Abstract
Dechlorination patterns of three tetrachlorobenzene isomers, 1,2,3,4-, 1,2,3,5-, and 1,2,4,5-TeCB, were studied in anoxic microcosms derived from contaminated harbor sludge. The removal of doubly, singly, and un-flanked chlorine atoms was noted in 1,2,3,4- and 1,2,3,5-TeCB fed microcosms, whereas only singly flanked chlorine was removed in 1,2,4,5-TeCB microcosms. The thermodynamically more favorable reactions were selectively followed by the enriched cultures with di- and/or mono-chlorobenzene as the main end products of the reductive dechlorination of all three isomers. Based on quantitative PCR analysis targeting 16S rRNA genes of known organohalide-respiring bacteria, the growth of Dehalococcoides was found to be associated with the reductive dechlorination of all three isomers, while growth of Dehalobacter, another known TeCB dechlorinator, was only observed in one 1,2,3,5-TeCB enriched microcosm among biological triplicates. Numbers of Desulfitobacterium and Geobacter as facultative dechlorinators were rather stable suggesting that they were not (directly) involved in the observed TeCB dechlorination. Bacterial community profiling suggested bacteria belonging to the phylum Bacteroidetes and the order Clostridiales as well as sulfate-reducing members of the class Deltaproteobacteria as putative stimulating guilds that provide electron donor and/or organic cofactors to fastidious dechlorinators. Our results provide a better understanding of thermodynamically preferred TeCB dechlorinating pathways in harbor environments and microbial guilds enriched and active in anoxic TeCB dechlorinating microcosms.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-016-8004-8) contains supplementary material, which is available to authorized users.
Keywords: Tetrachlorobenzenes, Organohalide respiration, Dehalococcoides mccartyi, Dehalobacter, Thermodynamically favorable reactions
Introduction
Chlorobenzenes (CBs) are aromatic chlorinated compounds with a benzene ring substituted with one to six chlorine atoms. CBs are important intermediates in industrial synthesis of pesticides and bulk chemicals (Barber et al. 2005; Malcolm et al. 2004). As a result of massive use and uncontrolled disposal on one hand and high persistence and bioaccumulation in ecosystems on the other hand, broad distribution of these pollutants has been found in a range of different environments (Barber et al. 2005; Malcolm et al. 2004; Wang et al. 2010). Partial reductive dechlorination of highly chlorinated benzenes has been shown in a broad range of sub-oxic/anoxic environments such as soil (Jiang et al. 2015; Ramanand et al. 1993), riverbed sediment (Nowak et al. 1996; Taş et al. 2010; Taş et al. 2011), sewage sludge (Fathepure et al. 1988; Fennell et al. 2004), and drainage ditch (Nelson et al. 2014) with lower chlorinated benzenes as end products. Even though monochlorobenzene (MCB) was considered to be recalcitrant to biotransformation in anoxic environments (Field and Sierra-Alvarez 2008), several studies reported further dechlorination of MCB to benzene in anaerobic enrichment cultures originating from freshwater sediment samples (Fung et al. 2009; Nowak et al. 1996; Zhou et al. 2015). TeCBs are important intermediates of HCB dechlorination, and include three isomers, i.e., 1,2,3,4-, 1,2,3,5-, and 1,2,4,5-TeCB. TeCBs can be reductively dechlorinated following diverse pathways (Adrian and Görisch 2002; Field and Sierra-Alvarez 2008). Preference for thermodynamically more favorable CB reductive dechlorination pathways was shown before by a microbial consortium originating from lake sediment (Beurskens et al. 1994), but this selectivity was not observed in other microcosms derived from contaminated freshwater sediment samples (Vandermeeren et al. 2014; Zhou et al. 2015).
Organohalide respiration (OHR), i.e., energy conservation from respiratory reductive dechlorination of organohalides as the final electron acceptors, is currently the only known metabolic microbial process for the partial/complete dechlorination of highly chlorinated benzenes like hexachlorobenzene (HCB), pentachlorobenzene (QCB), and tetrachlorobenzenes (TeCBs) under anoxic conditions (Adrian and Görisch 2002; Field and Sierra-Alvarez 2008). Therefore, the presence and activity of organohalide respiring bacteria (OHRB) is pivotal for bioremediation of anoxic CB-contaminated environments. Currently known bacterial isolates able to dechlorinate TeCB isomers belong to Dehalococcoides mccartyi in the phylum Chloroflexi (Adrian et al. 2000; Fennell et al. 2004) and Dehalobacter in the phylum Firmicutes (Nelson et al. 2014). D. mccartyi strains CBDB1 (Adrian et al. 2000), 195 (Fennell et al. 2004) and DCMB5 (Pöritz et al. 2015) dehalogenate CB congeners with four or more chlorine substitutes to trichlorobenzenes (TCBs) or dichlorobenzenes (DCBs). Besides, Dehalobacter spp. strains 12DCB1 and 13DCB1 and a highly enriched culture containing Dehalobacter sp. 14DCB1 were shown to dechlorinate highly chlorinated CBs to DCBs and MCB (Nelson et al. 2014). In addition to these obligate organohalide respiring isolates that as to the best of our knowledge, are restricted to OHR as the sole metabolism to conserve energy, evidence for the growth of members of metabolically versatile OHRB in the presence of CBs was reported. Bacteria of the genus Geobacter were found associated with HCB dechlorination in an anaerobic consortium also containing Dehalococcoides and originating from contaminated river sediment as determined by genus-specific quantitative PCR (qPCR), suggesting members of the genus Geobacter also may serve as CB dechlorinators (Zhou et al. 2015). Furthermore, a recently reported bacterial community analysis with 454 pyrosequencing of 16S ribosomal RNA (rRNA) gene fragments conducted on a range of CB-dechlorinating enrichments from contaminated river sludge revealed a high relative abundance (21–55%) of populations closely related to Desulfitobacterium, while 16S rRNA genes of Dehalococcoides and Dehalobacter together represented less than 0.53% of the bacterial community (Vandermeeren et al. 2014).
Up to this date, the fate of TeCBs in marine and estuarine environments is still not well known. 1,2,4-TCB dechlorination in sediment collected from Ise Bay (Japan) revealed that DCB production rates were related to anoxic plate count numbers and activity of sulfate reducing bacteria, although there was no solid evidence showing those sulfate reducers have direct involvement in 1,2,4-TCB dechlorination (Yonezawa et al. 1994). Furthermore, dechlorination of CBs was reported in enrichments derived from estuarine sediments collected in Tsurumi river (Japan) (Masunaga et al. 1996) and Bayou d’Inde (USA) (Pavlostathis and Prytula 2000), but the microorganisms responsible for dechlorination were not identified. To date, the only reported CB-dechlorinating bacterium isolated from an estuarine environment (Charleston Harbor, USA) is strain DF-1, later re-classified as Dehalobium chlorocoercia (Löffler et al. 2013), which was shown to dechlorinate HCB via 1,2,3,5-TeCB to 1,3,5-TCB (Wu et al. 2002). This pathway was also proposed as the main pathway of dechlorination of HCB and QCB in estuarine sediments with very low levels of other CBs detected (Pavlostathis and Prytula 2000; Wu et al. 2002). Overall, dechlorination of CBs and in particular of TeCB in marine and estuarine sediments remains poorly explored, and better understanding of the activity of CB-dehalogenating microbes and the microbial context they interact with in those habitats is required for their in situ bioremediation (Zanaroli et al. 2015).
The aim of this study was to assess the following: (i) the TeCB dechlorination potential of microbial communities present in an estuarine environment, (ii) microbial community dynamics during anoxic cultivation, and (iii) the preferred TeCB dechlorinating pathways. To this end, we set up TeCB-fed anoxic microcosms with sludge derived from a harbor contaminated with tributyltin, mineral oil, and polyaromatic hydrocarbons. A combination of bacterial community profiling and qPCR was applied to gain better insight into TeCB dechlorinators and co-existing, non-dechlorinating bacterial guilds that may be important for sustained dechlorination and therefore aid in improving in situ bioremediation.
Materials and methods
Chemicals
CBs, benzene, and 1,3,5-tribromobenzene (TBrB) of analytical grade and high purity were purchased from Sigma Aldrich. Benzene, MCB, 1,2-DCB, 1,4-DCB, 1,2,3-TCB, 1,2,4-TCB, 1,3,5-TCB, and 1,2,3,5-TeCB had a purity of 99%; 1,3-DCB, 1,2,4,5-TeCB and 1,3,5-TBrB 98%; and 1,2,3,4-TeCB 96%.
Microcosm setup
Harbor sludge from Zeebrugge, Belgium was sampled (Fig. 1, step 1) to prepare anoxic microcosms. All samples were collected in September 2008, and stored for 3 months at 4 °C until they were used in this study. The microcosms were set up in triplicates as described elsewhere (Vandermeeren et al. 2014) with 10 g of sludge and 40 ml of an anoxic and sterilized medium with 2 g/L yeast extract (Holliger et al. 1998). One spike of lactate (20 mM) was added as electron donor and two spikes of TeCB isomers were added from acetone stocks to final concentrations of 50 μM for each isomer where necessary and microcosms were incubated non-shaking at 25 °C in the dark for a period of 93 days (Fig. 1, step 2). After dechlorination of the first TeCB spike to TCBs and DCBs, a second spike (50 μM) of the corresponding TeCB isomer was supplied to the cultures. As residual lactate concentrations were considered sufficient, lactate was not replenished with the second TeCB spike. Abiotic controls were prepared by addition of 0.5 ml of 37% formaldehyde. Cultures were subsequently kept for 5 years at 4 °C, after which a subset (single 1,2,3,4- and 1,2,4,5-TeCB enrichment cultures and triplicate 1,2,3,5-TeCB cultures) was transferred to Wageningen, the Netherlands, for further characterization of TeCB dechlorination. To revive TeCB dechlorination in the original cultures, 20 mM lactate and 30 ml of fresh anoxic medium was added to the original bottles. Bottles were sealed with a viton stopper, the headspace was exchanged with N2/CO2 (80:20 v/v), and the respective TeCBs (50 μM/each in acetone) were added. The dechlorinating cultures were amended with the respective TeCBs and lactate as mentioned on day 35, 96, and 134 of incubation. All cultures were maintained actively dechlorinating for over 200 days without shaking at 25 °C in the dark (Fig. 1, step 3). To ensure efficient electron donor and micronutrients supply, 20 mM lactate and 42 ml fresh anoxic medium were added to the cultures on day 245 of incubation.
Fig. 1.
Schematic representation of the experimental strategy
To study the dynamics of degradation pathways and microbial communities and to obtain sediment-free TeCB-enriched cultures, serial transfers were performed. A 5% inoculum from the original TeCB-fed microcosms was transferred into anoxic bottles containing growth medium, fed with 50 μM of the same TeCB isomer as the initial inoculum and cultivated for 66 days (Fig. 1, step 4). Further transfers were conducted with the 1,2,4,5-TeCB dechlorinating culture enriched from harbor sludge (culture A in Fig. 1, step 5), and their performance was compared to that of transfers of two sediment-free 1,2,4,5-TeCB-enriching cultures (cultures B and C in Fig. 1, step 5) that were previously obtained from chlorinated aromatics-contaminated river sludge collected in Kanaal Ieper-Ijzer, Belgium (Vandermeeren et al. 2014). A 5% inoculum was taken for serial transfers (Fig. 1, step 6).
Effects of vancomycin addition and/or vitamin B12 starvation on the dechlorination activity of the sediment-free 1,2,4,5-TeCB-fed enrichments were investigated with a 2 × 2 factorial design experiment (Fig. 1, step 7) that was set up in duplicate. Twelve microgram per liter of vancomycin and 0.03 μM of vitamin B12 were added as required.
Detection of benzene and chlorinated benzenes
Benzene and CBs were quantified on a GC-2000 equipped with flame ion detector (FID) (Shimadzu, Japan) and an Rxi-5Sil capillary column (RESTEK, USA). 0.2 ml headspace samples were used for benzene measurement as described before (Liang et al. 2011), and 1 ml liquid samples were used for the detection of CBs. Liquid samples were extracted with 400 μl hexane by overnight shaking at 600 rpm followed by 100 μl hexane phase transfer to HPLC vials containing 5 μl of 1 mM 1,3,5-tribromobenzene dissolved in hexane as an internal standard. The column temperature program was 60 °C held for 4 min, followed by a gradient of 5 °C min−1 to 180 °C and another held for 3 min. The carrier gas was nitrogen at a flow of 1 ml min−1, a split flow of 12 ml min−1 with a split ratio of 1:10. Injection was performed by an auto-sampler (Shimadzu, Japan) injecting 2 μl of the sample into a split injector held at 250 °C. Standards with a mixture of benzene and CBs at 10, 20, 40, 60, 100, and 200 μM were set up in 125 ml serum bottles with the same headspace-liquid phase ratio of treatment cultures.
Sampling for molecular analysis and DNA extraction
Cultures were sampled for molecular analysis as shown in Fig. 1. The samples taken before adding TeCBs to the original bottles were indicated as ‘Before reviving’ (Fig. 1, step 3). Two milliliters of slurry or liquid samples were withdrawn from each bottle, centrifuged (5417R, Eppendorf, Germany) at 6010 g for 5 min at 4 °C, and pellets were stored at −20 °C. DNA isolation was performed using the PowerSoil DNA isolation kit (MO-BIO, USA) according to the manufacturer’s instructions except that three 45 s bead beating (5.5 m/s) steps using a Fastprep Instrument (MP Biomedicals, USA) were included instead of the horizontally vortexing step suggested in the manual. The quality and quantity of DNA was checked using a Nanodrop 2000c Spectrophotometer (Thermo Fisher Scientific, USA) and agarose gel electrophoresis.
Illumina MiSeq sequencing and data analysis
A 2-step PCR strategy was used to generate barcoded amplicons from the V1-V2 region of the 16S rRNA gene. The first PCR (50 μl) contained 1× HF buffer (Thermo Scientific™, The Netherlands), 1 μl dNTP Mix (10 mM; Promega, The Netherlands), 1 U of Phusion® Hot Start II High-Fidelity DNA polymerase (Thermo Scientific™), 500 nM of 27F–DegS forward primer, 500 nM of 338R I and II reverse primers (Table S1), and 1 μL template DNA (15–20 ng/μl). The forward and reverse primers were appended at the 5′ end with 18 bp Universal Tag (Unitag) 1 and 2, respectively (Table S1). PCR conditions were 98 °C for 30 s, 25 cycles of denaturation at 98 °C for 10 s, annealing at 56 °C for 20 s and elongation at 72 °C for 20 s, and a final extension at 72 °C for 10 min. The PCR product was examined by gel electrophoresis. The second PCR (100 μl) contained 1× HF buffer, 2 μl dNTP Mix, 2 U of Phusion® Hot Start II High-Fidelity DNA polymerase, 500 nM of a forward and reverse primer (equivalent to the Unitag1 and Unitag2 sequences, respectively) that were each appended with an 8 nt sample specific barcode (Ramiro-Garcia et al. 2016) at the 5′ end, and 5 μl PCR product of the first reaction. PCR conditions were similar to those used for the first PCR except for an annealing temperature of 52 °C and a reduced number of 5 cycles. The PCR product was examined by gel electrophoresis, and amplicons were purified with HighPrep™ (Magbio Genomics, USA) and quantified using a Qubit in combination with the dsDNA BR Assay Kit (Invitrogen, USA). Purified PCR products were pooled, underwent adaptor ligation, and were sequenced on a MiSeq platform (GATC-Biotech, Germany). Sequence analysis was performed with NG-Tax, an in-house pipeline (Ramiro-Garcia et al. 2016) in which operational taxonomic units (OTUs) were assigned to taxonomy using uclust (Edgar 2010) in an open reference approach against the SILVA 16S rRNA gene reference database (Quast et al. 2013). Finally, a biom file was generated and sequence data were further analyzed using Quantitative Insights Into Microbial Ecology (QIIME) v1.2 (Caporaso et al. 2010).
Quantitative PCR
The abundance of the 16S rRNA genes of Dehalococcoides, Dehalobacter, Desulfitobacterium, Geobacter, and total bacteria was determined by qPCR performed on a CFX384 Real-Time system with C1000 Thermal Cycler (Bio-Rad Laboratories, USA) with iQ™ SYBR Green Supermix (Bio-Rad Laboratories, USA). Each DNA sample was analyzed in triplicates. The primers and qPCR amplification programs used in this study were described before for Dehalobacter (Atashgahi et al. 2013) and other targets (Sutton et al. 2015). The relative abundance of Dehalococcoides in qPCR data was calculated by dividing the number of Dehalococcoides 16S rRNA gene copies by the copy number of bacterial 16S rRNA gene copies, multiplied by 100.
Nucleotide sequences
16S rRNA gene sequences were deposited in the European Nucleotide Archive (ENA) with accession number ERS1082969-ERS1083005 under study PRJEB13024.
Results
TeCB dechlorination in sediment microcosms
During 93 days of anoxic cultivation (Fig. 1, step 2), complete depletion of two spikes of 50 μM of the added TeCBs, i.e., either 1,2,3,4-, 1,2,3,5-, or 1,2,4,5-TeCB, was observed in all sediment microcosms with TCBs and DCBs as products (data not shown). In contrast, no decrease of TeCBs was observed and no dechlorination products were detected in the abiotic controls (data not shown).
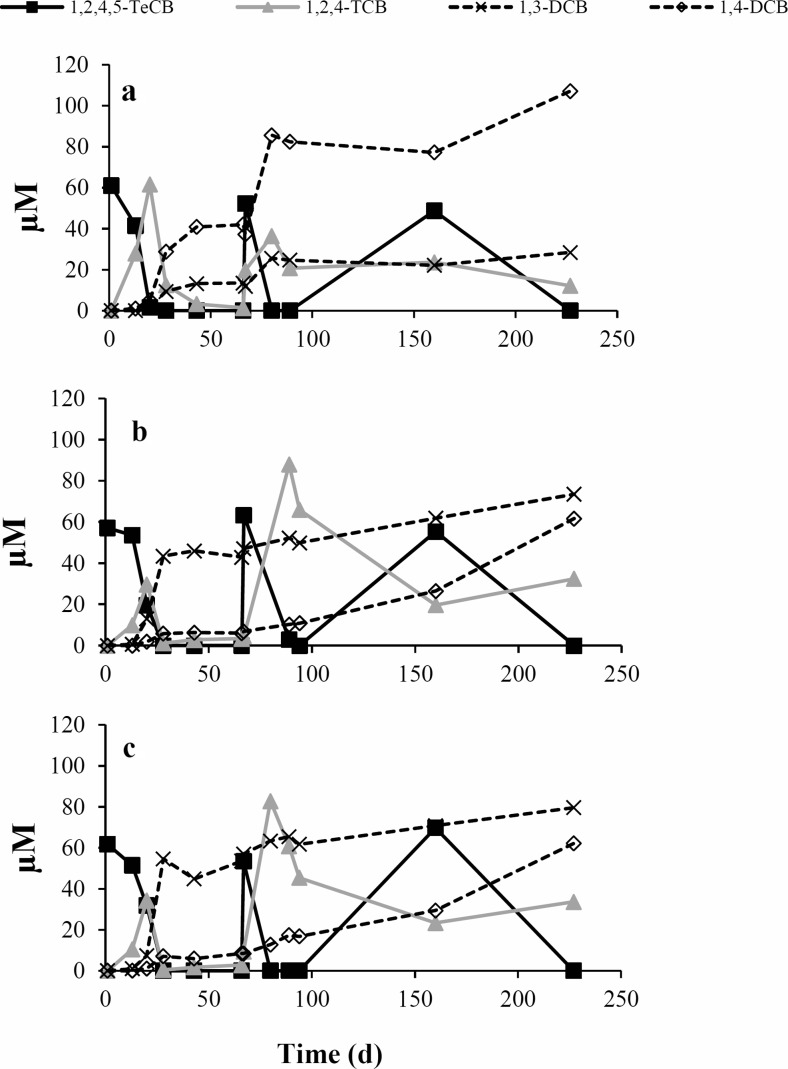
After these microcosms had been stored for 5 years at 4 °C, new spikes of TeCBs were added to recover dechlorination activity. After a short lag phase of 1–8 days, dechlorination of TeCBs was observed in all microcosms (Fig. 2) indicating that long-term storage at cold temperature did not negatively affect the dechlorination activity of the microcosms. 1,2,4-TCB was observed as a transient intermediate during the dechlorination of all three TeCB isomers, whereas 1,3,5-TCB was only detected as an intermediate product of 1,2,3,5-TeCB-dechlorination. All microcosms showed MCB accumulation except for the 1,2,4,5-TeCB-fed microcosm in which 1,4-DCB was detected as the main end product (Fig. 2e). 1,2,3-TCB, 1,2-DCB, and benzene were not detected in any of the microcosms during 161 days of incubation.
Fig. 2.
Dechlorination of 1,2,3,4-TeCB (a) 1,2,3,5-TeCB (b–d, triplicate bottles), and 1,2,4,5-TeCB (e) in original microcosms derived from harbor sludge. 50 μM spikes of the respective TeCBs are indicated by vertical dashed lines. Concentrations of TeCBs are not shown for the clarity of the figure. Mass balance between parent and daughter products during dechlorination is shown in Table S2
1,2,4,5-TeCB dechlorination in sediment-free cultures
To achieve sediment-free TeCB-enriched cultures, a 5% inoculum was directly transferred from original TeCB-fed microcosms (Fig. 1, step 4). Dechlorination was observed in the transferred 1,2,4,5-TeCB-fed enrichments (culture A in Fig. 1, step 5) with 1,3- and 1,4-DCB as end products after 66 days of anoxic cultivation, but not in the 1,2,3,4- and 1,2,3,5-TeCB-fed cultures (data not shown).
In the following, the dechlorination performance of the newly obtained transfer cultures derived from the harbor environment was compared with that of another two sediment-free 1,2,4,5-TeCB-enriching cultures that were previously enriched from river sludge in Kanaal Ieper-Ijzer (Belgium) (Vandermeeren et al. 2014). During incubation of the second transfer, three spikes of 1,2,4,5-TeCB were completely depleted in all transferred enrichments, concomitant with the production of 1,2,4-TCB, 1,4-DCB, and 1,3-DCB (Fig. 3). 1,4-DCB was the dominant end product in harbor derived enrichment (Fig. 3a), which was in line with metabolites observed in the original 1,2,4,5-TeCB-fed microcosm (Fig. 2e). In contrast, slightly more 1,3-DCB was detected in river sludge-derived enrichments (Fig. 3b, c).
Fig. 3.
Dechlorination of 1,2,4,5-TeCB in second transfers (Fig. 1, step 6) from the 1,2,4,5-TeCB dechlorinating enrichment derived from harbor sludge (a) and 1,2,4,5-TeCB dechlorinating sediment-free enrichments (culture B and C in Fig. 1) derived from river sludge (b–c). 50 μM spikes of 1,2,4,5-TeCBs were added at the beginning and on day 66 and 159 of incubation. Data from an individual bottle is shown here as a representative of duplicate cultures, which both followed the same overall dechlorination pattern
Dechlorination pattern of TeCB isomers and selectivity of the reactions
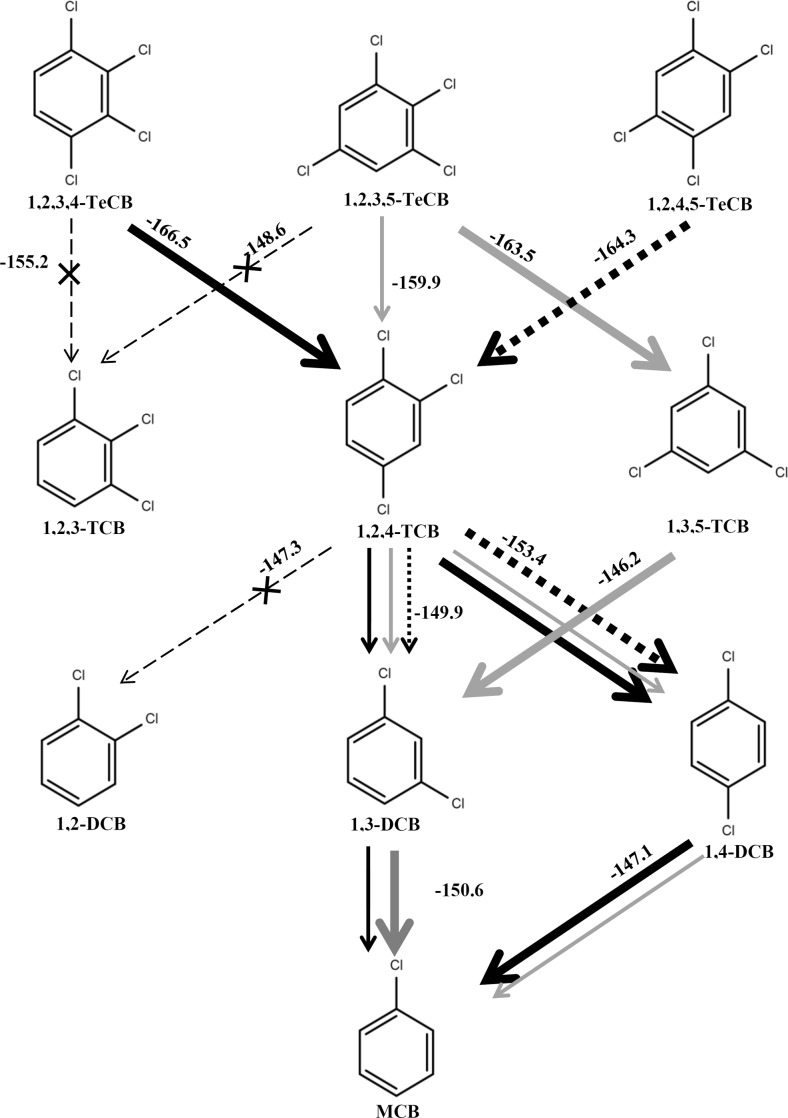
The reductive removal of doubly, singly, and non-flanked chlorine atoms was noted in the 1,2,3,4- and 1,2,3,5-TeCB-fed microcosms derived from contaminated harbor sludge. In contrast, only singly flanked, but not the non-flanked chlorine, was removed during the dechlorination of 1,2,4,5-TeCB. Based on Gibbs free-energies (ΔG0′ under standard condition, kJ/mol) taken from Dolfing and Keith Harrison (1993) calculated with hydrogen as electron donor, the harbor sludge-derived microcosms showed a preference to follow the thermodynamically most favorable dechlorination pathways. For example, 1,2,3,5-TeCB was mainly dechlorinated to 1,3,5-TCB as an intermediate rather than 1,2,4-TCB, and 1,2,3-TCB was not detected, in line with the fact that the latter reaction theoretically yields the least negative free energy (Fig. 4). A similar pattern was observed for the dechlorination of 1,2,3,4-TeCB to 1,2,4-TCB instead of 1,2,3-TCB and further dechlorination of 1,2,4-TCB to 1,4-DCB instead of 1,3-DCB as the main product (Fig. 4).
Fig. 4.
Dechlorination patterns of 1,2,3,4- (black solid line), 1,2,3,5- (gray solid line) and 1,2,4,5- (black dotted line) TeCB in microcosms derived from harbor sludge. Thin arrows represent reactions that were detected but were not dominant. Reactions catalyzed preferentially are indicated with bold arrows, and reactions not observed in our study are indicated by dashed lines with a cross. The ΔG0′ (kJ/mol) indicated for each dechlorination reaction is based on previously reported calculations with hydrogen as the electron donor (Dolfing and Keith Harrison 1993)
Effect of vancomycin addition and vitamin B12 starvation
Vancomycin, an inhibitor of the peptidoglycan synthesis of Gram-positive bacteria, was added to harbor sludge-derived, sediment-free 1,2,4,5-TeCB enrichments to test its effect on TeCB dechlorination. After 21 days of anoxic cultivation, the amended 1,2,4,5-TeCB was completely depleted in control microcosms (Fig. S1a) with 1,4-DCB and 1,3-DCB as the main products. In contrast, in vancomycin treated microcosms, only 36–59% of the 1,2,4,5-TeCB was dechlorinated within the same incubation time, with 1,2,4-TCB as the main product and trace amounts of 1,4-DCB and 1,3-DCB (Fig. S1b). The delayed dechlorination in enrichments supplied with vancomycin suggested (in)direct involvement of vancomycin-sensitive Gram-positive bacteria in 1,2,4,5-TeCB dechlorination. Additionally, vitamin B12 (cofactor of reductive dehalogenase enzymes) starvation was investigated in the presence or absence of vancomycin (Fig. S1c, d). However, no effect was observed on TeCB dechlorination activity, suggesting that the 1,2,4,5-TeCB-enriched microbial communities tested here were capable of de novo vitamin B12 biosynthesis.
Dynamics of bacterial communities
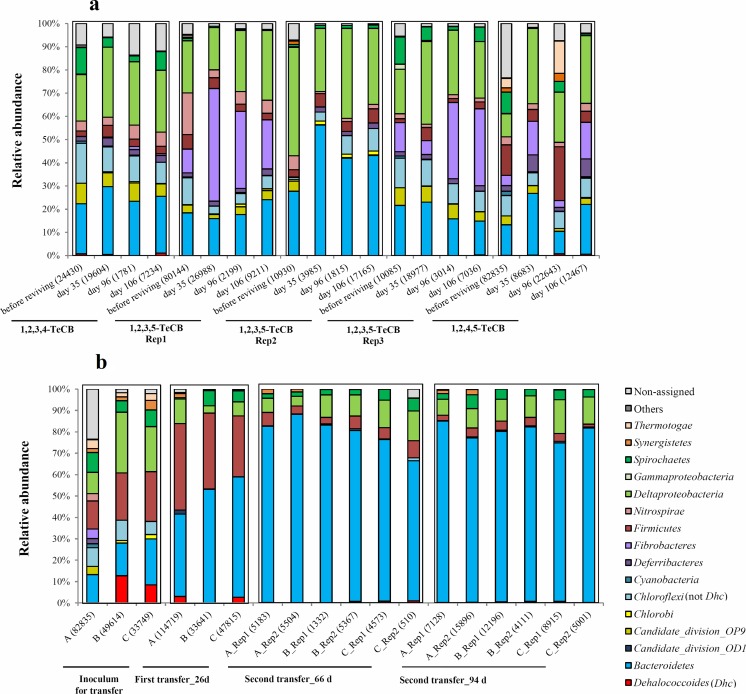
During 161 days of cultivation of original TeCB microcosms, Dehalococcoides as a potential TeCB dechlorinator was detected at relative abundances below 2% in 1,2,3,4- and 1,2,4,5-TeCB fed microcosms and one (Rep3) of the triplicate 1,2,3,5-TeCB fed microcosms (Fig. 5a). The detected sequence of Dehalococcoides (200 bp) showed 100% identity with that of D. mccartyi strains CBDB1 and 195 (data not shown). Additionally, diverse bacteria were found in the bacterial community of TeCB-fed microcosms, especially from the phyla Bacteroidetes and Fibrobacteres and the class Deltaproteobacteria (Fig. 5a) that have not reported as OHRB except the deltaproteobacterial Geobacter lovelyi that did not show appreciable increase using qPCR analysis (Fig. 6). 16S rRNA genes of Bacteroidetes and Deltaproteobacteria were detected in all microcosms in the range of 10–56% and 10–47%, respectively. Fibrobacteres was mainly enriched in the 1,2,4,5-TeCB-fed microcosm and two of the three replicate 1,2,3,5-TeCB fed microcosms (Rep1, Rep3), whereas it was not detected in the third 1,2,3,5-TeCB fed microcosm during cultivation (Fig. 5a). Differences in bacterial community composition observed among the three replicate 1,2,3,5-TeCB-fed microcosms may have been caused by the heterogeneity of the sediment used for initial inoculation.
Fig. 5.
Bacterial 16S rRNA gene-based community dynamics of TeCB-fed microcosms during 161 days of cultivation (a) and 1,2,4,5-TeCB dechlorinating enrichments during serial transfers (b). a. Reps 1, 2, and 3 are triplicate original bottles fed with 1,2,3,5-TeCB. Sampling times are labeled as in Fig. 1 (step 3). b. Rep1 and 2 are duplicates. Inoculum and sampling time are labeled as in Fig. 1 (step 6). Data are shown at phylum level, except Delta- and Gammaproteobacteria that are presented at the class level and the genus Dehalococcoides that is shown separately from its corresponding phylum (Chloroflexi). The number of reads of each sample is given in brackets. Phyla with relative abundance lower than 1% in all samples are summed up as ‘rest’
Fig. 6.
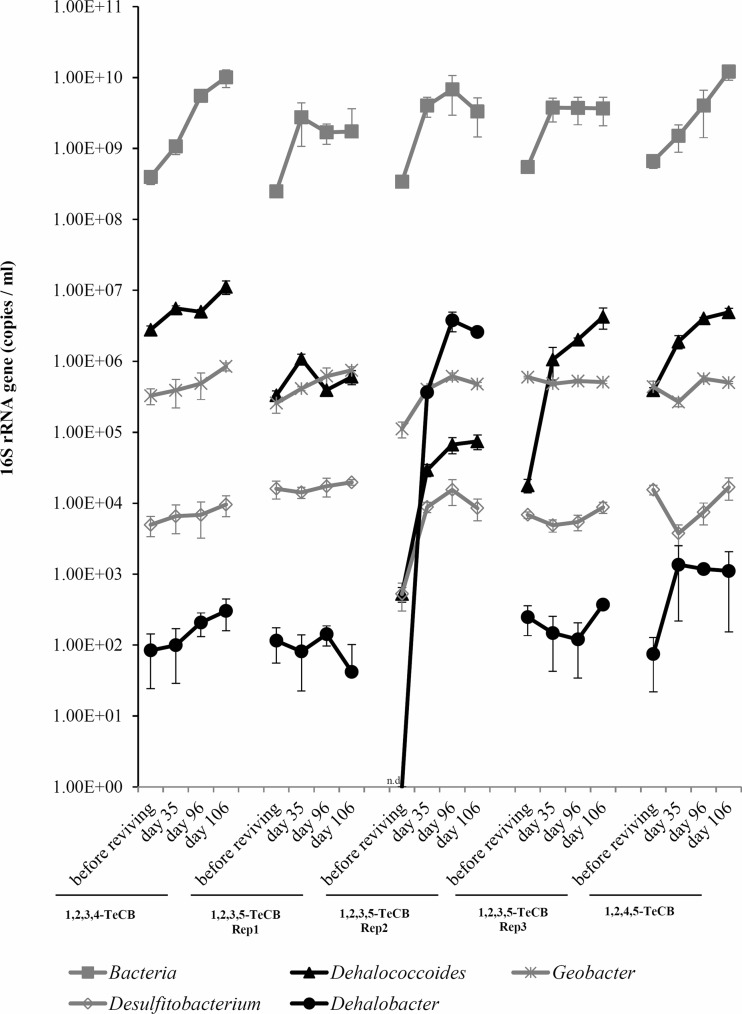
Quantitative PCR (qPCR) targeting 16S rRNA genes of Dehalobacter, Desulfitobacterium, Geobacter and Dehalococcoides, and of total bacteria during 161 days of cultivation of TeCB-fed microcosms derived from harbor sludge. Microcosm set up and sampling times are labeled as Fig. 1 (step3). Reps 1, 2, and 3 are triplicate original bottles fed with 1,2,3,5-TeCB. Error bars indicate standard deviations of qPCR technical triplicates. n.d. nondetectable
During serial transfers of the different 1,2,4,5-TeCB-enriched cultures, members of the Bacteroidetes phylum became strongly enriched with an average relative abundance of 79.7% (SD = 3.8, Table S3) at the end of the second transfer (Fig. 5b). Bacteroidetes detected in the MiSeq dataset were mainly from the classes Bacteroidia (especially from the genus Petrimonas) and Sphingobacteriia (Table S3). Petrimonas was not detected in the TeCB-enriched cultures derived from harbor sludge, in which the Sphingobacteriia dominated the bacterial community (Table S3).
Quantification of potential dechlorinators during TeCB dechlorination
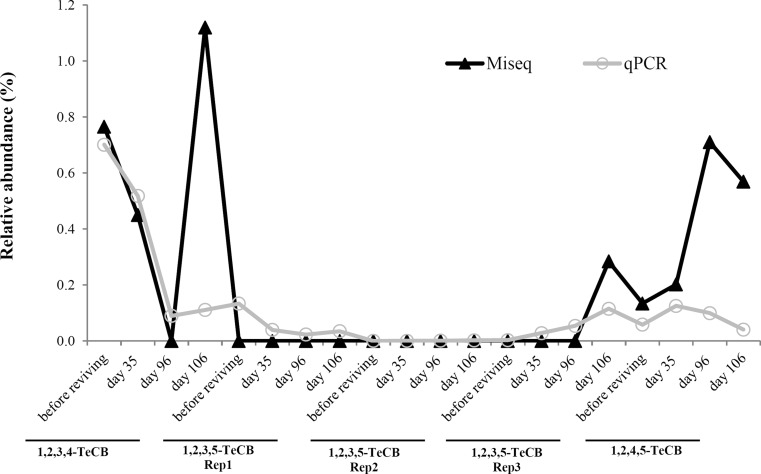
Bacterial growth, as measured by bacterial 16S rRNA gene-specific qPCR, was observed in all TeCBs fed microcosms (Fig. 6). Interestingly, in one out of three 1,2,3,5-TeCB fed microcosms (Rep2), the abundance of the genus Dehalobacter increased considerably from non-detectable levels before reviving to 2.59E + 06 copies/ml (SD = 3.48E + 05) at day 106 of cultivation, which was higher than the abundance of Dehalococcoides 16S rRNA genes with 7.41E + 04 copies/ml (SD = 1.74E + 04). This indicated that in this microcosm, Dehalobacter rather than Dehalococcoides was the main TeCB dechlorinator. In all other microcosms, a clear dominance of Dehalococcoides during cultivation was observed with an average abundance in the range of 3.93E + 05 to 1.12E + 07 copies/ml at the end of the cultivation (Fig. 6). In contrast, 16S rRNA gene copy numbers of Desulfitobacterium and Geobacter were rather stable (Fig. 6). In all of the 1,2,4,5-TeCB-fed transferred cultures, Dehalococcoides was detected with abundances that were more than three orders of magnitude higher than those of Dehalobacter (Fig. S2), suggesting a more important role of Dehalococcoides in 1,2,4,5-TeCB dechlorination in these enrichments. Overall, the relative abundance of Dehalococcoides calculated from qPCR data generally matched with the results derived from the MiSeq dataset, although in several samples higher relative abundance of Dehalococcoides was detected via MiSeq (Fig. 7, Fig. S3). Both qPCR and MiSeq results showed decreased relative abundance of Dehalococcoides during transfer, which was mainly caused by the increase of average bacterial numbers from 3.05E + 08 copies/ml in the inoculum to 1.98E + 09 copies/ml at 94 days, while the absolute numbers of Dehalococcoides remained around 1.0E + 07 copies/ml during serial transfer (Fig. S2).
Fig. 7.
Relative abundance of Dehalococcoides in TeCB-fed microcosms during 161 days of cultivation. Reps 1, 2 and 3 are triplicate original bottles fed with 1,2,3,5-TeCB. Sampling times are labeled the same as Fig. 1 (step 3)
Discussion
Dechlorination of 1,2,3,4-, 1,2,3,5-, and 1,2,4,5-TeCB isomers to DCB and/or MCB was observed in anoxic microcosms derived from harbor sludge, and enriched consortia showed selectivity in mediating thermodynamically more favorable steps in the reductive dechlorination of TeCB isomers (Fig. 4). Such preference for thermodynamically more favorable CB dechlorination pathways was shown before in an anaerobic microbial consortium originated from lake sediment (Beurskens et al. 1994), however, in that report, the TeCB isomers were dechlorinated with 1,3,5-TCB or 1,2,4-TCB as the end products. In our study, 1,2,3,5-TeCB was mainly dechlorinated to 1,3,5-TCB with the removal of doubly flanked chlorine, and subsequent removal of the unflanked meta-substituted chlorines to produce 1,3-DCB (Fig. 2b) and MCB (Fig. 2c, d). This implies that the enriched consortia from harbor sludge host CB dechlorinator(s) with complementary dechlorination functions. In line with our data, the dechlorination of HCB via 1,2,3,5-TeCB was found to predominate in cultures derived from estuarine sediment with 1,3,5-TCB (Wu et al. 2002) and 1,3-DCB (Pavlostathis and Prytula 2000) as the end products. Furthermore, 1,3,5-TCB production from 1,2,3,5-TeCB was reported before for D. mccartyi strains (Adrian et al. 2000; Fennell et al. 2004; Pöritz et al. 2015), however, no further dechlorination of 1,3,5-TCB was observed with those isolates, which might be due to the lack of respective dehalogenases. A different dechlorination pattern of 1,2,3,5-TeCB, with even less favorable thermodynamics, was found in Dehalobacter sp. 13DCB1, which predominantly dechlorinated singly flanked ortho-substituted chlorine with 1,2,4-TCB as the dominant intermediate leading to the formation of 1,4-DCB as the main end product and small amounts of MCB (Nelson et al. 2014). 1,2,4-TCB and 1,4-DCB were also detected in our 1,2,3,5-TeCB amended microcosms, albeit not as dominant dechlorination products (Fig. 4). Up to date, the dechlorination of 1,2,3,5-TeCB with the removal of unflanked chlorine to produce 1,2,3-TCB has not been reported.
1,2,3-TCB was shown to be the only product of 1,2,3,4-TeCB dechlorination by Dehalobacter strain 12DCB1 and 14DCB1 (Nelson et al. 2014) and could be further dechlorinated to 1,3-DCB by D. mccartyi strain DCMB5 (Pöritz et al. 2015). However, those metabolites were not detected in our study (Fig. 2a), probably because 1,2,3-TCB production was thermodynamically less favorable compared to the reductive removal of doubly flanked chlorine from 1,2,3,4-TeCB that leads to 1,2,4-TCB formation (Fig. 4).
Furthermore, 1,2,4-TCB was the only TCB detected in the 1,2,4,5-TeCB amended microcosms that was predominantly dechlorinated to 1,4-DCB with slightly more negative change of Gibbs free energy than formation of 1,3-DCB by the microcosm originated from contaminated harbor sludge (Fig. 4). In contrast, more 1,3-DCB than 1,4-DCB was produced via 1,2,4-TCB in the transferred cultures derived from Kanaal Ieper-Ijzer sludge (Fig. 3b, c), indicating the impact of inoculum source on the preference for the prevailing biotransformation pathway. The formation of 1,2-DCB from 1,2,4-TCB with the removal of unflanked meta-chlorine and lowest energy release was neither observed in our microcosms (Fig. 4) nor in other reported enrichment cultures (Middeldorp et al. 1997; Ramanand et al. 1993; Vandermeeren et al. 2014; Zhou et al. 2015) or in studies using CB dechlorinating isolates (Adrian et al. 2000; Fennell et al. 2004; Nelson et al. 2014; Pöritz et al. 2015).
We detected Dehalococcoides and Dehalobacter in all TeCB-fed microcosms as putative TeCB dechlorinators. In most of the microcosms, the growth of Dehalococcoides during 161 days of cultivation was associated with the addition of new spikes of the different TeCB substrates and subsequent TeCB dechlorination (Figs 2 and 6). However, no Dehalococcoides isolates have been previously reported to dechlorinate 1,2,3,5-TeCB to 1,4-DCB via 1,2,4-TCB or to dechlorinate any DCBs to MCB. Hence, the detected 1,2,4-TCB and 1,4-DCB in one of the 1,2,3,5-TeCB-fed microcosm among triplicates and the MCB increase in all the 1,2,3,4- and 1,2,3,5-TeCB-fed microcosms were possibly due to the dechlorination activity of Dehalobacter or other unknown dechlorinators. Except for one replicate microcosm, the obtained growth yields of Dehalococcoides by reductive dechlorination of amended TeCB isomers in original microcosms derived from harbor sludge (1.7 × 1010 to 2.4 × 1010 cells per mmol chloride released, Table S4) were comparable to the values reported for reductive dehalogenation of 2,3-dichlorophenol (7.6 × 1010 cells per mmol chloride released) (Adrian et al. 2007) and tetra- and trichloroethene (3.9 × 109 cells per mmol chloride released) (Marco-Urrea et al. 2011) by a pure culture of Dehalococcoides mccartyi strain CBDB1. This indicates that Dehalococcoides was mainly responsible for the reductive dechlorination patterns observed in these microcosms. However, Dehalococcoides growth yield was lower in one of the 1,2,3,5-TeCB dechlorinating microcosms (7.9 × 108 cells per mmol chloride released). Such low growth yields were previously reported from biostimulated aquifer microcosms dechlorinating trichloroethene (Schneidewind et al. 2014). Hence, we cannot exclude reductive dechlorination by other OHRB not quantified in this study or even co-metabolic reductive dechlorination (Assafanid et al. 1992, Gantzer and Wackett 1991).
Dehalococcoides and Dehalobacter both use hydrogen as the sole electron donor for reductive dehalogenation and are corrinoid auxotrophs (Holliger et al. 1998; Löffler et al. 2013; Nelson et al. 2014; Rupakula et al. 2015). Hence, the lack of a negative effect of vitamin B12 starvation on the 1,2,4,5-TeCB dechlorination capability of the transferred cultures indicated the presence of corrinoid producers in the microbial community that fulfill nutritional requirements of the TeCB dechlorinators. Considerable inhibition of dechlorination activity by vancomycin was observed in our study, and the 1,2,4,5-TeCB dechlorination pathway was changed in transferred cultures (Fig. S1). This sensitivity to vancomycin was observed before in HCB and 1,3,5-TCB fed microcosms derived from anoxic sediments (Duan and Adrian 2013), and therefore indicated the (in)direct involvement of vancomycin-sensitive bacteria in 1,2,4,5-TeCB dechlorination in transferred cultures. Dehalococcoides members lack a typical bacterial cell wall and therefore are not sensitive to vancomycin (Löffler et al. 2013), and members of Dehalobacter were found resistant to vancomycin before (Nelson et al. 2011) which might explain why vancomycin treatment did not fully abolish dechlorination activity.
Bacteria, belonging to the genera Desulfitobacterium and Geobacter, were also detected in the TeCB-fed microcosms by qPCR (Fig. 6). Although high abundance of Geobacter was previously found in HCB dechlorinating consortia (Zhou et al. 2015), and Desulfitobacterium-like populations predominated CBs-dechlorinating bacterial communities (Vandermeeren et al. 2014), these two genera have not been previously reported to dechlorinate CB, and hence no conclusion can be drawn on their (in)direct role in dechlorination of TeCB isomers and/or corresponding daughter products. Moreover, Desulfitobacterium and Geobacter are known as versatile facultative OHRB (Lovley et al. 2011; Villemur et al. 2006) that can use lactate as electron donor and other electron acceptors like sulfate that was also included in the media used in our study.
In our study, members of the phylum Fibrobacteres were detected in all TeCB-fed microcosms except one of three 1,2,3,5-TeCB-fed microcosms, with relative abundances ranging up to 49.5% (Fig. 5). Members of this phylum are known as plant polymer-degrading bacteria and were reported before from anoxic cellulose-fed digesters and herbivore guts (Rahman et al. 2015). Even though no evidence has been shown for their (in)direct involvement in reductive dechlorination of organohalides, they were detected before in contaminated sites, including a hexahydro-1,3,5-trinitro-1.3.5-triazine (RDX) contaminted vadose zone (Cupples 2013; Ronen et al. 2008) and soils contaminated with cadmium (Jeong et al. 2013) and crude oil (Abbasian et al. 2016). High relative abundance of sulfate-reducing Deltaproteobacteria, mainly from the Desulfobacteraceae family, with relative abundances ranging between 5 and 25.9% was detected in all TeCB-fed microcosms during 161 days of cultivation (Fig. 5a). Sulfate reducing members of the Deltaproteobacteria have previously been suggested to support Dehalococcoides by providing essential corrinoids at batch (Hug et al. 2012) and field (Atashgahi et al. 2016) scales and therefore may have a stimulatory effect on dechlorination (Sutton et al. 2015).
Increase of Bacteroidetes abundance has previously been reported after in situ bioaugmentation to treat trichloroethene (TCE)-contaminated groundwater, which indicated a potential syntrophy and/or synergistic relationship between Bacteroidetes and chlorinated ethene dechlorinators (Pérez-de-Mora et al. 2014). Bacteroidetes were also found highly enriched in our study, especially from the genus Petrimonas and the class Sphingobacteriia. Members of the genus Petrimonas are known as anaerobic fermentative bacteria and were previously isolated from a biodegraded oil reservoir and confirmed as lactate fermentator (Grabowski et al. 2005). Furthermore, Petrimonas was found predominant in a laboratory upflow anoxic sludge blanket (UASB) reactor treating TCE containing wastewater supplied with glucose and lactate as substates, and Petrimonas showed a positive correlation with TCE removal efficiency (Zhang et al. 2015). In our study, Petrimonas was mainly detected in the transferred cultures derived from river sludge collected in Kanaal Ieper-Ijzer (Table S3), but not from transfers derived from the harbor sludge, in which Sphingobacteriia dominated the bacterial community (Table S3). Sphingobacteriia were reported before to use lactate as carbon source (Shivaji et al. 1992), which might explain their considerable growth found in our study. Bacterial clones affiliated with Sphingobacteriia were detected before in a TCE-degrading community after in situ biostimulation with lactate (Macbeth et al. 2004), and sequences of Sphingobacteriia (in family WCHB1–69) were detected in a dioxin-dehalogenating enrichment derived from contaminated freshwater sediment with acetate as carbon source (Bunge et al. 2008) and in RDX-contaminated groundwater after biostimulation with acetate (Livermore et al. 2013). In our study, a strong increase of the Bacteroidetes population in the transferred cultures suggested a potential link (e.g., syntrophy and/or synergy) between lactate-utilizing Bacteroidetes and CB dechlorinators. Additionally, Firmicutes, all from the Clostridiales order, were detected in the transferred cultures, and especially in the first transfer (Fig. 5b). In addition to known organohalide-respiring Clostridiales belonging to the genera Dehalobacter and Desulfitobacterium, other Clostridiales were reported at high relative abundances in microcosms enriched with CBs (Vandermeeren et al. 2014), chlorinated ethane (Merlino et al. 2015), chlorinated ethene (Cichocka et al. 2010), and chlorinated phenols (Zhang et al. 2012) as electron acceptor and lactate as the electron donor, which suggested their role as important co-existing microbial guilds in organohalide-contaminated environments and a potential partnership between corrinoid-producing Clostridiales that are most probably non-dechlorinating and CBs dechlorinators (Men et al. 2013). One of the genera within the Clostridiales order found in this study, Sedimentibacter, was previously reported in co-culture with Dehalobacter sp. E1 and was postulated to support β-hexachlorocyclohexane (HCH) dechlorination by providing essential co-factors (e.g., corrinoid) (Maphosa et al. 2012; van Doesburg et al. 2005). Sedimentibacter alone was not able to survive in HCH-fed media (van Doesburg et al. 2005), and therefore, OHRB as detoxificator of chlorinated contaminants may have stimulatory impact on the growth of other anaerobic community members.
In summary, in this study we report the reductive dechlorination of three TeCB isomers in anoxic microcosms enriched from contaminated harbor sludge. Thermodynamically favorable dechlorinating pathways were found to be selectively followed by the TeCB-fed microcosms. Additionally, growth of Dehalobacter and Dehalococcoides was found to be associated with CB dechlorination based on 16S rRNA gene-targeted qPCR supporting their role in the observed reductive dehalogenation. Bacterial community profiling showed diverse putative non-dechlorinating bacteria, some of which may have stimulated TeCB dechlorination by providing essential nutrients, including electron donor and cofactors. Overall, these results provide further insight into the potential CB dechlorination in contaminated harbor environments and the interaction between TeCB dechlorinators and co-existing non-dechlorinators.
Electronic supplementary material
(PDF 201 kb)
Acknowledgments
We thank Shangwei Zhang and the members of the lab of Dr. Lorenz Adrian in UFZ, Germany for technical assistance in CB measurements, and acknowledge the China Scholarship Council for the support to Yue Lu. This work was furthermore supported by a grant of BE-Basic-FES funds from the Dutch Ministry of Economic Affairs.
Compliance with ethical standards
The work described in this article did not involve any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abbasian F, Lockington R, Megharaj M, Naidu R (2016) The biodiversity changes in the microbial population of soils contaminated with crude oil. Curr Microbiol 72:663–670 [DOI] [PubMed]
- Adrian L, Görisch H. Microbial transformation of chlorinated benzenes under anaerobic conditions. Res Microbiol. 2002;153:131–137. doi: 10.1016/S0923-2508(02)01298-6. [DOI] [PubMed] [Google Scholar]
- Adrian L, Szewzyk U, Wecke J, Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Adrian L, Hansen SK, Fung JM, Görisch H, Zinder SH. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ Sci Technol. 2007;41:2318–2323. doi: 10.1021/es062076m. [DOI] [PubMed] [Google Scholar]
- Assafanid N, Nies L, Vogel TM. Reductive dechlorination of a polychlorinated biphenyl congener and hexachlorobenzene by vitamin B12. Appl Environ Microbiol. 1992;58:1057–1060. doi: 10.1128/aem.58.3.1057-1060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashgahi S, Maphosa F, Doğan E, Smidt H, Springael D, Dejonghe W. Small-scale oxygen distribution determines the vinyl chloride biodegradation pathway in surficial sediments of riverbed hyporheic zones. FEMS Microbiol Ecol. 2013;84:133–142. doi: 10.1111/1574-6941.12044. [DOI] [PubMed] [Google Scholar]
- Atashgahi S, Lu Y, Zheng Y, Saccenti E, Suarez-Diez M, Ramiro-Garcia J, Eisenmann H, Elsner M, Stams AJM, Springael D, Dejonghe W, Smidt H. Geochemical and microbial community determinants of reductive dechlorination at a site biostimulated with glycerol. Environ Microbiol. 2016 doi: 10.1111/1462-2920.13531. [DOI] [PubMed] [Google Scholar]
- Barber JL, Sweetman AJ, Van Wijk D, Jones KC. Hexachlorobenzene in the global environment: emissions, levels, distribution, trends and processes. Sci Total Environ. 2005;349:1–44. doi: 10.1016/j.scitotenv.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Beurskens JE, Dekker CG, Van Den Heuvel H, Swart M, de Wolf J, Dolfing J. Dechlorination of chlorinated benzenes by an anaerobic microbial consortium that selectively mediates the thermodynamic most favorable reactions. Environ Sci Technol. 1994;28:701–706. doi: 10.1021/es00053a026. [DOI] [PubMed] [Google Scholar]
- Bunge M, Wagner A, Fischer M, Andreesen JR, Lechner U. Enrichment of a dioxin-dehalogenating Dehalococcoides species in two-liquid phase cultures. Environ Microbiol. 2008;10:2670–2683. doi: 10.1111/j.1462-2920.2008.01688.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocka D, Nikolausz M, Haest PJ, Nijenhuis I. Tetrachloroethene conversion to ethene by a Dehalococcoides-containing enrichment culture from Bitterfeld. FEMS Microbiol Ecol. 2010;72:297–310. doi: 10.1111/j.1574-6941.2010.00845.x. [DOI] [PubMed] [Google Scholar]
- Cupples AM. RDX degrading microbial communities and the prediction of microorganisms responsible for RDX bioremediation. Int Biodeterior Biodegradation. 2013;85:260–270. doi: 10.1016/j.ibiod.2013.08.002. [DOI] [Google Scholar]
- Dolfing J, Keith Harrison B. Redox and reduction potentials as parameters to predict the degradation pathway of chlorinated benzenes in anaerobic environments. FEMS Microbiol Ecol. 1993;13:23–29. doi: 10.1111/j.1574-6941.1993.tb00047.x. [DOI] [Google Scholar]
- Duan TH, Adrian L. Enrichment of hexachlorobenzene and 1, 3, 5-trichlorobenzene transforming bacteria from sediments in Germany and Vietnam. Biodegradation. 2013;24:513–520. doi: 10.1007/s10532-012-9607-0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fathepure BZ, Tiedje JM, Boyd SA. Reductive dechlorination of hexachlorobenzene to tri-and dichlorobenzenes in anaerobic sewage sludge. Appl Environ Microbiol. 1988;54:327–330. doi: 10.1128/aem.54.2.327-330.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol. 2004;38:2075–2081. doi: 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- Field JA, Sierra-Alvarez R. Microbial degradation of chlorinated benzenes. Biodegradation. 2008;19:463–480. doi: 10.1007/s10532-007-9155-1. [DOI] [PubMed] [Google Scholar]
- Fung JM, Weisenstein BP, Mack EE, Vidumsky JE, Ei TA, Zinder SH. Reductive dehalogenation of dichlorobenzenes and monochlorobenzene to benzene in microcosms. Environ Sci Technol. 2009;43:2302–2307. doi: 10.1021/es802131d. [DOI] [PubMed] [Google Scholar]
- Gantzer CJ, Wackett LP. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ Sci Technol. 1991;25:715–722. doi: 10.1021/es00016a017. [DOI] [Google Scholar]
- Grabowski A, Tindall BJ, Bardin V, Blanchet D, Jeanthon C. Petrimonas sulfuriphila gen. nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int J Syst Evol Microbiol. 2005;55:1113–1121. doi: 10.1099/ijs.0.63426-0. [DOI] [PubMed] [Google Scholar]
- Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJB. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- Hug LA, Beiko RG, Rowe AR, Richardson RE, Edwards EA. Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics. 2012;13:327. doi: 10.1186/1471-2164-13-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Moon HS, Shin D, Nam K. Survival of introduced phosphate-solubilizing bacteria (PSB) and their impact on microbial community structure during the phytoextraction of Cd-contaminated soil. J Hazard Mater. 2013;263:441–449. doi: 10.1016/j.jhazmat.2013.09.062. [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Q, Liu H, Yao J. Influence of degradation behavior of coexisting chlorobenzene congeners pentachlorobenzene, 1,2,4,5-tetrachlorobenzene, and 1,2,4-trichlorobenzene on the anaerobic reductive dechlorination of hexachlorobenzene in dye plant contaminated soil. Water Air Soil Pollut. 2015;226:1–9. doi: 10.1007/s11270-015-2559-3. [DOI] [Google Scholar]
- Liang X, Howlett MR, Nelson JL, Grant G, Dworatzek S, Lacrampe-Couloume G, Zinder SH, Edwards EA, Sherwood Lollar B. Pathway-dependent isotope fractionation during aerobic and anaerobic degradation of monochlorobenzene and 1,2,4-trichlorobenzene. Environ Sci Technol. 2011;45:8321–8327. doi: 10.1021/es201224x. [DOI] [PubMed] [Google Scholar]
- Livermore JA, Jin YO, Arnseth RW, LePuil M, Mattes TE. Microbial community dynamics during acetate biostimulation of RDX-contaminated groundwater. Environ Sci Technol. 2013;47:7672–7678. doi: 10.1021/es4012788. [DOI] [PubMed] [Google Scholar]
- Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol. 2013;63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP (2011) Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100 [DOI] [PubMed]
- Macbeth TW, Cummings DE, Spring S, Petzke LM, Sorenson KS. Molecular characterization of a dechlorinating community resulting from in situ biostimulation in a trichloroethene-contaminated deep, fractured basalt aquifer and comparison to a derivative laboratory culture. Appl Environ Microbiol. 2004;70:7329–7341. doi: 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm H, Howe P, Dobson S (2004) Chlorobenzenes other than hexachlorobenzene: environmental aspects. Concise International Chemical Assessment Document 60. World Health Organization, Geneva
- Maphosa F, van Passel MWJ, de Vos WM, Smidt H. Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ Microbiol Rep. 2012;4:604–616. doi: 10.1111/j.1758-2229.2012.00376.x. [DOI] [PubMed] [Google Scholar]
- Marco-Urrea E, Nijenhuis I, Adrian L. Transformation and carbon isotope fractionation of tetra- and trichloroethene to trans-dichloroethene by Dehalococcoides sp. strain CBDB1. Environ Sci Technol. 2011;45:1555–1562. doi: 10.1021/es1023459. [DOI] [PubMed] [Google Scholar]
- Masunaga S, Susarla S, Yonezawa Y. Dechlorination of chlorobenzenes in anaerobic estuarine sediment. Wat Sci Tech. 1996;33:173–180. doi: 10.1016/0273-1223(96)00290-9. [DOI] [Google Scholar]
- Men Y, Lee PK, Harding KC, Alvarez-Cohen L. Characterization of four TCE-dechlorinating microbial enrichments grown with different cobalamin stress and methanogenic conditions. Appl Microbiol Biotechnol. 2013;97:6439–6450. doi: 10.1007/s00253-013-4896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlino G, Balloi A, Marzorati M, Mapelli F, Rizzi A, Lavazza D, de Ferra F, Carpani G, Daffonchio D (2015) Diverse reductive dehalogenases are associated with Clostridiales-enriched microcosms dechlorinating 1, 2-dichloroethane. BioMed Res Int 2015:242856 [DOI] [PMC free article] [PubMed]
- Middeldorp P, De Wolf J, Zehnder A, Schraa G. Enrichment and properties of a 1,2,4-trichlorobenzene-dechlorinating methanogenic microbial consortium. Appl Environ Microbiol. 1997;63:1225–1229. doi: 10.1128/aem.63.4.1225-1229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JL, Fung JM, Cadillo-Quiroz H, Cheng X, Zinder SH. A role for Dehalobacter spp. in the reductive dehalogenation of dichlorobenzenes and monochlorobenzene. Environ Sci Technol. 2011;45:6806–6813. doi: 10.1021/es200480k. [DOI] [PubMed] [Google Scholar]
- Nelson JL, Jiang J, Zinder SH. Dehalogenation of chlorobenzenes, dichlorotoluene, and tetrachloroethene by three Dehalobacter spp. Environ Sci Technol. 2014;48:3776–3782. doi: 10.1021/es4044769. [DOI] [PubMed] [Google Scholar]
- Nowak J, Kirsch N, Hegemann W, Stan H-J. Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture enriched from Saale river sediment. Appl Microbiol Biotechnol. 1996;45:700–709. doi: 10.1007/s002530050751. [DOI] [Google Scholar]
- Pavlostathis SG, Prytula MT. Kinetics of the sequential microbial reductive dechlorination of hexachlorobenzene. Environ Sci Technol. 2000;34:4001–4009. doi: 10.1021/es991214n. [DOI] [Google Scholar]
- Pérez-de-Mora A, Zila A, McMaster ML, Edwards EA. Bioremediation of chlorinated ethenes in fractured bedrock and associated changes in dechlorinating and nondechlorinating microbial populations. Environ Sci Technol. 2014;48:5770–5779. doi: 10.1021/es404122y. [DOI] [PubMed] [Google Scholar]
- Pöritz M, Schiffmann CL, Hause G, Heinemann U, Seifert J, Jehmlich N, von Bergen M, Nijenhuis I, Lechner U. Dehalococcoides mccartyi strain DCMB5 respires a broad spectrum of chlorinated aromatic compounds. Appl Environ Microbiol. 2015;81:587–596. doi: 10.1128/AEM.02597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman NA, Parks DH, Vanwonterghem I, Morrison M, Tyson GW, Hugenholtz P (2015) A phylogenomic analysis of the bacterial phylum Fibrobacteres. Front Microbiol 6:1469 [DOI] [PMC free article] [PubMed]
- Ramanand K, Balba M, Duffy J. Reductive dehalogenation of chlorinated benzenes and toluenes under methanogenic conditions. Appl Environ Microbiol. 1993;59:3266–3272. doi: 10.1128/aem.59.10.3266-3272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro-Garcia J, Hermes G, Giatsis C, Sipkema D, Zoetendal EG, Schaap PJ, Smidt H. NG-tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Res. 2016;5:1791. doi: 10.12688/f1000research.9227.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen Z, Yanovich Y, Goldin R, Adar E. Metabolism of the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in a contaminated vadose zone. Chemosphere. 2008;73:1492–1498. doi: 10.1016/j.chemosphere.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Rupakula A, Lu Y, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J. Functional genomics of corrinoid starvation in the organohalide-respiring bacterium Dehalobacter restrictus strain PER-K23. Front Microbiol. 2015;5:751. doi: 10.3389/fmicb.2014.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind U, Haest PJ, Atashgahi S, Maphosa F, Hamonts K, Maesen M, Calderer M, Seuntjens P, Smidt H, Springael D, Dejonghe W. Kinetics of dechlorination by Dehalococcoides mccartyi using different carbon sources. J Contam Hydrol. 2014;157:25–36. doi: 10.1016/j.jconhyd.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Shivaji S, Ray M, Rao NS, Saisree L, Jagannadham M, Kumar GS, Reddy G, Bhargava PM. Sphingobacterium antarcticus sp. nov., a psychrotrophic bacterium from the soils of Schirmacher Oasis, Antarctica. Int J Syst Bacteriol. 1992;42:102–106. doi: 10.1099/00207713-42-1-102. [DOI] [Google Scholar]
- Sutton NB, Atashgahi S, Saccenti E, Grotenhuis T, Smidt H, Rijnaarts HH. Microbial community response of an organohalide respiring enrichment culture to permanganate oxidation. PLoS One. 2015;10:e0134615. doi: 10.1371/journal.pone.0134615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N, Heilig HG, Eekert V, Miriam H, Schraa G, De Vos WM, Smidt H. Concurrent hexachlorobenzene and chloroethene transformation by endogenous dechlorinating microorganisms in the Ebro River sediment. FEMS Microbiol Ecol. 2010;74:682–692. doi: 10.1111/j.1574-6941.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Taş N, van Eekert MH, Wagner A, Schraa G, de Vos WM, Smidt H. Role of “Dehalococcoides” spp. in the anaerobic transformation of hexachlorobenzene in European rivers. Appl Environ Microbiol. 2011;77:4437–4445. doi: 10.1128/AEM.01940-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doesburg W, Eekert MH, Middeldorp PJ, Balk M, Schraa G, Stams AJ. Reductive dechlorination of β-hexachlorocyclohexane (β-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol Ecol. 2005;54:87–95. doi: 10.1016/j.femsec.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Vandermeeren P, Herrmann S, Cichocka D, Busschaert P, Lievens B, Richnow H-H, Springael D (2014) Diversity of dechlorination pathways and organohalide respiring bacteria in chlorobenzene dechlorinating enrichment cultures originating from river sludge. Biodegradation 25:1–20 [DOI] [PubMed]
- Villemur R, Lanthier M, Beaudet R, Lépine F. The Desulfitobacterium genus. FEMS Microbiol Rev. 2006;30:706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Lu Y, Han J, Luo W, Shi Y, Wang T, Sun Y. Hexachlorobenzene sources, levels and human exposure in the environment of China. Environ Int. 2010;36:122–130. doi: 10.1016/j.envint.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Wu Q, Milliken CE, Meier GP, Watts JE, Sowers KR, May HD. Dechlorination of chlorobenzenes by a culture containing bacterium DF-1, a PCB dechlorinating microorganism. Environ Sci Technol. 2002;36:3290–3294. doi: 10.1021/es0158612. [DOI] [PubMed] [Google Scholar]
- Yonezawa Y, Fukui M, Masunaga S, Urushigawa Y. Dechlorination of 1, 2, 4-trichlorobenzene in the sediment of Ise Bay. Chemosphere. 1994;28:2179–2184. doi: 10.1016/0045-6535(94)90185-6. [DOI] [Google Scholar]
- Zanaroli G, Negroni A, Häggblom MM, Fava F. Microbial dehalogenation of organohalides in marine and estuarine environments. Curr Opin Biotech. 2015;33:287–295. doi: 10.1016/j.copbio.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Zhang C, Suzuki D, Li Z, Ye L, Katayama A. Polyphasic characterization of two microbial consortia with wide dechlorination spectra for chlorophenols. J Biosci Bioeng. 2012;114:512–517. doi: 10.1016/j.jbiosc.2012.05.025. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hu M, Li P, Wang X, Meng Q (2015) Trichloroethylene removal and bacterial variations in the up-flow anaerobic sludge blanket reactor in response to temperature shifts. Appl Microbiol Biotechnol 99:1–12 [DOI] [PubMed]
- Zhou X, Zhang C, Zhang D, Awata T, Xiao Z, Yang Q, Katayama A. Polyphasic characterization of an anaerobic hexachlorobenzene-dechlorinating microbial consortium with a wide dechlorination spectrum for chlorobenzenes. J Biosci Bioeng. 2015;120:62–68. doi: 10.1016/j.jbiosc.2014.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 201 kb)