Abstract
The regulation and timing of spindle pole body (SPB) duplication and maturation in fission yeast was examined by transmission electron microscopy. When cells are arrested at G1 by nitrogen starvation, the SPB is unduplicated. On release from G1, the SPBs were duplicated after 1–2 h. In cells arrested at S by hydroxyurea, SPBs are duplicated but not mature. In G1 arrest/release experiments with cdc2.33 cells at the restrictive temperature, SPBs remained single, whereas in cells at the permissive temperature, SPBs were duplicated. In cdc10 mutant cells, the SPBs seem not only to be duplicated but also to undergo partial maturation, including invagination of the nuclear envelope underneath the SPB. There may be an S-phase–specific inhibitor of SPB maturation whose expression is under control of cdc10+. This model was examined by induction of overreplication of the genome by overexpression of rum1p or cdc18p. In cdc18p-overexpressing cells, the SPBs are duplicated but not mature, suggesting that cdc18p is one component of this feedback mechanism. In contrast, cells overexpressing rum1p have large, deformed SPBs accompanied by other features of maturation and duplication. We propose a feedback mechanism for maturation of the SPB that is coupled with exit from S to trigger morphological changes.
INTRODUCTION
The duplication of the centrosome, like replication of DNA, is under precise cell cycle control and occurs once in each eukaryotic cell cycle (Kallenbach and Mazia, 1982). Centrosome duplication occurs at the G1/S boundary in the cells of many organisms, although splitting of the centrosome complex, for example, as monitored by the separation of the centrioles that are embedded in the centrosome in animal cells, does not occur until later in the cell cycle (Hinchcliffe and Sluder, 2001). Recent studies of centrosome duplication in mammalian tissue culture cells or in Xenopus egg extracts demonstrate the centrosome duplication is driven by the cdk2–cyclin E complex (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999). In budding yeast cells, duplication of the spindle pole body (SPB), the yeast equivalent of the centrosome, is under the control of Cdc28/Clns complex, the G1 cyclins, and occurs late in G1 and spindle formation occurs in S phase (Haase et al., 2001).
The reported cycle of SPB duplication and maturation in the fission yeast Schizosaccharomyces pombe differs from that in budding yeast and other organisms. The fission yeast SPB, a laminar body, spends most of interphase in the cytoplasm adjacent to the nuclear envelope. After the formation of a half bridge a second laminar body forms adjacent to the first. As the SPB matures, osmophilic material accumulates in a pocket in the nuclear envelope that forms as the nuclear envelope invaginates. Subsequently, as the cell enters mitosis the two laminar bodies separate as the mitotic spindle forms, giving rise to a bipolar spindle (Ding et al., 1997; Tanaka et al., 2000). The timing of SPB duplication in the fission yeast cell cycle is controversial. Although the latest reports place duplication, maturation, and separation at late G2 (Ding et al., 1997), earlier reports suggest that duplication occurs upon entry into mitosis (McCully and Robinow, 1971) or anaphase (Kanbe et al., 1990) or that it is independent of the DNA replication cycle (King et al., 1982).
We wanted to reexamine the timing of SPB duplication in fission yeast because it is the only organism that is reported to initiate centrosome duplication at a point in the cell cycle other than G1/S. Although we have shown in permeabilized cells that the SPB becomes competent to nucleate microtubules at G2/M (Masuda et al., 1992), it is possible that other changes in SPB structure and function, including SPB duplication occur earlier in the cell cycle. For example, two fission yeast SPB components, alp4p and alp6p, which are homologous to the γ-tubulin binding proteins, Sc. Spc97/h.GCP2 and Sc. Spc98/h.GCP3, respectively, have an essential role that is required earlier in the cell cycle than M, i.e., during G1 (Vardy and Toda, 2000). Their essential function may be associated with a step in the duplication of the SPB. Different steps in duplication and maturation of the Drosophila centrosome happen at different stages in the cell cycle (Vidwans et al., 1999). A similar cell cycle-dependent separation of SPB duplication and maturation may occur during the fission yeast cell cycle.
To accurately place the events of fission yeast duplication and maturation in the cell cycle, we have monitored changes in SPB morphology in high-pressure fast frozen, freeze-substituted cells followed either by serial thin section analysis or tomography. Various cell cycle arrest and release techniques such as nitrogen starvation/release and hydroyxurea treatment allowed us to determine whether SPB duplication is associated with crossing the G1/S boundary. The large collection of cell cycle arrest mutants gave us further insight into the cell cycle-dependent control of SPB duplication and maturation. We find that duplication of the SPB occurs at the G1/S boundary and that maturation of the SPB occurs later in the cell cycle and requires exit from S.
MATERIALS AND METHODS
Yeast Strains and Media
The cells were cultured in rich medium (YES) for drug treatment and for temperature arrest of ts– mutants. For G1 arrest, the cells were cultured in minimal medium (PM) and then starved for nitrogen and carbon sources with PM-ND (PM without ammonium chloride or dextrose) (Horie et al., 1998). The wild-type strain used was 972L(h–). For G1 arrest and release by nitrogen starvation experiments, autotroph strains were used. Cells were cultured at 25°C unless otherwise stated. Temperature-sensitive strains cdc2.33 (h–) and cdc10.v50 (h–) were a kind gift of Paul Nurse (Imperial Cancer Research Fund, London, United Kingdom). The integrant strains of cdc18+ and rum1+ genes under nmt1+ promoter also were obtained from Paul Nurse. The strain FY1166 h+ mcm4::[mcm4HA::leu1+] ura4-D18 leu1–32 ade6-M210 used in the in situ mcm4p binding assay was a kind gift of Susan Forsburg (University of Southern California).
Electron Microscopy
The cells were high-pressure frozen in a Bal-tec high-pressure freezer as described previously (Ding et al., 1997). The frozen cells were substituted in acetone with 2% osmium oxide and 0.1% uranyl acetate for 3 d at –90°C and then warmed to 20°C at 10°C/h. The cells were embedded in Epon 812 resin, sectioned at a thickness of ∼40–50 nm, and poststained in lead citrate and uranyl acetate. The pictures of SPBs were taken with either a JEOL 100CX transmission electron microscope at 80 keV, a JEOL 1200EX at 100 keV, or a Philips TECNAI 12 at 100 keV. For each SPB found, serial sections were followed, including two sections outside the ends of the SPB in both directions. For each section, a low-magnification image for the entire cell (3300–16,000×) and a close-up image of the SPB (26,000–50,000×) were taken. For selected samples, the stage was tilted to obtain images at different angles for a clearer view of the internal structure of the SPB. The SPBs were scored as single or duplicated judging from their shape. All of the SPBs without complete lamellae were scored as single, including all the putative duplication intermediates. With only a few exceptions, >10 complete serial section images of SPBs were taken for each time point or condition in each experiment. The total number of electron microscope (EM) negatives used in this study is >1800.
EM Tomography
EM tomographic reconstruction was performed as described previously (Braunfeld et al., 1994). Briefly, after fixation, processing, and embedding in Epon 812, cells were sectioned to a thickness of ∼0.4 μm and placed on a 50 × 200 mesh grid coated with Formvar. The sections were scanned for SPBs on either Philips TECNAI 12 electron microscope at 100 keV or a Philips EM430 at 300 keV. The position of SPBs was recorded for later reference. The scanned grids were then treated with poly-l-lysine followed by coating with 15-nm gold beads. The poly-l-lysine treatment and gold bead coating were repeated until sufficient density of gold beads was obtained around the SPB and then the grids were carbon coated. The SPBs were preexposed for 15–20 min to minimize shrinkage during tilting. The tilt data sets were taken on a Philips CM430 automated data correction system at 300 keV. The system is fully automated with a Philips C400 interface and a SGI OCTANE workstation. Data were collected on a Gatan 676 cooled slow-scan charge-coupled device camera with 2× binning (512 × 512 pixels) at a magnification of 21,200× (1.68 nm/pixel) with a tilting angle ranging from –70 to +70° at 1.25° intervals except for a few samples for which a high tilting angle >60° was not available in one tilting direction. Each data set was processed for reconstruction by using alignment of gold beads on the surface of the section, mass normalization, and calculation of the tomographic alternating projection iterative reconstruction in Priism software. The reconstructed images were displayed, analyzed, and modeled with Priism or DeltaVision software (Chen et al., 1996).
Cell Cycle Arrest and Release
For G1 arrest experiments with nitrogen starvation, logarithmic cultures were centrifuged and cells were washed three times in PM-ND. Then, the cells were cultured in PM-ND for 2–2.5 h followed by addition of glucose to a final concentration of 2% (Horie et al., 1998). The culture was subsequently incubated for 6–7.5 h and was subjected to high-pressure freezing. The arrest was monitored with FACscan analysis for DNA content according to the literature (Moreno and Nurse, 1994; Labib et al., 1995). For arrest/release experiments, G1 arrested cultures were refed with ammonium chloride and cultured for an hour before high-pressure freezing. For cdc2.33 strain with G1 arrest/release experiment, the G1 arrested culture cdc2.33 strain was split into two halves, and one was shifted up to the nonpermissive temperature (35°C) and the other was kept at the permissive temperature (25°C). Ten minutes later, prewarmed nitrogen source (ammonium chloride, final 1%) was added to both cultures. The fixation of cells was started between 60 and 90 min after the addition of nitrogen source and took 15–20 min for one culture condition typically. For hydroxyurea (HU) treatment, the cells were cultured in YES to mid-log phase and then HU was added to 10 mM, and cells were further incubated for 3.5 h followed by high-pressure freezing. The cdc10.v50 strain was cultured in YES to mid-log phase and then shifted up the nonpermissive temperature (35°C) for 3.5 h before fixation. Overexpression of rum1p or cdc18p under the control of the nmt1+ promoter was done according to Moreno and Nurse (1994) or Nishitani and Nurse (1995). Cells were cultured in YES to mid-log phase. The cells were washed three times in PM followed by diluting the culture in PM for 30-fold. The cells were then cultured for 17 h before the fixation to allow overexpression of the proteins by depletion of thiamine. Most of the cells in these populations had more than 4C DNA content as monitored by flow cytometry.
Fluorescence-activated cell sorting (FACS) analysis of the DNA content of cell populations in cell cycle arrest/release experiments were done according to Moreno et al. (Moreno and Nurse, 1994; Labib et al., 1995). The DNA content of cells stained with SYTOX Green (Molecular Probes, Eugene, OR) were analyzed using a Beckman Coulter EPICS XL-MCL flow cytometer at the Flow Cytometry Facility (Cancer Research Laboratory, University of California, Berkeley, CA). To confirm the stage of cell cycle arrest of the various populations of cells analyzed by flow cytometry, we used an in situ chromatin binding assay developed by Kearsey et al. (2000) as modified by Gomez and Forsburg (2004), to study the cell cycle-dependent binding of mcm4p to chromatin in individual cells. As described previously, after Zymolyase 20T treatment, cells were washed with low (0.025%) or high (1%) Triton X-100 to determine whether HA-tagged mcm4p was retained in the nucleus bound to chromatin after high-detergent treatment. Cells were examined by indirect immunofluoresence and photographed using the same exposure times.
RESULTS
Serial Section EM Analysis Is Required to Determine Whether a SPB Is Duplicated
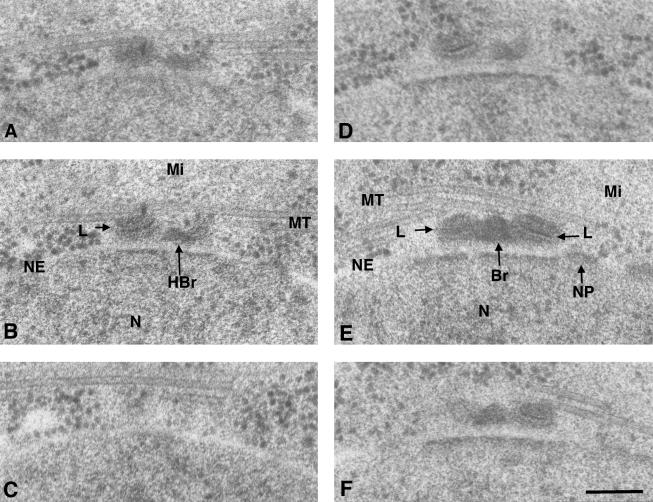
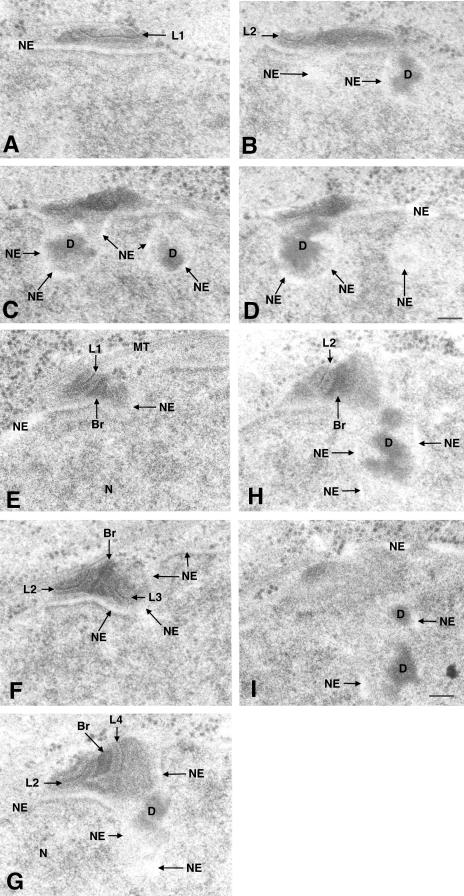
Our criteria for identifying SPBs at different stages of duplication were based on changes in SPB morphology and required a complete set of serial sections through the SPB to distinguish unduplicated from duplicated SPB. As shown in Figure 1, A–C, unduplicated SPBs are a single laminated body with a small well-stained appendage. This appendage was named the half bridge by Ding et al. (1997), because of the structural resemblance to the half bridges found in unduplicated SPBs of Saccharomyces cerevisiae. After duplication, the SPB is two laminated bodies interconnected by a densely stained pyramid shaped bridge (Figure 1, D–F). However, this structure does not show the morphological changes associated with maturation. During maturation, the nuclear envelope becomes invaginated, and dark staining material accumulates in the pocket that forms beneath the duplicated SPB (see Figure 5 for an example of a maturing SPB). Subsequently, the two daughter SPBs are separated by microtubules as a bipolar mitotic spindle is formed between them (Ding et al., 1997). For clarity of analysis, we have divided SPB maturation into two stages, e.g., early and late maturation. Early maturation is marked by growth in size of the lamellae bodies, invagination of the nuclear envelope, and accumulation of darkly stained material between the nuclear envelope and the SPB, whereas late maturation is marked by the physical separation of the two daughter SPBs and formation of the mitotic spindle.
Figure 1.
Serial sections through two SPBs comparing the structure of an unduplicated (A–C) versus a duplicated but immature (D–F) SPB. In A–C, the nitrogen-starved cell is arrested in early G1. The SPB in this cell consists of a single laminar structure (L) and a half bridge (HBr), which lies adjacent to an intact NE. In D–F, the cdc10-arrested cell at the nonpermissive temperature is arrested at the G1/S boundary. The SPB is duplicated and has two laminar structures separated by a dark staining elipsoid bridge (Br). In both cells, the nuclear envelope is continuous and unfenestrated and shows no signs of invagination, although dark material has accumulated on the nuclear but not the cytoplasmic face of the nuclear envelope adjacent to the SPB. Several microtubules (MT) are in proximity to the cytoplasmic face of the SPB in each cell, accompanied by mitochondria. A nuclear pore (NP) is always found near the SPB. Note the image of the unduplicated SPB in section B superficially resembles the image of the duplicated but immature SPB in section D. The size of each linear structure is similar (ranging from 70 to 90 nm) in unduplicated and duplicated SPBs. Bar, 100 nm.
Figure 5.
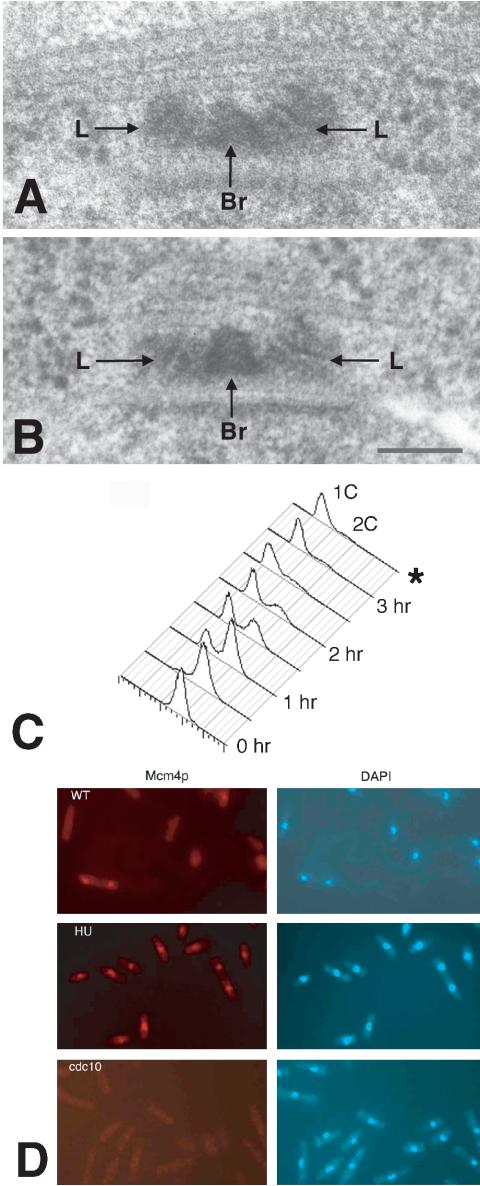
(A) 3D tomographic reconstruction of a SPB in a cdc10-arrested cell. The 3D volume of the tomographic reconstruction is shown as consecutive two-dimensional projections (A–L). Each projection is 16 slices, which is the equivalent of an image of a serial section 27 nm in thickness. The SPB shows invagination of the NE, accumulation of dark material in the membrane pocket (D), microtubule (MT) in the cytoplasm associated with SPB, and expansion in overall size of the SPB (L, laminar structure). The invaginated nuclear envelope is continuous and is not fenestrated. Bar, 100 nm. A movie of the whole volume can be found in Supplemental Data. (B) Schematic DNA histograms of cells after switching cdc10 cells to the nonpermissive temperature (35°C) at time 0, as determined by flow cytometry. The first peak represents cells with 1C DNA content; the second peak cells with 2C DNA content. At zero time, almost all cells had 2C DNA content, but by 3 h almost all cells had 1C DNA content. Cells were fixed for electron microscopy after 3.5 h as indicated by the *.
Because of the contradictory literature and the difficulty in analyzing SPB morphology as it duplicates and matures, a large data set consisting of >120 complete sets of serial sections containing SPBs were obtained and analyzed for this study. When the lamellae were not clearly visible in the sections, the sections were examined with various tilting angles, and images were obtained at the best tilting angle for visualization of the lamellae. We examined complete serial sections of SPBs running from two sections outside of the boundary of the SPB and scored the SPBs as single or duplicated only when the complete sets of sections were recorded on film. Any ambiguous SPBs, including putative duplication intermediates, were scored as duplicated. In addition, three tomographic reconstructions of SPBs at different stages of duplication were made. As shown in Figure 1, the images of an unduplicated (section B) and a duplicated but immature (section D) SPB can seem similar when comparing two single sections, but they can be distinguished from each other in serial section analysis.
The SPB Is Duplicated in Cells Arrested at S Phase by HU
Although Ding et al. (1997) concluded that the SPB undergoes duplication and maturation at the G2/M boundary, other accounts in the literature (Vardy and Toda, 2000; Garcia et al., 2001) and our own observations of SPB morphology in log phase culture (S.U., unpublished data) suggest that SPB duplication was initiated at G1-S phase. To further study the timing of SPB duplication, we examined the morphology of SPBs in cells arrested at various stages of the cell cycle around the G1-to-S phase transition. First, we examined the SPB morphology in cells arrested at S phase by hydroxyurea. HU is an inhibitor of ribonucleotide reductase (RNR) and delays the progression of S phase by depleting the dNTP pool required for efficient DNA synthesis (Kim and Huberman, 2001). Figure 2, A and B, shows SPBs from wild-type cells treated with HU for 3.5 h at 25°C. All 11 SPBs scored were duplicated according to our criteria. It is striking that the most of the SPBs found in HU-treated cells are uniform in shape and length as shown in Figure 2. SPBs contained a pair of lamellar bodies; one was smaller and at an angle relative to the nuclear envelope, and the other larger and slightly more extended. They are interconnected by a pyramid-shaped bridge that is on top of a continuous nuclear envelope. The nuclear envelope beneath the SPB was straight and closely oppressed to the SPB. Electron-dense material was found in the nucleus adjacent to the nuclear envelope beneath the SPB at the site where the centromere cluster is located (Tanaka and Kanbe, 1986; Nishimoto et al., 1992; Uzawa and Yanagida, 1992; Funabiki et al., 1993; Kniola et al., 2001). Because all SPB scored had a duplicated SPB, we conclude that the SPB duplication is completed before or during S phase. Moreover, because 100% of the cells have duplicated SPBs, it is not possible that SPB duplication in HU-arrested cells is actually a later cell cycle event unaffected by HU treatment because the cell population is variable in the length of time spent arrested in S. If the HU-treated cells had progressed further in the cell cycle with respect to SPB duplication, then only a fraction of the cells would have had duplicated SPBs.
Figure 2.
(A and B) The images of the duplicated but immature SPBs in two hydroxurea-treated cells arrested early in S. The SPB consists of two laminar structures (L) separated by a bridge (Br). For both examples, only the center section is presented. The SPB has not undergone maturation as shown by the small size of the laminar structures (50–60 nm) and the absence of any nuclear envelope invagination or fenestration. The laminations of one of the two structures (left-hand side) lie oblique to the nuclear envelope relative to the other laminated structure. Due to the angle and the size of the smaller laminar body, it was essential to examine images of tilted specimen in series to find it reliably. Bar, 100 nm. (C) Schematic DNA histograms of cells arrested early in S after addition of hydroxyurea at time 0 as determined by flow cytometry. The first peak represents cells with 1C DNA content; the second peak cells with 2C DNA content. At zero time, almost all cells had 2C DNA content but by 3 h posthydroxyurea addition all cells had 1C DNA content. Cells were fixed for electron microscopy after 3.5 h in hydroxyurea as indicated by the *. (D) In situ chromatin binding assay for HA-tagged mcm4p proteins in cells permeabilized with 1.0% Triton X-100. HA-labeled proteins were visualized in left panels (red) by using HA antibody and a secondary conjugated to Cys3 and right (blue) panels display 4,6-diamidino-2-phenylindole staining of DNA in the same cells. In the wild-type population (top), most cells are in G2 and do not retain mcm4p in the nucleus after permeabilization. The only cell that retained mcm4p protein is a binucleate cell at the G1/S boundary that has not completed cytokinesis (lower left). After 3.5-h treatment with hydroxurea, all cells (middle) have retained mcm4p after permeabilization. No mcm4p protein was retained in the nuclei of cdc10-arrested cells grown for 3.5 h at the nonpermissive temperature (bottom).
As shown by flow cytometry (Figure 2C), cells fixed after 3.5 h in HU for electron microscopy were arrested in early S phase with unreplicated DNA. At the beginning of S phase, the mcm4p/mcm6p complex is bound to chromatin and is then displaced from chromatin as DNA replication occurs and becomes soluble in the nucleus (Kearsey et al., 2000). As shown in Figure 2D by using an in situ binding assay for chromatin bound mcm4p, HA-tagged mcm4p is retained in the nucleus after high-detergent treatment in the HU-treated cells. This result, consistent with that shown previously by Kearsey et al. (2000), confirms that SPB duplication can occur in cells blocked in DNA replication during early S phase. By way of contrast in the wild-type, non-HU–treated population where most cells are in G2, the only cells observed that have retained mcm4p after detergent treatment are binucleate cells undergoing septation that are at the G1/S boundary.
The SPB Is Not Duplicated in Cells Arrested at G1 Phase by Nitrogen Starvation
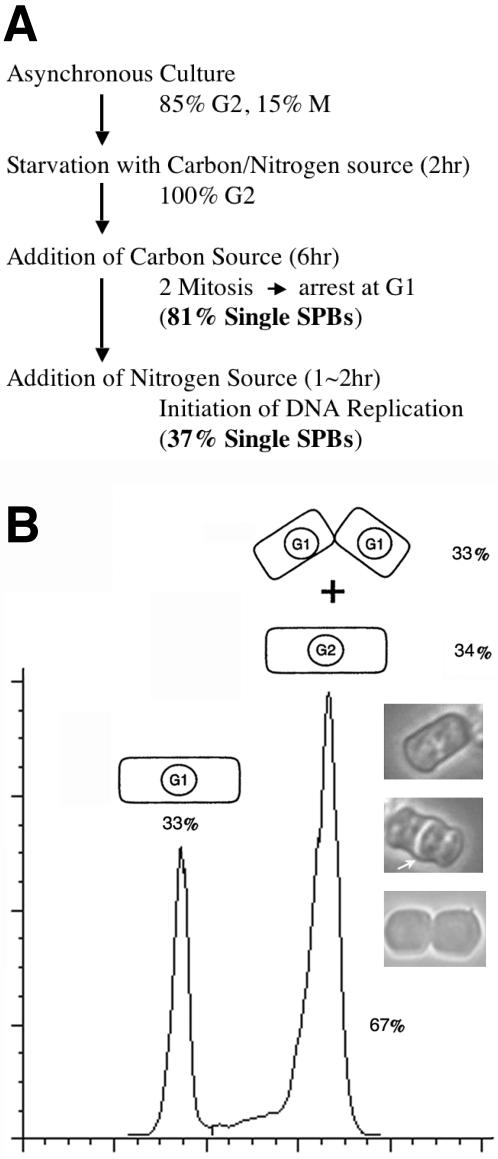
To further study the timing of SPB duplication, we examined the morphology of SPBs in cells arrested at various stages of the cell cycle around the G1/S boundary. G1 arrest by nitrogen starvation is before cdc2 arrest and clearly is preStart. For an unknown reason, the conventional method for G1 arrest (nitrogen starvation for 16 h or more) yielded very poor quality specimens for EM. An alternative protocol developed by Shimoda (Horie et al., 1998) yielded cells that had well preserved ultrastructure. By this method, cells in log phase are arrested at G2 by starving with both nitrogen and carbon sources (Figure 3A). On addition of a carbon source, the cells undergo two rapid successive cell divisions and then arrest at G1. Although many cells in this population are multiply septated and daughters have not separated (Figure 3B), only cells lacking a septum were used for EM. Flow cytometry of the arrested population combined together with a septation index suggested that >66% of the cells were arrested in G1 by this nitrogen starvation protocol (Figure 3B). A single laminated structure with two layers and a half bridge (Figure 1, A–C) was found in most of the cells examined (81%; 13/16), demonstrating that the SPB is not duplicated in cells arrested at G1. The nuclear membrane running under the SPB seems to be straight and continuous. The nuclear membrane had no indication of early maturation events, such as fenestration or invagination adjacent to the SPB. A bundle of cytoplasmic microtubules running parallel to the longitudinal axis of the cells were found at the cytoplasmic side of SPBs. Four to seven microtubules were usually found within the bundle; a number that matches our live data analysis for cytoplasmic microtubule behavior (Sagolla et al., 2003). Mitochondria were always associated with the microtubule bundles near the SPB and extended most of the length of the cytoplasmic bundle. This observation is consistent with genetic studies that demonstrate that microtubules are involved in mitochondrial partitioning to daughter cells during cell division (Yaffe et al., 1996). The three cells that had duplicated SPBs may be the result of an incomplete arrest by nitrogen starvation as suggested by flow cytometry (Figure 3B). This observation clearly demonstrates that the SPB is not duplicated at an early G1 arrest point.
Figure 3.
(A) Flow diagram shows the method used for G1 arrest in this study (Horie et al., 1998). The cells cultured in rich medium were transferred to a synthetic medium lacking both carbon source and nitrogen source (PM-ND). The cells were cultured for 2 h to allow them to arrest in G2 phase followed by addition of carbon source (2% dextrose final). After two rapid sequential divisions, cells arrest at G1. For arrest/release experiments, nitrogen source (ammonium chloride, 1% final) was added back. The percentage of unduplicated versus duplicated SPBs is shown in bold in parentheses. (B) Schematic DNA histogram of cells arrested in G1 as determined by flow cytometry. The square represents the cells where the cell cycle phase is indicated. The highest peak represents cells with 2C DNA content, including G1 cells with incomplete separation after septation, and G2 cells. The percentage of cells that have not separated is 33%. Therefore, 66% of the cells were in G1. The lesser, left-hand peak (33%) is the cells that have 1C DNA content and have undergone septation and separation. The phase micrographs show three cells from the 2C peak; the top cell has 2C DNA, and the middle and bottom binucleate cells have 1C nuclear DNA content. The lower two cells have formed septa (arrow), and the bottom cell shows partial separation of the two daughters. The bar to the right of the histograms indicates times when cells were fixed for electron microscopy.
SPB Duplicates upon Release from Nitrogen Starvation
To determine whether SPBs are duplicated at a stage in the cell cycle that falls between arrest by nitrogen starvation and HU arrest, we investigated the timing of SPB duplication by releasing cells from nitrogen starvation and fixing the cells shortly after release. The method of starvation and release is shown in the flow diagram in Figure 3A (Horie et al., 1998). One to 1.5 h after the addition of a nitrogen source, when cells are at the onset of S phase or in early S phase, as shown by flow cytometry (Figure 4B), the cells were fixed and the SPB morphology was examined. Where possible, only cells that had no septa were examined. The flow cytometry data suggest that these cells at 25°C have not completed DNA replication. Of the eight cells examined, five cells had duplicated SPBs (63%) in contrast to the 81% that had single SPBs in nitrogen-starved G1-arrested cells (Figure 1, A–C). Two of the three cells scored as having unduplicated SPBs contained an unknown structure associated with the half bridge, which may be a duplication intermediate (our unpublished data). None showed any signs of maturation as defined previously. Nor had any of the cells entered mitosis. This result, together with the results of the HU arrest experiment, strongly suggests that the duplication of the SPB occurs at the G1/S boundary instead of at the G2/M boundary.
Figure 4.
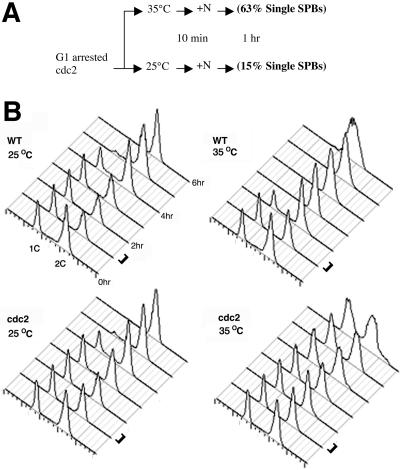
(A) Flow diagram showing the procedure used in the cdc2 arrest/release experiments. One hour after the release from G1 arrest, either at the permissive temperature or restrictive temperature, the cells were fixed and examined. At the nonpermissive temperature for cdc2 (35°C), 83% of the cells had unduplicated SPBs, but at the permissive temperature (25°C) only 13% of the cells examined had unduplicated SPBs. (B) Cell cycle arrest and release were analyzed by flow cytometry. Samples were taken of the cells after release from G1 arrest for FACS analysis at the times indicated. The bar to the right of the histograms indicates times when cells were fixed for electron microscopy. DNA histograms are shown for WT and cdc2.33 strains at permissive temperature (25°C) and nonpermissive temperature (35°C). These populations were grown in parallel. The highest peak is cells with 2C DNA content, including G1 cells that have undergone septation but not separated, and G2 cells. The percentage of cells that are incompletely separated was 25% for cdc2.33 cells and 33% for the wild-type control. The left-hand peak is the cells that have 1C DNA content and have undergone both septation and separation. In wild-type cells, the G1 peak decreases over time; the largest changes are observed at 35°C. In the cdc2.33 strain, there is no change in the G1 peak at the nonpermissive temperature (35°C) and only about a 10% decrease in the G1 peak at the permissive temperature (25°C) at 2 h. At 0 h of cell release by addition of the nitrogen source, the percentage of cdc2.33 G1 cells that have undergone both septation and separation is 35% and the percentage of G1 cells that have undergone septation but not separation is 25% (also see Figure 3B). The total percentage of cdc2.33 cells in G1 before cell release (0 h) is 60%, and the percentage of G2 cells is 40%.
The Duplication of SPB Is Dependent on cdc2p Kinase at the G1/S Boundary
To investigate the role of cdc2p kinase during SPB duplication, a nitrogen starvation arrest/release experiment was carried out with a temperature-sensitive allele of cdc2+ gene. Wild-type cdc2p kinase activity is required both for entrance into S and into M. Normally, if a log phase population of cdc2.33 cells were shifted to the nonpermissive temperature, almost all cells arrest at the G2/M boundary because most cells in a log phase population are in G2 (King and Hyams, 1982). In this experiment, cells were arrested at G1 by nitrogen starvation and then released from arrest by addition of the nitrogen source. The population of nitrogen-starved, G1-arrested cdc2.33 cells was divided into two cultures, and one-half was shifted up to 35°C, the nonpermissive temperature (Figure 4A, diagram). Ten minutes later, the G1 arrest was released by addition of a prewarmed nitrogen source to both cultures. The cells were fixed 1–1.5 h after the addition of the nitrogen source (Figure 4B, bar), as in the nitrogen arrest/release experiment. At the nonpermissive temperature, 83% (10/12) of cells had a single, unduplicated SPB. In contrast, 13% of the cells (2/16 cells) at the permissive temperature (25°C) had unduplicated SPBs. These results clearly demonstrate that the duplication of the SPB occurs at the G1/S boundary in fission yeast and is downstream of G1/S cdc2p kinase activity. As is shown by the FACS analysis of the cdc2.33 populations of cells (Figure 4B), at 25°C the cells progressed through the cell cycle with kinetics similar to wild-type cells at the same temperature, whereas at 35°C after being released from nitrogen starvation the cdc2.33 cells remain arrested at the G1/S boundary with unreplicated DNA.
The SPB Is Duplicated and Undergoes Early Maturation in cdc10-arrested Cells
To further characterize regulation of SPB duplication timing, we investigated the morphology of SPBs in G1-arrested cells with the cdc10.v50 ts– mutation, which arrested at a step in the cell cycle downstream from cdc2p kinase but earlier than HU arrest. cdc10p is a component of (MluI binding factor transcription complex, which regulates the transcription of genes required for S phase, including the large subunit of RNR and cdc18p (Tanaka and Okayama, 2000). cdc10p activity is regulated through phosphorylation and is a down-stream target of cdc2p/G1 cyclin complex. The execution point of the cdc10+ gene is commonly used to define Start in the fission yeast cell cycle (Nurse and Bissett, 1981)
As shown by flow cytometry, cdc10 cells after 3 h at the nonpermissive temperature are arrested with a 1C DNA content (Figure 5B). mcm4 p is not retained in the nucleus in permeabilized cells at the nonpermissive temperature (Figure 2D), presumably because it cannot be loaded onto chromosomes in the absence of cdc18p expression (Kearsey et al., 2000). Ogawa et al. (1999) have previously demonstrated that the mcm4p/mcm6p complex is not bound to chromatin in the absence of cdc10p. Cells were fixed for EM after 3.5 h at the nonpermissive temperature, at a time when all cells remain arrested at the G1/S boundary with 1C DNA content. The SPBs were found to be duplicated in cdc10-arrested cells as shown in Figure 1, D–F and Figure 5, A–L, and the Supplemental Movie. Two laminated bodies were found in 97% (26/27 cells) of the SPBs examined, demonstrating that they have already undergone duplication. This result shows that SPB duplication has happened before the initiation of S phase. The SPB scored as “single” in the cdc10-arrested culture had an appendage at the tip of half bridge, indicating that it is actually a duplication intermediate. Although to be consistent we scored this SPB as single, this unusual morphology suggests that none of the SPBs in cdc10-arrested cells possess a single SPB.
SPB morphology was heterogeneous in cdc10-arrested cells. We conducted a reconstruction of three-dimensional (3D) structure of SPBs in several cells by using EM tomography (Figure 5, A–L, and Supplemental Figure) (Braunfeld et al., 1994). We found duplicated SPBs with an unmodified nuclear membrane underneath them (Figure 1, D–F) duplicated SPBs with a small invagination in the nuclear membrane underneath the bridge (our unpublished data) or duplicated SPBs with a large invagination of the nuclear membrane and associated dark staining material underneath the SPB (Figure 5, A–L, and Supplemental Movie). The SPB shown in Figure 5 bears a close resemblance to the SPB morphology described by Ding et al. (1997) as a premitotic SPB. Our results demonstrate that after cdc10 arrest, duplication and subsequent early maturation of the SPB could be initiated without exiting from G1 or entering S phase. However, late-stage maturation, as defined by separation of the SPBs and nucleation of microtubules on the nucleoplasmic face of the SPB for the assembly of the mitotic spindle, was not observed.
Overexpression of cdc18p Is Sufficient to Block the Early Maturation
Cdc10p is a subunit of the transcription factor required for the progression through G1-S boundary. Because the block of early maturation is cdc10p dependent, it is natural to make the assumption that one of the cdc10+-regulated genes is responsible for the blockage of the early maturation. We, by accident, found that the overexpression of cdc18p, a protein whose expression is regulated by cdc10+, by itself can induce the blockage of early maturation. Overexpression (OE) of cdc18p is known to induce repeated initiations of DNA replication at various sites in the genome independent of cell cycle phase. These cells initiate DNA replication without going through a corresponding G1, G2, or M phase (Figure 6A, diagram) (Moreno and Nurse, 1994; Nishitani and Nurse, 1995). Cdc18p is directly involved in initiation of DNA replication (Nishitani and Nurse, 1995). It is a down-stream target of the res1p-cdc10p transcription activator (complex) and when overexpressed, there is repeated initiation of DNA replication bypassing a requirement for cdc10+ (Martín-Castellanos et al., 2000).
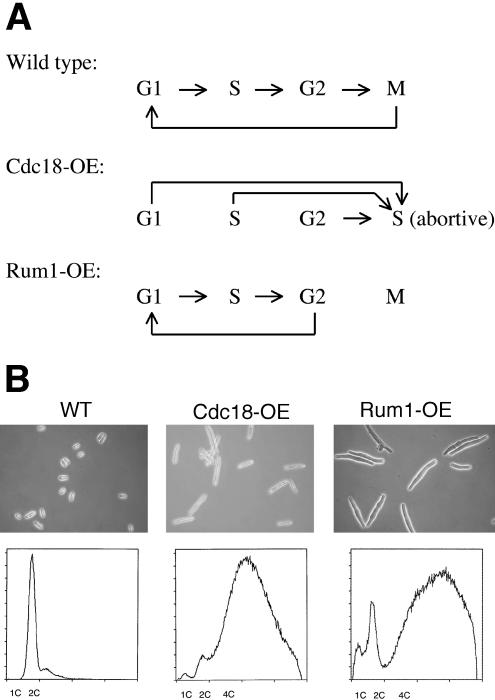
Figure 6.
(A) Diagram showing the impact of rum1 or cdc18p overexpression on the cell cycle (based on Moreno and Nurse, 1994; Nishitani and Nurse, 1995). When cdc18p is overexpressed, the cells start DNA replication regardless of their position in the cell cycle, and the cells remain in S. For cells overexpressing rum1p, cells in G1, S, or G2 continue to progress toward the G2/M boundary but skip M and reenter G1.(B) Phase micrographs of wt cells, cdc18p-overexpressing cells, and rum1p-overexpressing cells with schematic DNA histograms of the same cell populations as determined by flow cytometry. Wild-type cells have a 2C DNA content, whereas the other two populations of cells have the equivalent of 4C or greater DNA content.
We investigated SPB morphology in cells overexpressing cdc18p. The DNA content of these cells at the time of fixation for EM was roughly equivalent to 4C, and their morphology was abnormally long because they continued to grow without undergoing mitosis and cytokinesis (Figure 6B). As shown in Figure 7, cells overexpressing cdc18p have duplicated SPBs, but the SPBs have not undergone any maturation except for a minor fraction of the cells (2 of the 20 cells examined; out unpublished data). As in the case of cells treated with HU, SPB morphology is mostly uniform. The two cells that exhibited some signs of SPB maturation may already have initiated maturation during the induction period for cdc18p-OE. If the frequency of SPBs with early stage maturation in the cdc18p-OE population (10%) reflects the execution point for this process, then early stage maturation must be initiated very close to the boundary of the G2/M transition. This estimate is consistent with the previous observation that the maturation does not start until the later stages of G2 (Ding et al., 1997). All of the SPBs observed in cdc18p-OE cells seemed normal in terms of size and morphology, in contrast to the SPBs found in rum1p-OE cells (described below). The observation that most of the SPBs show no signs of maturation demonstrates that the block of early maturation during S phase is dependent on either the cdc18p or an S-phase event such as formation of replication forks.
Figure 7.
Serial sections of a SPB in a cell overexpressing cdc18p. The SPB has duplicated but shows no signs of maturation. The laminar bodies (L), separated by a bridge (Br), on the cytoplasmic face of the NE adjacent to a microtubule (MT), are the same size (50 nm) as those in HU-arrested cells. Bar, 100 nm.
Duplication and Early Maturation of SPB Can Occur without Preceding M Phase
We further investigated the requirement of previous cell cycle events for the initiation of SPB duplication. rum1p is an inhibitor of the mitotic form of cdc2p kinase and prevents mitosis from occurring prematurely. Overexpression of rum1p directs cells to go through G1, S, and G2 and reenter G1 before Start without going through M, and as a result cells end up with a higher DNA content (Figure 6A, diagram). As shown in Figure 6B, these cells have a 4C or greater DNA content and are abnormally long because mitosis and cytokinesis has not occurred. In cells overexpressing rum1p, overreplication of the genome is dependent on cdc2p kinase and the G1 cyclins pcl1p, cig1p, and partially cig2p and requires activation of the res2p–cdc10 p complex. (Fisher and Nurse, 1995; Martín-Castellanos and Moreno, 1996; Tanaka and Okayama, 2000). Rum1p-OE affects pre-Start events, whereas cdc18p-OE affects post-Start events but only with rum1p-OE does the cell actually leave S and enter G2. Analysis of SPB maturation in cells overexpressing rum1p allowed us to determine whether maturation requires a cdc10+-dependent passage through Start and exit from S.
The SPBs in cells (13 cells examined) overexpressing rum1p having gone through the equivalent of two or more S phases (Figure 6B) and have a morphology consistent not only with duplication but also with multiple early maturations (Figure 8). In the example shown in Figure 8, A–D, two pairs of laminated structures, one member of the pair more than twofold larger than the other in length, can be distinguished in the serial sections. The consistent size difference between the two lamellae bodies in each pair, observed in all cells examined, may reflect the difference between new and old SPBs as described by Grallert et al. (2004). However, SPB maturation, as determined by extent of membrane invagination and deposition of darkly staining material, was more variable in the cell population. For example, in a second cell (Figure 8, E–I), although the SPB has a similar morphology to that observed in Figure 8, A–D, several invaginations in the nuclear envelope are present beneath this complex structure. However, the morphology is also consistent with two rounds of duplication of the laminated structure, and several early maturations, because the laminated structures have not physically moved far apart, and the nuclear envelope (NE) seems to be intact. As shown by examining serial sections (our unpublished data), the lamellar bodies are discrete structures and the laminations in the two structures are at different angles with respect to each other. These results demonstrate that duplication of SPB and partial maturation of SPBs can occur even in the absence of a previous M phase, provided that cells enter S phase in a cdc2-dependent manner and leave S (Figure 6A, diagram). Also, these observations demonstrate that SPB duplication can occur in the absence of late-stage maturation or separation of the SPBs by mitosis.
Figure 8.
SPBs in two cells after rum1p overexpression. In A–D (the sections are not a continuous series), the SPB has undergone duplication and maturation. The nuclear envelope has invaginated, and several dense masses of dark staining material (D) have accumulated in the pockets in the NE beneath the SPB. Only one of the two pairs of laminated structures (labeled L1 and L2) is displayed, and it is accompanied by two discrete invaginations in the nuclear envelope beneath the SPB. The small one is comparable in size to ones found in wild-type, partially matured SPB (measured 80 nm) and lies at an oblique angle relative to its large partner. The larger ones were 230 nm in length. In serial sections E–I, part of another SPB in serial section is shown. In this cell, the nuclear envelope invagination is larger and the whole SPB is within it. The SPB consists of four laminated structures; two pairs, one large and one small, each labeled as L1 through L4. Careful examination of serial sections revealed that these laminated structures are not continuous but are separate structures. Two large laminated structures were connected by a bridge (Br), which has also grown in size (180 nm) compared with the ones found in other duplicated SPBs (70 nm). Bar, 100 nm.
DISCUSSION
In most organisms investigated to date, centrosomes are duplicated during S phase in parallel with the replication of the genomic DNA, and the process is initiated near the G1/S boundary. However, it has previously been reported that in fission yeast SPB duplication occurs at the G2/M boundary (Ding et al., 1997). We have investigated the cell cycle-dependent regulation and timing of SPB duplication in fission yeast by monitoring the morphology of SPBs in cells during cell cycle arrest/release experiments and summarize our results in Figure 9. We show that duplication of the SPB is initiated near the G1/S boundary and is under the regulation of cdc2p kinase but is independent of cdc10+. SPB maturation occurs in two phases; early maturation leading to NE invagination and deposition of material under the SPB requires exit from S, whereas late maturation, leading to fenestration of the NE and SPB separation as the spindle forms requires entrance into M. Expression of cdc18p is sufficient for inhibition of early maturation and highly likely to be required for it, too. We suggest that in S. pombe cells there is a feedback inhibition mechanism downstream from cdc10+ that delays early SPB maturation until S phase is complete (Figure 9B, model). To test this model we analyzed SPB duplication in cells with prolonged or repeated S phases (Figure 6A). Provided that cells never left S, maturation was inhibited. However, cells that repeatedly left and reentered S, even if they never transited M, underwent both duplication and maturation, although maturation was not complete. Our observation suggests that once the two major events of S phase, DNA replication and SPB duplication, are triggered by cdc2p/G1 cyclin complexes, they are independent of each other. A feedback control of SPB maturation may serve as a mechanism to coordinate these two independent pathways and prevent accidental spindle formation during S phase, an event which leads to catastrophic cell death.
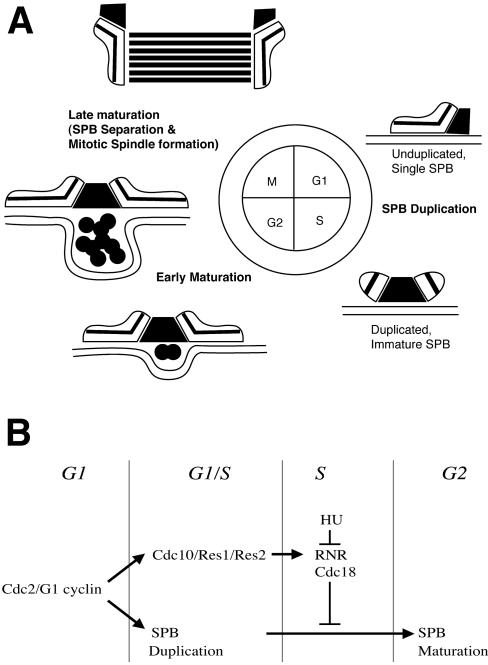
Figure 9.
Cartoon in A shows the changes in SPB morphology that occur as the cell progresses through the cell cycle. In G1, the lamellar body is associated with a half bridge outside the nuclear envelope. The lamellar bodies are duplicated at the G1/S boundary. After exiting S, the SPB undergoes early maturation, including an increase in size of the lamellar bodies, invagination of the nuclear envelope, and accumulation of material in a pocket underneath the SPB. As the cell enters M, the nuclear envelope becomes fenestrated and SPBs separate and enter the nucleus, giving rise to the spindle (late maturation). The diagram in B illustrates the regulatory mechanism that coordinates SPB duplication and maturation with the fission yeast cell cycle. Both DNA replication and SPB duplication are triggered by cdcp kinase at the G1/S boundary but follow different pathways. During S phase, early stage maturation is blocked either by cdc18p or downstream events.
To interpret our studies properly, it is necessary to make a distinction between the unduplicated and duplicated SPB and the subsequent changes in morphology as it matures. As described previously (Ding et al., 1997), the morphology of the fission yeast SPB immediately after duplication consists of two laminated structures connected by an ellipsoid bridge. Because this structure sits on the outside of the NE it is difficult to distinguish it from unduplicated SPBs and may have been misidentified in cells grown in log phase (Ding et al., 1997; Kniola et al., 2001). To avoid this problem, in all experiments only complete serial sections running through the SPB were scored, and in several experiments tomographic reconstructions were made to confirm our interpretation of the serial sectioned SPBs. Our sample size was also large—in all, 123 SPBs were reconstructed in this study.
Even though cdc10+ was originally used to define Start in S. pombe (Nurse and Bissett, 1981), as we show in our results, some events down stream from the G1/S transition occur in cdc10-arrested cells. For this discussion, rather than defining Start as the cdc10+ execution point, we will define Start as being under cdc2p kinase control and cdc10p as downstream from that event. This is in agreement with the definition of START in budding yeast and many other organisms (Nurse and Bissett, 1981).
cdc2p Kinase Control of SPB Duplication at the G1/S Boundary
Using nitrogen starvation to arrest cells in G1, and then releasing them from the block by using a ts– allele of cdc2+, we demonstrate that SPB duplication in S. pombe, like in S. cerevisiae and mammalian cells, is under control of cdc2p kinase and occurs at the G1/S boundary. It is likely that they are under a direct control and in a pathway independent of cdc10+ because duplication does not require cdc10+, which is immediately down stream of cdc2+ (Labib et al., 1995). There are three G1 cyclins in fission yeast, namely, cig1p, cig2p, and puc1p (Fisher and Nurse, 1995; Martin-Castellanos et al., 1996; Martín-Castellanos et al., 2000; Tanaka and Okayama, 2000). In addition to these characterized cyclins, there are other putative G1 cyclins in the genome database (S. pombe genome database, Sanger Institute, Cambridge, United Kingdom). One or more of these G1 CDK complexes may be responsible for controlling SPB duplication. Our conclusions placing the timing of duplication at the G1/S boundary are also supported by evidence for SPB duplication in HU-treated cells and cdc10 ts– cells at the nonpermissive temperature. After HU treatment, cells enter S but proceed very slowly if all to replicate their DNA. The cdc10 mutants cells never start DNA replication. Under both conditions, the SPBs are duplicated. These experiments show that SPB duplication has been uncoupled from DNA replication and is consistent with SPB duplication happening at the G1/S boundary. In mammalian cells, it has been demonstrated that centrosome duplication occurs at the G1/S boundary and is under control of cdk2/cyclin E complex, or in some cases cyclin A (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999). Similarily in S. cerevisiae, SPB duplication is regulated by Cdc28 and the three G1 cyclins Cln1, 2, and 3 (Haase et al., 2001). Thus, the timing of centrosome duplication at the G1/S boundary seems to be an evolutionary conserved event.
SPB Maturation Occurs in Two Phases, Probably under Different Cell Cycle Control
SPB maturation involves different events in fission yeast than in budding yeast and is not completed until the G2/M boundary. In budding yeast, SPB maturation, defined morphologically as SPB separation, is dependent on the S phase and M phase cyclins. Maturation is completed in S, and a small central spindle is present in S phase budding yeast cells (Haase et al., 2001). In fission yeast, there is no spindle formation in S-phase cells, and there must be a mechanism, either negative or positive, to delay the onset of maturation until S is complete. As shown by the rum1p-OE results, exit from S and/or transit through G2 may be required for early stage maturation. A similar timing of initiation of centrosome maturation and hence the need for a similar S phase inhibitory mechanism is observed in many mammalian cells (Balczon et al., 1995; reviewed in Hinchcliffe and Sluder, 2001).
SPB maturation occurs in several steps. After duplication the SPB is associated with the cytoplasmic face of the NE (Tanaka and Kanbe, 1986; Ding et al., 1997). The laminated bodies increase in length and the bridge structure between them becomes attenuated, and dark staining material accumulates underneath these structures on the nucleoplasmic face of the NE as the NE invaginates. We call these events the early phase of maturation, and they do not occur until after cells exit S. Eventually, the laminated structures begin to nucleate microtubules and then separate, forming the spindle (Ding et al., 1997). We have called these events the late phase of maturation.
We suggest that late stage maturation is under a different cell cycle regulation than early phase maturation. In log growth phase cells, early and late phase maturation events seem to occur in rapid succession; early phase events were seldom observed in these cultures (Ding et al., 1997). However, in the cell cycle arrest/release experiments described here, maturation when observed was incomplete and seemed not to progress beyond the early stage. Using log phase cells, Ding et al., 1997 demonstrated that maturation occurred in G2. We concur and think that both stages of maturation occur after leaving S, but late stage maturation requires crossing the G2/M boundary for its completion. Because the cells used in our experiments were never allowed to cross the G2/M boundary, the conditions for late stage maturation were not met in our arrest/release experiments. In cells overexpressing rum1p, the cells in question cross the G1/S boundary, initiate DNA replication in a cdc10+-dependent manner, exit into G2, and then reenter G1 without ever going through M (Moreno and Nurse, 1994; Nishitani and Nurse, 1995). rum1p-overexpressing cells show early stage maturation. In contrast, cdc18p-overexpressing cells bypass a requirement for cdc2p/G1 cyclins and res1p-cdc10p to initiate DNA replication and never leave S. Although cdc18p-overexpressing cells undergo SPB duplication, SPB maturation is not initiated. This is consistent with a model that exit from S is required for the early stage maturation to occur and that late stage maturation requires entrance into M (Figure 9B).
S Phase-dependent Feedback Inhibition of Early SPB Maturation
We invoke a feedback inhibition mechanism downstream of cdc10+ to block the early phase of maturation until completion of S, i.e., completion of DNA synthesis. Although it is possible that early stage maturation is under positive cell cycle and only G2 is permissive for early stage maturation to occur, this simple model is contradicted by our observations of SPB duplication and early stage maturation in cdc10+-inactive cells. When cells initiated SPB duplication in the absence of normal cdc10+, they also underwent early stage maturation, even though these cells never entered S or G2. Maturation was blocked in cells overexpressing cdc18p and in HU-treated cells, conditions that place these cells down-stream of a requirement for cdc10+. The simplest explanation of these results is that during S-phase early stage maturation is blocked by a feedback inhibition mechanism that is cdc10+ dependent. These results also demonstrate that early stage maturation cannot be under control of the cdc2p/mitotic cyclin complex because only the cdc2p/S phase cyclin complex is active under these conditions (Labib et al., 1995; Tanaka and Okayama, 2000). The absence of SPB maturation in HU-treated cells and in cells overexpressing cdc18p suggests that the mechanism of feedback inhibition is coupled to an event involved in DNA synthesis, for example, the formation of replication forks or that it requires cdc18p itself.
We propose that cdc18p is the protein blocking the early maturation of duplicated SPBs until the completion of the S phase. Cdc18p has been shown to be required for maintenance of the replication fork and it activates the S phase DNA replication checkpoint pathway (Murakami et al., 2002), The presence of replication forks maintained by cdc18p could send signals to the cell to inhibit early phase SPB maturation and thus coordinate SPB duplication/early maturation with DNA replication. This model is consistent with our observation of early stage maturation in cells overexpressing rum1p, because rum1p OE triggers entrance into S upstream of cdc10+, but these cells subsequently exit from S. In the future, it will be important to identify other proteins involved in the SPB duplication/maturation checkpoint pathway and to show whether it shares some common components with the DNA replication checkpoint pathway.
Supplementary Material
Acknowledgments
This study was inspired by Takashi Toda (Cancer Research, London, United Kingdom) who also helped us through various discussions and provided careful reading of the manuscript. We also thank Susan Forsburg for comments on the manuscript and for strains and protocols needed for the in situ mcm4p binding assay. We thank members of the Cande laboratory for reading drafts of this manuscript. Leah Kenaga helped with the early stages of electron microscopy. This research was supported by grants from National Institutes of Health and from Torry Mesa Research Institute, Syngenta Research and Technology, San Diego, CA. S.U. was supported by a fellowship from Keiyu Medical Research Foundation (Tatebayashi, Japan).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0255. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0255.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Balczon, R., Bao, L., Zimmer, W.E., Brown, K., Zinkowski, R.P., and Brinkley, B.R. (1995). Dissociation of centrosome duplication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunfeld, M.B., Koster, A.J., Sedat, J.W., and Agard, D.A. (1994). Cryo automated electron tomography: towards high-resolution reconstructions of plastic-embedded structures. J. Microsc. 174, 75–84. [DOI] [PubMed] [Google Scholar]
- Chen, H., Hughes, D.D., Chan, T.A., Sedat, J.W., and Agard, D.A. (1996). IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116, 56–60. [DOI] [PubMed] [Google Scholar]
- Ding, R., West, R.R., Morphew, D.M., Oakley, B.R., and McIntosh, J.R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D., and Nurse, P. (1995). Cyclins of the fission yeast Schizosaccharomyces pombe. Semin. Cell Biol. 6, 73–78. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., Hagan, I., Uzawa, S., and Yanagida, M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.A., Vardy, L., Koonrugsa, N., and Toda, T. (2001). Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 20, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, E.B., and Forsburg, S.L. (2004). Analysis of the fission yeast Schizosaccharomyces pombe cell cycle. Methods Mol. Biol. 241, 93–111. [DOI] [PubMed] [Google Scholar]
- Grallert, A., Krapp, A., Bagley, S., Simanis, V., and Hagan, I.M. (2004). Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 18, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, S.B., Winey, M., and Reed, S.I. (2001). Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 3, 38–42. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., Li, C., Thompson, E.A., Maller, J.L., and Sluder, G. (1999). Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283, 851–854. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., and Sluder, G. (2001). “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Horie, S., Watanabe, Y., Tanaka, K., Nishiwaki, S., Fujioka, H., Abe, H., Yamamoto, M., and Shimoda, C. (1998). The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18, 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach, R.J., and Mazia, D. (1982). Origin and maturation of centrioles in association with the nuclear envelope in hypertonic-stressed sea urchin eggs. Eur. J. Cell Biol. 28, 68–76. [PubMed] [Google Scholar]
- Kanbe, T., Hiraoka, Y., Tanaka, K., and Yanagida, M. (1990). The transition of cells of the fission yeast beta-tubulin mutant nda3-311 as seen by freeze-substitution electron microscopy. Requirement of functional tubulin for spindle pole body duplication. J. Cell Sci. 96, 275–282. [DOI] [PubMed] [Google Scholar]
- Kearsey, S.E., Montgomery, S., Labib, K., and Lindner, K. (2000). Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.M., and Huberman, J.A. (2001). Regulation of replication timing in fission yeast. EMBO J. 20, 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., and Hyams, J.S. (1982). Interdependence of cell cycle events in Schizosaccharomyces pombe. Terminal phenotype of cell division cycle mutants arrested during DNA synthesis and nuclear division. Protoplasma 110, 54–62. [Google Scholar]
- King, S.M., Hyams, J.S., and Luba, A. (1982). Absence of microtubule sliding and an analysis of spindle formation and elongation in isolated mitotic spindles from the yeast Saccharomyces cerevisiae. J. Cell Biol. 94, 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola, B., O'Toole, E., McIntosh, J.R., Mellone, B., Allshire, R., Mengarelli, S., Hultenby, K., and Ekwall, K. (2001). The domain structure of centrosomes is conserved from fission yeast to Humans. Mol. Biol. Cell 12, 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Moreno, S., and Nurse, P. (1995). Interaction of cdc2 and rum1 regulates Start and S-phase in fission yeast. J. Cell Sci. 108, 3285–3294. [DOI] [PubMed] [Google Scholar]
- Lacey, K.R., Jackson, P.K., and Stearns, T. (1999). Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 96, 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Castellanos, C., Blanco, M.A., de Prada, J.M., and Moreno, S. (2000). The puc1 cyclin regulates the G1 phase of the fission yeast cell cycle in response to cell size. Mol. Biol. Cell 11, 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos, C., Labib, K., and Moreno, S. (1996). B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 15, 839–849. [PMC free article] [PubMed] [Google Scholar]
- Martín-Castellanos, C., and Moreno, S. (1996). Regulation of G1 progression in fission yeast by the rum1+ gene product. Prog. Cell Cycle Res. 2, 29–35. [DOI] [PubMed] [Google Scholar]
- Masuda, H., Sevik, M., and Cande, W.Z. (1992). In vitro microtubule-nucleating activity of spindle pole bodies in fission yeast Schizosaccharomyces pombe: cell cycle-dependent activation in Xenopus cell-free extracts. J. Cell Biol. 117, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, Y., Hayashi, K., and Nishida, E. (1999). Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 9, 429–432. [DOI] [PubMed] [Google Scholar]
- McCully, E.K., and Robinow, C.F. (1971). Mitosis in the fission yeast Schizosaccharomyces pombe: a comparative study with light and electron microscopy. J. Cell Sci. 9, 475–507. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Lukas, J., Fry, A.M., Bartek, J., and Nigg, E.A. (1999). Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1, 88–93. [DOI] [PubMed] [Google Scholar]
- Moreno, S., and Nurse, P. (1994). Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature 367, 236–242. [DOI] [PubMed] [Google Scholar]
- Murakami, H., Yanow, S.K., Griffiths, D., Nakanishi, M., and Nurse, P. (2002). Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nat. Cell Biol. 4, 384–388. [DOI] [PubMed] [Google Scholar]
- Nishimoto, T., Uzawa, S., and Schlegel, R. (1992). Mitotic checkpoints. Curr. Opin. Cell Biol. 4, 174–179. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., and Nurse, P. (1995). p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83, 397–405. [DOI] [PubMed] [Google Scholar]
- Nurse, P., and Bissett, Y. (1981). Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292, 558–560. [DOI] [PubMed] [Google Scholar]
- Ogawa, Y., Takahashi, T., and Masukata, H. (1999). Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla, M.J., Uzawa, S., and Cande, W.Z. (2003). Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116, 4891–4903. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Fuchs, J., Loidl, J., and Nasmyth, K. (2000). Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and Kanbe, T. (1986). Mitosis in the fission yeast Schizosaccharomyces pombe as revealed by freeze-substitution electron microscopy. J. Cell Sci. 80, 253–268. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and Okayama, H. (2000). A pcl-like cyclin activates the Res2p-Cdc10p cell cycle “start” transcriptional factor complex in fission yeast. Mol. Biol. Cell 11, 2845–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa, S., and Yanagida, M. (1992). Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101, 267–275. [DOI] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast gamma-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans, S.J., Wong, M.L., and O'Farrell, P.H. (1999). Mitotic regulators govern progress through steps in the centrosome duplication cycle. J. Cell Biol. 147, 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P., Hirata, D., Verde, F., Eddison, M., Toda, T., and Nurse, P. (1996). Microtubules mediate mitochondrial distribution in fission yeast. Proc. Natl. Acad. Sci. USA 93, 11664–11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.