Abstract
Glycoprotein gp120 is a surface antigen and virulence factor of human immunodeficiency virus 1. Broadly neutralizing antibodies (bNAbs) that react to gp120 from a variety of HIV isolates offer hope for the development of broadly effective immunogens for vaccination purposes, if the interactions between gp120 and bNAbs can be understood. From a structural perspective, gp120 is a particularly difficult system because of its size, the presence of multiple flexible regions, and the large amount of glycosylation, all of which are important in gp120–bNAb interactions. Here, the interaction of full-length, glycosylated gp120 with bNAb b12 is probed using high-resolution hydroxyl radical protein footprinting (HR-HRPF) by fast photochemical oxidation of proteins. HR-HRPF allows for the measurement of changes in the average solvent accessible surface area of multiple amino acids without the need for measures that might alter the protein conformation, such as mutagenesis. HR-HRPF of the gp120–b12 complex coupled with computational modeling shows a novel extensive interaction of the V1/V2 domain, probably with the light chain of b12. Our data also reveal HR-HRPF protection in the C3 domain caused by interaction of the N330 glycan with the b12 light chain. In addition to providing information about the interactions of full-length, glycosylated gp120 with b12, this work serves as a template for the structural interrogation of full-length glycosylated gp120 with other bNAbs to better characterize the interactions that drive the broad specificity of the bNAb.
The human immunodeficiency virus 1 (HIV-1) gp120 envelope glycoprotein is the major target of neutralizing antibodies.1,2 The gp120 molecule consists of a polypeptide core of roughly 60 kDa. Extensive modification by N-linked glycosylation increases the molecular weight of the molecule to ∼120 kDa.3 The amino acid sequence of gp120 is composed of five conserved regions (C1–C5) and five variable regions (V1–V5), many of which are highly flexible. The majority of antibodies raised against gp120 have very narrow ranges of effectiveness and are eventually evaded by the virus. However, a subset of raised antibodies have been found to be effective against a broader array of isolates. The development of a vaccine immunogen that elicits these broadly neutralizing antibodies (bNAbs) and confers protective immunity remains a challenge. Improved knowledge of the Env structure and what constitutes a full neutralization epitope will aid in rational immunogen design to elicit potent bNAbs. However, gp120 is a very challenging molecule for structural biology. The extensive glycosylation, diversity of isoforms, and broad conformational flexibility of gp120 pose formidable barriers for crystallization. To surmount these difficulties and construct a crystal structure of gp120, sources of likely conformational heterogeneity such as N-linked carbohydrates, flexible or mobile N- and C-termini, and variable internal loops (like V1/V2 and/or V3) are often reduced or eliminated, and ligands such as CD4 are used to restrict conformational mobility and to alter the crystallization surface.4−12 These stabilized structures provide valuable information at high resolution, but at the cost of eliminating regions that have been shown to be important for many gp120–antibody interactions.13
The first broadly neutralizing human monoclonal antibody (mAb), b12, was isolated from clade B-infected patients and binds to gp120 at and near its CD4 binding site (CD4bs).7,10,14 Binding of b12 to the surface of gp120 blocks attachment of CD4 and thus prevents the entry of HIV-1 into a target cell.7,10 Therefore, gp120 appears to present the b12 epitope in conjunction with several other weakly neutralizing and overlapping epitopes. However, while several other CD4bs antibodies with potency and breadth greater than those of b12 have been discovered since then, b12 remains a valuable model for anti-CD4bs bNAbs because of its history of experimental study.15−17 A crystal structure of b12 in complex with a truncated, deglycosylated, and mutationally stabilized gp120 core [Protein Data Bank (PDB) entry 2NY7] has revealed that the contacts between b12 and gp120 are centered around the CD4 binding loop spanning residues 364–373 but involves many other residues.10 The truncated, deglycosylated, and mutationally stabilized gp120 core is different from its mature counterpart in important ways, including a fully truncated V1/V2 domain. It was found that the removal of V1/V2 loops significantly weakens the binding of b12 to gp120.17 The removal of a single N-linked glycosylation site at the V3 loop increased the neutralization sensitivity of CD4bs antibodies.18 Because of their absence in the crystal structure of the gp120 core in complex with b12, it remains unclear how the V1/V2 and V3 loops interact with b12. The characterization of the contact sites between mature gp120 and b12 will provide a better understanding of the specific broadly neutralizing activity of b12 against gp120. As there is no crystal structure available of an intact, glycosylated gp120 in complex with b12, molecular modeling has been used to predict the interface between b12 and the gp120 core in the truncated area by using available crystal structures as the basis for homology modeling.5,7,19 However, such molecular modeling has limitations in terms of accuracy and reliability and should ideally be coupled with experimental testing and constraints.20
In this study, we used high-resolution hydroxyl radical protein footprinting (HR-HRPF) by fast photochemical oxidation of proteins (FPOP) coupled with mass spectrometry (MS) to characterize the binding interface between mature HIV-1 JR-FL gp120 and b12. Hydroxyl radical protein footprinting is a relatively new method for probing changes in the topography of a protein. The measured rate of the reaction of hydroxyl radicals at any particular amino acid side chain depends primarily upon two factors: the chemical reactivity of the side chain (which is invariant between conformations) and the accessibility of the side chain to the radical. Therefore, by monitoring changes in the HR-HRPF reactivity, we can determine changes in the average solvent accessibility caused by changes in conformation and/or dynamics or direct shielding by protein–protein or protein–ligand binding. MS-based hydroxyl radical protein footprinting has shown great promise in the measurement of protein conformational changes, protein–protein interactions, and protein–ligand binding events.21−27 HR-HRPF by certain techniques, including FPOP28 and electron accelerator radiolysis,29 stably modifies solvent accessible amino acid side chains without deforming the protein structure during the time scale of modification, allowing for heavy surface labeling of the native structure.
To measure changes in HR-HRPF reactivity, the stable modifications to the protein side chains are analyzed by liquid chromatography and mass spectrometry (LC–MS), relatively quantifying modified and unmodified peptides, and the MS signal of the oxidized version(s) of each peptide is compared to the MS signal of the unoxidized version of the same peptide to quantify oxidation at the peptide level. Via measurement of the rate of reaction of each amino acid side chain of a protein under two different structural conditions (in this case, b12-bound vs free gp120), relative changes in the accessibility of each amino acid can be measured.25 Previous work from our group has shown that, using ETD-based methods for quantifying multiple adjacent sites of isomeric oxidation products, we can accurately quantify changes in the hydroxyl radical footprint down to single-amino acid spatial resolution,30,31 providing structural information with higher spatial resolution and more accuracy. This method offers an important alternative method for characterizing protein–protein and protein–ligand interactions in cases in which it is not possible to determine high-resolution three-dimensional structures of complexes because of the heterogeneity, flexibility, and/or size of the target(s).
A glycosylated, full-length homology model of JR-FL gp120 was constructed, and a 500 ns molecular dynamics simulation was performed. The resulting simulation was aligned to the available structure of gp120 HXBc2 in complex with b12, allowing an estimation of the contact area between b12 and glycosylated JR-FL. This provided a structure-based rationalization of the experimentally determined protection sites as well as detailed insights into the regions of JR-FL affected by complexation with b12.
Materials and Methods
Materials and Reagents
Hydrogen peroxide (30%) was purchased from J. T. Baker (Phillipsburg, NJ). Dithiothreitol (DTT) and high-performance liquid chromatography-grade acetonitrile were purchased from Fisher Scientific (Fair Lawn, NJ). Methionine amide was purchased from Bachem (Torrance, CA). Catalase, adenine, formic acid, l-glutamine, and phosphate-buffered saline (PBS) were obtained from Sigma-Aldrich Corp. (St. Louis, MO). Sequencing-grade modified trypsin and Glu-C were purchased from Promega Corp. (Madison, WI). PNGase F (500000 units/mL) was purchased from New England Biolabs (Ipswich, MA). All reagents were used without further purification. Purified water (18 MΩ) was obtained from an in-house Milli-Q Synthesis system (Millipore, Billerica, MA).
Construction, Expression, and Purification of gp120 and Human mAbs IgG1 b12
Recombinant HIV-1 gp120 protein vaccine was produced from gp120-JR-FL DNA vaccine-transfected 293F cells. Codon-optimized gp120-JR-FL DNA vaccine was constructed in the pJW4303 DNA vaccine vector as previously reported.32 Seventy-two hours after the transient transfection of 293F cells, the culture supernatant was harvested and the secreted gp120 protein purified using a lectin column. The purified gp120 protein was verified and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blot analysis (data not shown). Monoclonal antibody (mAb) IgG1 b12, specific for the gp120 CD4 binding site (CD4bs), was purchased from Polymun Scientific (Klosterneuburg, Austria).
HR-HRPF by FPOP
Six microliters of gp120 JR-FL (25 μM) was incubated with 6 μL of PBS either with or without 25 μM b12 at 37 °C for 1 h. HR-HRPF by FPOP was then performed in triplicate as previously described. Briefly, 4 μL of glutamine (100 mM) and 2 μL of adenine (200 μg/mL) were mixed with 12 μL of gp120 alone or the gp120–b12 reaction solution. Adenine was added as a radical dosimeter to monitor the available radical dose in each sample. Two microliters of 3% hydrogen peroxide (1 M, freshly prepared) was added to the mixture immediately prior to irradiation. FPOP labeling of the total volume of a 20 μL mixture was immediately performed. The KrF excimer laser power (GAM Laser Inc., Orlando, FL) was adjusted to 75 mJ/pulse and its pulse frequency set to 5 Hz. The flow rate was set to 12.19 μL/min to ensure a 10% exclusion volume to prevent repeat hydroxyl radical exposure.28 Each replicate was collected in a microcentrifuge tube containing a quench mixture of 10 μL of 140 mM H-Met-NH2 and 10 μL of 2 μM catalase to eliminate secondary oxidants such as remaining hydrogen peroxide, protein peroxides, superoxide, etc. The samples were incubated in the quench solution for 30 min at room temperature with pipet mixing. Control samples were handled in the same manner as those submitted to FPOP, but they were not laser irradiated; instead, they were incubated for 2 min with H2O2, after which the solution was added to the collection solution containing catalase and H-Met-NH2. All experiments were performed in triplicate for statistical analysis.
Following irradiation, 2 μL of the sample (40 μL) was analyzed to assess the oxidation of adenine after HR-HRPF by UV at 260 nm (Thermo Scientific NanoDrop 2000c UV–vis spectrophotometer) to ensure comparable amounts of available hydroxyl radical for oxidizing the analyte, as previously reported.33 The amounts of adenine oxidation are similar among all tested samples with an average of around 53% oxidized adenine and a CV of <5% (data not shown), which indicates the same level of hydroxyl radical available among all samples.
The lyophilized powder was resuspended in 50 μL of 100 mM ammonium bicarbonate buffer. DTT (10 μL, 50 mM) was added and incubated at 65 °C for 30 min to denature and reduce the protein. After the mixture had been cooled to room temperature, a 1:20 Glu-C:protein weight ratio was added to the protein sample and the mixture incubated at room temperature for 2 h. Glu-C was deactivated by heating the sample to 95 °C for 10 min; then after the samples had been cooled to room temperature, a 1:20 trypsin weight ratio was added to the samples for overnight digestion at 37 °C while the sample was being rotated. Digestion was terminated by adding 12 μL of DTT (50 mM) and heating the samples to 95 °C for 10 min. Finally, when the samples were cooled to room temperature, 150 units of PNGase F was added to the digested samples and the samples were incubated at 37 °C for 16 h. The reaction was terminated by adding 12 μL of DTT (50 mM) and heating the sample to 95 °C for 10 min. Samples were stored at −20 °C until nanoLC–MS/MS analysis.
HR-HRPF samples were analyzed on an LTQ Orbitrap XL instrument (Thermo Fisher Scientific) controlled with Xcalibur version 2.0.7 (Thermo Fisher, San Jose, CA). Samples were loaded on a 10 cm column with a PicoFrit tip, bomb-packed with C18 reverse phase material (Magic, 0.075 mm × 100 mm, 5 μm, 300 Å, Michrom, Auburn, CA). Chromatography was performed using mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in 80% acetonitrile) with a 160 min gradient consisting of 0 to 60% solvent B over 60 min at a rate of 135 μL/min, ramped to 100% B over 20 min, held for 9 min, and then returned to 0% B over 1 min and held for 70 min. Peptides were eluted directly into the nanospray source of an LTQ Orbitrap XL instrument using the bomb-packed column as the nanospray emitter. All data were acquired in positive ion mode. CID (collision-induced dissociation) and/or ETD (electron transfer dissociation) was used to fragment peptides. The spray voltage was set to 2.0 kV, and the temperature of the heated capillary was set to 200 °C. In CID mode, full MS scans were acquired from m/z 400 to 1500 followed by eight subsequent MS2 scans on the top eight most abundant peptide ions. In ETD mode, the parent ions of all identified peptides under CID mode were listed in the parent ion mass list. ETD-based precursor activation was performed for 100 ms, including charge state-dependent with supplemental activation enabled. Precursor ions were isolated with width of 3 m/z units.
HR-HRPF Data Analysis
Unoxidized gp120 controls, oxidized gp120, and oxidized gp120–b12 complex peptide sequences were initially identified using ByOnic version 1.2-250 (Protein Metrics). The enzyme was defined to cleave the protein after lysine, arginine, asparatic acid, and glutamine. Deamination on both asparagine and glutamine and all possible major oxidation modifications25 were included as variable modifications for database searches. All tandem mass spectral assignments and sites of oxidation were verified manually via the peptide modification complexity. The LC–MS peak intensities of the digested peptides and corresponding oxidation products were used to calculate the average oxidation events per peptide in the sample. Peptide level quantitation is calculated by summing the ion intensities of all the oxidized peptides multiplied by the number of oxidation events required for the mass shift (e.g., one event for +16, two events for +32) and then divided by the sum of the ion intensities of all unoxidized and oxidized peptide masses (eq 1):
| 1 |
where P denotes the oxidation events at the peptide level and I values are the peak intensities of oxidized and unoxidized peptides.
gp120 control samples unilluminated by the UV laser, but otherwise identical to the experimental samples, were analyzed to ensure that background oxidation would not be intense enough to interfere with HR-HRPF data. Illumination-free controls showed <10% of the oxidation extent of the gp120 sample with UV laser for peptides 19–31, 37–63, 64–69, 178–197, 261–274, 386–396, and 397–404 (data not shown). The small amount of oxidation happening occurred at M or C because of the sample preparation process (especially for residue M), protein expression, and purification. The interference of this background is not significant enough to affect the comparison of the oxidation extent between gp120 and gp120–b12 samples and was subtracted from experimental samples.
In cases in which oxidation at specific sites can be identified on the basis of either the mass differences between nonisomeric oxidation products or the presence of only a single oxidation site within a peptide, residue level quantitation is calculated from the LC–MS signal intensities of each peptide containing a specific oxidized amino acid (Ioxidized), relative to the total of all intensities associated with that peptide sequence (Ioxidized + Iunoxidized) using the following straightforward relationship:24
| 2 |
In cases in which the peptide isomers are oxidized at multiple adjacent sites in a single sequence that results in an identical mass shift, residue level quantitation is calculated from the fragment ion intensities from ETD to determine the oxidation extent at a specific residue site based on our previous studies.30,31 Briefly, an oxidized peptide with multiple sites of oxidation can generate both oxidized and unoxidized sequence ions in its tandem mass spectrum. The fractional oxidation of a given sequence ion is defined as the ratio between the oxidized sequence ion intensity and the sum of the intensity of the corresponding oxidized and unoxidized sequence ion (eq 3).
| 3 |
where f(Ci) denotes the fractional oxidization of sequence ion i (e.g., oxidized C3 ions generated by ETD) and I(Ci) denotes the intensity of sequence ion i, whether it is the oxidized or unoxidized form. The absolute level of oxidation for a given amino acid residue i is based on both the average oxidation event of the peptide and the fractional oxidation of the corresponding sequence ions, as shown in eq 4:
| 4 |
where P is the average oxidation events per peptide as derived from eq 1 and the term in brackets is the fractional oxidation difference of two adjacent sequence ions i and i – 1. Multiplying the average number of oxidations per peptide by the fraction of that oxidation that occurs on a given amino acid residue yields the average oxidation events per residue. In cases in which ETD did not yield sufficient product ions for quantification, CID was used to provide semiquantitative information about changes in the footprint at a residue level.
The protection by b12 binding is defined as the ratio of the difference in oxidation extent between the gp120 sample alone and the gp120–b12 binding sample to the oxidation extent of the gp120 sample alone, as shown in eq 5:
| 5 |
Glycosylation Analysis
An aliquot of each sample was denatured by being incubated with 10 mM dithiothreitol at 56 °C for 1 h and alkylated with 55 mM iodoacetamide for 45 min in the dark prior to digestion with proteases optimized on the basis of the amino acid sequence of each target protein. Following digestion, the samples were deglycosylated by PNGaseF in the presence of H218O that coverts Asn residues carrying N-linked glycans to [18O]Asp residues. The resulting peptides were separated on a 75 μm (inside diameter) × 15 cm C18 capillary column (packed in house, YMC GEL ODS-AQ120 ÅS-5, Waters) and eluted into the nanoelectrospray ion source of an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) with a 180 min linear gradient consisting of 0.5 to 100% solvent B over 150 min at a flow rate of 200 nL/min. The spray voltage was set to 2.2 kV, and the temperature of the heated capillary was set to 280 °C. Full MS scans were acquired from m/z 300 to 2000 at 120K resolution, and MS2 scans following collision-induced fragmentation were collected in the ion trap for the most intense ions in the Top-Speed mode within a 3 s cycle using Fusion instrument software (version 1.1, Thermo Fisher Scientific). The resulting spectra were analyzed using SEQUEST (Proteome Discoverer 1.4, Thermo Fisher Scientific) with a full MS peptide tolerance of 20 ppm and a MS2 peptide fragment tolerance of 0.5 Da and filtered using ProteoIQ (version 2.7, Premier Biosoft) at the protein level to generate a 1% false discovery rate for protein assignments. Site occupancy was calculated using spectral counts of [18O]Asp peptides to the identical peptide containing Asn. The sole exception is N88, which was wholly unobserved as glycosylated or nonglycosylated by this method. Glycosylation of this site was assigned on the basis of the high abundance of the N → D substitution of this site after PNGase F treatment during HR-HRPF data acquisition and analysis, as detailed above.
A site was assigned as occupied for the purpose of modeling if the site was predominantly glycosylated across all peptides in which the site was observed. Notably, sites N241 and N262 co-reside on a single peptide and were detected as individually glycosylated 56% of the time, but simultaneous glycosylation was detected for only 6% of observed peptides. Thus, the predominant state was for only one site to be glycosylated. Given their equal rate of occupancy, we arbitrarily chose N241 for glycosylation in the model.
Model Building
Blastp34 was used to find three-dimensional (3D) structures from the PDB with sequences similar to that of JR-FL. A modified JR-FL has been crystallized previously (PDB entry 2B4C); however, the N- and C-termini were truncated, and the V1/V2 region was substituted with a GAG tripeptide. The model used here is based on 2B4C and employs additional crystal structures (PDB entries 4NCO, 3JWD, 4HPO, 4R4H, 4TVP, and 9B4C) to model a complete JR-FL. Modeler35 incorporated each of these structures (see alignments in Figure S1) to generate five models of complete gp120.
Model Glycosylation
Previous work revealed that expression of gp120 from Ba-L (a clade B isolate) in 293F cells resulted in a mixture of a complex and high-mannose glycans and that the level of interaction of the B clade gp120 with the b12 bNAb was increased by expression in cells that increased the level of high-mannose glycosylation.36 Because the full diversity of possible glycan structures is impractical to simulate even for individual compositions of complex glycans, Man9GlcNAc2 structures were modeled into all sites assigned as glycosylated, as this glycan is a single, biologically relevant structure that explores the extended reach of glycans in examining both glycan–antibody interactions and glycan shielding of the antibody binding surface. GLYCAM-Web (www.glycam.org) was used to generate 3D structures of the common rotamers of Man9GlcNAc2. These structures were superimposed onto the Asn side chain at each assigned glycosylation site. The glycan structure was adapted to the local protein environment and any previous glycans in the following manner: the four most populated conformations of N-linked Asn side chains found in the PDB were assessed in series by altering the χ, ϕ, and ψ angles of the N-linked glycan. Any van der Waals (vdW) overlaps between the glycan and the glycoprotein were minimized by altering the torsion angles of the Asn-GlcNAc and interglycosidic linkages by ±20° in 5° increments. The first glycan rotamer and Asn side chain conformer that allowed the glycan to fit without large vdW overlaps was added to the glycoprotein structure.
Model Selection
A single model of JR-FL was chosen on the basis of the number of experimentally determined glycosylation sites that were accessible for glycosylation and the DOPE score provided by Modeler. A recent crystal structure of a complete JR-FL (PDB entry 5FYK),37 released during the preparation of this work, allowed validation of the model generated here. A Needleman–Wunsch alignment using a BLOSUM-62 matrix38 gave a relatively large root-mean-square deviation (RMSD) of 5.2 Å over 450 atom pairs. However, most of the deviation is in the flexible N- and C-terminal domains that are ordered by the presence of gp41 in the crystal structure. When these regions are discounted, the RMSD drops to 1.0 Å over 303 atom pairs, indicating that the homology model successfully predicted the remaining 3D structure.
Molecular Dynamics Simulation
All simulations were performed with the CUDA implementation of PMEMD39,40 in the Amber14 software suite.41 The carbohydrate was modeled using the GLYCAM06j-1 force field,42 while the Amber ff14SB force field43 was employed for the protein. A Berendsen barostat with a time constant of 1 ps was employed for pressure regulation, while a Langevin thermostat with a collision frequency of 2 ps–1 was employed for temperature regulation. A nonbonded interaction cutoff of 8 Å was employed. Long-range electrostatics were treated with the particle mesh Ewald (PME) method.44 Covalent bonds involving hydrogen were constrained with the SHAKE algorithm allowing a time step of 2 fs.45
Each system was placed in a cubic box of TIP5P water46 with an 8 Å buffer between the solute and the periodic boundary. Five sodium ions were added to neutralize the overall charge. Energy minimization was performed for 20000 steps (10000 steepest decent, followed by 10000 conjugant gradient) with Cartesian restraints (5 kcal mol–1 Å–2) on solute heavy atoms. This was followed by a 400 ps solvent equilibration phase at 300 K under nPT conditions. Cartesian restraints (5 kcal mol–1 Å–2) were employed on the Cα atoms of the first and last three residues (N- and C-terminal) during a 1 ns structural equilibration phase and 500 ns production phase (nPT).
Structure of gp120 (JR-FL) in Complex with the b12 Antibody
A structure-based sequence alignment of gp120 (HXBc2) in complex with the b12 Ab (PDB entry 2NY7) with 100 snapshots from the MD simulation of JR-FL was performed using Sequoia,47 with an RMSD of 4.14 Å for 245 equivalent matches based on alignment with the original model, with a Sequoia alignment score of 0.08. On the basis of each alignment, an estimation of the contact area between b12 and JR-FL was obtained by averaging the percentage change in the respective solvent accessibilities48 for each snapshot in the presence and absence of the b12 structure.
Results
Glycosylation Analysis of gp120 and the gp120–b12 Complex
The recombinant glycoprotein gp120 used in this study was expressed as a 477-amino acid mature protein containing intact N- and C-termini, five variable regions (V1/V2, V3, V4, and V5) and five conserved regions (C1–C5). Following Glu-C and trypsin digests of gp120, 32 unique peptides were identified by CID-based MS/MS data, covering 72% of the overall sequence. Eighteen highly occupied N-linked glycosylation sites were identified. The details of the oxidation sites and glycosylation sites are listed in Figure 1.
Figure 1.
Sequence of HIV-1 gp120 (JR-FL). The sequences covered by HR-HRPF experiments are overlined with green. The identified oxidation sites are shown in red text with protected sites underlined and the sole exposed site in italics. The heavily occupied N-linked glycosylation sites are shown in green text. The variable domains are labeled above the sequence.
HR-HRPF of the gp120–b12 Complex
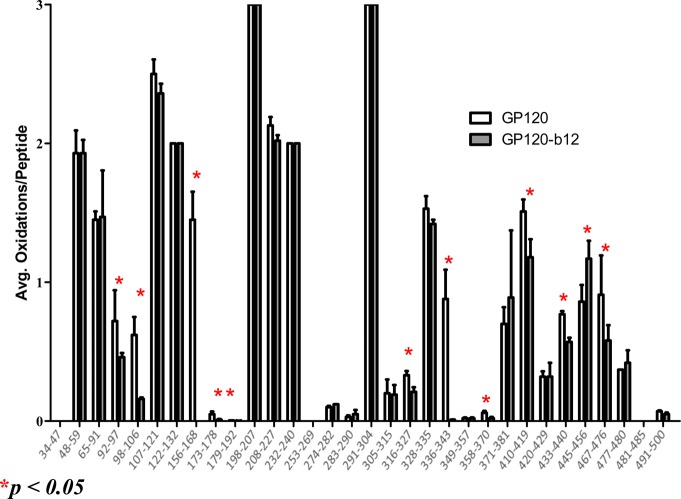
Twenty-nine peptides were seen to be modified by FPOP footprinting. Sixty-one oxidized residues were identified in these modified peptides on the basis of a Byonic database search and verified manually from the CID and/or ETD MS/MS spectra. Almost one in five residues in the peptides detected was modified, indicating a high yield of protein labeling through the short FPOP reaction window. The oxidation extent of the 29 modified peptides was determined at the peptide level by measuring the intensity of the oxidized peptide ion compared to the sum of oxidized and unoxidized peptide ions as calculated by eq 1. The side-by-side comparison of the oxidation extent of sequenced peptides in gp120 and gp120–b12 samples is shown in Figure 2. The overall number of average oxidations per peptide for different peptides varies significantly from 0.003 for peptide 179–192 to 3 for peptide 198–207. Most peptides show comparable amounts of oxidation between binding and nonbinding samples. Eleven peptides, including 92–97, 98–106, 156–168, 173–178, 179–192, 316–327, 336–343, 358–370, 410–419, 433–440, and 467–476, showed a significant decrease in oxidation extent upon b12 binding, indicating a structural event that shielded the sites from the radical.
Figure 2.
Peptide level HRPF of gp120 footprinting for gp120 alone (white) and the gp120–b12 complex (gray) (mean ± standard deviation; n = 3). Peptides highlighted with a red star showed a statistically significant change in oxidation extent upon gp120–b12 binding (α ≤ 0.05).
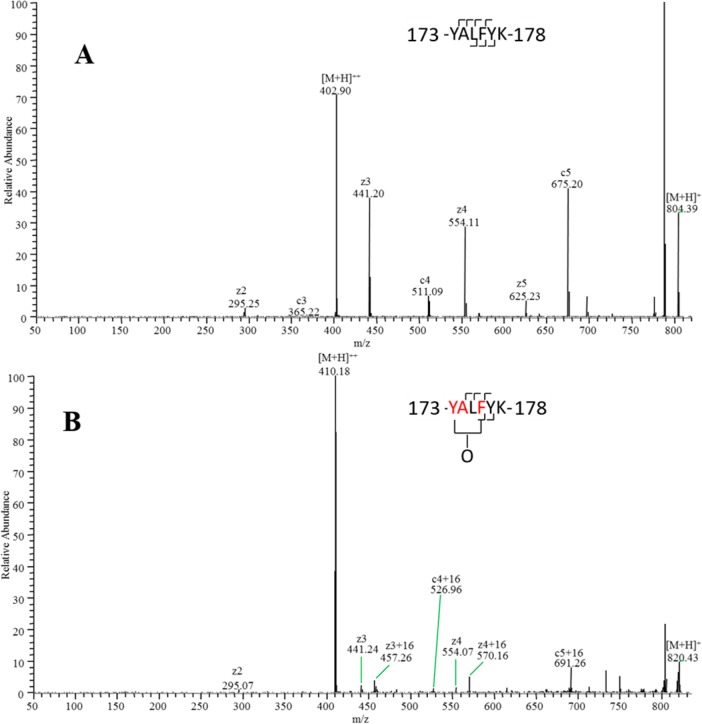
To investigate structural changes upon binding between gp120 and b12 at a high spatial resolution, the CID- and ETD-based tandem mass spectra of these 11 peptides were analyzed. A representative ETD spectrum is shown in Figure 3. On the basis of the CID/ETD fragmentation ions, most of the 11 peptides were found to have multiple oxidation sites except peptides 156–168 and 336–343. For peptides with only one oxidation site, the oxidation extent at the residue level was calculated by eq 2. For oxidation isomers having multiple oxidation sites on a single sequence (peptides 143–148, 149–162, 284–296, 326–338, 375–383, and 397–404), the oxidation extent at residue level was calculated by eqs 3 and 4. Oxidized residues that exhibit HR-HRPF protection upon b12 binding are listed in Table 1. Because of the semiquantitative nature of CID-based oxidation quantification,30 CID-based data were selected only when ETD data were not available (residues M72, M76, and C127).
Figure 3.
ETD spectrum of peptide 143-YALFYK-148 and its oxidation products. (A) Unoxidized peptide 173-YALFYK-178. (B) Mixture of singly oxidized isomers. By measuring the ratio of oxidized product ions to total product ions for each fragment, oxidation can be quantified to occur on F176 and the Y173-A174 fragment (on the basis of CID data and the relative reactivities of alanine and tyrosine, almost no oxidation occurs on A174).
Table 1. Protection of Residues from HR-HRPF upon b12 Binding.
| residuea | % HR-HRPF protection upon b12 bindingb | direct protection in MD upon b12 binding (Δ%)c |
|---|---|---|
| M95 | 31.9 | 0% |
| W96 | 81.8 | 0% |
| M100 | 84.6d | 0% |
| M104 | 66.0d | 0% |
| C157 | 100d | 64% |
| Y173, A174 | 100 | 53%, 76% |
| F176 | 100 | 70% |
| D180 | 64.5 | 35% |
| V182, P183 | 21.3 | 32%, 54% |
| I322a | 43.6 | 0% |
| I323 | 28.2 | 14% |
| W338 | 100 | 0% |
| V360 | 72.4 | 0% |
| F361 | 73.3 | 0% |
| H363 | 70.4 | 0% |
| P417, C418, R419 | 45 | 0%, 0%, 0% |
| M434 | 28.5 | 0% |
| Y435 | 28.2 | 0% |
| C445 | –35.7 | 0% |
| M475 | 11.3 | 0% |
The residue numbering used is based on the HXB2 numbering system.
The protection upon binding for each residue was calculated by the changesin oxidation extent between gp120 and the gp120–b12 complex divided by the oxidation extent of gp120.
The direct protection is calculated by measuring the change in solvent accessibility after alignment of b12. The values are averages from the MD simulation.
CID-based data were used in HR-HRPF results and should be considered semiquantitative.
It should be noted that the HR-HRPF technique cannot differentiate changes in solvent accessibility as a result of direct binding from those due to conformational or dynamic perturbations. Therefore, even a preliminary 3D model is valuable for interpretation of the HR-HRPF data, especially when dealing with a protein with as many dynamic domains as full-length, glycosylated gp120. With the exception of the small V5 domain, we were able to probe at least one amino acid in each domain by HR-HRPF. Residues that show significant differences in oxidation due to altered average solvent accessibility after complex formation can be directly involved in b12 binding or experiencing conformational and/or dynamic changes that occur upon b12 binding. Residues that were oxidized, yet showed no protection upon b12 binding, experienced no net change in the average solvent accessible surface area of the side chains probed.
Alignment of b12
The 3D structure of gp120 (HXBc2) in complex with b12 (PDB entry 2NY7) was aligned with the gp120 (JR-FL) model generated here. Residues that experienced protection from HR-HRPF oxidation upon b12 binding are plotted against a full-length model of glycosylated gp120 aligned with the stabilized gp120–b12 complex structure (RMSD of 2.3 Å) and shown in Figure 4. The alignment provided an estimation of the direct contact area between b12 and gp120 (JR-FL), and changes in solvent accessibility due to direct interaction between b12 and gp120 were calculated (Table 1). During the MD simulation, the V1/V2 region samples conformations that overlap with the area occupied by a bound b12, indicating that b12 binding would impact the ensemble of conformations adopted by the V1/V2 region, either by inducing new conformations or by just limiting the available conformational space.
Figure 4.
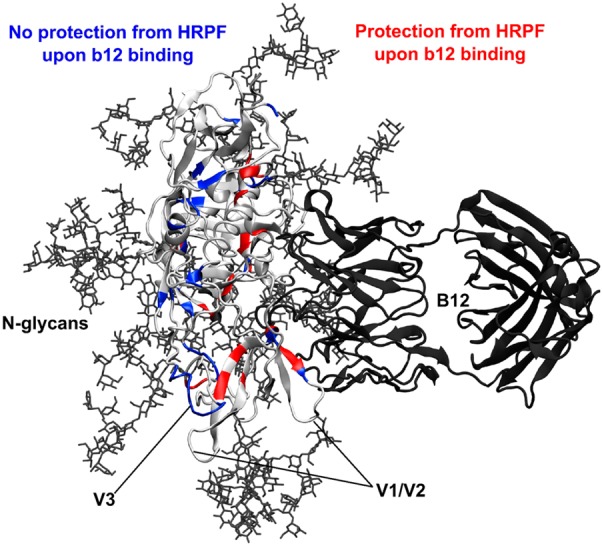
Glycosylation and HRPF of gp120. A model of full-length glycosylated gp120 (ribbon protein in light gray, licorice Man9GlcNAc2 glycans in dark gray) was generated and relaxed by MD simulation and then aligned with the crystal structure of stabilized gp120 bound to the b12 Fab (aligned gp120 not shown, ribbon b12 in dark gray) (PDB entry 2NY7). Residues that showed protection from HRPF upon b12 binding are colored red. Residues that showed no protection from HRPF are colored blue. Residue C445 (also blue) showed an exposure to HRPF upon b12.
The combination of HR-HRPF data and modeling allows us to predict which of those residues are directly protected from oxidation upon complexation (residues D180, V182, P183, P417, R419, and M426) and which become protected via induced conformational and/or dynamic changes in the gp120 (JR-FL) structure (residues M95, W96, M100, M104, F176, C157, Y173, I322a, I323, W338, M434, Y435, V360, F361, and H363). The combination also allows us to determine residues at which the model disagrees with the HR-HRPF data, indicating regions of either insufficient modeling or unreliable experimental results to be probed by further experimentation (L179 and I184).
Discussion
The X-ray crystal structure of monomeric gp120 in complex with the b12 Fab includes only 62% of our intact gp120 polypeptide sequence and does not include glycosylation.10 The structure lacks most of the residues in the V1/V2 loops (residues 121–203), V3 loop (residues 298–329), V4 loop (residues 393–404), and portions of the amino and carboxy termini (residues 31–82 and 493–507, respectively). In this reported crystal structure, a total of 33 residues were defined as contact sites. In the current HR-HRPF study, 22 residues were identified as being protected from hydroxyl radicals upon b12 binding. It is clear that the V1/V2 loops and V3 loop can affect binding of the antibody to the CD4bs either by direct contact or by transmitted conformational effects.49 The deletion of the V1/V2 and V3 loops also has an adverse effect on the binding of b12 to gp120.50 The application of a full-length, glycosylated model of gp120 allows us to probe and try to explain the observed roles of these and other gp120 domains in b12 binding.
V1/V2 Domain
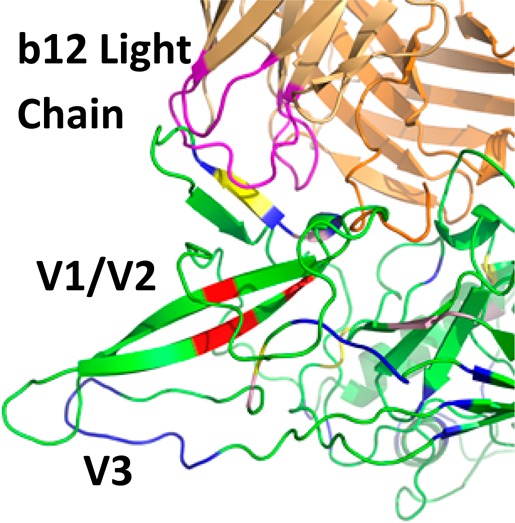
The V1/V2 domain is highly dynamic and is excised from the published X-ray crystal structure of gp120 bound to b12 to promote crystallization.10 However, alanine scanning mutagenesis data have shown that several amino acids in the V2 region are important for b12 binding.50 Similarly, the crystal structure shows no direct interaction between the light chain of the b12 antibody and gp120; however, mutagenesis found that several amino acids in the L1 and L3 CDR loops were important for binding of b12 to gp120.51 HR-HRPF results of the V1/V2 domain within full-length, glycosylated gp120 are shown in Figure 5, with glycans labeled and shown as 3D SNFG symbols52 positioned at each residue’s ring center. These results revealed several amino acids that were protected from HR-HRPF upon b12 binding. C157 and Y173 are two residues on adjacent strands of an antiparallel β-sheet that both experience 100% protection from HR-HRPF upon b12 binding. F176 resides on the C-terminal end of the second strand of that same β-sheet and also exhibits 100% protection from HR-HRPF upon b12 binding. F176 and C157 present their side chains to the opposite face of the β-sheet from Y173 (Figure 5, inset), suggesting that the protection observed by these residues is probably not all from direct interaction with the b12 antibody but rather from stabilization of this β-sheet upon antibody binding. We also observe modest protection of D180 and V182/P183 upon b12 binding, but neighboring residues L179, V181, and I184 are not protected upon b12 binding. This targeted protection is more in line with the effects of direct interaction, as a change in dynamics or gross conformation is likely to affect all amino acids in a region.
Figure 5.
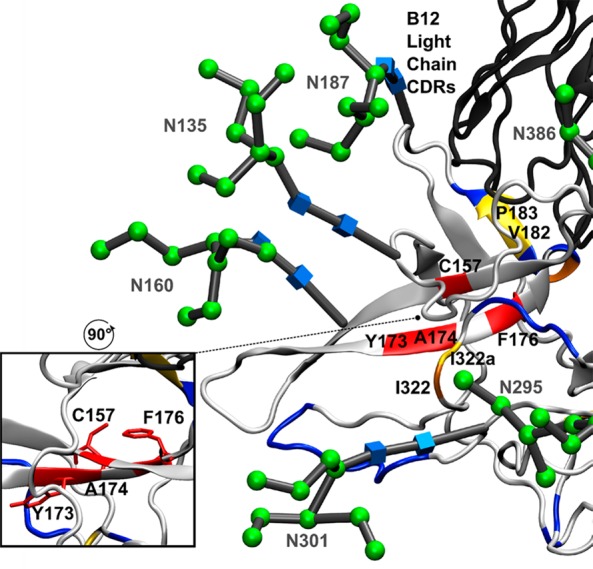
HR-HRPF and glycosylation of the V1/V2 and V3 domains. Aligned model of full-length glycosylated gp120 after MD simulation (light gray ribbon) bound to the b12 Fab (dark gray ribbon) (PDB entry 2NY7). Residues that showed >80% protection from HR-HRPF upon b12 binding are colored red. Residues that showed between 40 and 80% protection from HR-HRPF upon b12 binding are colored orange. Residues that showed statistically significant protection from HR-HRPF of <40% are colored yellow. Residues showing no protection from HR-HRPF upon b12 binding are colored blue. The CDR loops of the light chain modeled to potentially interact with the V1/V2 domain are shown. The inset shows the orientation of the C157, F176, and Y173 side chains relative to the β-sheet.
Modeling of the glycosylated V1/V2 domain into the gp120 structure revealed a potential interaction between the V1/V2 domain and the CDR loops of the b12 light chain, as well as a loop of the b12 heavy chain. However, both L179 and I184 are outliers as their oxidation was not affected by b12 binding but were predicted to be in direct contact with b12. These outliers are within the flexible V1/V2 region, and the inconsistency with the footprinting data may be the result of the dynamics of this region. Previously, alanine scanning mutagenesis was performed on monomeric gp120 (JR-CSF) to define in more detail which residues on gp120 influence or modulate b12 reactivity. The outcome indicated that in the V2 loop, the F176A, D180A, I184A, and D185A mutations significantly reduced the binding affinity of b12, which supports our finding in regard to the contact sites identified in the V2 loop.50
The oxidation extents of residues M95 and W96 were decreased by 41.9 and 71.3%, respectively, upon b12 binding and are likely involved in the binding interface with b12 based on the docking model of the crystal structure of b12 and the CD4-complexed gp120 core structure of HXB2. However, the previous observation that the M95A mutation causes a very modest decrease in b12 binding affinity for JR-CSF gp120 (75% of WT binding) suggests that this residue, while protected by the gp120–b12 interface, is probably not energetically involved in stabilizing the binding to any significant extent.7,50
Hydrogen–deuterium exchange (HDX) studies were previously reported for the KNH1144 SOSIP.664 trimer structure bound to b12. These studies largely monitor changes in secondary structure dynamics that alter backbone hydrogen bonding stability, serving as complementary information for the side chain-oriented HR-HRPF data presented here. HDX protection profiles of the V1/V2 domain upon b12 binding were complex, with some peptides showing a stabilized secondary structure and others showing a destabilized secondary structure, and with changes in stability differing over time. The fact that HDX data are usually limited in spatial resolution to the peptide level further increases the difficulty in interpreting the data. The V1/V2 region is clearly structurally altered by b12 binding in complex ways, with the region of residues ∼100–175 being destabilized by b12 binding and residues 176–179 being slightly stabilized.53 Combined with our HR-HRPF data, this suggests that HDX protection of this region is largely limited to the area of direct contact with the b12 antibody, with regions affected conformationally by b12 binding in this region exhibiting increased HDX rates.
V3 Domain
The V3 domain is a highly flexible loop that is missing from the stabilized gp120–b12 crystal structure. The V3 loop is heavily modified by HR-HRPF, suggesting a high degree of solvent accessibility. The vast majority of residues in the V3 domain (C296, K305–R313, D324, I325, R326, and Q327) show no protection upon b12 binding, indicating that the domain remains highly flexible in the b12-bound form. However, slight to moderate protection is observed for I322a and I323. Via examination of the location of these residues in the MD simulation of full-length glycosylated gp120 (Figure 5), I322a and I323 are adjacent to Y173, an amino acid in the V1/V2 domain that shows complete protection upon b12 binding. We suggest that the b12-induced conformational change in the V1/V2 domain allows for interaction between I322a and I323 and the V1/V2 domain, perhaps through Y173. While the modest extent of protection of I322a and I323 suggests that this interaction is not completely protecting, the protection is both significant and unique in the V3 domain. This suggests that interactions between V3 and other molecules (e.g., heparan sulfate54) could modulate the affinity of the b12 antibody. This observation is supported by the fact that the V3 domain of KNH1144 SOSIP.664 shows a marked increase in HDX rates upon b12 binding, and the V3 peptides most markedly affected are those containing residues 322 and 323,53 as well as by mutational and recombinant analysis yielding evidence that the V3 region interacts directly with the V1/V2 domain in DH102 gp120.55 Though the deletion of the V3 loop (residues N302–D324) was reported to reduce the binding affinity of b12, it is currently unknown which particular residues mediate the interaction.50
C1 and C5 Domains
Ten residues were observed as oxidized in the C1 domain, four of which experience protection upon b12 binding: M95, W96, M100, and M104. These residues cluster at a loop and the N-terminal region of the large central helix of the C1 domain. Alanine scanning mutagenesis of binding of b12 to JR-CSF gp120 did not cover this end of the helix thoroughly and did not directly test any of the residues protected by HRPF; K97A caused a modest increase in apparent affinity, while E102A caused a modest decrease. Mutagenesis at the C-terminal end of this helix showed an almost complete loss of binding affinity, suggesting a role for the helix in the interaction.50 Hydrogen–deuterium exchange of the KNH1144 SOSIP.664 trimer with b12 indicated this region exhibits increased exchange rates upon antibody binding, suggesting that antibody binding destabilizes the secondary structure of the N-terminal end of the C1 helix.53 The HDX data (which largely probe backbone secondary structure stability) combined with our HR-HRPF data (which probe side chain solvent accessibility) suggest that antibody binding increases the amount of side chain packing not by stabilizing the helical structure, but by destabilizing the helical structure and granting the side chains additional flexibility to maximize interactions. In our model, the protected N-terminal end of the C1 helix packs against a helix in the C5 domain that contains the only protected residue in C5, M475 (Figure 6). Residues G471–M475 show substantial contact with the heavy chain of b12, with M475 showing a 29.5 Å2 interaction surface with the b12 heavy chain.10 Mutagenesis data revealed that mutation of M475 to alanine increased the apparent affinity of b12 for JR-CSF gp120 by a very modest amount, while mutation of the nearby G473 to alanine increased affinity almost 10-fold. However, quadruple mutation of G473, D474, M475, and R476 to alanine increased the apparent relative affinity for b12 only 2.5-fold, and GM and DMR multiple mutations increased the apparent affinity to an extent similar to that seen for the M475A mutant alone, suggesting that the large effect of G473A may be due to stabilization of the helix, with the interactions mediated by the amino acids in the helix.50 On the basis of our data and the glycosylated homology model, the binding of the b12 heavy chain fixes the C5 domain helix by direct contact, shielding M475. The binding simultaneously destabilizes the backbone of the N-terminus of the C1 helix as shown by HDX,53 with the destabilized helix packing against the fixed C5 helix, providing the observed shielding of M95, W96, M100, and M104. Residues in domains C1 and C5 that are not protected (F53, C54, H66, H72, and C74 in domain C1 and L494 and K500 in the C-terminal tail of domain C5) are distal to this interaction interface and do not experience any direct contact or significant stabilization by b12 binding, showed no change in HDX experiments,53 and were not in the covered regions of the previously reported alanine mutagenesis work.50
Figure 6.

HR-HRPF protection suggests stabilization of the helix α1–helix α5 interaction upon binding of b12 to helix α5. Helix α1 of the C1 domain (green) shows protection of four residues: M95 and W96 in the loop at the N-terminus of helix α1 and M99 and M104 within helix α1. The sole residue probed in helix α5, M475, interacts directly with a CDR loop of the b12 heavy chain (dark gray), with helix α5 (light gray) packing against the N-terminus of helix α1. Residues that showed >80% protection from HR-HRPF upon b12 binding are colored red. Residues that showed between 40 and 80% protection from HR-HRPF upon b12 binding are colored orange. Residues that showed statistically significant protection from HR-HRPF of <40% are colored yellow. Residues showing no protection from HR-HRPF upon b12 binding are colored blue.
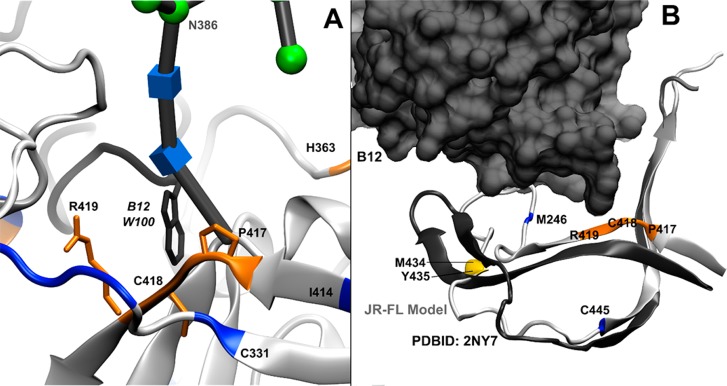
C4 Domain. The C4 domain contains three sites that exhibited modest protection upon b12 binding (P417-C418-R419, M434, and Y435) as well as one highly reactive cysteine that showed modest exposure upon b12 binding (C445). The P417-C418-R419 region interacts directly with W100 from CDR H3 of the b12 antibody (Figure 7A). While the P417-C418-R419 region was not analyzed by alanine mutagenesis, W100 of b12 was shown to be essential for b12 binding,51 as was K421 of JR-CSF gp120;50 this interaction explains the protection observed here.
Figure 7.
b12 both interacts directly with domain C4 and alters its dynamics. Residues that showed >80% protection from HR-HRPF upon b12 binding are colored red. Residues that showed between 40 and 80% protection from HR-HRPF upon b12 binding are colored orange. Residues that showed statistically significant protection from HR-HRPF of <40% are colored yellow. Residues showing no protection from HR-HRPF upon b12 binding are colored blue. Residue C445 (also blue) showed an increase in the level of oxidation upon b12 binding. (A) P417, C418, and R419 of strand β19 (orange ribbon and licorice) interact directly with W100 (dark gray ribbon and licorice) of the CDR H3 loop of the b12 heavy chain, which extends out from the bulk of the b12 IgG fold. (B) Model of full-length, glycosylated gp120 (light gray) aligned with the crystal structure of stabilized, b12-bound gp120 (dark gray), with only the C4 domain backbone shown for the sake of clarity and the b12 antibody shown as a space-filling model. Strand β22/β23 remains largely unperturbed; however, strands β20 and β21 are much more flexible in the full-length, glycosylated, unbound gp120 MD simulation. HR-HRPF data of M426 show no protection upon b12 binding, suggesting that there is no appreciable change in the conformation or dynamics of this residue in strand β20. However, the protection of M434 and Y435 suggests that strand β21 does experience a stabilization upon b12 binding.
The slight observed protection of M434 and Y435 and the modest exposure observed by C445 are more difficult to understand. This region does not appear to directly interact with b12 in our MD model or in the crystal structure. This region shows considerable flexibility in the MD simulations of free gp120 as the C-terminal end of the β-strand of residues 413–426, as well as the entire β-strand of residues 432–436, completely unravels, deviating significantly from the crystal structure of the stabilized gp120–b12 complex (Figure 7B). Upon b12 binding, M434 and Y435 in the small β-strand show protection, suggesting this β-strand may be stabilized by b12 binding. This hypothesis is supported by HDX analysis of KNH1144 SOSIP.664 bound to b12, where binding shows slight protection of peptide 427–433 and very little change in peptide 435–445,53 indicating that the protection is largely occurring in residues associated with this strand. However, M426 at the extreme C-terminal end of the β-strand of residues 413–426 shows no protection yet shows considerable flexibility in the MD simulation of free gp120 as the C-terminal of its β-strand unravels. C445 in the C4 domain exhibits modest exposure upon b12 binding but shows little deviation between the MD simulation of free gp120 and the X-ray crystal structure of the stabilized gp120–b12 complex, remaining in a β-strand configuration. While HDX analysis suggests no significant change in the secondary structure stability at C44553 and our MD simulation indicates that the displacement of this residue in the strand is small, b12 binding does result in statistically significant exposure of C445. Of note, previous HR-HRPF results have indicated that cysteine oxidation is sensitive to relatively small changes in solvent accessibility;31 it is probable that the modest exposure observed in C445 indicates a small change in the average solvent accessibility of this residue.
Previous alanine scanning mutagenesis data support the importance of this region to b12 binding: mutations K421A and M426A resulted in large losses of binding affinity of b12 for JR-CSF gp120, while mutation N425A resulted in a very slight loss of affinity. Similarly, mutation K432A (the only mutated residue in the strand of residues 432–436) resulted in a large loss of affinity. Interestingly, the V430A mutant in the turn between these two strands showed a >5-fold increase in apparent affinity,50 suggesting the reduced bulk of the alanine residue in this turn improves the interaction efficiency, perhaps by improving stabilization of these two β-strands. On the basis of these data and the MD simulation, we hypothesize that b12 binding stabilizes the C-terminus of the strand of residues 413–426 and the paired 432–436 strand, but the extreme C-terminal end of the strand of residues 413–426 in the stabilized crystal structure remains sufficiently disordered to not register topographical protection in HR-HRPF. The β-strand starting with C445 appears to undergo a slight displacement upon b12 binding by MD simulation, which we can detect only because of the sensitivity of cysteine residues to SASA changes in HR-HRPF.
C3 Domain
Seven residues in the C3 domain were oxidized. Three residues at the edges of the folded domain (C331, F353, and C377) experienced no protection upon b12 binding. These residues mostly exist in loops between secondary structure elements, and these loops are distal to the b12 interaction elements; a lack of protection of these residues upon b12 binding would be anticipated.
Four residues experience strong protection upon b12 binding. Three of these residues cluster around glysocylation site N362: V360, F361, and H363. The Man9GlcNAc2 glycan modeled on the N362 site is poised to interact with the b12 light chain (Figure 8). On the basis of the glycosylated gp120 model and our HR-HRPF data, it appears that the interaction of the N362 glycan with the light chain of b12 protects the region around N362. Examination of the CATNAP database56 suggests that an intact glycosylation sequon at N362 is not correlated with b12 binding across various gp120 sequences, suggesting that while interaction with b12 may stabilize the topography around this glycosylation site, the interaction is not energetically important for b12 binding.
Figure 8.
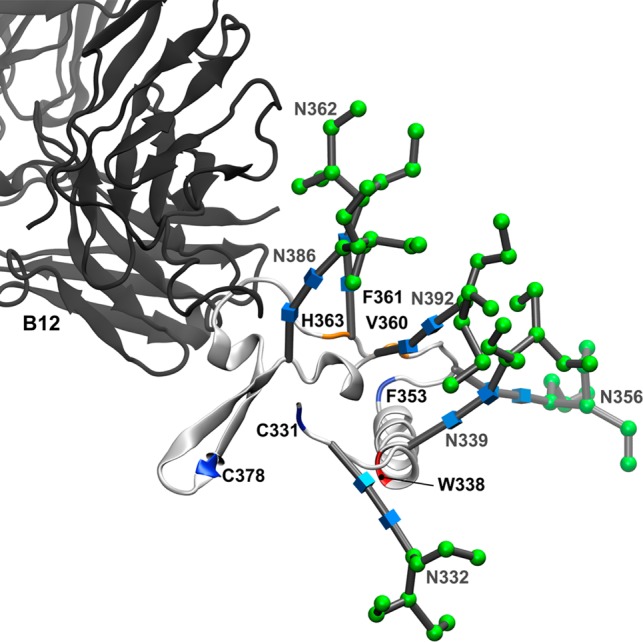
Interaction of b12 with the C3 domain of gp120 mediated by N-linked glycosylation. Backbone and N-linked glycans of the C3 domain from the full-length, glycosylated gp120 MD simulation (light gray ribbon) aligned with the b12 Fab fragment (dark gray ribbon). All other gp120 domains have been excluded for the sake of clarity. Residues that showed >80% protection from HR-HRPF upon b12 binding are colored red. Residues that showed between 40 and 80% protection from HR-HRPF upon b12 binding are colored orange. Residues showing no protection from HR-HRPF upon b12 binding are colored blue. Glycans are labeled and shown as 3D-SNFG symbols, which are positioned at each residue’s ring center.
W338 in the C3 domain shows complete protection from HR-HRPF upon b12 binding. W338 exists at the N-terminal end of the large α2 helix of the C3 domain. This helix does not interact with b12 in either the X-ray crystal structure or the full-length glycosylated model. W338 is directly adjacent to the N339 glycosylation site. However, in our model, the N339 Man9GlcNAc2 glycan does not directly interact with the b12 antibody. It is possible that b12 binding triggers conformational changes in the core of the C3 domain or in the C3–C4 domain–domain interactions (namely, between helix α2 of the C3 domain and strand β22/β23 of the C4 domain) that are observed in the strong HR-HRPF protection of W338 observed. Mutagenesis of the K343 residue in helix α2 and T450 in the center of strand β22/β23 showed a greatly decreased affinity for the b12 antibody,50 supporting a potential impact of b12 binding on this region.
It is also possible that the N339 glycan is involved in interactions that are not properly captured by our model. Interestingly, the CATNAP database56 shows a strong positive correlation between an intact N339 glycosylation sequon and b12 binding, suggesting that glycosylation of N339 is involved, directly or indirectly, in the energetics of b12 binding. Previous deglycosylation studies by Koch and co-workers did not examine elimination of the N339 or N362 glycosylation site.18 While it is not apparent how the N339 glycan could interact directly with the b12 antibody, perhaps the N339 glycan contributes indirectly via another glycan, which interacts directly with b12.
Unprotected Domains
The C2 and V4 domains both had at least one residue probed by HR-HRPF, but no residues that showed protection upon b12 binding. For the C2 domain, eight residues were probed by HR-HRPF (C205, C218, C239, S274, D275, F277, L288, and K289), including three cysteines that are highly sensitive to changes in solvent accessibility. This domain is distal to the b12 antibody binding interface and largely consists of flexible loops and small antiparallel β-sheets. All of the HR-HRPF-probed residues are found on regions lacking secondary structure in both the MD model and the X-ray crystal structure of the stabilized gp120–b12 complex, indicating that the binding of b12 does not appreciably change the structure or dynamics of the loops. The residues in the β-sheets were silent in this HR-HRPF experiment. Interestingly, several residues in the C2 domain were found by mutagenesis to affect b12 binding affinity, mostly among residues that are conserved among most gp120 sequences.50 Unfortunately, in no case did we receive HR-HRPF data directly on a residue mutated by Pantophlet and co-workers, and the pattern of mutations that changed binding affinity is by no means clear; several of the mutations that decreased b12 binding affinity occurred on the stretch of residues 223–239 of the C2 domain that was unoxidized by HR-HRPF (Figure 2).
The V4 domain had one residue at its extreme C-terminus probed by HR-HRPF, I414, which can be seen near the N-terminal end of strand β19 in Figure 7A. The interaction with b12 occurs at the C-terminal end of the long β19 strand, and the lack of HR-HRPF protection of I414 upon b12 binding suggests that the N-terminal end of this strand remains stable in the absence of b12 binding. No mutagenesis data for this residue have been reported, and CATNAP database analysis showed no correlation between this amino acid identity and b12 affinity.
Conclusion
We demonstrate the ability of the hydroxyl radical-mediated protein footprinting combined with molecular modeling to examine the protein–protein interactions between a highly glycosylated mature gp120 and its bNAb b12. The data presented here identified the highly occupied glycosylation sites and probed the solvent accessibility of 61 residues in full-length, glycosylated JR-FL gp120 and identified 23 residues protected upon b12 binding. Analysis of our results in the context of a model of full-length, glycosylated gp120 indicates a prominent role of interactions of the V1/V2 domain with the light chain of the b12 antibody, as well as potential roles for the N362 glycan in interacting directly with b12. HR-HRPF was able to detect both direct interactions and conformational changes upon b12 binding without the need to introduce mutations or site-specific chemical labels that could alter the native conformation we seek to probe. The combination of HR-HRPF for providing biophysical data on the full-length glycosylated gp120 protein and molecular modeling for providing a context to interpret changes in HR-HRPF protection is powerful. Further studies examining the interactions of gp120 with other bNAbs are underway.
Acknowledgments
Figures depicting 3D structures were created using Visual Molecular Dynamics (VMD).57
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00888.
A table of glycosylation sequon occupancy for gp120 samples used for this study and a figure showing the gp120 sequence alignments used by Modeler to generate the homology model (PDF)
Author Contributions
X.L. and O.C.G. contributed equally to this work.
This research is supported by the National Institute of General Medical Sciences (1R01GM096049-01A1 and R01GM100058). Instrumentation used for analysis was funded in part by the National Institute of General Medical Sciences-funded “Research Resource for Integrated Glycotechnology” (P41 GM103390) from the National Institutes of Health. Glycosylation occupancy identification by L.W. was supported in part by the National Center for Biomedical Glycomics (P41 GM103490). S.L. acknowledges funding from a Gates Foundation grant (OPP1033112) and the National Institute of Allergy and Infectious Diseases (AI082274 and AI082676). R.J.W. was supported by the National Institutes of Health (U01 CA207824 and P41 GM103390).
The authors declare the following competing financial interest(s): J.S.S. discloses a significant ownership share of Photochem Technologies, LLC, a small company active in the area of hydroxyl radical protein footprinting.
Supplementary Material
References
- Kowalski M.; Potz J.; Basiripour L.; Dorfman T.; Goh W. C.; Terwilliger E.; Dayton A.; Rosen C.; Haseltine W.; Sodroski J. (1987) Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237, 1351–1355. 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- Nara P. L.; Garrity R. R.; Goudsmit J. (1991) Neutralization of HIV-1: a paradox of humoral proportions. FASEB J. 5, 2437–2455. [DOI] [PubMed] [Google Scholar]
- Lasky L. A.; Groopman J. E.; Fennie C. W.; Benz P. M.; Capon D. J.; Dowbenko D. J.; Nakamura G. R.; Nunes W. M.; Renz M. E.; Berman P. W. (1986) Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233, 209–212. 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- Kwong P. D.; Wyatt R.; Robinson J.; Sweet R. W.; Sodroski J.; Hendrickson W. A. (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R.; Kwong P. D.; Desjardins E.; Sweet R. W.; Robinson J.; Hendrickson W. A.; Sodroski J. G. (1998) The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711. 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Kwong P. D.; Wyatt R.; Desjardins E.; Robinson J.; Culp J. S.; Hellmig B. D.; Sweet R. W.; Sodroski J.; Hendrickson W. A. (1999) Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274, 4115–4123. 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- Saphire E. O.; Parren P. W.; Pantophlet R.; Zwick M. B.; Morris G. M.; Rudd P. M.; Dwek R. A.; Stanfield R. L.; Burton D. R.; Wilson I. A. (2001) Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293, 1155–1159. 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Chen B.; Vogan E. M.; Gong H.; Skehel J. J.; Wiley D. C.; Harrison S. C. (2005) Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841. 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Huang C. C.; Tang M.; Zhang M. Y.; Majeed S.; Montabana E.; Stanfield R. L.; Dimitrov D. S.; Korber B.; Sodroski J.; Wilson I. A.; Wyatt R.; Kwong P. D. (2005) Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028. 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T.; Xu L.; Dey B.; Hessell A. J.; Van Ryk D.; Xiang S. H.; Yang X.; Zhang M. Y.; Zwick M. B.; Arthos J.; Burton D. R.; Dimitrov D. S.; Sodroski J.; Wyatt R.; Nabel G. J.; Kwong P. D. (2007) Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737. 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R.; Marcovecchio P. M.; Bjorkman P. J. (2010) Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17, 608–613. 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J. S.; Pancera M.; Carrico C.; Gorman J.; Julien J. P.; Khayat R.; Louder R.; Pejchal R.; Sastry M.; Dai K.; O’Dell S.; Patel N.; Shahzad-ul-Hussan S.; Yang Y.; Zhang B.; Zhou T.; Zhu J.; Boyington J. C.; Chuang G. Y.; Diwanji D.; Georgiev I.; Do Kwon Y.; Lee D.; Louder M. K.; Moquin S.; Schmidt S. D.; Yang Z. Y.; Bonsignori M.; Crump J. A.; Kapiga S. H.; Sam N. E.; Haynes B. F.; Burton D. R.; Koff W. C.; Walker L. M.; Phogat S.; Wyatt R.; Orwenyo J.; Wang L. X.; Arthos J.; Bewley C. A.; Mascola J. R.; Nabel G. J.; Schief W. R.; Ward A. B.; Wilson I. A.; Kwong P. D. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343. 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P. D.; Mascola J. R.; Nabel G. J. (2013) Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701. 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Burton D. R.; Pyati J.; Koduri R.; Sharp S. J.; Thornton G. B.; Parren P. W.; Sawyer L. S.; Hendry R. M.; Dunlop N.; Nara P. L.; Lamacchia M.; Garratty E.; Stiehm E. R.; Bryson Y. J.; Cao Y.; Moore J. P.; Ho D. D.; Barbas C. F. III (1994) Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027. 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Barbas C. F. 3rd; Bjorling E.; Chiodi F.; Dunlop N.; Cababa D.; Jones T. M.; Zebedee S. L.; Persson M. A.; Nara P. L.; Norrby E.; Burton D. R. (1992) Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 89, 9339–9343. 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C. F. 3rd; Collet T. A.; Amberg W.; Roben P.; Binley J. M.; Hoekstra D.; Cababa D.; Jones T. M.; Williamson R. A.; Pilkington G. R.; Haigwood N. L.; Cabezas E.; Satterthwait A. C.; Sanz I.; Burton D. R. (1993) Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230, 812–823. 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- Roben P.; Moore J. P.; Thali M.; Sodroski J.; Barbas C. F. 3rd; Burton D. R. (1994) Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68, 4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.; Pancera M.; Kwong P. D.; Kolchinsky P.; Grundner C.; Wang L.; Hendrickson W. A.; Sodroski J.; Wyatt R. (2003) Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313, 387–400. 10.1016/S0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Liu J.; Bartesaghi A.; Borgnia M. J.; Sapiro G.; Subramaniam S. (2008) Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113. 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. J.; Tessier M. B. (2010) Computational glycoscience: characterizing the spatial and temporal properties of glycans and glycan-protein complexes. Curr. Opin. Struct. Biol. 20, 575–583. 10.1016/j.sbi.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye T. T.; Low T. Y.; Sze S. K. (2005) Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal. Chem. 77, 5814–5822. 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- Chance M. R. (2001) Unfolding of apomyoglobin examined by synchrotron footprinting. Biochem. Biophys. Res. Commun. 287, 614–621. 10.1006/bbrc.2001.5628. [DOI] [PubMed] [Google Scholar]
- Goldsmith S. C.; Guan J. Q.; Almo S.; Chance M. (2001) Synchrotron protein footprinting: a technique to investigate protein-protein interactions. J. Biomol. Struct. Dyn. 19, 405–418. 10.1080/07391102.2001.10506750. [DOI] [PubMed] [Google Scholar]
- Charvatova O.; Foley B. L.; Bern M. W.; Sharp J. S.; Orlando R.; Woods R. J. (2008) Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J. Am. Soc. Mass Spectrom. 19, 1692–1705. 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.; Chance M. R. (2007) Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543. 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- Kiselar J. G.; Maleknia S. D.; Sullivan M.; Downard K. M.; Chance M. R. (2002) Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int. J. Radiat. Biol. 78, 101–114. 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- Kiselar J. G.; Mahaffy R.; Pollard T. D.; Almo S. C.; Chance M. R. (2007) Visualizing Arp2/3 complex activation mediated by binding of ATP and WASp using structural mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 104, 1552–1557. 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau B. C.; Sharp J. S.; Rempel D. L.; Gross M. L. (2009) Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 81, 6563–6571. 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C.; Janik I.; Zhuang T.; Charvatova O.; Woods R. J.; Sharp J. S. (2009) Pulsed electron beam water radiolysis for submicrosecond hydroxyl radical protein footprinting. Anal. Chem. 81, 2496–2505. 10.1021/ac802252y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Li Z.; Xie B.; Sharp J. S. (2013) Improved identification and relative quantification of sites of peptide and protein oxidation for hydroxyl radical footprinting. J. Am. Soc. Mass Spectrom. 24, 1767–1776. 10.1007/s13361-013-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Moniz H.; Wang S.; Ramiah A.; Zhang F.; Moremen K. W.; Linhardt R. J.; Sharp J. S. (2015) High structural resolution hydroxyl radical protein footprinting reveals an extended robo1-heparin binding interface. J. Biol. Chem. 290, 10729–10740. 10.1074/jbc.M115.648410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Arthos J.; Lawrence J. M.; Van Ryk D.; Mboudjeka I.; Shen S.; Chou T. H.; Montefiori D. C.; Lu S. (2005) Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. Journal of virology 79, 7933–7937. 10.1128/JVI.79.12.7933-7937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B.; Sharp J. S. (2015) Hydroxyl Radical Dosimetry for High Flux Hydroxyl Radical Protein Footprinting Applications Using a Simple Optical Detection Method. Anal. Chem. 87, 10719–10723. 10.1021/acs.analchem.5b02865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden T. (2002) in The BLAST Sequence Analysis Tool (McEntyre J., and Ostell J., Eds.) National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- Sali A.; Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Kong L.; Sheppard N. C.; Stewart-Jones G. B.; Robson C. L.; Chen H.; Xu X.; Krashias G.; Bonomelli C.; Scanlan C. N.; Kwong P. D.; Jeffs S. A.; Jones I. M.; Sattentau Q. J. (2010) Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 403, 131–147. 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G. B.; Soto C.; Lemmin T.; Chuang G. Y.; Druz A.; Kong R.; Thomas P. V.; Wagh K.; Zhou T.; Behrens A. J.; Bylund T.; Choi C. W.; Davison J. R.; Georgiev I. S.; Joyce M. G.; Kwon Y. D.; Pancera M.; Taft J.; Yang Y.; Zhang B.; Shivatare S. S.; Shivatare V. S.; Lee C. C.; Wu C. Y.; Bewley C. A.; Burton D. R.; Koff W. C.; Connors M.; Crispin M.; Baxa U.; Korber B. T.; Wong C. H.; Mascola J. R.; Kwong P. D. (2016) Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 165, 813–826. 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R.; Götz A. W.; Poole D.; Le Grand S.; Walker R. C. (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Gotz A. W.; Williamson M. J.; Xu D.; Poole D.; Le Grand S.; Walker R. C. (2012) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555. 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Babin V., Berryman J. T., Betz R. M., Cai Q., Cerutti D. S., Cheatham T. E. III, Darden T. A., Duke R. E., Gohlke H., Goetz A. W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossváry I., Kovalenko A., Lee T. S., LeGrand S., Luchko T., Luo R., Madej B., Merz K. M., Paesani F., Roe D. R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C. L., Smith W., Swails J., Walker R. C., Wang J., Wolf R. M., Wu X., and Kollman P. A. (2014) AMBER 14, University of California, San Francisco. [Google Scholar]
- Kirschner K. N.; Yongye A. B.; Tschampel S. M.; Gonzalez-Outeirino J.; Daniels C. R.; Foley B. L.; Woods R. J. (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655. 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Babin V., Berryman J. T., Betz R. M., Cai Q., Cerutti D. S., Cheatham T. E. III, Darden T. A., Duke R. E., Gohlke H., Goetz A. W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossvary I., Kovalenko A., Lee T. S., LeGrand S., Luchko T., Luo R., Madej B., Merz K. M. Jr., Roe D. R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C. L., Smith W. L., Swails J., Walker R. C., Wang J., Wolf R. M., Wu X., and Kollman P. A. (2014) The FF14SB force field. AMBER 14 Reference Manual, pp 29–31, University of California, San Francisco. [Google Scholar]
- Darden T.; York D.; Pedersen L. (1993) Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089. 10.1063/1.464397. [DOI] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. C. (1977) Numerical integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Mahoney M. W.; Jorgensen W. L. (2000) A Five-site Model for Liquid Water and the Reproduction of the Density Anomoly by Rigid, Non-polarizible Potential Functions. J. Chem. Phys. 112, 8910–8922. 10.1063/1.481505. [DOI] [Google Scholar]
- Bruns C. M.; Hubatsch I.; Ridderstrom M.; Mannervik B.; Tainer J. A. (1999) Human glutathione transferase A4–4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J. Mol. Biol. 288, 427–439. 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
- Hubbard S. J., and Thornton J. M. (1993) NACCESS, Department of Biochemistry and Molecular Biology, University College London, London. [Google Scholar]
- Zwick M. B.; Kelleher R.; Jensen R.; Labrijn A. F.; Wang M.; Quinnan G. V. Jr.; Parren P. W.; Burton D. R. (2003) A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. Journal of virology 77, 6965–6978. 10.1128/JVI.77.12.6965-6978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R.; Ollmann Saphire E.; Poignard P.; Parren P. W.; Wilson I. A.; Burton D. R. (2003) Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. Journal of virology 77, 642–658. 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M. B.; Parren P. W.; Saphire E. O.; Church S.; Wang M.; Scott J. K.; Dawson P. E.; Wilson I. A.; Burton D. R. (2003) Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. Journal of virology 77, 5863–5876. 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieker D. F.; Hadden J. A.; Schulten K.; Woods R. J. (2016) 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology 26, 786–787. 10.1093/glycob/cww076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M.; Cupo A.; Julien J. P.; Sanders R. W.; Wilson I. A.; Moore J. P.; Lee K. K. (2015) Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat. Commun. 6, 6144. 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell B. J.; Lortat-Jacob H. (2013) Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 4, 385. 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. B.; Zhu L.; Holz-Smith S.; Matthews T. J.; Chen C. H. (2001) The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc. Natl. Acad. Sci. U. S. A. 98, 15227–15232. 10.1073/pnas.261359098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.; Macke J.; West A. P. Jr.; Foley B.; Bjorkman P. J.; Korber B.; Yusim K. (2015) CATNAP: a tool to compile, analyze and tally neutralizing antibody panels. Nucleic Acids Res. 43, W213–219. 10.1093/nar/gkv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 27–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.