Several lines of evidence support the role of Brain-Derived Neurotrophic Factor (BDNF) in the pathophysiology and pharmacotherapy of depression.1 The neurotrophin hypothesis of depression postulates that stress and depression are associated with decreased BDNF expression that can be reversed by antidepressant treatment.2 The goal of the present study was to investigate the effect of antidepressant treatment on the epigenetic regulation of BDNF in major depressive disorder (MDD).
The BDNF gene has distinct splice variants, each one regulated by a specific promoter region, that determine tissue-specific regulation of expression.3 Of these variants, BDNF-IV is the most commonly studied and its expression changes have been associated with behavioural phenotypes, psychiatric disorders and epigenetic modifications.4 The promoter region in exon-IV contains specific binding sites for the cyclic-AMP-responsive-element-binding protein (CREB)5 and the methyl-CpG-binding-protein-2 (MeCP2)6 making it a preferential candidate for epigenetic regulation.
Tsankova et al.7 reported that mice exposed to chronic social defeat stress displayed lower levels of BDNF-IV associated with a significant increase in histone H3 lysine 27 tri-methylation (H3K27me3), a modification associated with transcriptional repression. Long term imipramine treatment reversed BDNF-IV down-regulation to baseline levels.7 Studies by our group in postmortem brain of depressed subjects with or without history of antidepressant treatment compared to controls, showed an increased expression of BDNF-IV and a decrease of H3K27 tri-methylation levels in subjects treated with antidepressants only.8
In order to investigate the epigenetic regulation of BDNF in MDD patients according to antidepressant treatment, we conducted a prospective study in 25 treatment-naïve MDD patients. All patients had Hamilton Rating Scale for Depression [HAM-D] scores equal or above 24 at baseline (N=25, X=29.4 ± 1.2). All participants gave written informed consent for this study, approved by our Institutional Review Board.
Subjects were excluded from the study if they had comorbidity with other major psychiatric disorders, if they had positive tests for illicit drugs at any point during the study or if they had general medical illnesses. Patients (12 males and 13 females) were treated with citalopram, starting with an initial dose of 10mg die, which was titrated progressively to a maximum of 60mg die. All final doses were within the therapeutic range and blood levels of total BDNF and H3K27me3 were measured at baseline (T0) and after 8 weeks (T8) of treatment. Subject treatment compliance was assessed using high-performance liquid chromatography at the end of the trial. All subjects showed detectable plasma citalopram levels and we observed a significant correlation between citalopram dose and plasma concentration (Spearman r=0.54; p=0.005).
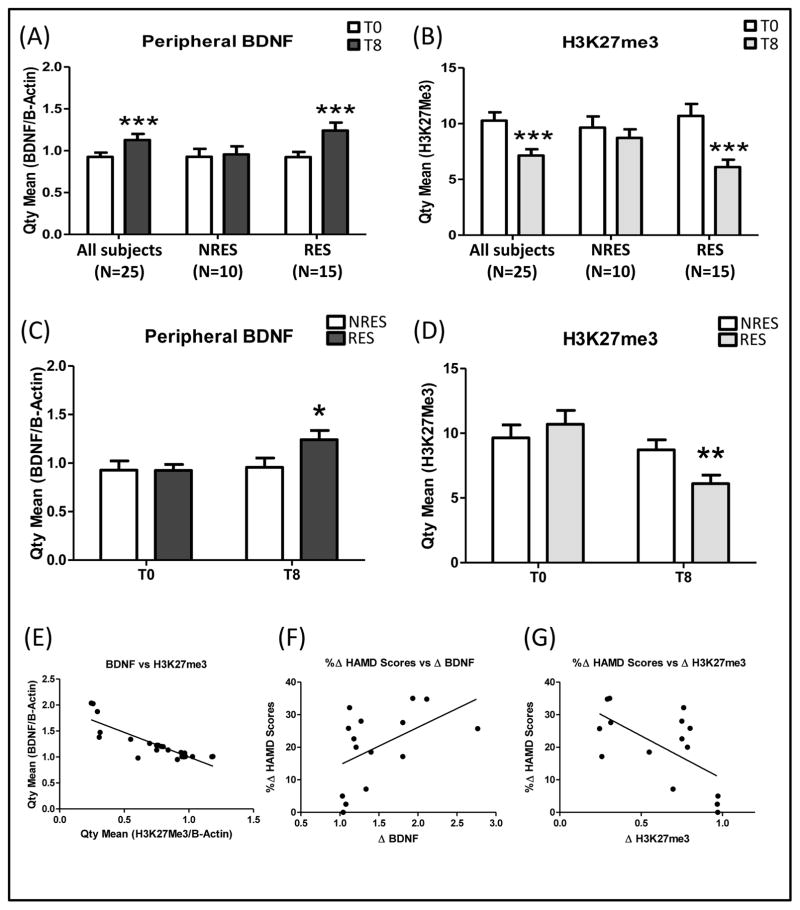
A repeated measures ANOVA with Bonferroni correction revealed that in line with previous findings, the expression of peripheral BDNF mRNA from depressed patients (N=25) was significantly elevated after 8-weeks of citalopram treatment (FC=34%; p<0.001; Fig 1A). Subjects were classified into responders (RES) and non-responders (NRES), based on changes to the HAM-D scores. We defined response as 8-week HAMD scores < 9, whereas non-response was defined as 8-week HAMD scores higher than 50% reduction in baseline HAMD scores (T8; RES =5.6 ± 0.7; N=15; NRES =17.0 ± 1.4; N=10). The RES group showed increased BDNF levels (T8-T0; FC=49%; p<0.001; Fig 1A) after treatment, while there was no significant difference in the NRES group (T8-T0; FC=3%; p>0.05; Fig 1A). Consistently, RES had higher T8 BDNF levels as compared to NRES (T-test p<0.05; Fig 1C), while there was no difference at T0 (T-test p=0.98; Fig 1C). Finally, we found a significant correlation between change in depression severity and change in BDNF expression (Pearson r=0.49; R2=0.25; p<0.05; Fig 1F). These findings indicated a relationship between peripheral BDNF expression and citalopram treatment response.
Figure 1. Epigenetic regulation of BDNF.
(A)(C) qRT-PCR AQ values of total BDNF and two endogenous controls (B-Actin and GAPDH). (B)(D) H3K27me3 levels at BDNF IV promoter normalized to input DNA and B-Actin as controls. (E) Correlation between total BDNF and H3K27me3 levels. (F) Correlation between change in depression severity and change in BDNF expression. (G) Correlation between change in depression severity and change in H3K27me3 expression. Depressed patients (N=25) were classified into responders (RES) and non-responders (NRES). The asterisks refer to p-values (*p≤0.05; **p≤0.01; ***p≤0.001). Variance bars represent standard error of the mean.
To investigate the role of chromatin modifications in BDNF expression changes, based on previous findings in rodents (7), we performed Chromatin Immunoprecipitation (ChIP) and found a significant decrease in H3K27me3 levels at promoter-IV of the BDNF gene after 8-weeks of citalopram treatment in all patients (N=25) according to a repeated measures ANOVA with Bonferroni correction (FC = 31%; p<0.001; Fig 1B). However, these results were explained primarily by changes in the RES group (FC=43%; p<0.001; Fig 1B), and there was no significant difference in the NRES group (p>0.05; Fig 1B). Consistently, we found a significant difference in H3K27me3 levels at T8 between groups (T-test p<0.01; Fig 1D), but no difference at T0 (T-test p>0.05; Fig 1D). Furthermore, we found a significant negative correlation between change in depression severity and change in H3K27me3 expression (Pearson r=−0.63; R2=0.39; p<0.01; Fig 1G). Finally, total BDNF and H3K27me3 levels were significantly negatively correlated (Pearson r=−0.86; R2=0.75; p<0.0001; Fig 1E).
To our knowledge, this is the first study that translates to human findings previously reported in an animal model of depression,7 reporting evidence suggesting that antidepressants regulate BDNF expression through alterations of promoter-IV H3K27me3 levels. Our results suggest that changes in H3K27 methylation state of the BDNF promoter IV and BDNF expression levels in peripheral tissue are biomarker correlates of antidepressant response. Finally, these findings are preliminary and await confirmation by larger samples and alternative designs. Moreover, additional work is necessary to better understand the relationship between epigenetic modifications and the exon-specific regulation of BDNF, as this has the potential to lead to new therapeutic options for MDD.
References
- 1.Dwivedi Y. Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatric disease and treatment. 2009;5:433–449. doi: 10.2147/ndt.s5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molendijk ML, Bus BA, Spinhoven P, Penninx BW, Kenis G, Prickaerts J, et al. Serum levels of brain-derived neurotrophic factor in major depressive disorder: state-trait issues, clinical features and pharmacological treatment. Molecular psychiatry. 2011;16(11):1088–1095. doi: 10.1038/mp.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90(3):397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Molecular psychiatry. 2011 doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- 5.Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33(3):383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- 6.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 7.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature neuroscience. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 8.Chen ES, Ernst C, Turecki G. The epigenetic effects of antidepressant treatment on human prefrontal cortex BDNF expression. Int J Neuropsychopharmacol. 2011;14(3):427–429. doi: 10.1017/S1461145710001422. [DOI] [PubMed] [Google Scholar]