Abstract
BACKGROUND
African-Americans (Afr-Amr) with head and neck squamous cell carcinoma (HNSCC) have a lower survival rate than Caucasians (Cau). This study investigates the functional importance of ancestry-informative SNPs in HNSCC and also examines the effect of functionally important genetic elements on racial disparities in HNSCC survival.
METHODS
Ancestry-informative SNPs, RNAseq, methylation, and copy number variation data for 316 oral cavity and laryngeal cancer patients were analyzed across 178 DNA repair genes. The results of eQTL analyses were also replicated using a Gene Expression Omnibus (GEO) dataset. The effects of eQTLs on overall survival (OS) and disease-free survival (DFS) were evaluated.
RESULTS
Five ancestry-related SNPs were identified as cis-eQTLs in the POLB gene (FDR<0.01). The homozygous/ heterozygous genotypes containing the Afr-allele showed higher POLB expression relative to the homozygous Cau-allele genotype (P<0.001). A replication study using a GEO dataset validated all five eQTLs, also showing a statistically significant difference in POLB expression based on genetic ancestry (P=0.002). An association was observed between these eQTLs and OS (P<0.037; FDR<0.0363) as well as DFS of oral cavity and laryngeal cancer patients treated with platinum-based chemotherapy and/or radiotherapy (P=0.018 to 0.0629; FDR<0.079). Genotypes containing the Afr-allele were associated with poor OS/DFS compared to homozygous genotypes harboring the Cau-allele.
CONCLUSIONS
Our analyses show that ancestry-related alleles could act as eQTLs in HNSCC and support the association of ancestry-related genetic factors with survival disparity in patients diagnosed with oral cavity and laryngeal cancer.
Keywords: Head and Neck Squamous Cell Carcinoma, survival disparity, genetic ancestry, eQTLs, DNA polymerase beta
INTRODUCTION
African-Americans (Afr-Amr) with Head and Neck Squamous Cell Carcinoma (HNSCC) have consistently lower survival rates compared to Caucasian (Cau) patients.1, 2 Previous studies have associated socio-economic status (SES) with these survival differences.3 However, recent literature suggests the contribution of genetic differences between populations to survival disparities in some cancer types.4
Survival from cancer depends on the success of common treatment methods such as chemotherapy, radiotherapy, and surgery. Treatment planning for potentially curative disease often requires a multidisciplinary approach.5-7 Ionizing radiation generates free radicals which damage cellular DNA, resulting in apoptosis.8 In chemotherapy, platinum-based drugs such as cisplatin and carboplatin are commonly used to treat HNSCC.9, 10 These platinum-based drugs bind to DNA, forming DNA adducts11, and lead to cell cycle arrest and cytotoxicity.12 In response to DNA lesions caused by chemotherapy and/or radiotherapy, the cellular DNA damage response system repairs DNA aberrations and can reduce treatment sensitivity in cancer patients.13, 14 Thus, DNA repair genes play a key role in the treatment outcome of many cancers including HNSCC.
Higher level expression of DNA repair genes have been observed in many cancer types15, 16, and increased expression levels of DNA repair genes are associated with reduced sensitivity to chemotherapy and radiation therapy.17, 18 The increased expression of DNA repair genes in some cancers arises from multiple factors, including signaling within the tumor or from the tumor microenvironment leading to epigenetic upregulation, and/or de novo somatic mutations. Some evidence suggests that host genomic factors, such as germline variants in DNA repair-associated genes, may be associated with individual differences in response. Previous studies have shown that germline variants can act as expression quantitative trait loci (eQTL) affecting the expression of genes in cancer.19 Such germline variants may be preferentially found in certain populations. Indeed, a number of studies have investigated the effect of population-specific genetic variants on gene expression in normal, non-cancer samples from various human populations and identified differential gene expression levels regulated by ancestry-related alleles.20
Afr-Amr and Cau HNSCC patients possess distinct genetic ancestries. Our recent genomic analysis of laryngeal cancer in Afr-Amr and Cau patients revealed that distinctive genetic ancestry corresponds to molecular differences in the laryngeal cancers arising in these two populations.21 To date, the functional role of population-specific genomics variants in HNSCC survival is unknown. In this study, the functional role of ancestry-related genomic factors on the expression of DNA damage response genes and the effect of ancestry-informative SNPs on racial disparity in HNSCC survival was investigated using an integrative genomics approach. We tested the hypothesis that ancestry-related genomic elements are associated with HNSCC survival disparity between Afr-Amr and Cau populations due to altered gene expression in DNA damage response genes, thereby affecting the sensitivity to chemotherapy and/or radiotherapy. This work, focusing on oral cavity and laryngeal cancer, is the first study to investigate the functional importance of ancestry-informative SNPs in HNSCC and to examine the effect of functionally important genetic elements on racial disparities in HNSCC survival.
MATERIALS AND METHODS
Data Source
The genotype data of 4,802 genome-wide ancestry-informative SNPs for 316 oral cavity and laryngeal cancer patients (30 Afr-Amr and 286 Cau) were retrieved from The Cancer Genome Atlas (TCGA) for eQTL analyses. Detailed methodologies explaining how 4,802 ancestry-informative SNPs were retrieved from TCGA are given in Supplementary Material 1. The raw genome-wide methylation data for ~485,000 CpG sites based on Infinium HumanMethylation450 BeadChip Kit (Illumina, Inc.) were retrieved for tumors of the 316 HNSCC patients from TCGA. The M-values (methylation signal) were calculated for each CpG site using the Bioconductor Minfi package.22 The somatic copy number variation (CNV) data and RNAseq based gene expression data (normalized RSEM values) for tumors of the 316 HNSCC patients in TCGA were also obtained from the Broad Institute (http://gdac.broadinstitute.org/). A gene was included in the analyses only when ≥ 50% of patients have expression values for that gene.
Expression quantitative trait loci (eQTL) analyses
The effect of each ancestry-informative SNP on the expression of nearby genes (± 1Mb from SNP) was analyzed to find potential ancestry-related eQTL candidates. A multivariable regression model was used to identify eQTLs (Equation 1). A negative binomial regression model was used based on the empirical mean-variance relationship for gene expression.
| (Equation 1) |
where, Expa denotes the expression of gene a,
GTa denotes the genotype of the SNP under study,
CNVa denotes the somatic copy number variation of gene a,
Ma denotes the methylation levels of CpG sites that are associated with gene a,
Popa denotes the top three principal component values to adjust for population stratification,
εa denotes the residual error
The regression analyses were carried out using the MASS package in R (version 3.0.2). All tests were two-sided and P-values were corrected for multiple tests following the Benjamini-Hochberg method (FDR). Potential ancestry-informative SNPs that affect the expression of DNA repair genes with FDR ≤ 0.01 were retrieved. A list of 178 DNA repair genes (updated on April 15th 2014) was obtained from the online resource of MD Anderson Cancer Center (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html) for our study. This comprehensive set of DNA repair genes has been used widely in several recent publications.23, 24 We also tested the effects of eQTLs (found in the pooled dataset (30 Afr-Amr and 286 Cau)) on the expression of DNA repair genes following the regression model (equation 1) using Cau patients (N=286) dataset.
Other statistical analyses
Age distribution differences between Afr-Amr and Cau patients were studied using a Mann-Whitney-Wilcoxon test. Differences in the proportions of current-smokers, ex-smokers, never-smokers, and each pathological stage between Afr-Amr and Cau patients were assessed using the test of proportions. Gene expression levels between Afr-Amr and Cau patients were compared using the Mann-Whitney-Wilcoxon test. The genotype data were retrieved for each eQTL based on ancestry and the allele frequencies of each eQTL were calculated. Also, the allele frequencies of each eQTL were retrieved for ASW (Americans with African ancestry in SW USA) and CEU (Americans with European ancestry) populations from the 1000 Genomes Project (1000G) data, and the proportions of allele frequencies were compared between TCGA and 1000G samples (ASW versus TCGA Afr-Amr and CEU versus TCGA Cau) using a test of proportions. All tests were two-sided and P-values ≤ 0.05 were considered to be statistically significant.
Replication of eQTL analyses
The TCGA eQTL analyses were replicated using a GEO dataset, GSE39368, generated by Walter et al.25 This dataset contains genome-wide SNP and CNV data for 99 HNSCC patients and gene expression data for 138 HNSCC patients. The SNP, CNV, and gene expression datasets were combined into a single dataset with missing values coded as “NA”. Data was extracted, if a patient's ancestry is either Afr-Amr or Cau, and the anatomical site represents oral cavity or larynx. After these filters, 96 HNSCC patients (73-Cau; 23-Afr-Amr) were retrieved for further analyses. In this dataset, the expression of each gene was measured by multiple probes and the mean of multiple probes of each gene was taken as the expression measure of that gene. The effect of each SNP, identified as an eQTL in the TCGA dataset, on gene expression was tested using the linear regression model after adjusting for CNV.
Linkage Disequilibrium (LD) analyses
Genomic data for all the eQTLs and SNPs within ± 50 kb distance from each eQTL were retrieved for the ASW population from the 1000G database and linkage disequilibrium between each eQTL and its nearby SNPs were analyzed using VCFtools.26 SNPs in strong LD (D’ ≥ 0.8) with eQTLs were identified for further analyses. The LD heatmap was generated using Haploview software.27
ENCODE functional analyses
The genome-wide DNAse-I sensitivity assay data and transcription factor sites were retrieved from ENCODE. 28 Each eQTL and LD SNP was intersected with the ENCODE database using custom perl and shell scripts. The eQTL/LD SNP is thought to be functionally important if the eQTL/SNP was found in the regulatory region of a gene, DNAse-I sensitivity and/or transcription factor binding site. The genomic position of functionally important eQTL/SNPs was visualized using the UCSC genome browser.29
Survival analyses based on eQTLs
The effect of eQTLs on overall survival (OS) and disease-free survival (DFS) in HNSCC patients with a history of platinum-based chemo and/or radiation therapy was investigated. First, Kaplan-Meier (KM) plots were generated for each eQTL to visualize the effect of eQTL genotypes using STATA v14.01. Secondly, Hazard Ratios (HR) for the risk of death (OS) according to the eQTL genotype was calculated using Cox proportional hazards (PH) regression models after adjusting for age and pathological stage using the survival package in R, and the goodness-of-fit test using Schoenfeld residuals was performed to test the appropriateness of the Cox PH model.30 All tests were two-sided and a P-value threshold of 0.05 was used to determine statistical significance. The P-values were corrected for multiple tests following the Benjamini-Hochberg method.
To validate the survival analyses results, existing germline DNA and clinical data for 20 additional oral cavity or laryngeal cancer patients of African-ancestry who had platinum-based chemo and/or radiation therapy were obtained.31 Germline DNA for all 20 patients was genotyped for one of the eQTLs, rs2272733, using a real-time PCR TaqMan assay (Life Technologies/ Thermo Fisher Scientific, Waltham, MA, USA). The genotype and clinical data for these 20 patients were combined with the 157 patient data of TCGA to generate an enriched dataset (Datset-2) containing 177 HNSCC patients (36 Afr-Amr and 141 Cau patients) with a history of platinum-based chemo and/or radiation therapy for survival analyses. All the human subject investigations were approved by Fox Chase Cancer Center's Institutional review boards.
Estimation of admixture proportions and survival analyses
Autosomal AIMs data for TCGA patients along with YRI, CEU, JPT, and CHB individuals from the 1000G were retrieved. The genetics admixture proportions for each individual including Afr-Amr and Cau patients from TCGA were estimated using a model-based clustering approach implemented in STRUCTURE V2.3.4.32 In STRUCTURE, the data were analyzed using different K (genetics clusters) values ranging from 3 to 10 under the admixture model. For each K, 10 runs were performed with 10,000 burn-in and an additional 20,000 replicates. The best K was estimated following the method of Evanno et al.33 as implemented in the Structure Harvester program.34 The output of STRUCTURE based on the best K was analyzed using CLUMPP.35 The Afr-admixed fraction of each HNSCC patient (N=157) with platinum-based chemo and/or radiotherapy history was obtained from the CLUMPP output and used for survival analyses. The effects of the Afr-admixed proportion on OS and DFS were analyzed using Cox PH regression models from the survival package in R after adjusting for age and pathological stage. Goodness-of-fit tests using Schoenfeld residuals were performed to evaluate the appropriateness of the Cox PH model. All tests were two-sided and a P-value threshold of 0.05 was used to determine statistical significance.
RESULTS
TCGA sample characteristics
Summary statistics of the 316 HNSCC patients included in this analysis are provided in Table 1. The differences in age, smoking status, and pathological stage between Afr-Amr and Cau patients were not significant.
Table 1.
Characteristics of TCGA HNSCC patients
| Characteristics | All | Afr-Amr | Caucasian |
|---|---|---|---|
| N | 316 | 30 | 286 |
| Age | 61.56±11.5 | 58±7.74 | 61.94±11.8 |
| Gender | |||
| Male | 226 | 24 (80.0%) | 202 (70.6%) |
| Female | 90 | 6 (20.0%) | 84 (29.4%) |
| Tumor site | |||
| Hypopharynx | 5 | 1 (3.3%) | 4 (1.4%) |
| Larynx | 81 | 11 (36.7%) | 70 (24.5%) |
| Oral cavitya | 230 | 18 (60.0%) | 212 (74.1%) |
| Smoking status | |||
| Current smoker | 115 | 16 (53.3%) | 99 (34.6%) |
| Ex-smoker | 128 | 9 (30.0%) | 119 (41.6%) |
| Never-smoker | 64 | 2 (6.7%) | 62 (21.7%) |
| Unknown | 9 | 3 (10.0%) | 6 (2.1%) |
| Pathologic_stage | |||
| Stage_I | 15 | 1 (3.3%) | 14 (4.9%) |
| Stage_II | 47 | 1 (3.3%) | 46 (16.1%) |
| Stage_III | 49 | 3 (10.0%) | 46 (16.1%) |
| Stage_IV | 177 | 22 (73.3%) | 155 (54.2%) |
| Unknown | 28 | 3 (10.0%) | 25 (8.7%) |
Oral cavity includes Alveolar ridge, Buccal mucosa, Floor of mouth, and Oral tongue.
Effect of ancestry-informative SNPs on expression of DNA repair genes (eQTL analysis)
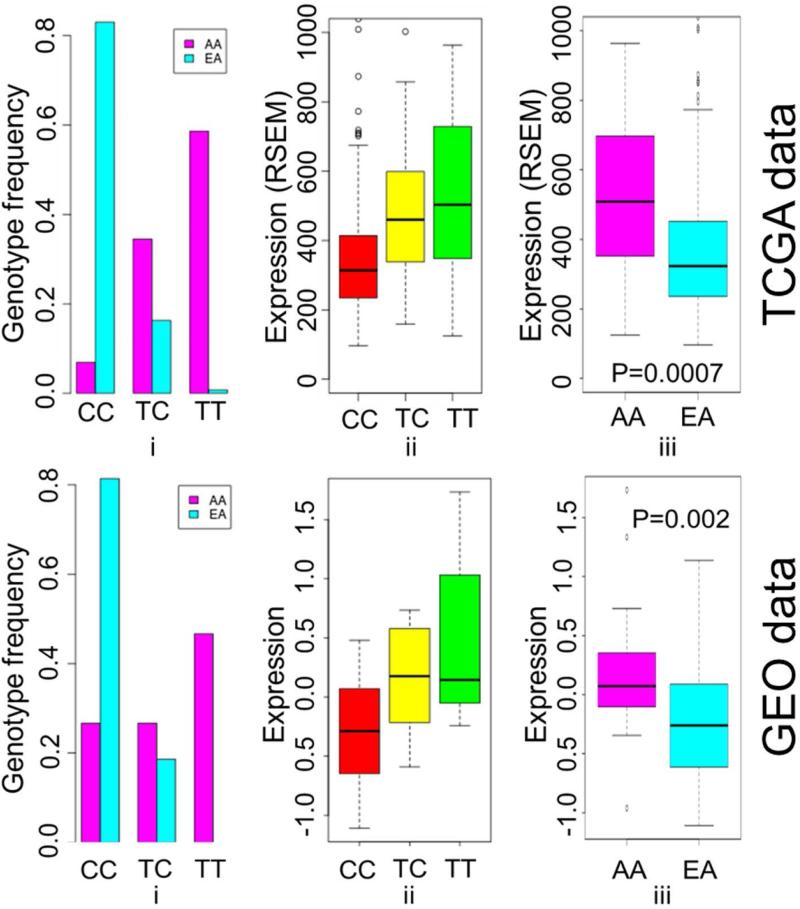
The focus of this study was to analyze the effect of ancestry-informative SNPs on an annotated set of 178 DNA repair genes. Our results showed that the expression of one DNA repair gene, DNA polymerase beta (POLB), was significantly affected by nearby ancestry-informative SNPs with FDR≤0.01. Of 4,802 ancestry-informative SNPs, five SNPs (rs2272733, rs3136790, rs6474387, rs2272732, and rs10096210) were found to be eQTLs that affect the expression of POLB (FDR≤0.01). Each of the five SNPs was also observed to be eQTLs when the dataset was limited to Caucasians only. The P-values for the five SNPs based on pooled (Afr-Amr and Cau patients) and Cau datasets are reported in Supplementary Table 1. As an illustration, the effects of rs2272732 on POLB are shown in Figure 1 (TCGA panel). The effects of the other four SNPs on POLB were similar to rs2272732 (Supplementary Figure 1). In this manuscript, we use “Afr-allele” and “Cau-allele” to denote the major-allele specific to Afr-Amr and Cau populations, respectively. Homozygous genotypes containing the Afr-allele were associated with higher levels of POLB while homozygous Cau-allele genotypes had decreased POLB expression. Heterozygous genotypes containing an Afr-allele and a Cau-allele were associated with moderately higher levels of POLB expression (Figure 1(ii)- TCGA panel). Comparison of POLB mRNA expression data showed higher level expression of POLB in Afr-Amr patients (Q1: 355.2; Median: 508.3; Q3: 693.7) compared to Cau patients (Q1: 236.1; Median: 323.2; Q3: 451.2). There was a statistically significant difference in POLB expression between Afr-Amr and Cau patients (P<0.001) (Figure 1(iii)–TCGA panel).
Figure 1.
Results of TCGA and GEO data analyses. (i) Genotype frequencies of rs2272732 for Afr-Amr (AA) and Caucasian patients (EA); (ii) Effect of rs2272732 on POLB gene expression; (iii) POLB gene expression for Af-Amr and Caucasian patients.
Replication of eQTL analyses
The results of eQTL analyses based on TCGA data were replicated using a GEO dataset and were consistent with the results observed from the TCGA data analyses. The regression analyses identified all five SNPs as eQTLs that affect the expression of the POLB gene (P<0.007). The effects of rs2272732 on POLB expression in the GEO dataset are shown in Figure 1 (GEO panel). As with the TCGA data, homozygous Afr-allele genotypes of all five SNPs were associated with higher expression levels of POLB compared to homozygous Cau-allele genotype from the GEO dataset. Also, the heterozygous genotypes had moderately higher levels of POLB expression as compared to homozygous genotypes of the Cau-allele. Evaluation of GEO data confirmed that Afr-Amr patients had a higher level of POLB expression (Q1: −0.10; median: 0.07; Q3: 0.35) compared to Cau patients (Q1: −0.61; median: −0.26; Q3: 0.09) with a significant difference in POLB expression levels between the two populations (P=0.002).
LD and ENCODE analyses
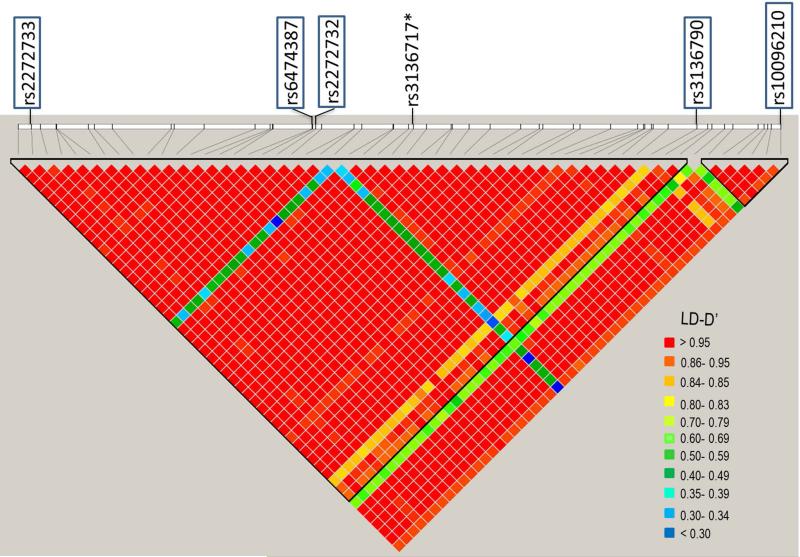
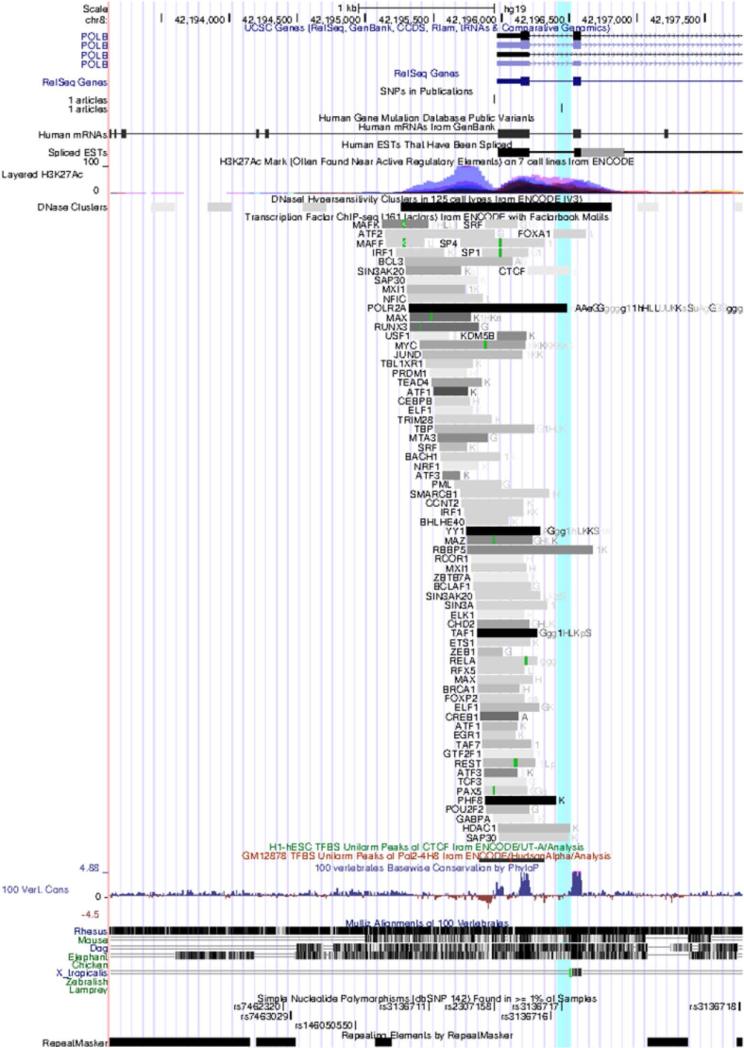
The results of the LD analyses are shown in Figure 2. Each of the five eQTLs are in strong LD (D’ ≥ 0.9) with each other. In addition, another ancestry-informative SNP, rs3136717 was in strong association with all five eQTLs (D’>0.8). The functional importance of the five eQTLs and rs3136717 was investigated using the ENCODE data. None of the five eQTLs were found to be on the POLB gene region with strong DNAse I sensitivity/ TF binding signals. However, the associated ancestry-informative SNP, rs3136717, is located in the regulatory region of POLB and in the DNAse I sensitivity region of POLB in all 125 cell lines assayed in the ENCODE project. In addition, rs3136717 is located in the binding site of several transcription factors, specifically polymerase (RNA) II subunit A (POLR2A). The genomic position and associated ENCODE annotations for rs3136717 are shown in Figure 3.
Figure 2.
LD analyses show all the five eQTLs are in strong LD. In addition, another ancestry-informative SNP, rs3136717 is in strong LD with all five eQTLs.
Figure 3.
Genomic position of rs3136717 based on ENCODE data. The SNP, rs3136717 (shaded in cyan,) is located in a known regulatory region, a DNAse I sensitivity region, and intersects several transcription factors binding sites of the POLB gene.
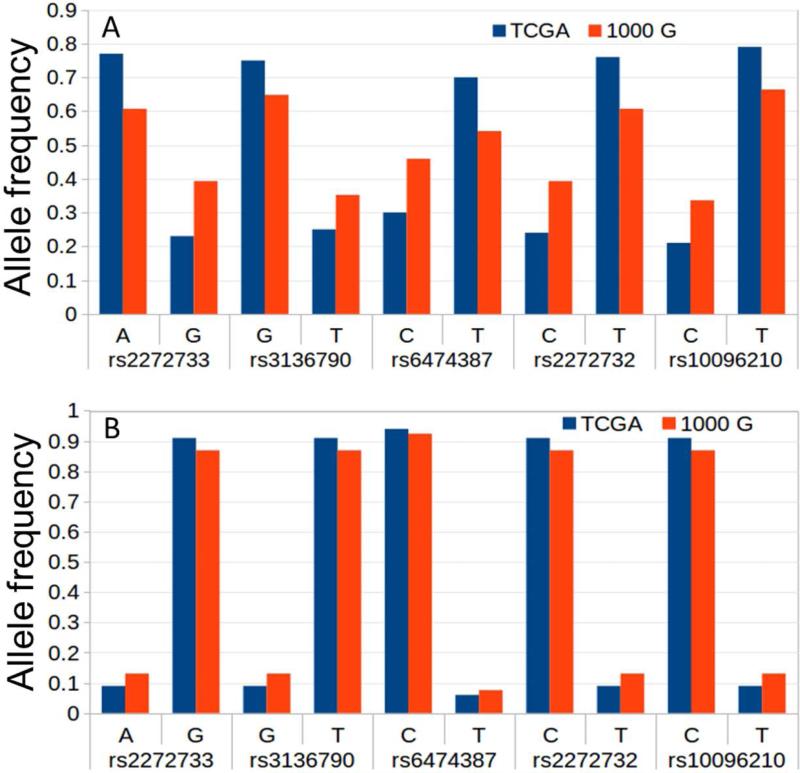
Allele frequencies of eQTLs between TCGA and 1000G data
Allele frequencies of the five eQTLs for Afr-Amr and Cau patients in TCGA were estimated, and compared with the allele frequencies of their respective populations (ASW for Afr-Amr patients (Figure 4A) and CEU for Cau patients (Figure 4B)) from the 1000G data. Allele frequency between the TCGA and the 1000G datasets were not significantly different.
Figure 4.
Allele frequency differences between TCGA and 1000G populations. (A) Allele frequency differences between TCGA Afr-Amr patients and 1000G ASW population data set. (B) Allele frequency differences between TCGA Caucasian patients and 1000G CEU population data set.
Survival analyses based on eQTLs
The effect of each eQTL on OS of HNSCC patients who were treated with platinum-based chemotherapy and/or radiotherapy (N=157; Afr-Amr-16; Cau-141) was examined. All five eQTLs were found to be significantly associated with OS (logrank test P<0.037; FDR<0.0363). The KM plot for a representative eQTL, rs2272733, is shown in Figure 5A. The KM plots for the other four eQTLs are shown in Supplementary Figure 2. The DFS analyses found rs2272732 and rs2272733 were significantly associated with DFS (P<0.05) while rs3136790 and rs10096210 were associated with DFS with moderate significance (P=0.0544 to 0.0629). The KM plots on DFS for each of the five eQTLs are shown in Supplementary Figure 3. The hazard ratios (HR) for each genotype of the five eQTLs on OS were calculated using the Cox PH model after adjusting for age and pathological stage (Table 2). The goodness-of-fit test confirmed the appropriateness of the Cox PH model for OS analyses. For four of the five eQTLs patients with the homozygous genotype of the Cau-allele had a significantly lower risk of death (P<0.0003; FDR<0.0008) compared to patients with the homozygous Afr-allele. Also, patients with the heterozygous genotypes for rs2272733 and rs3136790 were found to have a significantly lower risk of death (P<0.002; FDR<0.0016) compared to patients that had homozygous genotypes consisting of the Afr-Amr major-allele. The HR for the genotypes of rs6474387 was not found to be significant.
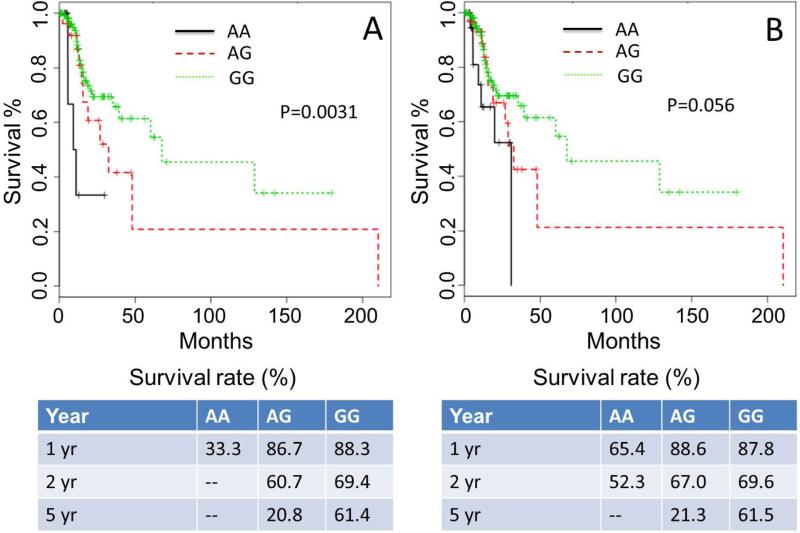
Figure 5.
Kaplan-Meier (KM) plot on overall survival of HNSCC patients with platinum-based chemo and/or radiation therapy based on rs2272733 genotypes. (A) KM plot for dataset-1 (N=157); (B) KM plot for dataset-2 (N=177).
Table 2.
Hazard ratio (HR) for five eQTLs based on overall survival after adjusting for age, and clinical pathological stage.
| eQTL | Afr-Amr Major-allele | Caucasian Major-allele | Genotype | HR | Lower .95 | Upper .95 | FDR |
|---|---|---|---|---|---|---|---|
| rs2272733 | A | G | AA | 1.00 (reference) | -- | -- | -- |
| AG | 0.079 | 0.016 | 0.380 | 0.0033 | |||
| GG | 0.075 | 0.018 | 0.310 | 0.0008 | |||
| rs3136790 | G | T | GG | 1.00 (reference) | -- | -- | -- |
| GT | 0.048 | 0.008 | 0.281 | 0.0016 | |||
| TT | 0.064 | 0.015 | 0.274 | 0.0008 | |||
| rs6474387 | T | C | CC | 1.00 (reference) | -- | -- | -- |
| TC | 1.869 | 0.695 | 5.026 | 0.27 | |||
| TT | 4.693 | 0.560 | 39.354 | 0.22 | |||
| rs2272732 | T | C | CC | 1.00 (reference) | -- | -- | -- |
| TC | 0.670 | 0.200 | 2.260 | 0.53 | |||
| TT | 13.500 | 3.260 | 55.876 | 0.0008 | |||
| rs10096210 | T | C | CC | 1.00 (reference) | -- | -- | -- |
| TC | 0.679 | 0.202 | 2.284 | 0.53 | |||
| TT | 13.478 | 3.258 | 55.755 | 0.0008 | |||
We also evaluated the effect of the five eQTLs on OS and DFS of patients not treated with cisplatin/carboplatin/radiotherapy. None of the eQTL genotypes were found to be significantly associated with OS (P>0.6) and DFS (P>0.8).
Validation of eQTL based survival analyses
The survival analyses results of Dataset-2 for SNP rs2272733 were consistent with the results of rs2272733 from the TCGA dataset. The KM plot and survival rates are shown in Figure 5B. The homozygous genotype of the Afr-allele, “AA”, is associated with lower OS as compared to the other two genotypes, “AG” and “GG” (logrank test: P=0.056). The Cox PH model, after adjusting for age, clinical stage, and cohort, revealed that patients with the heterozygous genotype “AG” had a significantly lower risk of death compared to patients with the homozygous Afr-allele genotype “AA” (AA vs AG: HR- 0.26; 95% CI- 0.08-0.84; P=0.024). Also, patients with the homozygous genotype containing the Cau-allele “GG” had a significantly lower risk of death compared to patients with the homozygous Afr-Amr genotype, “AA”, (AA vs GG: HR- 0.15; 95% CI- 0.04-0.48; P=0.0012).
Survival analyses based on genetics proportion
The main objective of this manuscript is to understand the effect of Afr ancestry on survival disparity in HNSCC. Thus, we analyzed the effect of Afr-admixture on survival in HNSCC patients with a history of platinum-based chemo and/or radiotherapy. The HR for OS and DFS were 8.99 (95% CI: 1.53- 52.95; P=0.015) and 7.12 (95% CI: 1.46- 34.77; P=0.015), respectively. The goodness-of-fit test confirmed the appropriateness of the Cox PH model for OS and DFS analyses.
DISCUSSION
Ancestry-informative SNPs act as eQTLs
Our stringent criteria (FDR≤0.01) found five ancestry-informative SNPs as eQTLs that affect POLB expression. This is the first study to demonstrate the impact of ancestry-informative SNPs on the expression of a gene involved in head and neck cancer tissues with a subsequent effect on survival disparity. The expression level of POLB significantly differs between Afr-Amr and Cau patients (P<0.001) with Afr-Amr patients possessing higher levels of POLB expression in their tumors compared to Cau patients due to ancestry-related eQTLs. In general, major factors that could alter gene expression are SNPs (eQTLs), CNVs, somatic point mutations, methylation, and population differences. In our analyses, we included CNV, methylation, and the top three principal components in our regression model. Therefore, the effects of CNV, methylation, and population structure on POLB expression were adjusted, suggesting an independent association of SNPs with gene expression. In addition, our candidate SNPs were also found to be eQTLs when tested exclusively on a Cau patients dataset. We have also checked somatic point mutations in our TCGA cohort based on exome data and did not find any somatic point mutations in the POLB gene.
Allele frequencies of the five eQTLs between Afr-Amr patients and the 1000G ASW population, and between Cau patients and the 1000G CEU population are not significantly different (Figure 2; P>0.6). Thus, we expect to observe the same effect for these eQTLs in any Afr-Amr and Cau populations as we found in TCGA patients. However, the effect of eQTLs in Afr-Amr and Cau controls (non-cancer samples) remain to be determined. Figure 4 reveals that candidate eQTL allele frequencies are different between Afr-Amr and Caucasian individuals, irrespective of which dataset (TCGA or / 1000G) was used. In addition, for all five eQTLs, each specific allele is enriched in Afr-Amr patients compared to Cau patients.
We also validated the results of TCGA dataset by replicating the eQTL analyses using a GEO dataset. If the results obtained from the TCGA dataset occurred by chance or due to some unknown factors associated with TCGA samples, we expect to see different results from the GEO dataset. Indeed, the results for the five ancestry-informative SNPs based on the GEO dataset are consistent with the results of TCGA data analyses. Each of the five eQTLs observed in the TCGA data were identified as eQTLs and affect POLB expression in the GEO dataset (Figure 1). Thus, we provide evidence that ancestry-informative SNPs could act as eQTLs and alter the expression of genes in HNSCC. While these results were observed based on RNA expression, further study is required to confirm the effect of eQTLs on protein expression.
Increased levels of POLB expression have been shown to be associated with tumorigenesis.36, 37 In addition, increased level of POLB expression has been observed in many cancer types38 suggesting that a higher level of POLB expression could be associated with the risk of HNSCC. Compared with other groups, Afr-Amr have a higher incidence of HNSCC, particularly in the larynx. These observations support the association between HNSCC incidence disparity and ancestry-informative SNPs. However, further studies are needed to confirm this association.
Identification of potentially functionally important eQTLs
All five eQTLs are located within or near (± 1Mb) the POLB gene region and all five eQTLs are in strong LD. In addition, we identify another ancestry-informative SNP, rs3136717, for which data are not available in TCGA and GEO datasets, in strong LD with the five eQTLs. It is expected that rs3136717 will be associated with similar levels of POLB expression as observed with the other five eQTLs. Unfortunately, we could not test the effect of rs3136717 on POLB due to the unavailability of genotype data for rs3136717 in TCGA. Our analyses showed that rs3136717 is found at the 5’ end of the POLB gene and in the DNAse I sensitivity region in all 125 surveyed cell lines. In addition, rs3136717 is located in the region where the transcription factor, POLR2A, binds to the POLB gene (Figure 4). Thus, rs3136717 is likely in an active regulatory region of the POLB gene. The POLR2A is the major subunit of RNA Polymerase II which is required for RNA transcription. Since rs3136717 is in an active regulatory region and on the POLR2A binding site, we speculate that the alternative alleles of rs3136717 could affect the binding affinity of POLR2A or other transcription factors to alter POLB gene expression. It is interesting to note that Figueroa el al39 has already identified rs3136717 to be associated with the risk of bladder cancer in a case-control study. Thus, rs3136717 could be functionally important and could be associated with the risk of many cancers including HNSCC. The role of rs3136717 in POLB expression in HNSCC needs to be further evaluated experimentally.
Effect of ancestry-related genomics variants on treatment outcome
Higher levels of POLB expression decrease the sensitivity of platinum-based chemotherapy and/or radiotherapy, and thus may be associated with poor survival.40, 41 Therefore, the five eQTLs shown to modulate POLB expression are expected to impact the outcome of patients treated with platinum-based chemo and/or radiation therapy. Among patients treated with platinum-based drugs and/or radiotherapy, the genotypes of eQTLs containing the Afr-allele (homozygous/heterozygous) were associated with poor OS and DFS as compared to the homozygous genotypes containing the Cau-allele. Even after adjusting for age and pathological stage, statistically significant associations persisted. However, the results of OS analyses were slightly different among eQTLs (Table 2), despite the fact that these eQTLs are in strong LD. In addition, the effect of these eQTLs on DFS differs among eQTLs. This is not surprising as it is known that independent cis-eQTLs in LD can have different functional effects.42 We also studied the effect of eQTLs among patients who were not treated with cisplatin/carboplatin/ radiotherapy and the results did not show any significant effect of eQTLs on OS (P>0.6) or DFS of these patients (P>0.8). These findings provide evidence that the associations of these five eQTLs with OS/DFS were limited to patients with cisplatin/carboplatin/radiotherapy treatment history and support the important role that POLB expression plays in treatment response.
We validated our survival analyses by analyzing one of five eQTLs, rs2272733, with 141 Cau and 36 HNSCC patients of African ancestry with a history of platinum-based chemo and/or radiation therapy (Dataset-2). The results of TCGA and Dataset-2 for rs2272733 were consistent. The genotype containing at least one Afr- allele (A) is associated with poorer OS compared to the homozygous Cau-allele (G) genotype.
To further test that Afr genetics ancestry is indeed related to survival disparity, we assayed the effect of Afr-admixed proportions on OS and DFS. The HR for OS and DFS are > 1.0 which indicates that higher Afr-admixture is associated with poorer OS and DFS in HNSCC patients with platinum-based chemo and/or radiotherapy history. These results are statistically significant; however, it is worth noting the wide confidence intervals partly due to the limited amount of data available on these endpoints. Moreover, a similar analysis using data from the entire cohort of subjects used in this study showed similar tendencies and indicated poorer OS and DFS in patients with increasing Afr genetic admixture (data not shown). Thus, our study reveals a clear association between African ancestry-related genetic factors and poor treatment outcome in Afr-Amr HNSCC patients who were treated with platinum-based chemo and/or radiotherapy. Validation of our findings in a larger, independent cohort of subjects would further help strengthen and establish their significance.
A limitation of this study is that SES and environmental factors were not included in the analyses due to such data not being available. However, we cannot ignore the effect of environmental and SES on survival disparity. Thus, this study needs to be extended further to analyze the interactions between genetics and environmental factors/ SES on survival disparity.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Biological Resource Center (CRB de Guadeloupe, Stanie Gaëte) as well as the Fox Chase Cancer Center Bio-repository Facility, Philadelphia, PA for managing and providing patient samples for this study. We thank Dr. Erica Golemis, Fox Chase Cancer Center for her valuable comments on this article. The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
FUNDING SUPPORT
This work was supported by the American Cancer Society (RSG-14-033-01-CPPB) to CR, and in part by the National Cancer Institute (CA006927). MPR is partially supported by the William J. Avery postdoctoral research fellowship of Fox Chase Cancer Center. DL acknowledges the support from the French National Cancer Institute (INCA).
Footnotes
AUTHOR CONTRIBUTIONS
Meganathan P. Ramakodi: Conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing – original draft, writing – review and editing, and visualization.
Karthik Devarajan: Methodology, formal analysis, writing – review and editing, and visualization.
Elizabeth Blackman: Data curation, writing – review and editing, and project administration.
Denise Gibbs: Validation, investigation, data curation, and writing – review and editing.
Danièle Luce: Resources, data curation and writing – review and editing.
Jacqueline Deloumeaux: Resources, data curation and writing – review and editing.
Suzy Duflo: Resources, data curation and writing – review and editing.
Jeffrey C. Liu: Writing – review and editing, and visualization.
Ranee Mehra: Writing – review and editing, and visualization.
Rob J. Kulathinal: Conceptualization, resources, writing – review and editing, and visualization.
Camille C. Ragin: Conceptualization, formal analysis, resources, writing – review and editing, visualization, supervision, and funding acquisition
CONFLICT OF INTEREST: None
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Ghafoor A, Jemal A, Cokkinides V, et al. Cancer statistics for African Americans. CA Cancer J Clin. 2002;52:326–341. doi: 10.3322/canjclin.52.6.326. [DOI] [PubMed] [Google Scholar]
- 4.Tan DS, Mok TS, Rebbeck TR. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. J Clin Oncol. 2016;34:91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 5.Vermorken JB, Specenier P. Optimal treatment for recurrent/metastatic head and neck cancer. Ann Oncol. 2010;21(Suppl 7):vii252–261. doi: 10.1093/annonc/mdq453. [DOI] [PubMed] [Google Scholar]
- 6.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moeller BJ, Yordy JS, Williams MD, et al. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clinical Cancer Research. 2011;17:2035–2043. doi: 10.1158/1078-0432.CCR-10-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borek C. Antioxidants and radiation therapy. J Nutr. 2004;134:3207S–3209S. doi: 10.1093/jn/134.11.3207S. [DOI] [PubMed] [Google Scholar]
- 9.Fung C, Grandis JR. Emerging drugs to treat squamous cell carcinomas of the head and neck. Expert Opin Emerg Drugs. 2010;15:355–373. doi: 10.1517/14728214.2010.497754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 11.Eastman A. The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol Ther. 1987;34:155–166. doi: 10.1016/0163-7258(87)90009-x. [DOI] [PubMed] [Google Scholar]
- 12.Jordan P, Carmo-Fonseca M. Molecular mechanisms involved in cisplatin cytotoxicity. Cell Mol Life Sci. 2000;57:1229–1235. doi: 10.1007/PL00000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell JS, Brady K, Burgan WE, et al. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 14.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clinical Cancer Research. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 15.Mathews LA, Cabarcas SM, Hurt EM, Zhang X, Jaffee EM, Farrar WL. Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas. 2011;40:730–739. doi: 10.1097/MPA.0b013e31821ae25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauffmann A, Rosselli F, Lazar V, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 17.Pitroda SP, Pashtan IM, Logan HL, et al. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci Transl Med. 2014;6:229ra242. doi: 10.1126/scitranslmed.3008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabholkar M, Vionnet J, Bostick-Bruton F, Yu JJ, Reed E. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest. 1994;94:703–708. doi: 10.1172/JCI117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Seo JH, Stranger B, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakodi MP, Kulathinal RJ, Chung Y, Serebriiskii I, Liu JC, Ragin CC. Ancestral-derived effects on the mutational landscape of laryngeal cancer. Genomics. 2015 doi: 10.1016/j.ygeno.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naccarati A, Pardini B, Stefano L, et al. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346–1351. doi: 10.1093/carcin/bgs172. [DOI] [PubMed] [Google Scholar]
- 24.Jacoby MA, De Jesus Pizarro RE, Shao J, et al. The DNA double-strand break response is abnormal in myeloblasts from patients with therapy-related acute myeloid leukemia. Leukemia. 2014;28:1242–1251. doi: 10.1038/leu.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter V, Yin X, Wilkerson MD, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grambsch PM, Therneau TM. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 31.Luce D. An epidemiological study on head and neck cancers in the French West Indies: rationale and study protocol.. The 5th International African-Caribbean Cancer Consortium Conference; Schoecher, Martinique. 2014. [Google Scholar]
- 32.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 34.Earl DA, Vonholdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- 35.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 36.Sweasy JB, Lang T, Starcevic D, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergoglio V, Pillaire MJ, Lacroix-Triki M, et al. Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–3514. [PubMed] [Google Scholar]
- 38.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 39.Figueroa JD, Malats N, Real FX, et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Human Genetics. 2007;121:233–242. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- 40.Vens C, Dahmen-Mooren E, Verwijs-Janssen M, et al. The role of DNA polymerase beta in determining sensitivity to ionizing radiation in human tumor cells. Nucleic Acids Res. 2002;30:2995–3004. doi: 10.1093/nar/gkf403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwatsuki M, Mimori K, Yokobori T, et al. A Platinum Agent Resistance Gene, POLB, Is a Prognostic Indicator in Colorectal Cancer. Journal of Surgical Oncology. 2009;100:261–266. doi: 10.1002/jso.21275. [DOI] [PubMed] [Google Scholar]
- 42.Bryois J, Buil A, Evans DM, et al. Cis and trans effects of human genomic variants on gene expression. Plos Genetics. 2014;10:e1004461. doi: 10.1371/journal.pgen.1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.