Summary
SIRT1, the most conserved mammalian NAD+-dependent protein deacetylase, plays a vital role in the regulation of metabolism, stress responses, and genome stability. However, the role of SIRT1 in the multi-step process leading to transformation and/or tumorigenesis, as either a tumor suppressor or tumor promoter, is complex and maybe dependent upon the context in which SIRT1 activity is altered, and the role of SIRT1 in tumor metabolism is unknown. Here we demonstrate that SIRT1 dose-dependently regulates cellular glutamine metabolism and apoptosis, which in turn differentially impact cell proliferation and cancer development. Heterozygous deletion of Sirt1 induces c-Myc expression, enhancing glutamine metabolism and subsequent proliferation, autophagy, stress resistance and cancer formation. In contrast, homozygous deletion of Sirt1 triggers cellular apoptotic pathways, increases cell death, diminishes autophagy, and reduces cancer formation. Consistent with the observed dose-dependence in cells, intestine-specific Sirt1 heterozygous mice have enhanced intestinal tumor formation, whereas intestine-specific Sirt1 homozygous knockout mice have reduced development of colon cancer. Furthermore, SIRT1 reduction but not deletion is associated with human colorectal tumors, and colorectal cancer patients with low protein expression of SIRT1 have a poor prognosis. Taken together, our findings indicate that the dose-dependent regulation of tumor metabolism and possibly apoptosis by SIRT1 mechanistically contributes to the observed dual roles of SIRT1 in tumorigenesis. Our study highlights the importance of maintenance of a suitable SIRT1 dosage for metabolic and tissue homeostasis, which will have important implications in SIRT1 small molecule activators/inhibitors based therapeutic strategies for cancers.
Keywords: SIRT1/Sirt1, glutaminolysis, inflammation, autophagy, colon cancer
Introduction
Tumor cells have unusually high demands of nutrients for survival and rapid growth compared to normal differentiated cells [1, 2]. Their preference for an extremely high rate of glycolysis in the presence of oxygen (aerobic glycolysis, “the Warburg effect” [3]) is considered a hallmark of cancer [4]. Many cancer cells also exhibit addiction to glutamine, a nonessential amino acid that is the most abundant in the body [5]. Both glucose and glutamine feed into multiple nodes of central metabolism, providing intermediates for synthesis of lipids, proteins and nucleic acids, thereby supporting cancer growth and proliferation [2]. Genetic factors, including Myc and p53, actively regulate cancer cell metabolic reprogramming. It has been increasingly recognized that a major impact of genetic mutations in cancer cells is to reprogram their metabolic pathways to maximize use of nutrients under various nutritional and stress conditions [2, 6, 7].
The most conserved mammalian NAD+-dependent protein deacetylase, SIRT1, is an important metabolic and stress sensor [8, 9]. However, its role in tumorigenesis remains unclear and inconclusive [10, 11]. SIRT1 promotes DNA repair through PARP1, XPC, and WRN, and represses inflammation and induces apoptosis through inhibition of NF-κB, STAT3, HIF-1α, and β-Catenin [12–14]. On the other hand, SIRT1 inhibits activities of tumor suppressors, such as p53, Rb, and PTEN, while stimulating oncogenic pathways including Myc, Ras, and Akt, thereby promoting growth and survival of cancer cells. As a result, whether SIRT1 is an oncogene or tumor suppressor remains controversial [15, 16].
In the present study, we explored the possibility that SIRT1 regulates cancer development in a quantitative dose-dependent manner and assessed the impact of this dosage-dependence on colorectal tumorigenesis. We hypothesized that different SIRT1-regulated pathways have distinct sensitivities to changes of SIRT1 dosages. Pathways that are sensitive to environmental alterations, such as metabolic cascades and inflammatory pathways, are also hypersensitive to fluctuations of SIRT1 expression/activity. In contrast, pathways involved in cell fate determination, including p53-mediated cell apoptosis, will only be activated upon drastic variations of SIRT1 expression/activity. The distinct sensitivities of different pathways to SIRT1 dosages differentiate the outcomes in cancer cell proliferation and growth, contributing to observed dual functions of SIRT1 in tumorigenesis.
Results
SIRT1 exerts a dose-dependent effect on cell proliferation and tumor formation
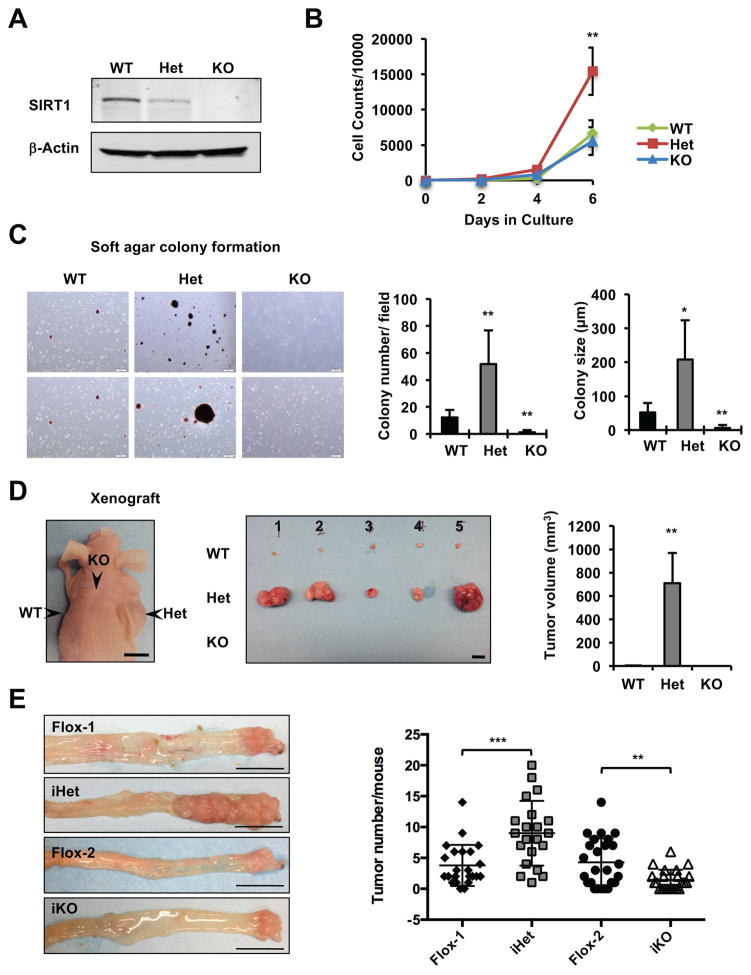
To test our hypothesis, we first analyzed immortalized Sirt1 wild-type (Sirt1+/+, WT), heterozygous (Sirt1+/−, Het), and knockout (Sirt1−/−, KO) mouse embryonic fibroblasts (MEFs) (Figure 1A). When cultured in complete DMEM medium containing 25 mM glucose and 4 mM glutamine, Het MEFs proliferated significantly faster than WT and KO MEFs (Figure 1B), and this increased proliferation was repressed (rescued) by putting back WT SIRT1 protein but not a catalytic inactive mutant of SIRT1 (Figure S1A and S1B). Het MEFs also formed significantly more and bigger colonies compared to WT MEFs in a soft-agar colony formation assay, whereas KO MEFs failed to form a significant number of colonies (Figure 1C). More strikingly, when injected subcutaneously into nude mice, Het MEFs readily developed into large tumors within 5–8 weeks, whereas tumor formation was minimal or undetectable from WT or KO MEFs (Figure 1D and S1C).
Figure 1. Dosage-dependent effects of SIRT1 on cell proliferation and tumorigenesis.
(A) Immortalized Sirt1 wild-type (WT), heterozygous (Het), and knockout (KO) MEFs.
(B) Het MEFs display an increased proliferation rate. Immortalized MEFs were cultured in complete DMEM medium, plated and counted (n=3 independent cell lines/genotype, **p<0.01, values are represented as mean + SEM).
(C) Het MEFs form more and larger colonies while KO MEFs fail to form any colonies in a soft agar colony formation assay (n=3 independent cell lines/genotype, *p<0.05, **p<0.01, values are represented as mean + SEM).
(D) Het MEFs readily form larger tumors in a xenograft model. Immortalized MEFs were injected subcutaneously into the flanks and back of host nude mice (Nu/J from Jax) and their tumorigenic potential was analyzed. n=3 independent cell lines/genotype, with total of 8 repeats/genotype. The tumor diameters were measured at the end of experiments and the volumes were calculated: tumor volume (mm3) = ½ × longest diameter2 × shortest diameter. Bars, 1 cm.
(E) Sirt iHet mice have enhanced while Sirt1 iKO mice have reduced formation of colon tumors in an AOM/DSS colon cancer model. Single-copy flox controls (Flox-1) and double-copy flox controls (Flox-2) were used as controls. Left, Representative images of colon tumors, bars, 1 cm. Right, Colon tumor numbers. n=23 (Flox-1), 21 (iHet), 25 (Flox-2), 23 (iKO) mice, **p<0.01, ***p<0.001.
Please also see Figure S1.
In line with above observations in vitro, in an Azoxymethane (AOM)/Dextran Sulfate Sodium (DSS)-induced colon cancer model, intestine-specific Sirt1 heterozygous deletion mice (Villin-Cre+/Sirt1flox/+, Sirt1 iHet) developed significantly more and larger tumors compared to those of control mice in colons, whereas Sirt1 homozygous deletion mice (Villin-Cre+/Sirt1flox/flox, Sirt1 iKO), consistent with a recent report [17], had a reduced number of colon tumors (Figure 1E). Histopathological analyses further revealed that 4 out of 12 analyzed control mice lacked a response to the AOM/DSS treatment at all. For those that developed tumors, some tumors had marked regenerative hyperplasia (Figure S1D). However, tumors from all analyzed iHet mice had clearly progressed beyond regenerative hyperplasia into neoplasia (Figure S1D). Most tumors from iKO mice, in contrast, were restricted to low-grade stages lacking regenerative hyperplasia (not shown). Taken together, our results confirmed our hypothesis that Sirt1 insufficiency but not complete deficiency enhances tumor development in vitro and in vivo.
Haploinsufficiency of Sirt1 promotes cell proliferation through enhanced glutamine metabolism
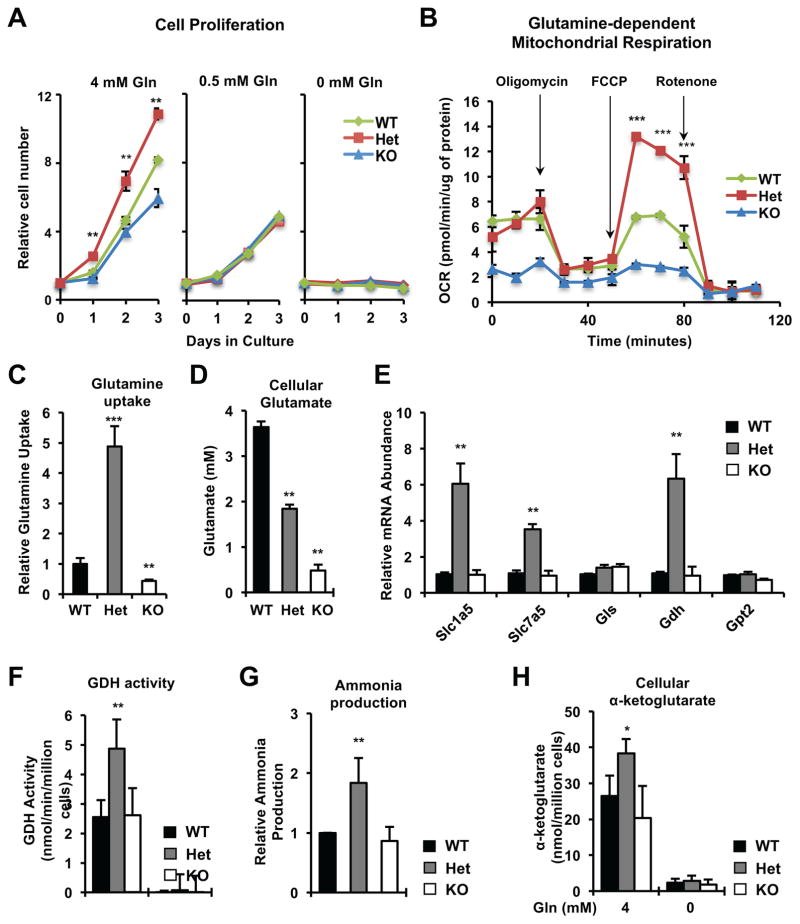
SIRT1 is a well-established metabolic regulator [8, 9], we therefore investigated whether metabolic dysregulation, particularly elevation of aerobic glycolysis, plays a role in Sirt1 haploinsufficiency-induced transformation. Indeed, compared to WT and KO MEFs, Het MEFs displayed increased glucose uptake/consumption, lactate production, and expression of several key genes involved in glycolysis and related pathways (Figure S2A–S2D). Furthermore, Het MEFs had an increased trend in their glycolytic capacity (Figure S2E) and significantly enhanced glucose-dependent mitochondrial respiration (Figure S2F). However, surprisingly, when cultured in a glucose-free medium, Het MEFs still proliferated significantly faster than WT and KO MEFs despite the reduction of proliferation of all three cells (Figure S2G), indicating that enhanced glucose metabolism is not essential for Sirt1 haploinsufficiency-induced cell hyper-proliferation.
In addition to glucose, most cancer cells have enhanced glutamine catabolism. Through the actions of glutaminase, glutamate dehydrogenase (Gdh), and aminotransferases, glutamine converts into α-ketoglutarate and feeds into the tricarboxylic acid cycle to support cell bioenergetics and anabolic reactions [2]. As shown in Figure 2A, restriction of medium glutamine from 4 mM to 0.5 mM eliminated the growth advantage of the Het MEFs vs WT and KO MEFs, suggesting that Het MEFs may sustain their high proliferation by increasing glutamine metabolism. Consistently, Het MEFs had significantly enhanced glutamine-dependent mitochondrial respiration (Figure 2B), a marked increase in their glutamine uptake but decrease in intracellular glutamate levels (Figure 2C and 2D), together with elevated levels of glutamine transporters Slc1a5 and Slc7a5, and Gdh (Figure 2E). Furthermore, compared to WT and KO MEFs, Het MEFs displayed an increased GDH activity (Figure 2F) and elevated cellular contents of two products of this reaction, ammonia (Figure 2G) and α-ketoglutarate (Figure 2H), suggesting that enhanced glutamine metabolism is the major underlying mechanism for SIRT1 haploinsufficiency induced cell hyper-proliferation.
Figure 2. Haploinsufficiency of Sirt1 promotes cell proliferation and glutamine metabolism.
(A) MEFs of all three genotypes display comparable proliferation rates upon glutamine restriction. Cells were plated and relative survived cell numbers were measured by the MTS/PMS reagents. n=3 independent cell lines/genotype, **p<0.01, values are represented as mean + SEM.
(B) Het MEFs have enhanced glutamine-dependent mitochondrial respiration. n=5 technical repeats, ***p<0.001, values are represented as mean + SEM.
(C) Het MEFs have increased uptake of glutamine. n=3 independent cell lines/genotype, ***p<0.001, **p<0.01, values are represented as mean + SEM.
(D) Cellular glutamate levels in WT, Het, and KO MEFs. n=3 independent cell lines/genotype, **p<0.01, values are represented as mean + SEM.
(E) Het MEFs have increased mRNA levels of key genes involved in glutamine metabolism in complete medium. Gls, Glutaminase; Gdh, glutamate dehydrogenase; Gpt2, glutamic pyruvate transaminase. n=3 independent cell lines/genotype, **p<0.01, values are represented as mean + SEM.
(F–H) Het MEFs have increased activity of GDH (F), ammonia production (G), and cellular α-ketoglutarate levels (H) when cultured in complete medium. n=3 independent cell lines/genotype, **p<0.01, *p<0.05, values are represented as mean + SEM.
Please also see Figure S2.
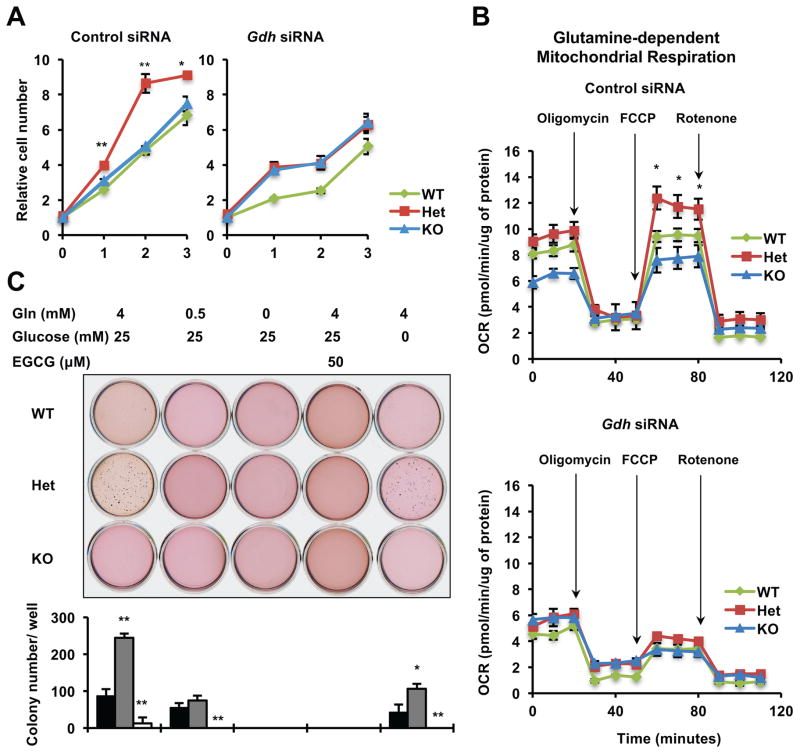
In support of the notion that an increase in glutaminolysis/glutamine metabolism rather than in glycolysis/glucose metabolism drives the hyper-proliferation phenotype of Het MEFs, depletion of GDH by siRNAs (Figure S3D) reduced the proliferation rates of Het and WT MEFs but not that of KO MEFs (Figure 3A), despite the fact that GDH depletion repressed and eliminated the difference of the glutamine-dependent mitochondrial respiration in WT, Het, and KO MEFs (Figure 3B). This observation suggested that enhanced glutamine metabolism significantly contributes to proliferation of Het and WT MEFs but not that of KO MEFs. Instead, KO MEFs appeared to maintain their proliferation by extremely high glycolysis as revealed by the ECAR profiling (Figure S2E, KO).
Figure 3. Haploinsufficiency of Sirt1 promotes proliferation and colony formation through enhanced glutamine metabolism.
(A) Knocking down Gdh represses hyper-proliferation of Het MEFs. Cells were plated and relative survived cell numbers were measured by the MTS/PMS reagents. n=3 technical repeats, **p<0.01, *p<0.05, values are represented as mean + SEM.
(B) Knocking down Gdh represses glutamine-dependent mitochondrial respiration. n=5 technical repeats with a different set of MEFs from those in Figure 2B, *p<0.05, values are represented as mean + SEM.
(C) The distinct colony formation abilities of WT, Het, and KO MEFs on soft agar are dependent on the status of glutamine metabolism. Representative images from one group of cells were shown. n=3 independent cell lines/genotype, **p<0.01, *p<0.05, values are represented as mean + SEM.
Please also see Figure S2 and S3.
Further colony formation assays (Figure 3C and S3A) confirmed that Het MEFs formed significantly more colonies while KO MEFs formed fewer colonies than WT MEFs in complete culture medium as observed previously (Figure 1C). Switching to a glutamine-restricted medium, a glutamine-free medium, or a complete medium containing 50 μM of a GDH inhibitor, EGCG, abolished the ability of Het MEFs to form more colonies than WT MEFs. In contrast, Het MEFs maintained their high colony-formation ability after glucose deprivation. Collectively, our findings demonstrate that haploinsufficiency of Sirt1 promotes glutamine metabolism, thereby inducing cell hyper-proliferation and colony formation.
Haploinsufficiency of Sirt1 regulates autophagy and apoptosis through enhanced glutamine metabolism
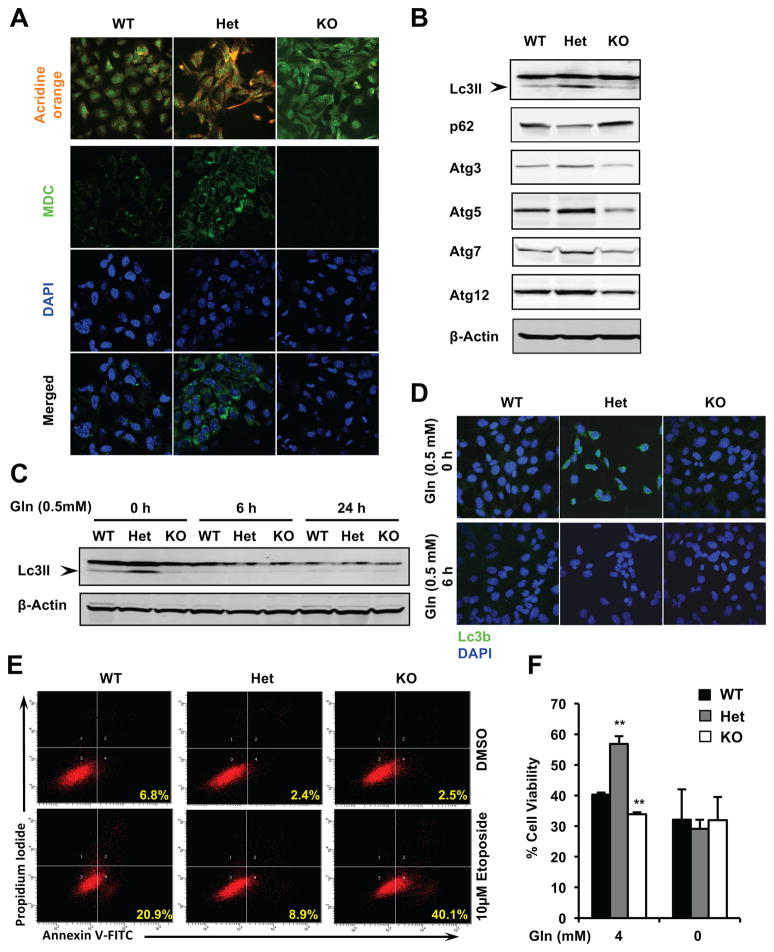
In addition to being a major nutrient source, glutamine also serves as a key regulator of autophagy. Glutamine-derived free ammonia has been reported to promote basal autophagy and protect cells from TNFα triggered cell death [18, 19]. Consistent with their enhanced glutamine metabolism (Figure 2 and Figure 3), Het MEFs displayed a striking enhancement in formation of autophagolysosomes (Acridine orange) and autolysosomes (MDC) (Figure 4A and S3B), along with increased levels of an autophagosomal marker Lc3II and several essential autophagy factors in complete glutamine-containing medium compared to WT MEFs (Figure 4B and S3C). Particularly, the increased formation of Lc3II-autophagosomes in Het MEFs was abolished upon glutamine restriction (Figure 4C and 4D) or GDH depletion (Figure S3D), supporting the notion that the enhancement of autophagy in Het MEFs is mediated by elevated glutamine metabolism. Furthermore, Het MEFs were protected from cell apoptosis induced by etoposide, a DNA damage reagent (Figure 4E), and displayed a higher survival rate in complete medium upon the etoposide treatment (Figure 4F). Depletion of glutamine abolished the protective effect provided by Sirt1 haploinsufficieny (Figure 4F) in spite of comparable degrees of DNA damage induced by etoposide in cells with the same genotype in both media (Figure S3E).
Figure 4. Haploinsufficiency of Sirt1 increases autophagy and stress resistance through glutamine metabolism.
(A) Het MEFs enhances yet KO MEFs reduces formation of acidic autophagolysosomes (acridine orange staining, top three panels) and autolysosomes (Monodansylcadaverine staining, MDC, bottom nine panels) in complete glutamine-containing medium.
(B) Het MEFs have increased levels of protein factors involved in autophagy.
(C–D) The enhanced autophagy in Het MEFs is dependent on glutamine. Immortalized MEFs were cultured in medium containing 0.5 mM glutamine for indicated times and expression of Lc3II was analyzed by immuno-blotting (C) or immune fluorescence (D).
(E) Het MEFs are resistant, whereas KO MEFs are sensitive to etoposide-induced apoptosis in normal culture medium. Immortalized MEFs cultured in complete glutamine-containing medium were treated with vehicle DMSO or 10 μM etoposide for 48 hours.
(F) Sirt1 dosage induced distinct sensitivities to etoposide are dependent on glutamine. Immortalized MEFs were treated with vehicle DMSO or 10 μM etoposide for 48 hours in DMEM medium containing 4 mM or 0 mM of glutamine. Cell survival ratios between Etoposide vs DMSO treated cells were analyzed as by the MTS/PMS reagents. n=3 technical repeats, **p<0.01, values are represented as mean + SEM.
Please also see Figure S3.
Sirt1 KO MEFs, on the other hand, had reduced formation of autophagolysosomes and autolysosomes (Figure 4A and S3B), reduced levels of autophagic protein factors (Figure 4B and S3C), and increased sensitivity to etoposide induced cell death (Figure 4E, 4F, and S3E). Together, our results indicate that loss of one copy versus both copies of Sirt1 gene differentially impact autophagy and apoptosis.
Dose-dependent regulation of the c-Myc pathway and p53 activity by SIRT1
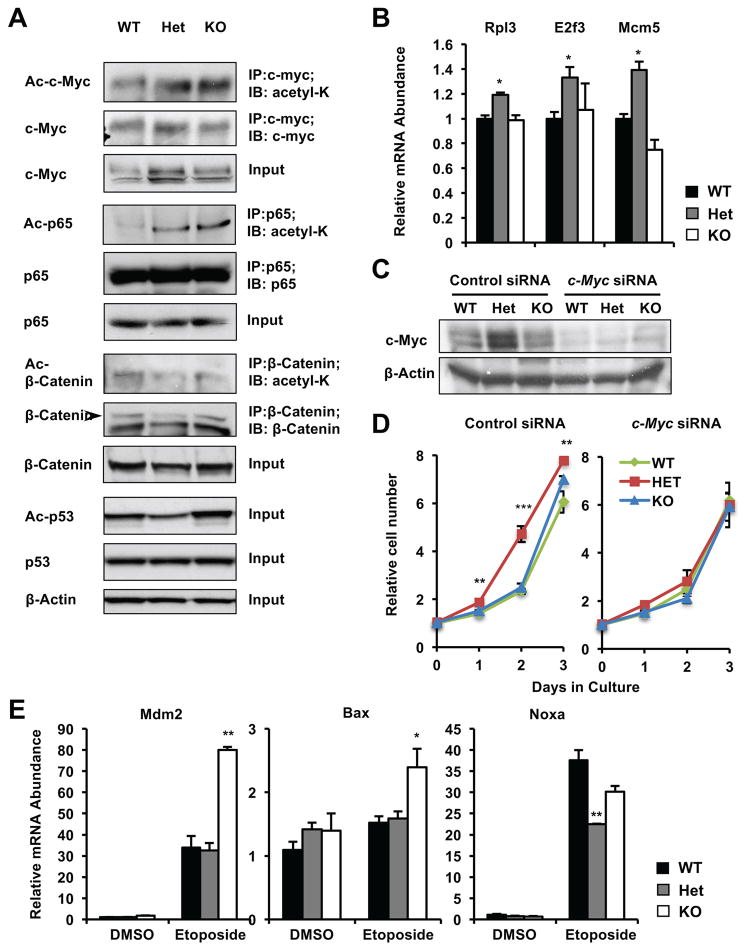
SIRT1 induce transcriptional reprogramming in response to environmental cues through modulating acetylation of transcription factors/cofactors [8, 9]. Microarray analysis of gene expression profiles from WT, Het, and KO MEFs suggested that the activities of several SIRT1 regulated factors/pathways, including p53, Wnt3a/β-catenin signaling, TNFα (NF-κB) activity, and the mTOR-c-Myc network, may be differentially altered by deletion of one copy or both copies of Sirt1 gene (Figure S4). p53, β-catenin, p65 subunit of NF-κB, and c-Myc are all known SIRT1 deacetylation substrates. While SIRT1 mediated deacetylation of p53, β-catenin, and p65 represses their transactivation, deacetylation of c-Myc increase its stability and activity [20].
Not surprisingly, the acetylation levels of c-Myc, p65, and p53 were all significantly increased in KO MEFs (Figure 5A and S3F). However, their acetylation levels were distinctly affected in Het MEFs. Single copy deletion of the Sirt1 gene failed to significantly induce the acetylation of p53, but it was sufficient to increase the acetylation of p65 (Figure 5A and S3F). Single copy deletion of the Sirt1 gene also increased the expression of c-Myc (Figure 5A and S3F), as well as a few c-Myc downstream target genes involved in cell growth and proliferation (Figure 5B). Additionally, several key metabolic genes that are transcriptional targets of c-Myc [6], including Glut1, Pdk1 Slc1a5, and Slc7a5, were highly induced in Het MEFs (Figure S2D and 2E). Consistent with the notion that increased c-Myc activities promote glutamine metabolism and subsequent cell proliferation [6, 21–23], knocking-down c-Myc in Het MEFs completely eliminated their growth advantage (Figure 5C and 5D), suggesting that SIRT1 haploinsufficiency induced cell hyper-proliferation is likely due to increased expression of c-Myc.
Figure 5. SIRT1 haploinsufficiency induces hyper-proliferation through c-Myc.
(A) Acetylation and expression of different substrates of SIRT1 display distinct sensitivities to the Sirt1 gene dosage. Immortalized MEFs cultured in complete medium were treated with 5 μM TSA for 30 minutes, and the total and acetylation levels of indicated proteins were analyzed by immuno-blotting or immunoprecipitation followed by immuno-blotting. Please also see Figure S3F.
(B) Deletion of one copy of Sirt1 stimulates the c-Myc pathway (n=3 technical repeats, *p<0.05, values are represented as mean + SEM).
(C–D) Knocking down c-Myc represses hyper-proliferation of Het MEFs. Relative survived cell numbers were measured by the MTS/PMS reagents (n=5 technical repeats, ***p<0.001, **p<0.01, values are represented as mean + SEM).
(E) Deletion of both alleles but not single allele of Sirt1 induces expression of p53 targets upon DNA damage. Immortalized MEFs cultured in complete medium were treated with DMSO or 10 μM etoposide for 6 hours, expression of indicated p53 targets were analyzed by q-PCR (n=3 technical repeats, *p<0.05, values are represented as mean + SEM).
Please also see Figure S3, S4 and S7.
Conversely, KO MEFs had increased formation of DNA double strand breaks (γ-H2AX) (Figure S3E), increased expression of apoptotic genes (Figure 5E), and elevated apoptosis (Figure 4E and 4F) upon etoposide treatment. We have previously shown that the enhanced sensitivity of KO MEFs to etoposide induced apoptosis is dependent on p53 [24]. This p53-mediated hypersensitivity to genotoxic stress may be responsible for observed reduced transformation of KO cells. Taken together, our observations demonstrate that distinct impacts of Sirt1 dosages on metabolic, proliferating, and apoptotic pathways contribute to the observed dose-dependent regulation of cell metabolism and proliferation by Sirt1.
Heterozygous deletion of Sirt1 enhances expression of glutamine metabolism genes and c-Myc signaling in AOM/DSS-induced colon tumors
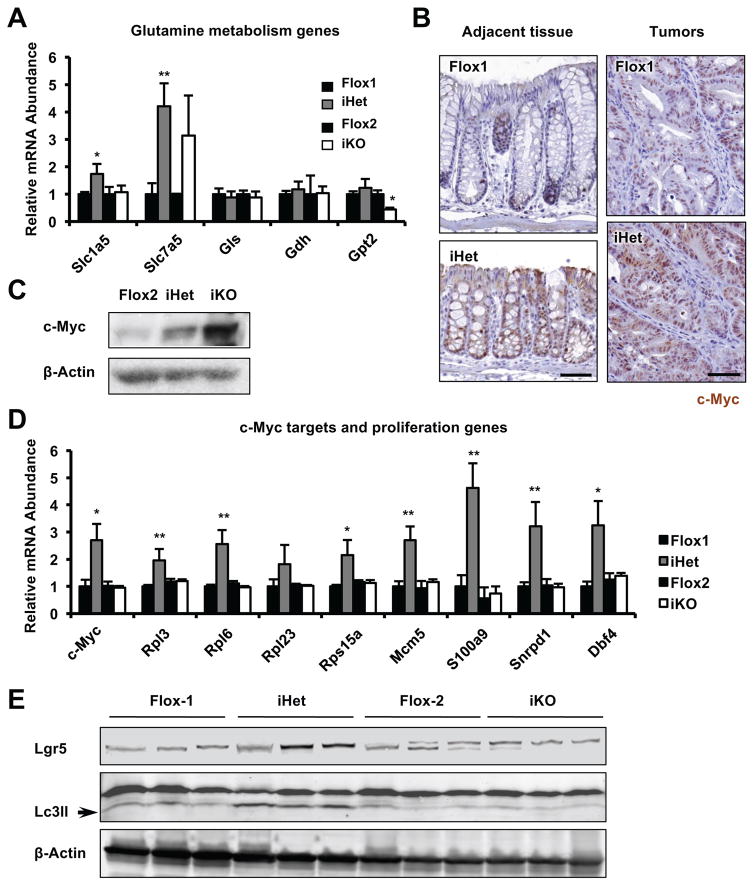
In agreement with our observations in immortalized MEFs (Figure 2 and 5), AOM/DSS treated colon tissues from Sirt1 iHet mice had significantly increased expression of genes involved in glutamine transport (Figure 6A), together with increased levels of c-Myc protein, mRNA, and target genes involved in cell growth and proliferation (Figure 6B–6D). Colon tumors from iKO mice, also consistent with observations in MEFs (Figure S3E), displayed enhanced apoptotic features (Figure S5A). Intriguingly, colon tumors from iHet mice also had higher levels of intestinal stem cell markers (Figure 6E and S5B), suggesting that colorectal tumorigenesis induced in iHet mice may also involve uncontrolled tumor stem cell expansion. Tumors from iHet mice also had increased levels of an autophagy marker Lc3II (Figure 6E) as observed in Het MEFs (Figure 4). Collectively, our observations suggest that heterozygous deletion of Sirt1 enhances glutamine metabolism and c-Myc signaling in mouse colon tumors.
Figure 6. Heterozygous deletion of Sirt1 in mouse intestine enhances glutamine metabolism, inflammation, and c-Myc signaling in AOM/DSS-colon tumors.
(A) Sirt1 iHet colon tumors have increased expression of glutamine transporters. n=6 mice/genotype, *p<0.05, **p<0.01, values are represented as mean + SEM.
(B–C) Sirt1 iHet colon tumors have increased c-Myc expression. c-Myc protein levels were analyzed by immunohistochemistry (B) or immune-blotting (C). Bars in (B), 50 μm. (D) Sirt1 iHet colon tumors have increased expression of c-Myc and cell proliferation genes. n=6 mice/genotype, *p<0.05, **p<0.01, values are represented as mean + SEM. (E) Sirt1 iHet colon tumors have increased protein levels of markers for intestinal stem cells and autophagy.
Please also see Figure S5.
Reduced expression of SIRT1 is associated with enhanced colorectal adenocarcinoma cell proliferation, colon cancer development, and poor prognosis in humans
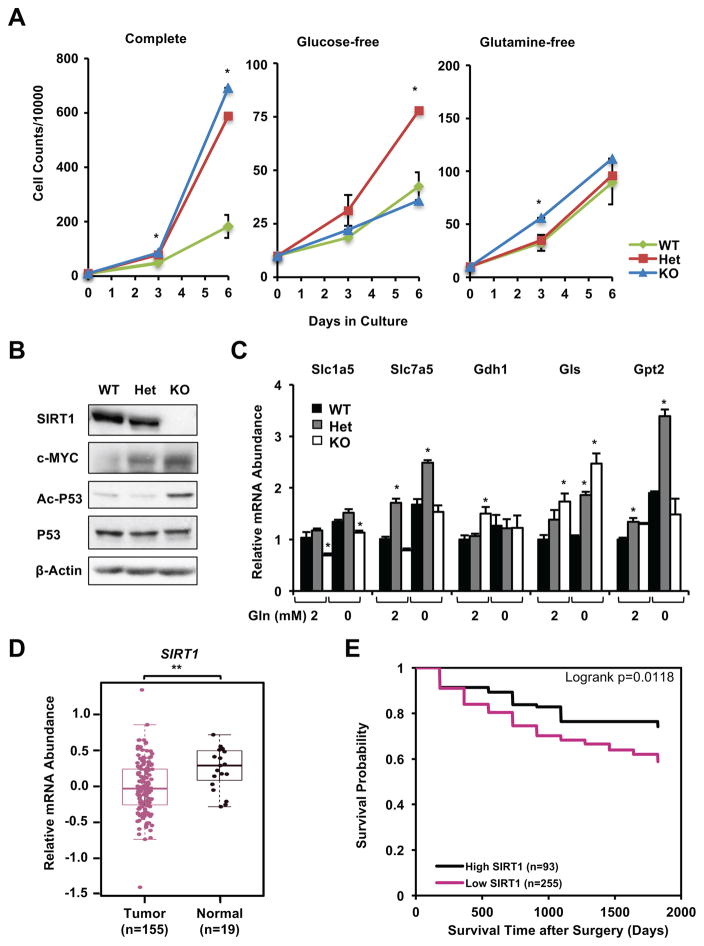
To further test our hypothesis, we generated human SIRT1 WT, Het, and KO colorectal adenocarcinoma cell lines using the Crispr/Cas9 technology (Figure S6A–S6D). Consistent with our observations in immortalized Het MEFs, deletion of one copy of SIRT1 gene in DLD1 cell line led to hyper-proliferation in a glutamine-dependent manner (Figure 7A), indicating that an increase in glutamine metabolism underlies the hyper-proliferation phenotype of Het DLD1 cells just like Het MEFs. In contrast to KO MEFs, however, KO DLD1 cells displayed a hyper-proliferation phenotype in a glucose-dependent manner (Figure 7A), possibly due to a S241F mutation thereby inactivation of p53 protein in these cells [25–27]. Moreover, deletion of single copy of SIRT1 gene in DLD1 cells was sufficient to increase the expression of c-MYC, whereas acetylation of P53 was elevated only when SIRT1 was completely deleted (Figure 7B). Additionally, Het DLD1 cells had increased expression of several genes involved in glutamine metabolism (Figure 7C), supporting the notion that haploinsufficiency of SIRT1 promotes glutamine metabolism and induces hyper-proliferation in cancer cells.
Figure 7. Reduced expression of SIRT1 is associated with enhanced colorectal adenocarcinoma cell proliferation, colon cancer development, and poor prognosis in humans.
(A) Haploinsufficiency of SIRT1 promotes proliferation of DLD1 cells in a glutamine-dependent manner. WT, Het, and KO DLD1 cells were cultured in complete RPMI1640 medium containing 11 mM glucose and 2 mM glutamine (Complete), no glucose (Glucose-free), or no glutamine (Glutamine-free), and cell numbers were counted as indicated times (n=2 independent clones/genotype, with 2 technical repeats each, *p<0.05, values are represented as mean + SEM).
(B) Different substrates of SIRT1 have distinct sensitivities to SIRT1 dosage in DLD1 cells. DLD1 cells cultured in complete medium were treated with 5 μM TSA for 30 minutes.
(C) SIRT1 Het DLD1 cells have increased expression of genes involved in glutamine metabolism. DLD1 cells were cultured in complete medium or in glutamine-free medium for 72 hours (n=3 technical repeats, *p<0.05, values are represented as mean + SEM).
(D) mRNA levels of human SIRT1 are reduced in colon tumor samples compared to normal colon samples from non-colorectal cancer individuals in TCGA database. The median, upper, and lower quartiles were plotted, and the Wilcox test is used to evaluate their significance (Tumor group n=155; Normal group n=19; **p=0.0018).
(E) Colorectal cancer patients with low expression of tumor SIRT1 have a poorer prognosis than those with high expression. Kaplan-Meier overall survival stratified by dichotomizing SIRT1 expression status in tumor tissues (n=255 low expression/93 high expression) on a continuous H-score scale of 0–300 at cut point with best model fit (234.415).
Please also see Figure S6 and Tables S1, S2, and S3.
Further analysis of SIRT1 gene alterations and expression in 1117 colorectal cancer samples collected in databases of The Cancer Genome Atlas (TCGA), Genentech, and Memorial Sloan Kettering Cancer Center (MSKCC) indicated that the vast majority (about 99%) of colorectal cancer patients had a normal SIRT1 gene, and no deletion of this gene was detected in any of these databases (Figure S6E and Table S1). Comparison of human SIRT1 mRNA levels between colon tumors and normal samples from non-colorectal cancer individuals in the TCGA revealed that human SIRT1 levels were modestly but significantly reduced in colon tumor samples (Figure 7D), consistent with the notion that SIRT1 insufficiency but not complete deficiency contributes to tumorigenesis.
Next we measured SIRT1 protein expression in colorectal tissue microarrays constructed for the North Carolina site of the Cancer Care Outcomes Research and Surveillance Consortium (CanCORS), a population-based prospective cohort study of colorectal and lung cancer [28–30]. We included 348 colorectal cancer subjects with medical records data available and at least one tumor core with a minimum of 50 epithelial cells and unambiguous histology (Table S2). SIRT1 proteins were detected in both tumor tissue and tumor-adjacent colorectal tissue (Figure S6F), and no statistically significant difference was detected between expression in tumor tissue and tumor-adjacent tissue after stratifying by sex, race, age, or tumor stage (Table S3). In tumor tissue on a continuous H-score scale of 0–300, the cut point distinguishing low SIRT1 expression from high expression with the best model fit in terms of association with all-cause mortality was observed at a value of 234.415. When dichotomized at this cut point, subjects with low expression of SIRT1 (n=255) showed significantly lower post-surgery survival rates than patients with high expression of SIRT1 (n=93) (Figure 7E, univariate analysis; multivariate adjusted HR 2.07, 95% CI 1.28, 3.35 in multivariate analysis). These results demonstrate that reduced tumor SIRT1 expression correlates with poorer prognosis in colorectal cancer patients.
Discussion
The association between SIRT1 dosages and distinct consequences in tumor development have been previously suggested [13, 31–33]. However, this dosage-dependence has not been systematically analyzed, and conflicting observations from mouse models with Sirt1 heterozygous deletion and homozygous deletion have resulted in inconsistent views on SIRT1’s roles in tumorigenesis [15, 16]. Our study showed that SIRT1 regulates colon cancer formation in a dosage-dependent manner, and this effect is dependent on glutamine metabolism. Intriguingly, the dosage-specific role of SIRT1 has also been observed in other SIRT1-regulated biological processes. For example, we and others have previously reported that whole-body Sirt1 heterozygous mice were sensitive to high-fat diet induced obesity and metabolic abnormalities [34, 35], while whole-body Sirt1 knockout mice were protected from high-fat diet induced body weight gain and obesity (unpublished observations). Likewise, the dosage-dependent effect also occurs in Sirt1 overexpressing models. Transgenic mice with modest overexpression of Sirt1 were protected from diet and aging induced metabolic dysfunctions and spontaneous tumorigenesis [36–39] and displayed a reduced incidence of tumors in several cancer models [12, 40]. On the other hand, massive overexpression or overactivation of SIRT1 resulting from loss of tumor suppressors, such as HIC1 and DBC1, promotes growth and survival of cancer [41, 42]. These observations were consistent with our hypothesis that different cellular pathways have distinct sensitivities to SIRT1 dosages, and strongly suggest that maintenance of a suitable SIRT1 dosage is critical for metabolic and tissue homeostasis.
In addition to activation of c-Myc signaling, Sirt1 Het MEFs and colon tumors from Sirt1 iHet mice also displayed an elevated NF-κB pathway (Figure 5A and S7A–D). Increased NF-κB has also been shown to promote glutamine metabolism and subsequent cell proliferation [43]. Although knocking-down p65 failed to completely eliminate the growth advantage of Het MEFs in culture (Figure S7E), we cannot exclude the possibility that the hyperactivated NF-κB pathway partially contributes to Sirt1 insufficiency induced transformation and tumor growth.
Experimental Procedures
Animal studies
The intestinal-specific Sirt1 homozygous knockout mice (iKO, Villin-Cre+/Sirt1flox/flox) and their littermate Flox-2 controls (Villin-Cre−/Sirt1flox/flox) on a C57BL/6 background were generated previously [44]. Sirt1 intestinal-specific heterozygous knockout mice (iHet, Villin-Cre+/Sirt1 flox/+) and their littermate Flox-1 controls (Villin-Cre−/Sirt1 flox/+) were generated by breeding Sirt1 iKO mice with WT C57BL/6J mice.
For AOM/DSS-colon cancer model, three-four month old mice injected with AOM (8 mg/Kg bwt). One week later, mice were fed 2.5% DSS in drinking water for 7 days followed by a 14-day of recovery with regular drinking water for three cycles. H&E stained slides of colons were evaluated by a designated study pathologist. c-Myc immunohistochemistry was performed with an anti-c-Myc specific antibody (Abcam, Cat# 39688, Cambridge, MA). All animal experiments were conducted in accordance with guidelines of NIEHS/NIH Animal Care and Use Committee.
Seahorse analysis
Seahorse analyses were performed on a Seahorse XF Analyzer according to manufacturer’s instructions (Agilent Technologies, Santa Clara, CA). Briefly, cells were plated in a 24-well assay plate the day before the analysis in the complete medium to ensure 80–90% confluent on next day, cells were then washed and incubated in a freshly prepared XF assay medium containing either 25 mM glucose, 0 mM glutamine, and 1 mM sodium pyruvate for measurement of glucose-dependent mitochondrial respiration, 0 mM glucose, 4 mM glutamine, and 1 mM sodium pyruvate for measurement of glutamine-dependent mitochondrial respiration, or 1 mM sodium pyruvate and 2 mM glutamate for measurement of glycolysis. Final mitochondrial respiration (OCR) and glycolysis (ECAR) were normalized to total cell protein contents.
Human studies
The analysis of human SIRT1 gene alterations in colorectal cancer was performed via http://www.cbioportal.org/index.do, from four independent studies with total of 1117 colorectal adenocarcinoma samples. The human SIRT1 gene expression data from normal control subjects and colon cancer patients was obtained from the TCGA Pan-Cancer Synapse (https://www.synapse.org/Portal.html#!Synapse:syn300013/wiki/), with 155 colorectal cancer patients and 19 normal controls.
To assess the relationship of SIRT1 expression to cancer patient prognosis, SIRT1 protein levels were analyzed by immunohistochemistry (cat #2493, Cell Signaling Technology, Danvers, MA) in tissue microarrays constructed for the North Carolina site of CanCORS [28–30], with total of 348 colorectal cancer patients. Cox proportional hazards models were used to estimate the association between SIRT1 expression and time from surgery to all-cause mortality adjusted for age, race, sex, tumor stage, and histologic grade, with censoring at 5 years after surgery. Multivariate models were run for both continuous and dichotomous SIRT1 expression. Multiple imputation was used to evaluate missing covariate data. A cut point to define dichotomous SIRT1 expression status was determined by a systematic search across continuous marker expression values for the cut point with the best model fit (lowest Bayesian Information Criterion statistic) as described previously [45]. The Institutional Review Board at the University of North Carolina at Chapel Hill approved the protocol. All subjects provided informed consent.
Statistical analysis
For clinical data, univariate survival was assessed using the Kaplan-Meier method, with survival of low and high SIRT1 expression groups compared using the logrank test. Univariate analyses were performed using SAS 9.3 (SAS Institute, Cary, NC). Other results are expressed as mean + standard error of mean (SEM) from at least three independent experiments or biological replicates, unless otherwise indicated in the figure legend. Significant differences between the means were analyzed by the two-tailed, unpaired, non-parametric Mann-Whitney test. For all statistical tests, p-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Anton Jetten and Dmitry Gordenin, and members of the Li laboratory for critical reading of the manuscript. We also thank Dr. Yi Fang and Ms. Qing Xu for technical support, Dr. E. Terence Adams from Integrated Laboratory Systems for histopathologic evaluation of colon tumors developed in mice, Stephanie Cohen (Image analysis), Yongjuan Xia and Gabriela De la Cruz (Staining) in the UNC Translational Pathology Laboratory (TPL) for expert technical assistance. This research was supported by the Intramural Research Program of National Institute of Environmental Health Sciences to Xiaoling Li (Z01 ES102205), and was also supported, in part, by extramural grants from the NIH (P30DK034987 and U01CA93326). Dr. Busch was supported in part by a grant from the National Cancer Institute (5T32CA009001). The UNC TPL is supported in part by grants from the National Cancer Institute (3P30CA016086), the UNC University Cancer Research Fund (UCRF).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Author Contributions
N. S. X. R. and M. J. designed experiments, carried out experiments, analyzed data, and prepared the manuscript. E. J. T. and D. L. carried out experiments. X. X., D. C. F., W. H., and L. L. analyzed the microarray data. Xiangchun Li and W. K. K. W. analyzed TCGA database. A. J. and Y. Z. provided equipment and technical supports in Seahorse analysis. E. L. B., T. O. K., and R. S. S. analyzed SIRT1 protein expression in colorectal tissue microarrays from CanCORS and prepared the manuscript. Xiaoling Li conceived the study, designed experiments, analyzed data, and prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 6.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 8.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Annals of Medicine. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Z, Fang D. The Roles of SIRT1 in Cancer. Genes Cancer. 2013;4:97–104. doi: 10.1177/1947601912475079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Sasso G, Ryu D, Mouchiroud L, Fernando SC, Anderson CL, Katsyuba E, Piersigilli A, Hottiger MO, Schoonjans K, Auwerx J. Loss of Sirt1 function improves intestinal anti-bacterial defense and protects from colitis-induced colorectal cancer. PLoS One. 2014;9:e102495. doi: 10.1371/journal.pone.0102495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 19.Eng CH, Abraham RT. Glutaminolysis yields a metabolic byproduct that stimulates autophagy. Autophagy. 2010;6:968–970. doi: 10.4161/auto.6.7.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;109:E187–196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, Lane DP. p53 mutations in colorectal cancer. Proc Natl Acad Sci U S A. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nazeer FI, Devany E, Mohammed S, Fonseca D, Akukwe B, Taveras C, Kleiman FE. p53 inhibits mRNA 3′ processing through its interaction with the CstF/BARD1 complex. Oncogene. 2011;30:3073–3083. doi: 10.1038/onc.2011.29. [DOI] [PubMed] [Google Scholar]
- 28.Ayanian JZ, Chrischilles EA, Fletcher RH, Fouad MN, Harrington DP, Kahn KL, Kiefe CI, Lipscomb J, Malin JL, Potosky AL, et al. Understanding cancer treatment and outcomes: the Cancer Care Outcomes Research and Surveillance Consortium. J Clin Oncol. 2004;22:2992–2996. doi: 10.1200/JCO.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Kang M, Shen XJ, Kim S, Araujo-Perez F, Galanko JA, Martin CF, Sandler RS, Keku TO. Somatic gene mutations in African Americans may predict worse outcomes in colorectal cancer. Cancer Biomark. 2013;13:359–366. doi: 10.3233/CBM-130366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malin JL, Ko C, Ayanian JZ, Harrington D, Nerenz DR, Kahn KL, Ganther-Urmie J, Catalano PJ, Zaslavsky AM, Wallace RB, et al. Understanding cancer patients’ experience and outcomes: development and pilot study of the Cancer Care Outcomes Research and Surveillance patient survey. Support Care Cancer. 2006;14:837–848. doi: 10.1007/s00520-005-0902-8. [DOI] [PubMed] [Google Scholar]
- 31.Ming M, Soltani K, Shea CR, Li X, He YY. Dual role of SIRT1 in UVB-induced skin tumorigenesis. Oncogene. 2015;34:357–363. doi: 10.1038/onc.2013.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purushotham A, Xu Q, Li X. Systemic SIRT1 insufficiency results in disruption of energy homeostasis and steroid hormone metabolism upon high-fat-diet feeding. FASEB J. 2012;26:656–667. doi: 10.1096/fj.11-195172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 37.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell metabolism. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:1–8. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Qiang L, Kon N, Zhao W, Jiang L, Knight CM, Welch C, Pajvani U, Gu W, Accili D. Hepatic SirT1-Dependent Gain of Function of Stearoyl-CoA Desaturase-1 Conveys Dysmetabolic and Tumor Progression Functions. Cell Rep. 2015;11:1797–1808. doi: 10.1016/j.celrep.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathore MG, Saumet A, Rossi JF, de Bettignies C, Tempe D, Lecellier CH, Villalba M. The NF-kappaB member p65 controls glutamine metabolism through miR-23a. Int J Biochem Cell Biol. 2012;44:1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Kazgan N, Metukuri MR, Purushotham A, Lu J, Rao A, Lee S, Pratt-Hyatt M, Lickteig A, Csanaky IL, Zhao Y, et al. Intestine-specific deletion of SIRT1 in mice impairs DCoH2-HNF-1alpha-FXR signaling and alters systemic bile acid homeostasis. Gastroenterology. 2014;146:1006–1016. doi: 10.1053/j.gastro.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busch EL, Keku TO, Richardson DB, Cohen SM, Eberhard DA, Avery CL, Sandler RS. Evaluating markers of epithelial-mesenchymal transition to identify cancer patients at risk for metastatic disease. Clin Exp Metastasis. 2015 doi: 10.1007/s10585-015-9757-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.