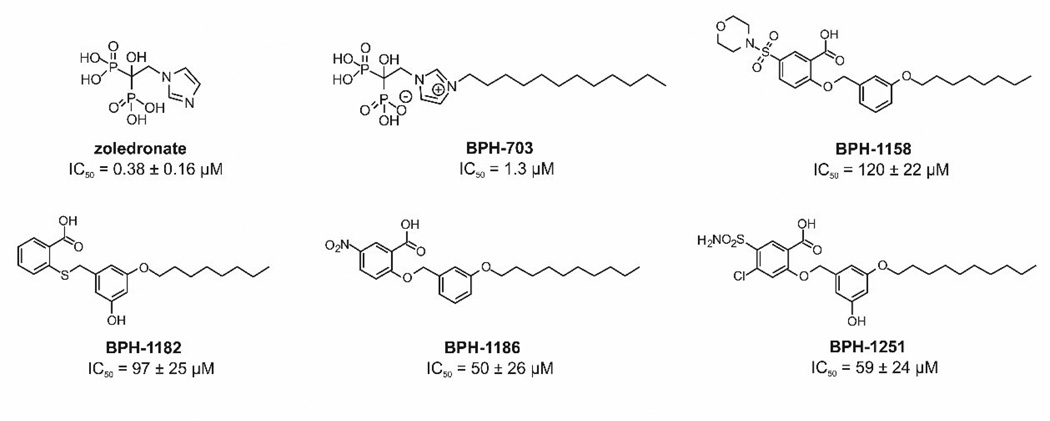
In this work, we report the discovery of several lipophilic, benzoic acid inhibitors of PvGGPPS; their X-ray crystallographic structures; their dynamic structures (from molecular dynamics [MD] simulations), as well as an MD investigation of a small bisphosphonate inhibitor, zoledronate (Chart 1), and a more lipophilic analog of zoledronate, BPH-703 (Chart 1), investigated previously both in vitro and in vivo in a mouse malaria model. Using MD simulations, we investigate apo, product-bound and inhibitor-bound structures, focusing on the motions that govern substrate binding and product release, as well as the key features that govern inhibitor binding.
Chart 1.
Plasmodium vivax GGPPS inhibitors investigated
Methods
Chemical Syntheses: General Methods
All chemicals were reagent grade. 1H and 13C NMR spectra were obtained on Varian (Palo Alto, CA) Unity spectrometers at 400 or 500 MHz for 1H and at 100 or 125 MHz for 13C. Elemental analyses were carried out in the University of Illinois Microanalysis Laboratory. HPLC-MS analyses were performed by using an Agilent LC/MSD Trap XCT Plus system (Agilent Technologies, Santa Clara, CA) with an 1100 series HPLC system including a degasser, an autosampler, a binary pump, and a multiple wavelength detector. All final compounds were ≥95% pure as determined by HPLC and structures were characterized by 1H NMR, LC-MS and HRMS. More detailed information on the synthesis of benzoic acids can be found in the Supporting Information.
Expression, purification and inhibition of PvGGPPS
The expression, purification and inhibition of P. vivax GGPPS was carried out as described previously.[9] Representative dose-response curves are in Figure S2.
X-Ray Crystallography
P. vivax crystals for soaking were obtained by using the hanging-drop method. Crystallization of PvGGPPS was carried out by co-crystallizing 1–2 mM of compound with 15 mg/mL PvGGPPS using 100 mM Tris (pH 8.5), 18~22% PEG 3, 350 and 200 mM Li2SO4. The drops were incubated at 18°C and single crystals formed overnight. X-ray diffraction data were collected at the Life Science Collaborative Access Team (LS-CAT) at the Advanced Photon Source of Argonne National Laboratory. Diffraction data were processed and scaled by using the program HKL2000 (HKL Research Inc., Charlottesville, VA, USA).[18] The statistics for data collection are given in Table 1. Model-building for PvGGPPS was performed by using molecular replacement with the PvGGPPS/GGPP complex structure (PDB ID code 3MYS). Structure refinements were carried out by using Refmac,[19] Phenix,[20] and Coot.[21]. Figure 3 was prepared by using PyMOL.[22]
Table 1.
Data collection and refinement statistics Crystals
| Crystals | BPH-1158 (5HN7) |
BPH-1182 (5HN8) |
BPH-1186 (5HN9) |
BPH-1251 (5HNA) |
|---|---|---|---|---|
| Data collection | ||||
| Space Group | P21 | P21 | P212121 | P212121 |
| Unit Cell Dimension a, b, c (Å) |
107.78, 108.38, 140.04 |
84.07, 117.22, 92.79 |
106.92, 108,29, 139,87 |
107.45, 108.62, 140.67 |
| X-ray source | APS 21-ID-F | APS 21-ID-F | APS 21-ID-F | APS 21-ID-F |
| Wavelength (Å) | 0.9787 | 0.9787 | 0.9787 | 0.9787 |
| Resolution (Å) | 50-2.15 (2.19-2.15) | 50-2.70 (2.75-2.70) | 50-2.10 (2.14-2.10) | 50-2.70 (2.75-2.70) |
| # of reflections observed |
865,103 | 231,318 | 689,414 | 442,309 |
| Unique | 170,179 (8,233) | 43,920 | 91,912 (4,507) | 46,114 (2,257) |
| Completeness (%) | 97.3 (94.3) | 99.5 (97.5) | 99.8 (99.5) | 100.0 (99.9) |
| R-merge | 0.126 (0.600) | 0.126 (0.561) | 0.094 (0.581) | 0.193 (0.777) |
| I/σI | 17.5 (2.2) | 18.3 (2.4) | 26.4 (2.7) | 11.16 (1.4) |
| Multiplicity | 5.1 (3.9) | 5.3 (3.9) | 7.5 (5.4) | 9.6 (5.1) |
| Refinement Statistics | ||||
| Resolution Range (Å) | 40-2.15 | 30-2.7 | 50-2.10 | 40-2.70 |
| R-work (%)/R-free (%) | 19.8/24.9 | 25.9/29.6 | 17.7/24.9 | 19.2/26.7 |
| RMSD (Å) | ||||
| Bond lengths | 0.006 | 0.013 | 0.01 | 0.009 |
| Bond angles | 0.953 | 1.407 | 0.882 | 1.262 |
| No. of atoms | ||||
| Protein | 22,371 | 10.497 | 11,136 | 11,304 |
| Ligand | 140 | 108 | 124 | 136 |
| B average (Å2) | ||||
| Protein | 25.18 | 46.81 | 29.58 | 40.11 |
| Ligand | 49.86 | 53.68 | 52.13 | 59.57 |
Values in parentheses are for the highest resolution shells.
Figure 3.
PvGGPPS complex structures. A, Stereo view of the four PvGGPPS inhibitors bound to the bc active site region of one monomer. Grey = BPH-1158; yellow = BPH-1182; pink = BPH-1186; blue = BPH-1251. B, Electron density maps contoured at 1.0σ (in gray) and 2.5σ (in blue) for the four PvGGPPS inhibitors. C, PvGGPPS-ligand structures and interactions. Note that these are "static" crystallographic structures and that the ligands move considerably in the MD simulations.
Molecular Dynamic Simulations
We performed molecular dynamics simulations for three of the four benzoic acid complexes reported herein: BPH-1158 (PDB ID code 5HN7), BPH-1182 (PBB ID code 5HN8), and BPH-1186 (PDB ID code 5HN9). We also used previously reported crystallographic structures to simulate GGPPS in its apo state (PDB ID code 3MAV),[8] in complex with the bisphosphonate inhibitors zoledronate (PDB ID code 3LDW)[8] and BPH-703 (PDB ID code 3RBM),[9] and also bound to its product, GGPP (PDB ID code 3PH7).[8] The zoledronate-GGPPS complex also contains IPP and Mg2+, so, to simulate apo-GGPPS in the presence of Mg2+, we used the zoledronate structure PDB ID code 3LDW, after (computational) removal of zoledronate and IPP.
Prior to MD simulations, ligands were geometry optimized by using the Gaussian-03 program[23] using the B3LYP functional and a 6-31G(d) basis set, then parameterized with Antechamber (in Amber 14)[24] using the Generalized Amber Force Field[25] and RESP charges.[26] The protonation states of ionizable amino acid residues were determined using the H++ server.[27] For each system, we used the tleap program in Amber 14 to build and neutralize simulation boxes containing TIP3P water models and Na+ counterions.[28–29] Proteins were solvated with a 12 Å water region.
MD equilibration was performed with the Sander module of Amber 14 and consisted of: i) 1000 steps of minimization with protein and ligands restrained (k = 500 kcal/mol/Å2); ii) 1000 steps of minimization with protein atoms restrained (k = 500 kcal/mol/Å2); iii) 2500 steps of minimization without position restraints; iv) 20 ps of NVT simulation, heating up the systems to 300 K with mild restraints applied to protein and ligands (k = 10 kcal/mol/Å2); v) 3 ns of NPT simulation with isotropic position scaling (with no position restraints), to equilibrate the density.
Subsequently, five independent 100 ns MD simulations were carried out for each system by using the pmemd.cuda module in Amber 14 and the ff14SB forcefield.[30] The simulations were kept at constant temperature using the Langevin thermostat with a collision rate of 5.0 ps−1. The PME method[31] was used to treat long-range electrostatic interactions, while short-range non-bonded interactions were truncated at 12 Å, employing periodic boundary conditions. All bonds involving hydrogen atoms were constrained with the SHAKE algorithm,[32] allowing for a time step of 2 fs.
MD analyses
Most of the MD analyses were performed with CPPTRAJ,[33] using functions rms, atomicfluct, contacts and hbond. Principal component analysis (PCA) was performed with a combination of the functions matrix, analyze and projection. To compare the dynamics of GGPPS in different apo and holo states, we stripped out the ligands and concatenated trajectories to create a common set of eigenvectors. Gas-phase interaction energies between protein and ligands were obtained by using the MM-PBSA method as implemented in pymdpbsa (Amber) with the option --solv=0 to deactivate the calculation of solvation effects.[34–35] Ligand occupancy maps were computed with the VolMap VMD plugin[36] after trajectories were aligned using the binding pocket as the reference.
Estimation of ligands conformational entropy
To estimate the entropy associated with ligand mobility in the bound state, we used a method based on conformational colony graphs, as proposed by Martinez et al.[37] We first assumed that a ligand's conformational ensemble is well represented by a 2D space formed by ligand mobility (RMSD) and interaction energy with the protein. We then divided the conformational colony space into an equally spaced grid, assuming each bin represents a thermodynamically relevant microstate (Figure S3). Next, we used a box-counting algorithm to count the number of bins that are occupied by each ligand in at least one snapshot of the MD simulations—that is, the area covered by each distribution of dots. The number of bins covered by ligand A, NA, can then be approximated as the number of microstates accessible to ligand A, ΩA, and the entropic contribution resulting from conformational mobility (Sconf,A) can be estimated by using the Boltzmann formula:
We tested different grid sizes and found that sizes of 100–300 bins best distinguished different conformational microstates, as also found by Martinez et al.[37] More details can be found in reference 37.
Results
Benzoic acid inhibitors occupy the product binding sites
We first screened a small in-house library of lipophilic benzoic acids finding four compounds (Chart 1) that had modest activity against PvGGPPS: BPH-1186, IC50=50 +/− 26 µM; BPH-1251, IC50=59 +/− 24 µM; BPH-1182, IC50=97 +/− 25 µM and BPH-1158; IC50=120 +/− 22 µM. We then obtained the x-ray crystallographic structures of these four compounds bound to PvGGPPS. Full crystallographic data collection and structure refinement statistics are shown in Table 1. All four ligands bind with their benzoic acid moieties in site c and their lipophilic side-chains in site b (Figure 3A). The ligand electron densities are quite well defined and 2Fo-Fc maps contoured at 1σ and 2σ are shown in Figure 3B. Protein-ligand interactions are shown in Figure 3C.
As noted above, in each of the four structures, the bound ligands occupy solely the GGPP binding sites, that is, sites bc. We thus next briefly consider the protein-ligand interactions suggested by these structures. As expected based on the adoption of the GGPP binding mode, the CO2− groups in BPH-1186 and BPH-1251 interact via electrostatic interactions with Lys-301 and Arg-135. In addition, Arg-135 side-chain interacts with the ligands phenyl group via a cation-π interaction, as does Lys-81 with the benzoate phenyl ring. Arg-136 appears to interact with the NO2 or SO2 groups, presumably due to a weak electrostatic interaction, and Lys-81 has a similar interaction with the NO2 group, in BPH-1186. The decrease in activity of BPH-1182 correlates with fewer interactions with these residues in the BPH-1182 structure, as it does in the BPH-1158 structure. These results suggest then that, just as we found with with inhibition of another prenyl transferase, undecaprenyl diphosphate synthase,[38] the presence of an electron-withdrawing group in the benzoate improves enzyme inhibition activity, presumably because this feature can facilitate a Coulombic or electrostatic interaction between the benzoic acid CO2− group and protein Lys/Arg residues, rather than simply a hydrogen bond interaction. That is, the benzoic acids with electron-withdrawing groups are stronger acids that more readily form the anionic species.
These binding poses in which the ligands occupy sites bc are to be compared with the binding poses for bisphosphonates, in which the highly polar bisphosphonate groups bind to site a and the alkyl groups bind to site b, that is, overall ligand binding is to sites ba (see Figure 2). Since these are the same sites that are occupied by the natural allylic substrate, we consider the bisphosphonates to be substrate-like inhibitors, as opposed to the benzoic acids, which display a product-like binding mode. The bisphosphonates are also more potent inhibitors than the best benzoate (zoledronate, IC50=0.38 µM; BPH-703, IC50=1.3 µM), presumably due in large part to the strong Coulombic interactions between the bisphosphonate groups and Mg2+. We next used molecular dynamics simulations to investigate in more detail how the benzoic acids and the bisphosphonates bind to and inhibit PvGGPPS, in addition to investigating how the GGPP product binds, in order to potentially provide new ideas to help guide inhibitor development.
9–10 loop stabilization correlates with inhibitor activity
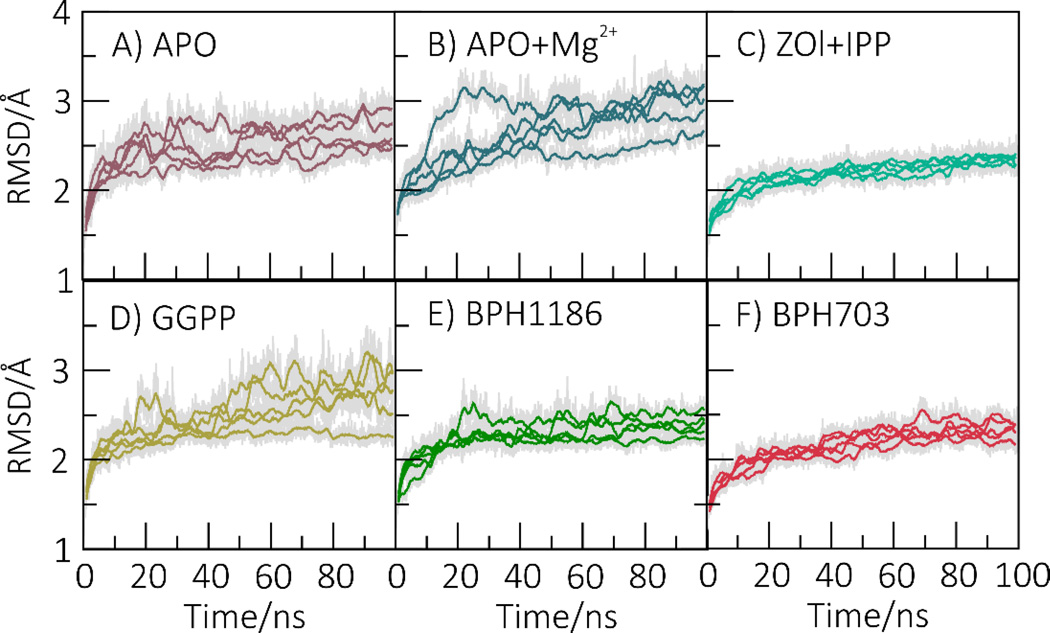
We carried out 6 sets of MD simulations in which we investigated the enzyme in its apo form (Figure 4A); +Mg2+ (Figure 4B); +zoledronate/IPP (Figure 4C); +GGPP (Figure 4D); +BPH-1186 (Figure 4E); and +BPH-703 (Figure 4F). We also simulated GGPPS+BPH-1182 and +BPH-1158; these results are shown in the Supporting Information (Figure S4). We find that GGPPS displays considerable mobility in the apo (Figure 4A), apo+Mg2+ (Figure 4B) and, to a lesser extent, product-bound states (Figure 4D). However, PvGGPPS is strongly stabilized by the three most potent inhibitors: zoledronate/IPP (Figure 4C), BPH-1186 (Figure 4E) and BPH-703 (Figure 4F).
Figure 4.
Mobility (RMSD) of GGPPS in different apo- and holo-states. Both bisphosphonate ligands (ZOL and BPH-703) and benzoic acid inhibitors (BPH-1186) stabilize the protein as compared to product-bound or apo-states. The RMSD values were computed for all heavy atoms with respect to the initial structure. The colored lines represent running averages of the original data (shown in grey).
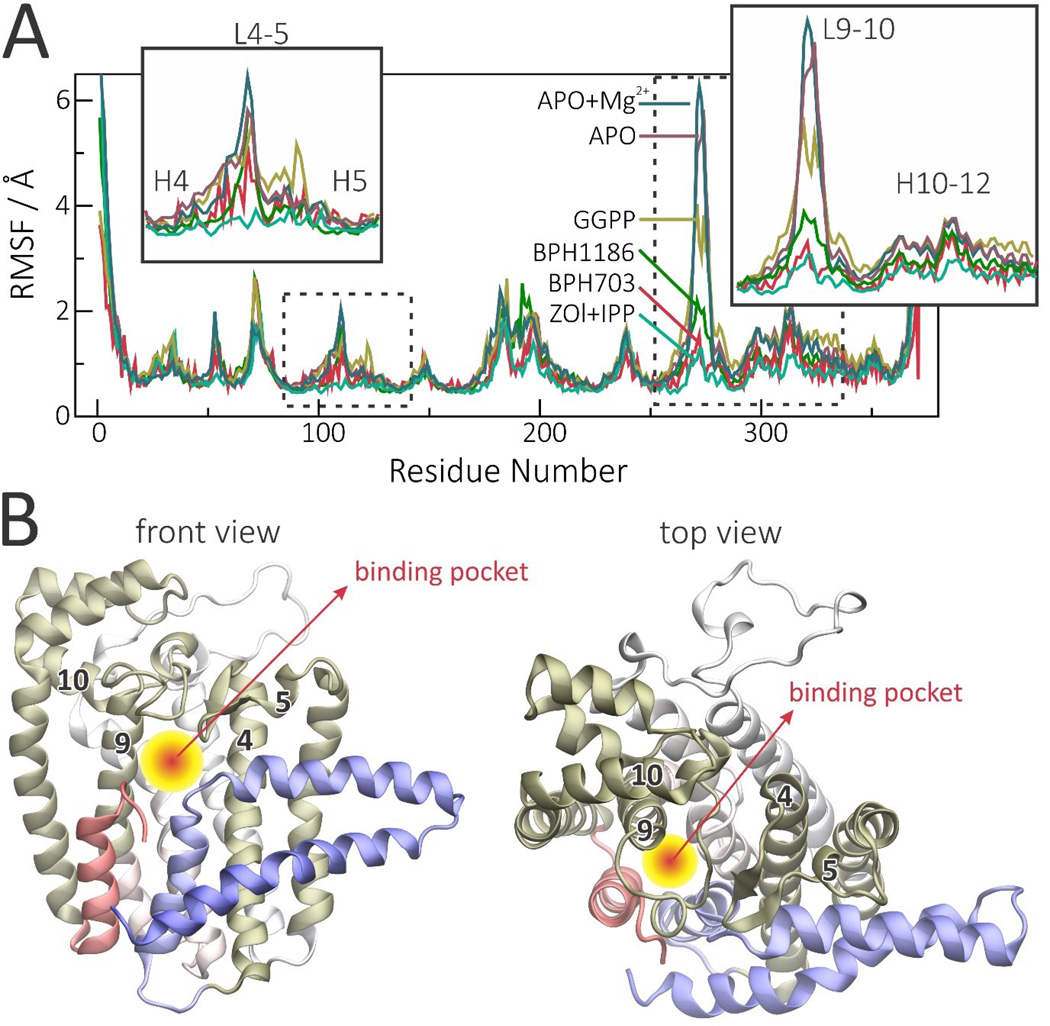
Such stabilization is particularly pronounced in the most flexible regions of the protein, primarily loops 9–10 and 4–5 (where the numbers refer to the helices shown in Figure 2A) with the product-bound GGPPS displaying intermediate mobility between apo- and inhibitor-bound states (Figure 5A). Adjacent helices are also stabilized, but to a lesser degree. Interestingly, loops 4–5 and 9–10 lie at the entrance of the binding cavity, while helices 4 and 9 form its walls (Figure 5B). Thus, changes in mobility or conformation of these regions can be expected to directly impact the accessibility, as well as the size, of the binding cavity. Figure 5 thus shows how different ligands stabilize different parts of the protein and how these ligands can affect the binding pocket.
Figure 5.
PvGGPPS Cα RMS fluctuation profiles (RMSF) in the presence or absence of ligands, and views of the binding cavity. A, RMSF profiles in different apo- and holo-states reveal that inhibitors stabilize the protein mainly around loops 4–5 and 9–10. B, Regions that are mostly stabilized by ligands are highlighted in gold in the monomer structure to illustrate how they can control binding cavity size and ligand accessibility. The RMSF profiles were computed for Cα with respect to the average structure and averaged over the two monomers.
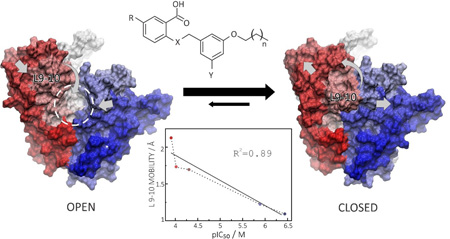
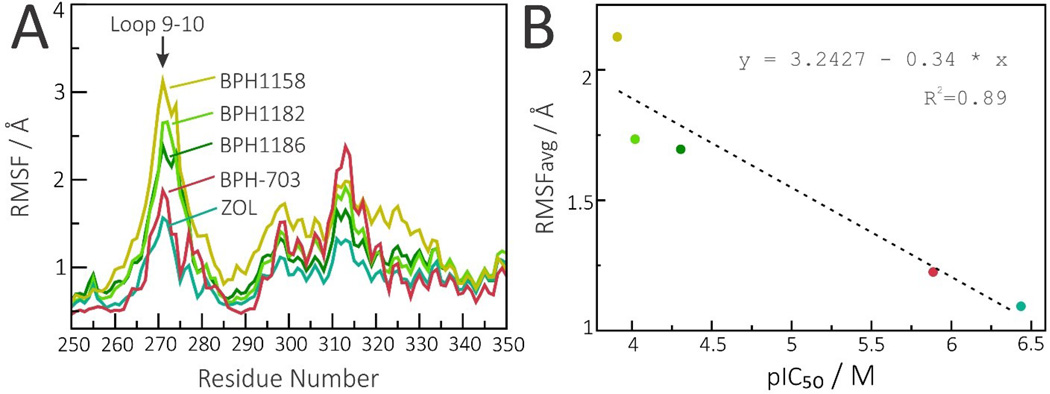
In Figure 6 we focus solely on the effects of inhibitors—the three benzoic acids, BPH-703 and zoledronate—on the loop 9–10. As can be seen in Figure 6, there is clearly a correlation between loop 9–10 stabilization and inhibitor efficiency (lower IC50 values) with all five inhibitors. More specifically, the pIC50 (=−log10 IC50 [M]) values anti-correlate with the loop 9–10 average fluctuation (average RMSF values computed for residues 266 to 280) with an R2=0.89.
Figure 6.
Effect of inhibitors on loop 9–10 mobility. A, Loop 9–10 RMSF profiles in the presence of all five inhibitors. B, Stabilization of the 9–10 loop is correlated with inhibitor efficiency (pIC50) for both bisphosphonate and benzoic acid inhibitors, with an R2=0.89. RMSFavg corresponds to average RMSF computed for residues 266 to 280.
PCA analysis of the effects of inhibitor binding on protein dynamics
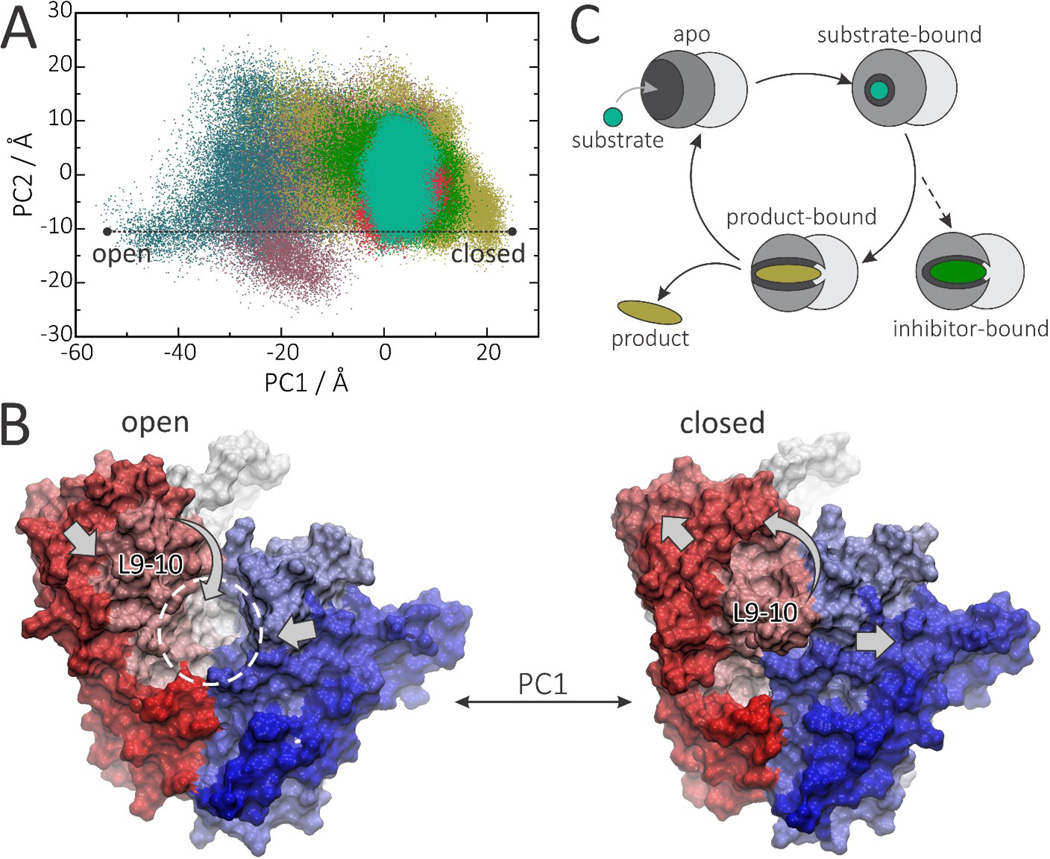
The results described above show that protein motion is strongly affected by ligand binding and that there are correlations between loop 9–10 dynamics and inhibitor activity. We thus next investigated this observation in more detail by using a principal component analysis (PCA). Projection of the GGPPS dynamics onto the subspace formed by the first and second principal components (PCs) shows that apo-states (especially in the presence of Mg2+) display significantly more conformational freedom than do the holo-states (Figure 7A). Whenever interacting with a ligand, GGPPS is stabilized in the same energy minimum, although the product-bound state retains significantly more conformational freedom than do the inhibitor-bound states. Similar conformational selection occurs with the other benzoic acid inhibitors (Figure S5). Conformational analysis of PC1 reveals that the main motion corresponds to a slight expansion (or compression) of the binding cavity together with a large rearrangement of loop 9–10 which opens (or closes) the entrance to the binding cavity (Figure 7B). Sampling of the essential subspace by different states shows that: 1) in the apo- state, the entrance to the binding site is more likely to be open, facilitating substrate binding; 2) after a substrate (or inhibitor) binds, it strongly stabilizes the closed conformation; 3) once formed, the product reduces the conformational selection, sampling semi-open conformations, which would be expected to facilitate product release (Figure 7C).
Figure 7.
Principal modes of motion control binding pocket accessibility in PvGGPPS. A, Sampling of essential subspace by apo (lilac), Mg2+-bound (blue), GGPP-bound (tan), BPH1186-bound (green), BPH-703-bound GGPPS (pink) or ZOL+IPP-bound (turquoise). B) Open and closed conformations corresponding to PC1 < −50 (left) and PC1 > 20 (right). C) Scheme of GGPPS dynamics as suggested by PCA analysis.
Substrate- and product-like inhibitors differ in terms of enthalpy-entropy balance
Initial inspection of the PvGGPPS X-ray structures discussed above suggested that bisphosphonate ligands form important hydrogen bonds with the entrance-loop residues displayed in Figure 8A. The MD simulations confirmed that zoledronate and BPH-703 form persistent hydrogen bond interactions with loops 2–3 (K81), 4–5 (R135 or R136) and 9–10 (K301), which lock these loops together, closing the entrance to the binding site (Figure 8B and Figure S6). The product, GGPP, is less efficient in forming these interactions, which is likely to be important in facilitating product release. None of these hydrogen bond interactions are persistently formed with BPH-1186 (or the other benzoic acid inhibitors), indicating that product-like inhibitors do not rely on the same mechanism as do substrate-like inhibitors, in "condensing" or stabilizing GGPPS in its closed conformations.
Figure 8.
Hydrogen bond interactions between ligands and PvGGPPS. A, residues K81 (loop 2–3), K135-K136 (loop 4–5) and K301 (loop 9–10), which can lock the entrance of the binding pocket. B, Frequency of hydrogen bond interactions formed during MD simulations. Zoledronate (in conjunction with IPP) and BPH-703 form persistent hydrogen bonds that are not so frequently formed with the product, GGPP, and even less so with benzoic inhibitors such as BPH-1186.
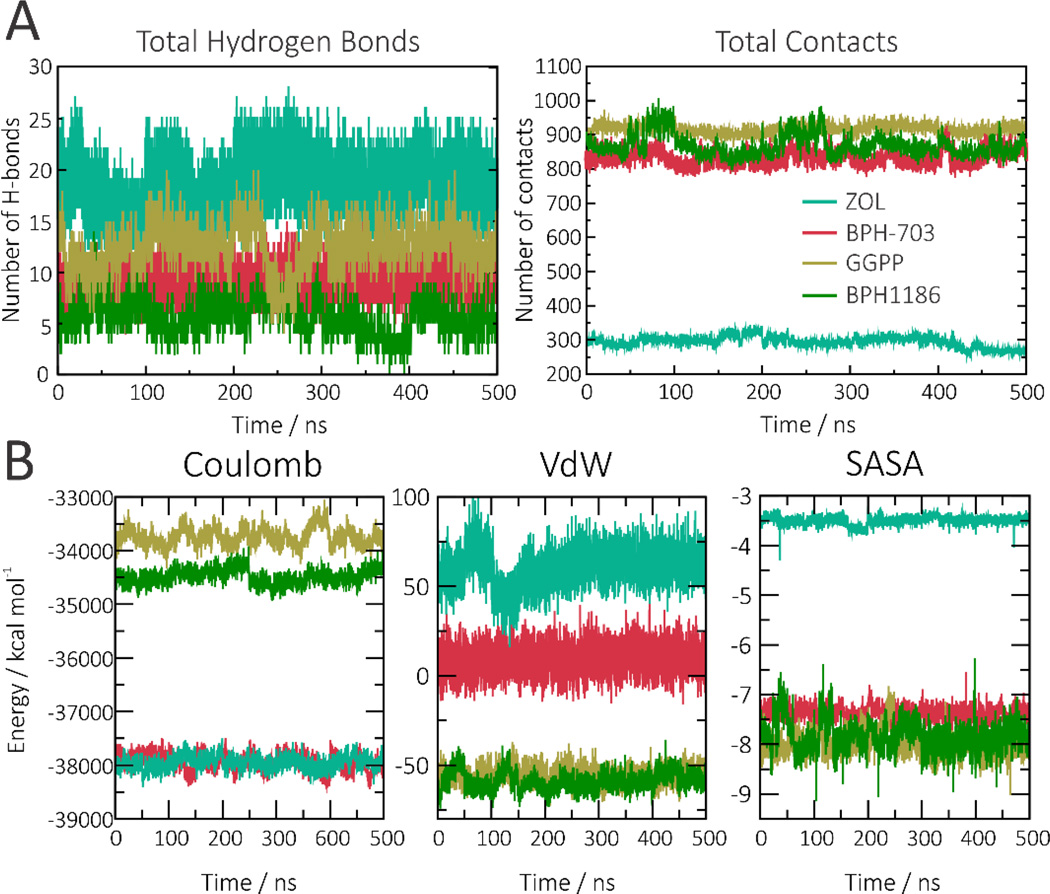
Instead, we found that the benzoic acid inhibitors compensate for the presence of fewer hydrogen bonds by having a larger number of hydrophobic interactions (Figure 9A and Figure S7). In addition to the presence of hydrophobic interactions in the b pocket, the MD results show that the benzoate and benzene rings in the benzoic acid inhibitors form transient π-stacking interactions with aromatic and arginine side-chains in the a and c pockets (Figure S8). Some of these were seen and briefly mentioned in the initial analysis of the X-ray structures discussed above, and are consistent with the gas-phase energetic results (arising from Coulombic and van der Waals interactions, and from the collapse of hydrophobic surfaces) shown in Figure 9B. All three benzoic acid inhibitors display rather similar energetic profiles to one another, so only the results for BPH-1186 are shown. Importantly, BPH-703 combines the favorable electrostatic interactions displayed by zoledronate with the favorable hydrophobic profile of BPH-1186. It should also be noted that BPH-703 is a slightly weaker GGPPS inhibitor than is zoledronate in vitro, but it is far more potent in cells and in vivo since its hydrophobic tail is expected to improve cell penetration.
Figure 9.
Protein-ligand interactions, and energetics. A, Total number of hydrogen bond interactions, and contacts, formed between ligands and GGPPS. A contact is formed whenever the ligand is less than 7Å from a protein atom. B, Coulombic, van der Waals and hydrophobic interaction energies (solvent-accessible surface area, SASA) computed for each ligand. Zoledronate in turquoise, BPH-703 in pink, GGPP in tan and BPH-1185 in green.
Because of the strength of electrostatic interactions, the larger number of primarily hydrophobic contacts found with the benzoic acid inhibitors does not compensate (enthalpically) for the lack of strong hydrogen bond/electrostatic interactions seen in bisphosphonate complexes. Thus, the overall (gas-phase) interaction energies of GGPPS with benzoic acid inhibitors are weaker than with the bisphosphonate ligands (see energy distributions in Figure 10A and Figure S9).
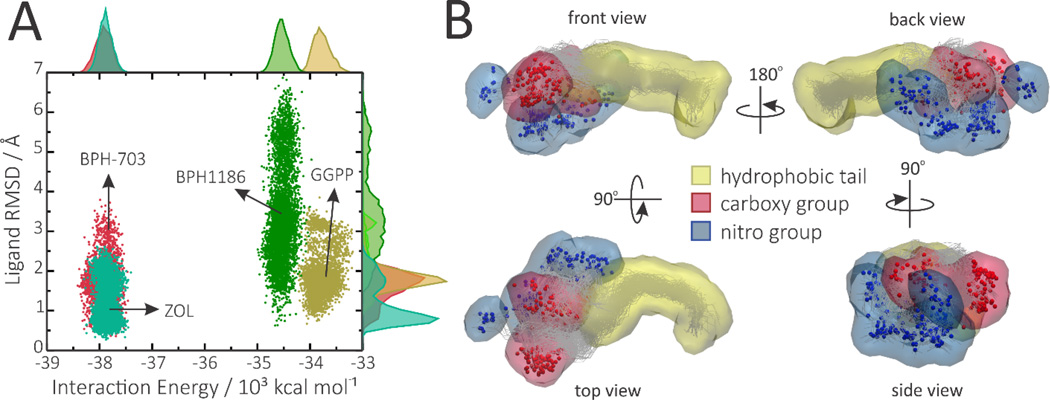
Figure 10.
The importance of conformational entropy for benzoic acid inhibitors. A, Projection of the simulations on the conformational colony space formed by ligand mobility (RMSD) and interaction energy with the protein, which provides a rough estimate of the number of thermodynamically relevant microstates accessible to each ligand in the bound state. B, Occupancy maps representing regions of the binding pocket that are occupied by BPH-1186 during at least 1% of the simulation time, showing occurrence of several major conformations, only one of which was seen in the average X-ray structure (PDB ID code 5HN9).
On the other hand, lacking persistent hydrogen bond interactions allows BPH-1186 to retain significant mobility and conformational freedom inside the binding pocket, as can be seen by the RMSD distributions in Figure 10A and by the multiple binding modes depicted in Figure 10B. The conformational entropy associated with this mobility can be roughly estimated by the area (or number of thermodynamically relevant microscopic states) covered by each ligand in the conformational space formed by the RMSD-versus-interaction energy, as shown in Figure 10A (and Figure S9).[37, 39] The predicted conformational entropies of BPH-1186, BPH-703, zoledronate and GGPP in the bound state are 7.76, 6.03, 6.55 and 6.81 cal K−1mol−1, respectively (Table S1). It should be noted that these are estimates for the bound state only. To account for the entropic gain associated with binding, one would need also to calculate the conformational entropies in the unbound states. Nevertheless, sampling of conformational space by the different ligands shows that they display very different binding profiles: the benzoic acid inhibitors (especially the most active BPH1186) take advantage of retaining a high conformational entropy in the bound state whereas the bisphosphonates rely on strong electrostatic interactions.
Discussion
In this work we employed molecular dynamics simulations to investigate the dynamic behavior of GGPPS in different apo- and holo-states. Holo-states included the complex with the enzyme's major product (GGPP), complexes with bisphosphonate inhibitors (zoledronate and BPH-703) as well as complexes with three benzoic acid inhibitors (BPH-1185, BPH-1182 and BPH-1158). The new crystallographic structures reveal that benzoic acid inhibitors bind in a product-like manner, different to the substrate-like binding mode adopted by bisphosphonate inhibitors such as zoledronate and its lipophilic analog, BPH-703.[8–9]
The molecular dynamics simulations reveal that GGPPS displays significant flexibility in the apo-states with its main motions consisting of large rearrangements of loop 9–10. It is also evident from the PCA analysis that loop 9–10 controls binding pocket accessibility, suggesting that these conformational changes may be an important part of the enzyme’s turnover kinetics—closing up after substrate binding and opening up for product release. In contrast, in human FPPS, it appears to be the C-terminal segment—not the 9–10 loop—that controls binding pocket accessibility in response to IPP binding.[40] Therefore, PvGGPPS and human FPPS do not appear to share the same opening/closing mechanism, which might be exploited to develop specific inhibitors.
In the apo-states, the 9–10 loop is found mostly open, which allows substrates to enter. In contrast, zoledronate (in conjunction with IPP) and BPH-703 both stabilize the 9–10 loop in a closed conformation by means of specific hydrogen bonds with residues lying at the entrance of the binding cavity. This loop stabilization mechanism is likely shared by the natural substrates to lock themselves inside the binding cavity and facilitate catalysis (in part, by shielding transition states/reactive intermediates from water). This stabilization is also in agreement with previous X-ray results in which the 9–10 loop changes from being disordered, in the apo-protein, to ordered, when the protein is co-crystallized with zoledronate and IPP.[8] The product, GGPP, promotes a 9–10 loop dynamics that is between that seen in the apo- and substrate-like states, in accord with the observation that there are fewer hydrogen bond interactions with the protein. Given that PvGGPPS is a multifunctional enzyme, such balanced open/closed behavior might enable intermediate products to be released, or else re-located for a second round of catalysis (GPP→FPP; FPP→GGPP).
The benzoic acid inhibitors are also capable of stabilizing the 9–10 loop, even though binding in a product-like manner with fewer and only transient hydrogen bonds. Such decrease in hydrogen bonding is partly compensated for by a larger number of transient π-stacking interactions and hydrophobic contacts. While the lack of persistent hydrogen bond interactions results in an overall less favorable interaction energy, it allows benzoic acid inhibitors to shift between several different binding modes. Though crystallographic structures of benzoic acids revealed a single average binding mode, diffuse B-factors suggested high mobility of the ligands, as confirmed by MD simulations. Thus, conformational entropy plays a role in the binding of these product-like inhibitors (benzoic acids), as opposed to substrate-like inhibitors (bisphosphonates), which rely on very specific hydrogen bonds.
Conformational entropy estimates confirm that the benzoic inhibitor BPH1186 displays a higher conformational entropy in the bound state as compared to the product, GGPP, or to the bisphosphonate ligands. The most flexible the ligand, the more dramatic is the entropy loss endured upon binding. Therefore, while retaining high conformational freedom inside the binding pocket might not be a crucial for binding of more intrinsically rigid ligands such as zoledronate, it can play a very important role for highly flexible ligands such as BPH1186. And finally, loop 9–10 stabilization is an important feature of the PvGGPPS inhibition mechanism since it was found to be highly correlated with inhibitor efficiency—regardless of inhibitor type.
In summary: by performing MD simulations on Plasmodium vivax GGPPS we provide interesting new insights into the conformational mechanisms required for the enzyme’s function, together with inhibition and binding mechanisms for different types of inhibitors. Our findings reinforce the important role of MD simulations in revealing molecular and entropic aspects not fully anticipated from crystallographic structures, and may help in the development of more potent GGPPS inhibitors as new malaria drug leads.
Supplementary Material
Acknowledgments
This work was supported in part by the United States Public Health Service (National Institutes of Health grants GM065307; CA158191), the National Science Foundation, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, the San Diego Supercomputer Center, by a Harriet A. Harlin Professorship (to E.O.), and by the University of Illinois Foundation/Oldfield Research Fund.
Abbreviations
- DMAPP
dimethylallyl diphosphate
- FPP
farnesyl diphosphate
- FPPS
farnesyl diphosphate synthase
- FTI
farnesyl transferase inhibitors
- GGPP
geranylgeranyl diphosphate
- IPP
isopentenyl diphosphate
- PfGGPPS
Plasmodium falciparum geranylgeranyl diphosphate synthase
- PvGGPPS
Plasmodium vivax geranylgeranyl diphosphate synthase
- ZOL
zoledronate
- MD
molecular dynamics
- PC
principal component
- PCA
principal component analysis
References
- 1.Who. World Malaria Report 2014. 2014:1–255.
- 2.Arnott A, Barry AE, Reeder JC. Malar. J. 2012;11 doi: 10.1186/1475-2875-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahimi BA, Thakkinstian A, White NJ, Sirivichayakul C, Dondorp AM, Chokejindachai W. Malar. J. 2014;13 doi: 10.1186/1475-2875-13-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. Nat. Rev. Microbiol. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 5.Noedl H. Curr. Pharm. Des. 2013;19:266–269. [PubMed] [Google Scholar]
- 6.Mukkamala D, No JH, Cass LA, Chang TK, Oldfield E. J. Med. Chem. 2008;51:7827–7833. doi: 10.1021/jm8009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh AP, Zhang YH, No JH, Docampo R, Nussenzweig V, Oldfield E. Antimicrob. Agents Chemother. 2010;54:2987–2993. doi: 10.1128/AAC.00198-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artz JD, Wernimont AK, Dunford JE, Schapira M, Dong A, Zhao Y, Lew J, Russell RG, Ebetino FH, Oppermann U, Hui R. J. Biol. Chem. 2011;286:3315–3322. doi: 10.1074/jbc.M109.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.No JH, de Macedo Dossin F, Zhang Y, Liu YL, Zhu W, Feng X, Yoo JA, Lee E, Wang K, Hui R, Freitas-Junior LH, Oldfield E. P Natl Acad Sci USA. 2012;109:4058–4063. doi: 10.1073/pnas.1118215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldfield E, Feng X. Trends Pharmacol. Sci. 2014;35:664–674. doi: 10.1016/j.tips.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahnke W, Rondeau JM, Cotesta S, Marzinzik A, Pelle X, Geiser M, Strauss A, Gotte M, Bitsch F, Hemmig R, Henry C, Lehmann S, Glickman JF, Roddy TP, Stout SJ, Green JR. Nat. Chem. Biol. 2010;6:660–666. doi: 10.1038/nchembio.421. [DOI] [PubMed] [Google Scholar]
- 12.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 13.Jordao FM, Gabriel HB, Alves JMP, Angeli CB, Bifano TD, Breda A, de Azevedo MF, Basso LA, Wunderlich G, Kimura EA, Katzin AM. Malar. J. 2013;12 doi: 10.1186/1475-2875-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruangweerayut R, Looareesuwan S, Hutchinson D, Chauemung A, Banmairuroi V, Na-Bangchang K. Malar. J. 2008;7 doi: 10.1186/1475-2875-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavanagh KL, Dunford JE, Bunkoczi G, Russell RGG, Oppermann U. J. Biol. Chem. 2006;281:22004–22012. doi: 10.1074/jbc.M602603200. [DOI] [PubMed] [Google Scholar]
- 16.Cao R, Chen CK, Guo RT, Wang AH, Oldfield E. Proteins. 2008;73:431–439. doi: 10.1002/prot.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C KMC, Hudock MP, Zhang Y, Guo RT, Cao R, No JH, Liang PH, Ko TP, Chang TH, Chang SC, Song Y, Axelson J, Kumar A, Wang AH, Oldfield E. J. Med. Chem. 2008;51:5594–5607. doi: 10.1021/jm800325y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. Acta. Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Acta. Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Acta. Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta. Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The PyMOL molecular graphics system (Schrödinger, LLC, New York) Version 1.3 [Google Scholar]
- 23.MJ F, GW T, HB S, GE S, MA R, JR C, JA MJ. Wallingford, CT: Gaussian Inc; 2004. [Google Scholar]
- 24.DA C, RM B, RM B, DS C, TE CI, TA D, TJ D. AMBER 2015. San Francisco: University of California; 2015. [Google Scholar]
- 25.Wang JM, Wang W, Kollman PA, Case DA. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Bayly CI, Cieplak P, Cornell WD, Kollman PA. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 27.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic. Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. J Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 30.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 32.JP R, G C, HJ B. J. Comput. Phys. 1976;23:327–341. [Google Scholar]
- 33.Roe DR, Cheatham TE. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 34.Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. J. Med. Chem. 2005;48:4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Martinez L, Nascimento AS, Nunes FM, Phillips K, Aparicio R, Dias SMG, Figueira ACM, Lin JH, Nguyen P, Apriletti JW, Neves FAR, Baxter JD, Webb P, Skaf MS, Polikarpov I. P Natl Acad Sci USA. 2009;106:20717–20722. doi: 10.1073/pnas.0911024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W, Zhang YH, Sinko W, Hensler ME, Olson J, Molohon KJ, Lindert S, Cao R, Li K, Wang K, Wang Y, Liu YL, Sankovsky A, de Oliveira CAF, Mitchell DA, Nizet V, McCammon JA, Oldfield E. P Natl Acad Sci USA. 2013;110:123–128. doi: 10.1073/pnas.1219899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang ZX, Soto CS, Honig B. P Natl Acad Sci USA. 2002;99:7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Lin YS, De Schutter JW, Tsantrizos YS, Berghuis AM. BMC Struct. Biol. 2012;12:32. doi: 10.1186/1472-6807-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.