Abstract
IMPORTANCE
Despite the moderate, well-demonstrated heritability of major depressive disorder (MDD), there has been limited success in identifying replicable genetic risk loci, suggesting a complex genetic architecture. Research is needed to quantify the relative contribution of classes of genetic variation across the genome to inform future genetic studies of MDD.
OBJECTIVES
To apply aggregate genetic risk methods to clarify the genetic architecture of MDD by estimating and partitioning heritability by chromosome, minor allele frequency, and functional annotations and to test for enrichment of rare deleterious variants.
DESIGN, SETTING, AND PARTICIPANTS
The CONVERGE (China, Oxford, and Virginia Commonwealth University Experimental Research on Genetic Epidemiology) study collected data on 5278 patients with recurrent MDD from 58 provincial mental health centers and psychiatric departments of general medical hospitals in 45 cities and 23 provinces of China. Screened controls (n = 5196) were recruited from a range of locations, including general hospitals and local community centers. Data were collected from August 1, 2008, to October 31, 2012.
MAIN OUTCOMES AND MEASURES
Genetic risk for liability to recurrent MDD was partitioned using sparse whole-genome sequencing.
RESULTS
In aggregate, common single-nucleotide polymorphisms (SNPs) explained between 20% and 29% of the variance in MDD risk, and the heritability in MDD explained by each chromosome was proportional to its length (r = 0.680; P = .0003), supporting a common polygenic etiology. Partitioning heritability by minor allele frequency indicated that the variance explained was distributed across the allelic frequency spectrum, although relatively common SNPs accounted for a disproportionate fraction of risk. Partitioning by genic annotation indicated a greater contribution of SNPs in protein-coding regions and within 3′-UTR regions of genes. Enrichment of SNPs associated with DNase I-hypersensitive sites was also found in many tissue types, including brain tissue. Examining burden scores from singleton exonic SNPs predicted to be deleterious indicated that cases had significantly more mutations than controls (odds ratio, 1.009; 95% CI, 1.003–1.014; P = .003), including those occurring in genes expressed in the brain (odds ratio, 1.011; 95% CI, 1.003–1.018; P = .004) and within nuclear-encoded genes with mitochondrial gene products (odds ratio, 1.075; 95% CI, 1.018–1.135; P = .009).
CONCLUSIONS AND RELEVANCE
Results support a complex etiology for MDD and highlight the value of analyzing components of heritability to clarify genetic architecture.
Major depressive disorder (MDD) is a common psychiatric disorder and a leading cause of disability worldwide.1 Global estimates of lifetime MDD prevalence range from 2.1% to 21.0%.2 The heritability of MDD is estimated as 37% from a meta-analysis of twin and family studies,3 supporting a complex etiology that includes both genetic and environmental factors. Identifying specific genetic variants that influence risk remains a challenge.
Genome-wide association studies (GWAS) have identified risk variants for many psychiatric disorders, but until recently, no replicated genome-wide significant loci had been identified for MDD, as clinically defined by the Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition, Text Revision).4,5 This lack of genome-wide significant loci may reflect the etiological heterogeneity of MDD, especially given the evidence that the genetic liability to MDD is only partially shared between the sexes.6,7 The CONVERGE (China, Oxford, and Virginia Commonwealth University Experimental Research on Genetic Epidemiology) study of MDD was designed to reduce phenotypic and genetic heterogeneity by examining only severe cases and carefully screened control patients, all of whom were female and of Han Chinese ancestry. Using sparse whole-genome sequencing, we detected and replicated 2 common variants that contribute to MDD risk.5 Not unexpectedly, these genome-wide significant loci accounted for only a small fraction of variance in MDD liability (approximately 0.6%). Given the polygenic nature of MDD, many additional loci likely contribute to disease risk but are of too small effect to attain genome-wide significance in our current sample.
However, aggregate analyses of single-nucleotide polymorphism (SNP) data have proven instrumental in furthering our understanding of complex trait genetics. For example, support for the polygenic basis of schizophrenia was demonstrated by the predictive value of polygenic risk scores.8 An alternative genome-wide approach derives narrow-sense heritability of quantitative traits by simultaneously considering all SNPs to estimate additive genetic variance.9 The Cross-Disorder Group of the Psychiatric Genomics Consortium used this approach to estimate SNP-based heritability of MDD at approximately 21%.10 In addition, significant associations with polygenic burden of private disruptive mutations from whole-exome sequencing have been reported for psychiatric disease, including schizophrenia.11
Here, we leverage advances in statistical methodologies to delineate the genetic architecture of MDD. Using genomic annotation databases, such as the Encyclopedia of DNA Elements, the enrichment of variants in regulatory elements and protein-coding regions can be assessed.12,13 Given our whole-genome sequencing data, enrichment of rare deleterious variants can also be tested. We apply an aggregate genetic risk method to estimate and partition heritability by chromosome, minor allele frequency (MAF), and various functional annotations as well as test for enrichment of rare deleterious variants. Our dense set of markers, which captures significantly more common and rare variation than is present on genotyping arrays, allows for a unique opportunity to add insight into the genetic architecture of this common and debilitating psychiatric disorder.
Key Points.
Question
What is the genetic architecture of recurrent major depressive disorder (MDD) in Han Chinese women?
Findings
In this case-control study of MDD, aggregate genetic risk accounted for 21.4% of the variance in MDD liability with significant heritability found across chromosomes and the allelic spectrum. Enrichment of variant associations was seen in protein-coding regions, 3′ UTR, and DNase I-hypersensitive sites, as was significant burden of singleton exonic variants in MDD, particularly in genes expressed in the brain or with mitochondrial gene products.
Meaning
Results confirm a complex genetic architecture for MDD, supporting etiological mechanisms for both common and rare genetic variation to MDD risk.
Methods
Sample Collection
Recurrent MDD cases were recruited from 58 provincial mental health centers and psychiatric departments of general medical hospitals in 45 cities and 23 provinces of China. Controls were recruited from several locations, including general hospitals and local community centers. All participants were Han Chinese women with 4 Han grand parents. Cases were aged between 30 and 60 years and had 2 or more episodes of MDD that met the criteria of the Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition, Text Revision),14 with the first episode occurring between ages 14 and 50 years; had not abused drugs or alcohol before their first depressive episode; and reported no history of schizophrenia or mania.
Data collection took place from August 1, 2008, to October 31, 2012. The study protocol was approved by the Ethical Review Board of Oxford University and the ethics committees of all participating hospitals in China. All participants provided written informed consent. Details on DNA sequencing and imputation of genotypes have been previously reported5 and are summarized in the eAppendix in the Supplement.
Population Stratification
To address population stratification, we constructed 10 ancestry principal components (PC) using EIGENSOFT 3.0 and smartpca (Harvard University).15,16 To circumvent overfitting, we used only PC1 and PC2, which distinguished north-south regional differences (eFigure 1 in the Supplement). Details appear in the eAppendix in the Supplement.
Estimation of SNP-Based Heritability of MDD
Single-nucleotide polymorphism-based heritability estimates were obtained using Genome-wide Complex Trait Analysis (GCTA), version 1.24.7,9 and Linkage Disequilibrium Adjusted Kinship (LDAK), version 5.9.17 Genetic relatedness matrices (GRMs) were constructed from 4.7M hard called SNPs that passed several quality control parameters: genotype probability (Pr[G]) of 0.9 or more, less than 1% missing rate, MAF of 0.005 or more, and Hardy-Weinberg equilibrium P > 10−6. To estimate the contribution of each chromosome to the total heritability as well as to test for inflation due to cryptic relatedness, we constructed GRMs for each chromosome and estimated per chromosome heritability using each GRM separately and all GRMs jointly. We partitioned SNPs into MAF quintiles (0.005–0.50) and estimated the proportion of variance contributed by each quintile using the multicomponent GREML approach. To assess the relative contribution of heritability of SNPs in functional categories, we partitioned SNPs into functional annotations (eg, exon, intron, or 3′UTR) using ANNOVAR, version 2015 (QIAGEN Bioinformatics).18 The functional classes were fitted jointly in a single GREML model.
To account for effects of uneven linkage disequilibrium, we applied the GCTA-LDMS19 and the LDAK17 approaches. In GCTA-LDMS, we calculated the linkage disequilibrium scores of all SNPs using a sliding-window approach (200 kB with 100-kB overlap between adjacent segments) and then partitioned them into linkage disequilibrium quartiles. Each linkage disequilibrium quartile was then partitioned into MAF quintiles, resulting in 20 GRMs that were fitted jointly. Using LDAK, we generated SNP weights that reflect a correlation with surrounding markers to construct GRMs adjusted for local linkage disequilibrium. For both methods, a relatedness filter (−grm-cutoff 0.05) was applied, giving a final sample of 10 474. We transformed the binary MDD disease status to the liability scale, assuming a prevalence of 8%(eAppendix in the Supplement). PC1 and PC2 were included as covariates.
Polygenic Risk Prediction
We constructed polygenic risk scores within CONVERGE by 2 methods. First, we randomly divided our sample (50–50 split) into independent subsets (sample 1 and sample 2). We conducted GWAS of each subset, subsequently performing linkage disequilibrium–based “clumping” to remove highly correlated markers (r2>0.1) while retaining the most significant SNP within 500-kB intervals. Using these linkage disequilibrium–independent SNPs, we computed per-individual polygenic scores on the basis of varying P value threshold signifying the proportion of SNPs with smaller P values in the training set; P value thresholds ranged from 0.001 to 1.8 Second, using the sample 1–sample 2 split, we also estimated SNP effects by the best linear unbiased prediction (BLUP) method implemented in GCTA.9 The latter scores were constructed with the profile option in PLINK,20 using SNP BLUP solutions as weights. We tested case-control differences by logistic regression with ancestry PC as covariates. The predictive value of these scores is reported in terms of Nagelkerke’s pseudo-R2 (fmsb package in R; package authored by Minato Nakasawa).
Enrichment of DNase I-Hypersensitive Sites
Studies have reported that SNPs with small P values, including those that do not reach genome-wide significance, are enriched for DNase I-hypersensitive sites (DHSs) in tissues related to the phenotype.21 We obtained DNase peaks from the Encyclopedia of DNA Elements project data website (https://genome.ucsc.edu/ENCODE/). We identified all SNPs with association P values with MDD less than threshold values (−log10[p] = 0, 0.5, 1, 1.5, ⋯), and then we computed the proportion of SNPs lying in DHSs. To determine the statistical significance of any particular enrichment curve (ie, how unlikely under the null hypothesis of no enrichment), we assessed the statistical significance of enrichment on the intervals between −log10(p), between 5 and 6, and separately upward of 6 by binomial tests, and then we combined these P values by the Fisher exact method. We determined 95% CIs for enrichment curves by bootstrapping and assessed significance by empirical null distributions (eAppendix in the Supplement).
Rare Variant Annotation and Enrichment Analysis
Methods for calling rare exonic variation from 1× sequencing appear in the eAppendix in the Supplement (eTable 5 and eFigures 4, 5, and 6). Exon coordinates contained 96 130 824 base pair positions in 254 986 exons in 21 946 genes. Singletons (both SNPs and INDELs [insertions and deletions])were called when 2 or more reads supported the same alternative allele in a single sample. All exonic SNPs were annotated using ANNOVAR.18 Variants of each annotation category and in each gene were aggregated for every individual and used in logistic regression as predictors of MDD, controlling for measures associated with sequencing runs, batch, read mapping quality, sequence coverage over the genome, GC (guanine-cytosine) content, PC from the common variant analysis, and city of origin. Permutations were performed to verify that P values were not inflated (eAppendix in the Supplement).
Results
MDD as a Polygenic Disease
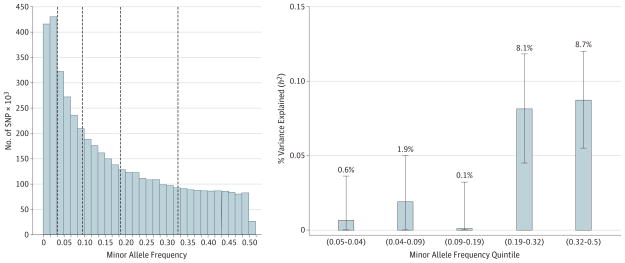
Using approximately 4.7M autosomal and X chromosome SNPs, we estimated that 21.4% of the variance in MDD risk (95% CI, 15.5–27.3; P < 1.0 × 10−16) is captured by genome-wide common variants (n = 10 474). eTable 1 in the Supplement shows heritability estimates based on varying MDD prevalence, which increases with higher prevalence rates. The variance in MDD explained by each chromosome was proportional to its length (r = 0.680; r2 = 0.463; P = .0003). Heritability estimates for separate vs joint analyses of all chromosomes indicated a negligible effect of confounding population structure (joint h2 = 21.4%; separate h2 = 23.6%) (eFigure 2 in the Supplement). To assess the relative contribution of MAF to heritability estimates, we partitioned SNPs into MAF quintiles. Higher-frequency SNPs (>19%) accounted for most of the heritability (Figure 1). As we are using imputed SNPs and therefore a denser set of markers than on genotyping arrays, we accounted for the biasing effect of uneven linkage disequilibrium by 2 methods. eTable 2 in the Supplement shows results for GCTA-LDMS, which partitioned heritability by linkage disequilibrium quartile and MAF to correct for region-specific linkage disequilibrium heterogeneity and indicated minimal bias in our unstratified heritability estimate of 21.4% vs 20.0%(SE = 3.4%) for LDMS. An alternative approach using LDAK, which accounts for local linkage disequilibrium by weighting all SNPs on the basis of correlations with surrounding SNPs, estimated heritability at 29.4% (SE = 4.6%; P = 9.09 × 10−11).
Figure 1. Variance in Major Depressive Disorder Risk Explained by Single-Nucleotide Polymorphisms (SNP) of Varying Minor Allele Frequency.
Genome-wide Complex Trait Analysis estimates of SNP h2 for major depressive disorder partitioned by minor allele frequency quintile. Error bars represent 95% CIs.
Polygenic Risk Prediction
Polygenic risk scores significantly predicted MDD disease status (eTables 3 and 4 in the Supplement). We attained the greatest predictive power using BLUP solutions (eTable 4 in the Supplement); this score was associated with MDD (P < 4.6 × 10−5), accounting for 1.1% of the variability in MDD risk. When applying the P value threshold method, we attained the greatest predictive ability using P(t)<0.4; this score was associated with MDD (P < 3.0 × 10−6), accounting for 0.55% of variability in MDD liability (eTable 3 in the Supplement).
Functional Variant Contribution to MDD
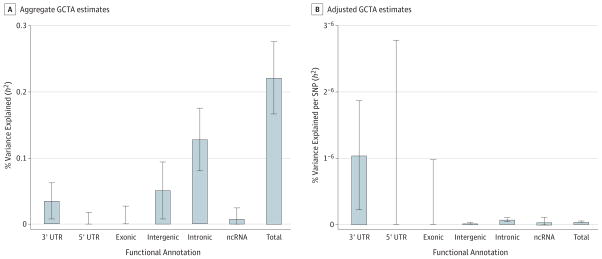
To assess the contribution of heritability due to SNPs in coding vs noncoding regions, we partitioned SNPs into their proposed genic annotations. When partitioning SNPs into 3′-UTR, 5′-UTR, exonic, and intronic regions, those in introns and 3′ UTR were significantly enriched for disease-relevant effects (Figure 2A). Considering the total number of SNPs in each functional category relative to the aggregate variance explained, the pattern of findings suggests that 3′-UTR effects may be particularly important to the etiology of MDD (Figure 2B).
Figure 2. Estimates of the Variance Explained by Genic and Intergenic Regions.
A, Aggregate Genome-wide Complex Trait Analysis (GCTA) estimates.
B, Adjusted (per SNP [single-nucleotide polymorphism]) GCTA estimates of SNP h2 partitioned by expected functional category. SNPs were mapped to 3′- or 5′-UTR, exonic, or intronic regions of known protein-coding genes or intergenic and ncRNA regions. Error bars represent 95% CIs, ncRNA, non-coding RNA.
Enrichment of SNPs in DHS
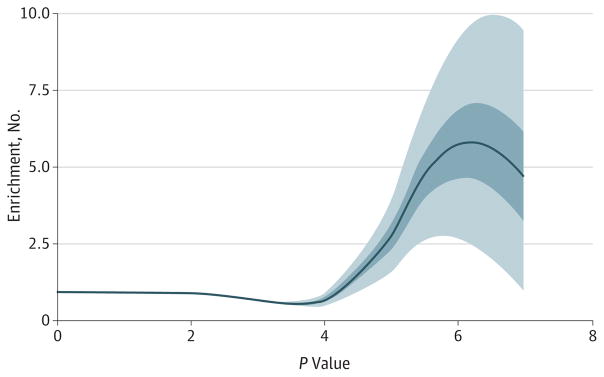
We find enrichment of SNPs with low P values associated with MDD in DHS of many cell types, including brain-related tissues. Figure 3 shows the enrichment curves for DHS annotated in 1 brain sample with bootstrap confidence intervals. Single-nucleotide polymorphisms with P values under 10−5 associated with MDD are 5 times as likely to lie in a DHS in this brain sample from the frontal cortex as are SNPs taken at random. The probability of an enrichment curve at least this high under random per mutations of DNase status of SNPs is less than 0.001. eFigure 3 in the Supplement shows enrichment of DHS in all of the samples available in July 2014 from the Encyclopedia of DNA Elements. While the 4 brains are among the most enriched tissues for MDD-associated SNPs in DHS, samples from the liver and pancreas also showed comparable enrichment.
Figure 3. Enrichment of Single-Nucleotide Polymorphisms With Small P Values in Major Depressive Disorder Analysis for DNase I-Hypersensitive Site in Frontal Cortex.
Enrichment curve for “Frontal Cortex OC” is a Loess curve interpolating the ratio of the number of single-nucleotide polymorphism (SNPs) whose association P value is smaller than various thresholds (x-axis) to the proportion of P values from all measured SNPs in DNase I-hypersensitive sites smaller than the same thresholds. The dark and light blue areas display 50% and 95% CIs, respectively, obtained by bootstrapping SNP sets.
Cumulative Burden of Private Deleterious Exonic Variants
We used low-coverage sequence data to test whether MDD cases have a polygenic burden of rare deleterious coding variants. For this analysis, we analyzed only SNPs. The Table shows a significant (odds ratio [OR], 1.011; 95% CI, 1.003–1.018; P = .004) excess of single ton deleterious mutations in brain-expressed genes in cases. In contrast, no significant enrichment was seen for associations between MDD and variants in genes not expressed in the brain (OR, 1.000; 95% CI, 0.994–1.014; P = .41). We have reported that CONVERGE cases have more mitochondrial DNA than controls.22 With our finding of loci near a gene with mitochondrial functions (SIRT1, an NAD+-dependent histone deacetylase and a mitochondrial ion transporter),5 we inquired whether singleton deleterious mutations would be enriched in nuclear-encoded genes with mitochondrial localized gene products. A significant enrichment in deleterious variants in nuclear-encoded mitochondrial genes (OR, 1.075; 95% CI, 1.018–1.135; P = .009) was found. We then applied a permutation-based method to investigate whether the ORs were significantly different from the average gene genome-wide value. We randomly selected an amount of coding DNA equal in length to that used when the analysis is restricted to genes expressed in the brain and in nuclear-encoded mitochondrial genes and then repeated the analyses 10000 times. The empirical P value was .04 for the OR observed in the brain-expressed gene set and was .02 for that in the nuclear-encoded mitochondrial genes (eFigure 7 in the Supplement). Because these tests explore whether the 2 ORs are significantly different, we applied a Bonferroni corrected threshold of 0.025 (0.05/2).
Table.
Polygenic Burden of Private Deleterious Exonic Variantsa
| Gene Set/Variant Type | No. of Variants | Odds Ratio (95% CI) | P Value |
|---|---|---|---|
| All (n = 18 169) | |||
| All variants | 302 838 | 1.005 (1.001–1.009) | .03 |
| Nonsynonymous | 179 546 | 1.008 (1.001–1.014) | .03 |
| Synonymous | 96 970 | 0.997 (0.986–1.008) | .61 |
| All deleterious | 202 374 | 1.009 (1.003–1.014) | .003 |
| Brain Expressed (n = 10 897) | |||
| All variants | 192 137 | 1.006 (0.999–1.011) | .07 |
| Nonsynonymous | 113 136 | 1.013 (1.004–1.023) | .006 |
| Synonymous | 62 882 | 0.992 (0.977–1.004) | .18 |
| All deleterious | 127 057 | 1.011 (1.003–1.018) | .004 |
| Non–Brain Expressed (n = 7272) | |||
| All variants | 110 701 | 0.999 (0.992–1.009) | .87 |
| Nonsynonymous | 66 410 | 0.996 (0.983–1.009) | .52 |
| Synonymous | 34 088 | 0.988 (0.970–1.008) | .26 |
| All deleterious | 75 317 | 1.000 (0.994–1.014) | .41 |
| Nuclear-Encoded Mitochondrial (n = 940) | |||
| All variants | 11 103 | 1.053 (1.001–1.096) | .04 |
| Nonsynonymous | 6536 | 1.063 (1.001–1.129) | .04 |
| Synonymous | 3226 | 1.004 (0.943–1.069) | .91 |
| All deleterious | 7414 | 1.075 (1.018–1.135) | .009 |
This table shows the increased risk of major depressive disorder with the number of rare coding variants (in which rare means that each variant is found only in a single individual in the CONVERGE [China, Oxford, and Virginia Commonwealth University Experimental Research on Genetic Epidemiology] sample). Numbers of variants are given for coding variants in all genes; for those expressed in the brain; for nuclear genes that have a role in mitochondrial function; and for brain-expressed genes, excluding mitochondrial genes. The table gives the odds ratio, the odds ratio 95% confidence intervals, and the P values of this analysis for the complete set of variants and for variants annotated by their predicted function. The category all deleterious includes nonsynonymous, stop-gain, stop-loss, and deletion frame shift variants.
Discussion
We extended our work in CONVERGE beyond identifying specific risk variants by evaluating aggregate contributions of molecular variation to risk for MDD. There are several noteworthy conclusions. First, we estimated the lower bound of narrow-sense heritability as between 20% and 29% depending on the method applied and the assumed MDD prevalence. Although these estimates were similar to those reported for populations of European descent (approximately 21%)10 but lower than the 37% reported by previous twin studies,3 heritability is a population-specific measure. Our results apply to Han Chinese women, aged between 30 and 60 years, with recurrent depression.
Second, our results support a substantial polygenic component to the risk of MDD involving many alleles of individually very small effect. Genome-wide polygenic risk scores constructed from SNPs were significantly associated with MDD liability, accounting for 1.1% of the variance in risk compared with 0.6% estimated by a similar method for European samples.23 Significant heritability was found across all chromosomes, with the amount of variance explained proportional to length, further demonstrating an underlying polygenic architecture of MDD.
It has been suggested that common variants have a smaller role in the etiology of MDD than originally posited by the common-disease–common-variant hypothesis because of the low proportion of variance explained by earlier GWAS.24,25 However, we found that the bulk of detectable heritability comes from common variants (MAF>0.19, the 2 topmost quintiles). This finding contrasts with the finding of a similar analysis carried out on a large schizophrenia cohort, in which heritability was distributed more evenly across the quintiles.26 The excess of heritability attributable to the most common MAFs is, in part, possibly because the small reduction in reproductive fitness associated with mood disorders exerts little selective power to drive risk variants to lower allele frequency.27,28
We found that particular functional categories of the genome contribute disproportionately to the heritability of MDD. Specifically, SNPs in genic regions, especially those in introns and 3′ UTR, explain more variance than in noncoding regions. We also found an enrichment of SNPs in DHSs, which mark transcriptionally active regions of the genome, in several tissue types, including brain tissue. Recently, Finucane et al29 have reported enrichment of functional elements in 17 complex traits and diseases, including 3 psychiatric disorders. They found significant enrichment in coding regions for schizophrenia and bipolar disorder as well as enrichment in 3′ UTR for schizophrenia. Performing a similar analysis on depressive symptoms, Okbay et al30 also reported enrichment of SNPs in DHSs but did not find enrichment of intron or 3′-UTR sites.
Regarding enrichment of DHSs, several other tissues, including the liver and pancreas, showed enrichment comparable to brain tissue. We propose 2 explanations for this finding. First, it is possible that DHSs are enriched in tissues other than brain tissue given that we have prior evidence of the role of genes with mitochondrial function in MDD,5 metabolism is regulated in many tissues, and many regulatory mechanisms are common to many tissues. Second, regulatory elements in brain cells are harder to identify by DHS because of greater cell-type heterogeneity than is found in most somatic tissues.
We report for the first time, to our knowledge, that, compared with controls, MDD cases had significantly more singleton deleterious SNPs in exons than controls. Similar results have been found for schizophrenia.11 We also showed that variation in nuclear-encoded mitochondrial genes contributes to the risk of MDD. Notably, MDD is reported as a comorbid illness in some human mitochondrial diseases, including those arising from mutations in genes that regulate mitochondrial DNA integrity; for example, depressive episodes are reported in patients who carry mutations in POLG1 (OMIM 174763).31 The identification of mitochondrial genes as risk factors for MDD might also explain some clinical features of the illness. For example, SIRT1 (OMIM 604479) influences processes that feature among the vegetative symptoms32 of MDD: alterations in food intake,33 wake fulness,34 and circadian rhythms.35 The involvement of mitochondrial genes might also explain why MDD increases the risk of cardiovascular disease.36
Design elements of CONVERGE sought to reduce genetic and phenotypic heterogeneity. Cases were recurrent and quite severe, with approximately 85% meeting the criteria for melancholia. An important theoretical question is the expected pattern of findings if we selected a more homogeneous and more severely ill cohort. We are guided by the only empirical study we know regarding this question. Using population-based female twins, Kendler37 tested a multiple-threshold model in which melancholia exists as a more severe form on the same continuum of liability as nonmelancholic MDD. This model fit the data well, and the heritability of melancholia was not different from nonmelancholic MDD, as expected under the liability threshold model. Based on these findings, we predict that the heritability of MDD in CONVERGE would not differ substantially from other samples, but the CONVERGE sample, in general, and our melancholic cases, on average, would have higher genetic liability. While SNP-based heritability estimates for melancholic and nonmelancholic MDD were not significantly different (eTable 6 in the Supplement), polygenic risk scores were more predictive of melancholic rather than nonmelancholic MDD (P = .002) (eTable 7 in the Supplement).
Conclusions
Our results are consistent with a polygenic architecture for MDD. A significant proportion of variance was due to common variants, although rare variation also appears to contribute to MDD disease liability. The genome partitioning results presented here provide direction for functional follow-up and will inform future studies. Taken together, our results support a complex etiology for MDD and highlight the value of partitioning heritability to better delineate the genetic architecture of this common, disabling psychiatric disorder.
Acknowledgments
Funding/Support: This work was funded by the Wellcome Trust WT090532/Z/09/Z, WT083573/Z/07/Z, and WT089269/Z/09/Z as well as by National Institutes of Health (NIH) grant MH100549. Dr Peterson is supported by NIH T32 grant MH020030. Dr Cai is supported by EBI-Sanger Postdoctoral Fellowship. Dr Bacanu is supported by NIH grants R21MH100560 and R21AA022717.
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: None reported.
Author Contributions: Drs Kendler and Flint had full access to all the data and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Peterson, Cai, and Bigdeli are first coauthors and contributed equally to this work. Drs Flint and Kendler are joint senior authors.
Study concept and design: Peterson, Cai, Bigdeli, Webb, Bacanu, Flint, Kendler.
Acquisition, analysis, or interpretation of data: Peterson, Cai, Bigdeli, Li, Reimers, Nikulova, Webb, Riley, Kendler.
Drafting of the manuscript: Peterson, Cai, Bigdeli, Reimers, Nikulova, Webb, Flint, Kendler.
Critical revision of the manuscript for important intellectual content: Peterson, Cai, Bigdeli, Li, Reimers, Webb, Bacanu, Riley, Kendler.
Statistical analysis: Peterson, Cai, Bigdeli, Li, Reimers, Nikulova, Webb, Bacanu, Flint.
Obtained funding: Webb, Riley, Flint, Kendler.
Administrative, technical, or material support: Peterson, Kendler.
Study supervision: Webb, Bacanu, Flint, Kendler.
Additional Contributions: All authors are part of the CONVERGE (China, Oxford, and Virginia Commonwealth University Experimental Research on Genetic Epidemiology) consortium and gratefully acknowledge the support of all CONVERGE partners in hospitals across China. Special thanks to all the CONVERGE collaborators and patients who made our work possible.
References
- 1.Ferrari AJ, Charlson FJ, Norman RE, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med. 2013;10(11):e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler RC, Bromet EJ. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119–138. doi: 10.1146/annurev-publhealth-031912-114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 4.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81(3):484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CONVERGE Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kendler KS, Gatz M, Gardner CO, Pedersen NL. A Swedish national twin study of lifetime major depression. Am J Psychiatry. 2006;163(1):109–114. doi: 10.1176/appi.ajp.163.1.109. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Gardner CO, Neale MC, Prescott CA. Genetic risk factors for major depression in men and women: similar or different heritabilities and same or partly distinct genes? Psychol Med. 2001;31(4):605–616. doi: 10.1017/s0033291701003907. [DOI] [PubMed] [Google Scholar]
- 8.Purcell SM, Wray NR, Stone JL, et al. International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SH, Ripke S, Neale BM, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium. A user’s guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusev A, Lee SH, Trynka G, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. AmJ Hum Genet. 2014;95(5):535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: 4th ed, text revision. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91(6):1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Bakshi A, Zhu Z, et al. Life Lines Cohort Study. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai N, Chang S, Li Y, et al. Molecular signatures of major depression. Curr Biol. 2015;25(9):1146–1156. doi: 10.1016/j.cub.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripke S, Wray NR, Lewis CM, et al. Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11(6):415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 26.Loh PR, Bhatia G, Gusev A, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47(12):1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry. 2009;14(12):1072–1082. doi: 10.1038/mp.2009.85. [DOI] [PubMed] [Google Scholar]
- 28.Williams KE, Marsh WK, Rasgon NL. Mood disorders and fertility in women: a critical review of the literature and implications for future research. Hum Reprod Update. 2007;13(6):607–616. doi: 10.1093/humupd/dmm019. [DOI] [PubMed] [Google Scholar]
- 29.Finucane HK, Bulik-Sullivan B, Gusev A, et al. Repro Gen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okbay A, Baselmans BML, De Neve JE, et al. Life Lines Cohort Study. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-γ and human disease. Hum Mol Genet. 2006;15(Spec2):R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 32.Kim HD, Hesterman J, Call T, et al. SIRT1 mediates depression-like behaviors in the nucleus accumbens. J Neurosci. 2016;36(32):8441–8452. doi: 10.1523/JNEUROSCI.0212-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoh A, Brace CS, Ben-Josef G, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30(30):10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panossian L, Fenik P, Zhu Y, Zhan G, McBurney MW, Veasey S. SIRT1 regulation of wakefulness and senescence-like phenotype in wake neurons. J Neurosci. 2011;31(11):4025–4036. doi: 10.1523/JNEUROSCI.5166-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson A, Kuper H, Hemingway H. Depression as an aetiologic and prognostic factor in coronary heart disease: a meta-analysis of 6362 events among 146 538 participants in 54 observational studies. Eur Heart J. 2006;27(23):2763–2774. doi: 10.1093/eurheartj/ehl338. [DOI] [PubMed] [Google Scholar]
- 37.Kendler KS. The diagnostic validity of melancholic major depression in a population-based sample of female twins. Arch Gen Psychiatry. 1997;54(4):299–304. doi: 10.1001/archpsyc.1997.01830160013002. [DOI] [PubMed] [Google Scholar]