Abstract
Most cells on earth exist in a quiescent state. In yeast, quiescence is induced by carbon starvation, and exit occurs when a carbon source becomes available. To understand how cells survive in, and exit from this state, mRNA abundance was examined using oligonucleotide-based microarrays and quantitative reverse transcription-polymerase chain reaction. Cells in stationary-phase cultures exhibited a coordinated response within 5–10 min of refeeding. Levels of >1800 mRNAs increased dramatically (≥64-fold), and a smaller group of stationary-phase mRNAs decreased in abundance. Motif analysis of sequences upstream of genes clustered by VxInsight identified an overrepresentation of Rap1p and BUF (RPA) binding sites in genes whose mRNA levels rapidly increased during exit. Examination of 95 strains carrying deletions in stationary-phase genes induced identified 32 genes essential for survival in stationary-phase at 37°C. Analysis of these genes suggests that mitochondrial function is critical for entry into stationary-phase and that posttranslational modifications and protection from oxidative stress become important later. The phylogenetic conservation of stationary-phase genes, and our findings that two-thirds of the essential stationary-phase genes have human homologues and of these, many have human homologues that are disease related, demonstrate that yeast is a bona fide model system for studying the quiescent state of eukaryotic cells.
INTRODUCTION
Quiescence is the most common state in which cells on earth exist (Lewis and Gattie, 1991). However, it has been a very difficult phase to study. Quiescent yeast cells (typically studied in stationary-phase [SP] cultures) exhibit very low metabolic activity, including low rates of protein synthesis (Fuge et al., 1994) and transcription (Choder, 1991; Jona et al., 2000). RNAses are abundant in these cells and have been found to copurify with poly(A)+ RNA (Gray et al., 2004). The thick cell walls of stationary-phase cells impede cell lysis, making isolation of proteins and native protein complexes difficult. Nevertheless, the importance of the quiescent state in all organisms makes this a compelling area of research.
The advent of genomics has made the study of quiescent cells much more tractable. Microarray analysis has allowed the characterization of global changes in transcript abundance that occurs as yeast cells enter the stationary-phase (Gasch et al., 2000). Nevertheless, major questions that remain unanswered include how the transitions into and out of the quiescent state are regulated and what mechanisms support the long-term survival in this state (Gray et al., 2004).
The quiescent state is highly regulated and programmed (Werner-Washburne et al., 1993; Herman, 2002; Gray et al., 2004). In multicelled eukaryotes, quiescence is regulated by hormones and growth factors. In microbes, the quiescent state is typically induced by nutrient limitation. During entry into the quiescent state, microorganisms undergo morphological and physiological changes that allow them to resist the effects of environmental stresses. In addition, both prokaryotic and eukaryotic cells, such as bacteria, yeast, and neuronal cells, can persist in the quiescent state for years (Werner-Washburne et al., 1993; Lewis, 2000). The signals for reentry into the cell cycle in eukaryotic cells are not completely known. For yeast, it includes the reintroduction of a carbon source (Granot and Snyder, 1993). For other cells, such as “unculturable” microbes (representing most of the microbes on earth) and neurons, the signals remain unknown.
The quiescent state is of major importance to both biomedical and environmental research. The persistence of pathogens, including Mycobacterium tuberculosis (Parrish et al., 1998) and Cryptococcus neoformans (Alexander and Perfect, 1997), in an antibiotic-resistant, quiescent state in the human host increases the difficulty and length of treatment. Thus, insights into the processes of survival in and exit from stationary-phase could lead to the development of treatment strategies that are independent of pathogen growth. Quiescent yeast cells also may provide an excellent model system for aging because cells in stationary-phase cultures were found to have a shorter replicative life span, mimicking aging in nondividing cells of other organisms (Ashrafi et al., 1999). Finally, most environmental microorganisms exist in an unculturable quiescent state, making it difficult to understand the contribution of these species to critical environmental processes, such as carbon fixation. The ability to induce reentry of these organisms into the cell cycle is an important beginning toward understanding their broad role in the environment.
Our long-term goal is to understand the regulation of and requirements for entry into and survival in the quiescent state, as well as for reentry into the mitotic cell cycle. Microarray analysis of entry into stationary-phase has been published (Gasch et al., 2000). Here, we report an analysis of gene expression in Saccharomyces cerevisiae cultures exiting stationary-phase for the first hour after transfer to rich, glucose-based medium. The most dramatic changes in mRNA abundance were observed within the first 5 min after refeeding. Phenotypic analysis of 95 genes induced in stationary-phase led to the identification of 32 genes required for survival in stationary-phase at 37°C. Many of these genes are involved in mitochondrial function, posttranslational protein modification, and resistance to oxidative stress. Several of these genes have human homologues, many of which are associated with diseases. These results provide a critical foundation that will have significant implications for understanding this important cellular state in all eukaryotic organisms.
MATERIALS AND METHODS
Cell Harvesting and RNA Preparation
Two independent cultures (time courses 1 and 2) of wild-type MATα S288C cells (American Type Culture Collection, Manassas, VA) were grown in 100 ml of YPD+A (1% yeast extract, 2% Bacto-peptone, 2% glucose, 0.04 mg/ml adenine) at 30°C for 7 d (stationary-phase) with aeration. For a reference standard, MATα S288C cells were grown to exponential phase in YPD+A with shaking (250 rpm) and harvested at 1 OD600. All methods and additional information are available on the Web Supplement (http://biology.unm.edu/biology/maggieww/Public_Html/SPexit.htm).
For time course 1, cells from a single 7-d culture were added to each of two 600-ml flasks with YPD+A prewarmed to 30°C. After refeeding, cells were harvested at T0 min, T1, T10, T20, T30, T40, from one flask and T50 and at T0, T5, T15, T25, T35, T45, and T55 from a second flask. These 14 time points plus 10 technical replicates were used for microarray analysis (see Web Supplement). For time course 2, which was used for quantitative reverse phase-polymerase chain reaction (qRT-PCR) analysis, cells were harvested from a single refed culture by using an air pressure-assisted sampling method (Quinones, unpublished data) at T0, T1, T2, T5, T10, T15, T20, T25, T30, T35, T40, T45, T50, T55, and T60. For all harvests, ∼40 ml of culture was added, in duplicate, to 50-ml Falcon tubes containing crushed ice. Cells were centrifuged at 2056 × g at 4°C for 3 min in a Beckman GPR Tabletop centrifuge. The supernatant was decanted, and the pellet was immediately frozen in liquid nitrogen before storage at –70°C.
RNA was isolated as described in the Web Supplement (http://biology.unm.edu/biology/maggieww/Public_Html/SPexit.htm). Briefly, cells were lysed using glass beads and a bead beater (Biospec, Bartlesville, OK). RNA reagents and initial protocols are based on the Purescript RNA purification kit for yeast (Gentra Systems, Minneapolis, MN). A phenol extraction was carried out before a final purification by using RNeasy columns (QIAGEN, Valenica, CA), which included the optional DNase I digestion. Each sample yielded ∼100–200 μg of RNA. Enough reference RNA was isolated and pooled at one time to provide a source for all microarrays of the entire time course experiment. RNA isolations were done in random order to avoid confounding preparation order with time of harvest. Purified total RNA was quantified using a Beckman DU640 spectrophotometer.
The percentage of budded cells (budding index) was determined by microscopic examination. Three separate counts were averaged to obtain the mean and SD. Cells began to bud at 60 min after refeeding and peak budding under these conditions was observed at 3–5 h.
Microarray Printing
Yeast 70-mer oligonucleotides (QIAGEN Operon, Alameda, CA) were resuspended to 40 μM in ArrayIt MicroSpotting Solution (TeleChem, Sunnyvale, CA). Before microarray printing, UltraGAPS glass slides (Corning Glassworks, Corning, NY) were incubated at 21°C in 48–52% humidity for 4–20 h (Martinez et al., 2003). The oligonucleotides were printed on slides by using a custom built arrayer (Pat Brown design) or an Omnigrid arrayer (GeneMachines) with SMP4 pins (TeleChem). After printing, slides were UV-cross-linked at 90 mJ in a UV-Stratalinker 1800 (Stratagene, La Jolla, CA) and baked for 2 h at 80°C. To remove spot-localized fluorescent background, slides were placed on a rocking platform and were given a postprinting treatment that included a 10-min wash in 0.1% SDS, a 2-min wash in 2× SSC, 3 min in boiling double distilled water, and 5 min in ice-cold 100% ethanol (Martinez et al., 2003). All slides were scanned before hybridization with a 4000B scanner (Axon Instruments, Union City, CA) to ensure the absence of spot-localized, contaminating fluorescence.
Preparation of Labeled cDNA
Direct Cy3- and Cy5-dCTP incorporation by first-strand cDNA synthesis was performed as described previously (Martinez et al., 2003). Twenty micrograms of total RNA was used for each labeling reaction. Cy3 and Cy5 (Amersham Biosciences, Piscataway, NJ) were incorporated into the experimental and reference cDNA, respectively. Samples were labeled in random avoid confounding factors. To normalize the arrays for labeling efficiency, equal amounts of mRNAs complementary to the Arabidopsis thaliana CAB, rbcL, and RCA genes (SpotReport-10; Stratagene) were added to each labeling reaction. Reference cDNA-Cy5 was synthesized in separate tubes and pooled before hybridization.
Microarray Hybridization
The entire labeling reaction of Cy3-cDNA generated from an aliquot obtained at a single time point was combined with an equivalent amount (20 μg of labeled total RNA) of the pooled reference Cy5-cDNA. The samples were dried using a SpeedVac (Savant Instruments, Holbrook, NY), resuspended in 30 μl of hybridization buffer containing 5% dextran sulfate, as described in the Web Supplement and Martinez et al. (2003), and applied to Lifter slips (Erie Scientific, Portsmouth, NH), which were then placed on the microarray. Slides were incubated in single-slide capacity chambers (Corning Glassworks) on a rotating platform at 42°C for 16–20 h. After hybridization, slides were washed as described previously (Martinez et al., 2003).
Microarray Data Acquisition and Processing
Slides were scanned with a 4000B scanner (Axon Instruments) with 100% laser power. Photo multiplier tube (PMT) settings were selected to maximize range of detection of fluorescence. Typically, PMT settings in the range of 500–700 were used for Cy3- and Cy5-labeled samples. Quantification and filtering were performed using Genepix 4.0 (Axon Instruments). Raw and analyzed data and grids are available in the Web Supplement. Spots were flagged as “good” if <10% of the pixels in one spot were saturated in either Cy3 (green, G) or Cy5 (red, R) channels. Spots were flagged as “bad” if the median pixel intensity of the feature varied from the mean pixel intensity by >20%. For each channel and each slide, the average background median pixel intensity (BG and BR) of the negative controls and a SD of this average (σBG and σBR, n ≥ 10) were calculated (Wu et al., 2001). BG and BR were used for background subtraction. A third precision filter was created, based on propagation of error, to flag spots as bad if the channel median intensities approached the background values (BG and BR), such that the error in measured G/R due to background variability would be outside the limits of a desired error (Ez). Specifically, the precision filter eliminated spots for which Ez2 < = [σBG2(R – BR)2 + σBR2(G – BG)2]/[(R – BR)2(G – BG)2], where σBG and σBR are the SD of measurements of the background in the green (Cy3) and the red (Cy5) channel, respectively (Wentzell, unpublished data). We chose Ez = 1 so our desired limit of error was 100%, or twofold. BG and BR values were then subtracted from the intensity value of each spot on each slide.
Because the amount of mRNA in the total RNA sample was less for cells in stationary-phase cultures than exponentially growing cells (the Cy5-reference sample), the Cy3/Cy5 ratio was normalized by setting the average ratios of median intensities of the good Arabidopsis gene control spots to 1. All spots flagged as bad or “not found” were excluded from the final data set.
Filtered data was converted into matrix format using MATLAB (Math-works, Natick, MA). MATLAB scripts were written (slide compare) that generated correlation plots and deviation histograms comparing any two experiments (time point to time point or replicate to replicate). Additional code was written (set compare) that generated a mesh plot to represent a pairwise comparison of each microarray with all of the other slides of the entire dataset. MATLAB code is available from Dr. P. D. Wentzell.
Evaluation of Microarray Reproducibility
To evaluate microarray reproducibility, the correlation between log2 ratios from the data of different array experiments was compared using slide compare (see above). Technical replicates performed by the same researcher were highly correlated with R2 = 0.985. The number of genes deviating from the normal distribution also was determined.
Clustering of Microarray Data
Normalized log2(G/R) data from a total of 23 time course 1 slides (13 time points plus 10 technical replicates) were clustered in VxOrd 1.58 (Viswave; www.viswave.com) and visualized in VxInsight 2.161 (Davidson et al., 2001). Genes were included in the clustering analysis only if data were available from >80% of the microarrays. Clustering was obtained using force-directed placement of the 20 nearest neighbors, i.e., the 20 genes whose expression pattern was most correlated, using the t-statistic of R (Werner-Washburne et al., 2002). The number of hills was not predetermined, nor was the number of genes in a hill. The total number of hills, however, was a function of the number of nearest neighbors that are used for clustering, i.e., genes whose Pearson's correlation coefficients were most similar. The VxInsight data set in a locked version of VxInsight can be downloaded from the Web Supplement (http://biology.unm.edu/biology/maggieww/Public_Html/SPexit.htm).
qRT-PCR Studies
For qRT-PCR analysis of mRNA abundance, samples of total RNA were reverse transcribed using random hexamers. Primers were designed using Primer Express (ABI, Foster City, CA). For qRT-PCR reactions, SYBR Green PCR Master Mix (ABI), 5 μM of both the forward and reverse primers, and 10 ng of cDNA template were used for each time point. PCR reactions for each sample were performed in triplicate by using a Prism 7000 real-time thermocycler (ABI), by using 18s rRNA as an internal control. Data was analyzed using the ABI Prism 7000 SDS software package, which determines the cycle threshold (CT) values for each reaction. The CT values were then used to calculate the mean fold change of the reactions via the 2–ΔΔCT method (Livak and Schmittgen, 2001). In addition to the qRT-PCR analyses presented in this article, results for 15 additional representative genes from several hills are provided on the Web Supplement.
Identification of Genes Induced in Stationary Phase
The list of 127 stationary-phase genes (or SP-expressed genes) to be evaluated was obtained in three ways (see Web Supplement). One hundred seven genes were identified from cluster lists from both glass-slide (this study) and membrane array data sets (Werner-Washburne et al., 2002). An additional 20 genes were manually “mined” from the glass array data set by using GeneSpring 4.0 (Silicon Genetics, Redwood City, CA).
Promoter Analysis
MOTIF REGRESSOR (Conlon et al., 2003) was used to evaluate the abundance of specific DNA-sequence motifs as a function of gene expression profiles.
Genes in the VxInsight cluster data set (n = 3995) for which values could be obtained in all three replicates of T0 (n = 3667) were selected for further analysis. For all genes, sequences for 1000 base pairs upstream of the initial ATG were obtained from the Stanford Genome Database (ftp://genome-ftp.stanford.edu/pub/yeast/). MOTIF REGRESSOR was then used to rank genes on the basis of their expression values and to find motifs from the most highly expressed or highly repressed genes based on gene expression values. MOTIF REGRESSOR returned a MR score for every gene based on the extent of identity between a motif and motifs in the promoter region of a gene. It also assigned a p value to every motif based on the correlation between motif identity and gene coexpression. Genes with p values ≤10–7 were identified as good hits. Consensus motifs with overlapping base sequences were then grouped, and the 100 genes with the highest MR scores were identified for each motif. Finally, VxInsight 2.161 (Davidson et al., 2001) was used to visualize the distribution of each group of 100 genes associated with specific motifs. Only genes represented in any of the fourteen hills shown in Figure 2 were used for the motif distribution analysis.
Figure 2.
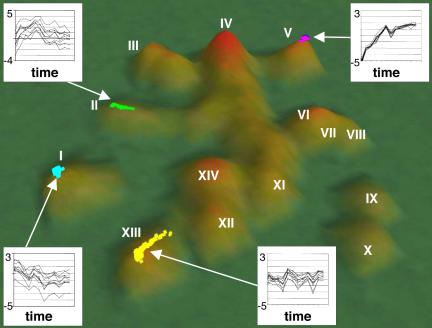
VxInsight (VxOrd-generated) topography from time course 1. Hills represent genes with similar expression profiles and are identified by Roman numerals. Height of hill (green to red) is a function of the number of clustered genes in the hill. The VxInsight dataset is available in the Web Supplement. Graphs of log2(G/R) as a function of time for ten representative genes in the hills I, II, V, and XIII illustrate major patterns of gene expression. Colored regions indicate clusters of genes (within particular hills) used for GO analysis (see Web Supplement).
Known binding factors for binding motifs identified by MOTIF REGRES-SOR were found using TRANSFAC database (http://www.gene-regulation.com). Additional details of this analysis are available on the Web Supplement. AlignACE and CompareACE (http://atlas.med.harvard.edu/) were used to produce a list of motifs and known binding factors from the upstream sequences of the selected 3667 genes, which was compared with the MOTIF REGRESSOR-generated list.
Gene Ontology (GO) Annotation of Gene Lists
Genes from each cluster identified by VxInsight were queried for annotations of molecular process using the GO Term Finder (The Gene Ontology Consortium, 2001) from the Saccharomyces Genome Database (SGD; http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermFinder). The most significant processes (those with the lowest p value as calculated by a GO algorithm) for each cluster were identified. The complete lists of genes, cluster lists, and all other annotations are provided in the Web Supplement.
Phylogenetic Distribution of Genes in the Yeast Genome
The Clusters of Orthologous Groups (COG, KOG, and TWOG) databases (http://www.ncbi.nlm.nih.gov/COG/) (Koonin et al., 2004) were used to study the phylogenetic distribution of S. cerevisiae open reading frames (ORFs) (n = 6702) obtained from SGD (http://www.yeastgenome.org). The COG database contains orthologous groups from 63 prokaryotic and three eukaryotic genomes. The KOG and TWOG database contains orthologous groups from seven eukaryotic organisms, including three also present in the COG database. The KOG and COG databases identify proteins with orthologues in at least three different organisms and the TWOG database identifies proteins with orthologues in two different organisms.
Genes were placed in categories, e.g., eukaryotes, prokaryotes, eukaryotes/archea, archea/prokaryotes, eukaryotes/prokaryotes, eukaryotes/archea/prokaryotes, or S. cerevisiae only, based on the phylogenetic distribution of one or more orthologues (Figure 6). For example, a gene was assigned to a category, such as eukaryotes or prokaryotes, if it had orthologues only in eukaryotic or prokaryotic organisms, in addition to S. cerevisiae. A gene was assigned into a combined category, such as eukaryotic/prokaryotic or eukaryotic/archea/prokaryotic, if it had orthologues in each phylogenetic domain. Individual genes were assigned to only one category.
Figure 6.
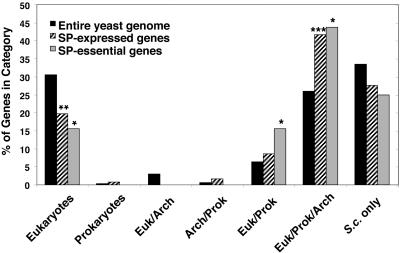
Comparison of the phylogenetic distribution of all yeast genes with SP-expressed and SP-essential genes. The phylogenetic distribution of genes identified in the yeast genome (n = 6702) (SGD), SP-expressed (n = 127), and SP-essential (n = 32) was determined using Clusters of Orthologous Groups Databases (COG, KOG, and TWOG) (see Materials and Methods). p values for SP-expressed and SP-essential genes were calculated using the phylogenetic distribution genes in the yeast genome: *p < 0.05; **p < 0.01; ***p < 0.001.
To determine the significance of the phylogenetic distribution of genes expressed in stationary-phase (SP-expressed) and genes essential for stationary-phase (SP-essential) with respect to genes in the yeast genome, a p value was calculated that reflects the probability of x or more genes out of n genes to be present in a category by random chance. The method for calculating this probability is described in more detail in http://www.yeastgenome.org/help/goTermFinder.html#method. A p value ≤0.05 was considered to be significant.
Phenotypic Analysis of Strains Carrying Deletions in Stationary Phase Genes
Strains carrying deletions in 95 of the 107 genes identified by clustering analysis were tested for their ability to survive in stationary-phase at 30 and 37°C. Strains with mutations in 12 genes could not be examined because they were not represented in the deletion library (including 4 genes that are essential for exponential growth) or could not be grown from the deletion library (Winzeler et al., 1999) (see Web Supplement).
MATα deletion mutants from the Yeast Deletion Consortium (Winzeler et al., 1999) were obtained from Research Genetics. Flasks containing 100 ml of YPD+A were inoculated from freshly streaked plates of each mutant and incubated for 2 d at 30°C with aeration (250 rpm). At day 2 (postdiauxic phase), cultures were divided into two 10-ml aliquots. One was incubated at 30°C and the other at 37°C, both with shaking. Cells were assayed for viability at 2, 6, 9, and 16 d after inoculation, i.e., at 0, 4, 7, and 14 d after the temperature shift. Serial 10-fold dilutions were made to produce samples with cell densities of 4 × 105, 4 × 104, 4 × 103, 4 × 102, and 4 × 101 cells/ml, respectively. Five microliters of each diluted sample was spotted onto a YPD+A plate and incubated at 30°C. Each spot was checked for growth after 2 d and was scored for the presence of colonies in any or all of the five spots. The last spot containing a colony indicated the number of colony-forming units per milliliter. Viability of each mutant was compared with the viability of the parental strain spotted on the same plate.
Strains were incubated at 37°C, even though it produced an added stress, because we had previously discovered that stationary-phase mutants often die more quickly, i.e., after 7 or 14 d compared with 30 d or longer, at 37°C (Werner-Washburne, unpublished data). This treatment is not a heat shock, which is caused by a rapid increase in temperature and measured over a period of typically 1 h. To ensure that strains were tested at 37°C only in the postdiauxic and stationary-phases, cultures were grown for 2 d at 30°C before the temperature shift. Because strains double only once between the diauxic shift and stationary-phase, if cells arrested at 37°C without dying, this would not be detected in our assay, because our assay is sensitive only to 10-fold changes in viability.
RESULTS
Gene expression in cells exiting stationary-phase over time after inoculation into fresh, glucose-based, rich medium was evaluated using total RNA samples collected at 13 time points spanning 55 min. Ten technical replicates also were analyzed, including two biological replicates of T0 (see Web Supplement). Of the ∼6200 ORFs represented in each microarray, 3995 genes passed all qualitative filters and contained enough data points (>80%) to allow further analysis (see Materials and Methods). A second, independent time course (time course 2) was generated for qRT-PCR and statistical analyses.
Correlation of Gene Expression over the Time Course
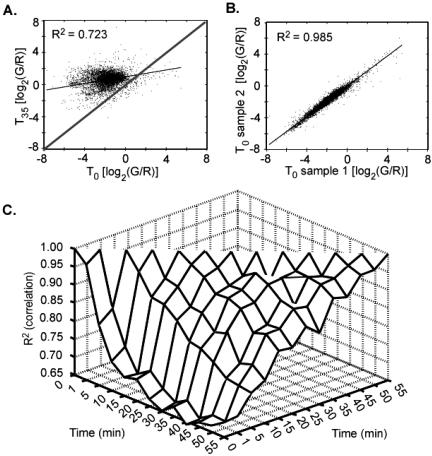
To understand the dynamics of gene expression during exit from stationary-phase, we evaluated the correlation between log2 gene-expression ratios for different time points. A comparison of log2 gene-expression ratios from T0 and T35 time points (0 and 35 min after refeeding) showed that gene expression changed dramatically over this time period, as indicated by loss of correlation (R2 = 0.698) (Figure 1A). A typical correlation plot for technical replicates is shown (Figure 1B). More than 1000 genes exhibited at least a twofold change in gene expression over this time period (Web Supplement), indicating a large, global accumulation of mRNAs in the first 35 min after refeeding. The position of the data points above the predicted regression for correlated experiments (Figure 1A) indicated that, for most genes, the change in gene expression between T0 and T35 was the result of increased mRNA abundance.
Figure 1.
Correlation plots of gene expression ratios for time course 1. Correlation (R2) was determined as described in Materials and Methods. (A) Correlation of log ratios of gene expression between T0 and T35. Gray line indicates the regression line for perfect correlation (slope = 1). (B) Correlation of log2 ratios of gene expression for technical replicates of T0. (C) Correlation map for pairwise R2 values obtained for all time-course 1 data points.
A map of pairwise correlations across the entire time course (by using set compare) allowed a more complete view of the dynamics of gene expression during exit from stationary-phase (Figure 1C). The correlation map showed sequential decreases in correlation (R2) between experiments as a function of time. However, almost half of the total decrease in correlation, i.e., changes in gene expression, occurred within the first 5 min after refeeding with additional, smaller decreases in correlation at 10 and 15 min. These results demonstrate that refeeding cells in stationary-phase cultures results in extremely rapid and global changes in transcript abundance.
Transcriptional Profile of Cells Exiting Stationary Phase
VxInsight (Viswave.com) was used for two-dimensional topographical clustering and visualization of the time-course data (Figure 2). In VxInsight topographies, hills are composed of clusters of genes with similar expression profiles. The height of the hill corresponds to the number of genes and the relative position of hills is related to the correlation between gene expression patterns (Werner-Washburne et al., 2002).
Approximately 14 major clusters were identified in the VxInsight topography, based on gene expression (Figure 2). Within the 14 clusters, three major gene-expression patterns were identified, involving dynamic changes in mRNA abundance as a function of time after refeeding (clusters I, II, and V). A fourth gene expression pattern was identified (cluster XIII), involving mRNAs whose abundance did not change dramatically over the time course. The biochemical processes that were significantly represented (GO term finder; www.yeastgenome.org) within the first five major clusters are shown in Table 1, and the processes for genes in the remaining nine clusters are shown in the Web Supplement.
Table 1.
Significant biological processes of clustered genes
| No. of genes (n)
|
||||
|---|---|---|---|---|
| Vx cluster | Process | Cluster | Genomea | p valuea |
| I (74) | Aerobic respiration | 11 | 58 | 3.74E-11 |
| Tricarboxylic acid cycle | 6 | 15 | 1.70E-08 | |
| Oxidative phosphorylation | 6 | 15 | 1.70E-08 | |
| II (54) | Amine/polyamine transport (amino acid transport) | 7 | 36 | 1.48E-08 |
| One-carbon compound metabolism | 3 | 11 | 2.92E-05 | |
| Invasive growth | 2 | 24 | 1.53E-02 | |
| III (79) | Ribosome biogenesis (20s pre-rRNA processing) | 7 | 149 | 1.62E-03 |
| Transcription from pol I promoter | 7 | 151 | 1.74E-03 | |
| Amino acid biosynthesis | 5 | 100 | 5.90E-03 | |
| IV (422) | Transcription from pol I promoter | 74 | 151 | 4.24E-43 |
| Ribosome biogenesis | 71 | 149 | 1.49E-40 | |
| Transcription from pol III promoter | 12 | 36 | 3.19E-06 | |
| V (128) | Protein biosynthesis | 100 | 754 | 3.39E-70 |
| Ribosome assembly | 14 | 51 | 1.12E-12 | |
| Regulation of translation | 8 | 25 | 2.85E-08 | |
GO Term Finder (SGD, June 2003, 6910 genes annotated).
A major expression pattern included mRNAs that were abundant in stationary-phase, i.e., at T0, and decreased as a function of time after refeeding. These are referred to as SP-expressed genes. Seventy-four genes with this profile were identified in cluster I (within hill I) (Figure 2 and Web Supplement). Messenger RNAs encoded by these genes decreased in abundance at least fourfold within 15–25 min of refeeding.
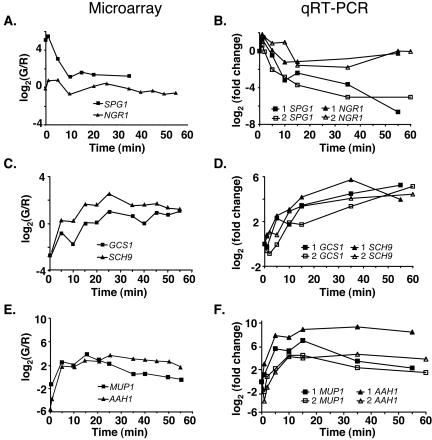
To determine the reproducibility of this expression pattern, representative genes were chosen for qRT-PCR analysis by using total RNA from both time course 1 and 2. The expression pattern of SPG1 and NGR1, two SP-expressed genes, were determined (Figure 3, A and B). The qRT-PCR results were essentially identical for time courses for both genes (Figure 3B), indicating that the patterns of gene expression during exit from stationary-phase were very robust. In addition, the relative changes over time observed by qRT-PCR, especially for SPG1, were essentially identical to that observed by microarray analysis (Figure 3A). Quantitative RT-PCR analysis also demonstrated that NGR1, originally identified as an SP-expressed gene from the membrane-array analysis of exit from stationary-phase (Werner-Washburne et al., 2002) but not from this analysis (Figure 3A), is, in fact, a stationary-phase gene (Figure 3B).
Figure 3.
Comparison of microarray data and quantitative RT-PCR measurements of abundance for representative mRNAs. Microarray expression data are presented for time course 1 and are plotted as log2(G/R) as a function of time after refeeding. Quantitative RT-PCR results for both time course 1(▪, ▴) and time course 2 (□, Δ) are plotted as log2(fold change) as a function of time after refeeding. Microarray analysis (A) and qRT-PCR analysis (B) of SPG1 and NRG1 expression (SP-expressed); microarray analysis (C) and qRT-PCR analysis (D) of GCS1 and SCH9 expression (showing rapid induction); microarray analysis (E) and qRT-PCR analysis (F) of MUP1 and AAH1 expression (showing increased then decreased expression).
A second expression pattern, found in the cluster II (Figure 2) included mRNAs that increased in abundance up to 16-fold in the first 5 min and then gradually decreased to a level similar to that found in exponentially growing cells by 35 min (Figures 2 and 3E). Genes in cluster II encode amino acid transporters, as well as proteins involved in adenine and methionine biosynthesis, sulfur metabolism, cell-wall biosynthesis, and gene regulation. Quantitative RT-PCR confirmed the expression pattern for a representative gene from cluster II, MUP1, in both time courses (Figure 3F). The relative change over time in mRNA abundance for MUP1 was essentially identical for qRT-PCR and microarray analyses (Figure 3E).
The third pattern of expression was exhibited by >1800 mRNAs. These transcripts, found in hills III–XII and XIV (Figure 2), accumulated rapidly and to very high levels upon refeeding. The most rapid and dynamic changes in mRNA abundance were observed for genes in hills III, IV, and V, with genes in hill III increasing as much as 64-fold within the first 10 min after refeeding (see Web Supplement). Cluster III (within hill III) contains genes encoding proteins involved in ribosomal biogenesis (Table 1), and genes encoding permeases and proteins involved in lipid metabolism, as well as the growth-related kinases Sch9p (Crauwels et al., 1997) and Sok1p (Ward and Garrett, 1994) (Web Supplement). Cluster IV was comprised of genes encoding ribosomal processing proteins. The extended cluster included TOR1 and TPK3, genes encoding kinases involved in regulation of translation and growth. Cluster V contains genes involved in protein biosynthesis and ribosome assembly and contained most (140) of the ribosomal protein genes (RPL and RPS genes). The rapid increase in transcript abundance of these groups of genes is consistent with an immediate requirement for ribosome assembly and protein synthesis after refeeding.
Slower, more linear increases in transcript abundance were observed with genes in hills VI, VII, and VIII. Many of these genes encode proteins involved in DNA repair, organelle biogenesis, and secretion. These hills also contain a relatively high number of genes with unknown functions (see Web Supplement). Expression patterns for AAH1 and SCH9 from hill III and GCS1 from hill VI were confirmed by qRT-PCR for using RNAs from both time courses (Figure 3, D and F) and were essentially identical to microarray results for these genes (Figure 3, C and E). We conclude from this analysis that cells in stationary-phase cultures respond rapidly and globally to refeeding by increasing mRNAs encoding proteins needed for growth and cell division.
Promoter Analysis of Coregulated Genes
To determine whether the promoters of genes that were coregulated during exit from stationary-phase shared transcription-factor binding motifs, the upstream regions (1000 base pairs) of genes analyzed in VxInsight (Figure 1) were examined with MOTIF REGRESSOR (Conlon et al., 2003), AlignACE, and CompareACE (Tavazoie et al., 1999).
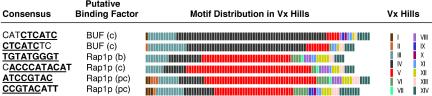
Although the SP-expressed genes were found to contain a variety of promoter motifs (see Web Supplement), these motifs were randomly distributed over the VxInsight topography. However, six motifs, which are reported to bind two different transcription factors, were found to be nonrandomly distributed in the VxInsight topography (Figure 4). Two motifs containing a sequence (GATGAG) complementary to a portion of a DNA fragment reported to be bound, in vitro, by BUF protein (also called RPA or RFA) (Luche et al., 1993) were found almost entirely in genes from hill IV (Figure 4), which contains a large percentage of ribosomal-processing genes. Four motifs, two resembling different Rap1p binding sites (TGTAT-GGGT), its complement, (ACCCATAC), a single mismatch (ATCCGTAC), and a partial sequence (CCGTAC) (Mizuta et al., 1998) were found in promoters of genes primarily in hill V, which contained a high proportion of ribosomal protein genes (Figure 2).
Figure 4.
Consensus sequences and VxInsight topographical distribution of genes with significant MR scores. The portion of the consensus sequence of a transcription factor-binding site (b), identified by MOTIF REGRESSOR, its complement (c), or partial complement (pc) are shown in the first column. The putative binding factor, as determined by the TRANSFAC database, is shown in the second column. Colored bars represent the genes with the highest MR scores for each motif and their distribution in VxInsight hills. The color code for the 14 VxInsight hills (indicated by Roman numerals and corresponding to the hills in Figure 2) is shown in the far right column.
Analysis of SP-expressed Genes
The function and phenotypic contribution of genes whose mRNAs accumulate in cells in stationary-phase cultures were examined to determine whether this information would provide additional insight into the quiescent state. One hundred twenty-seven stationary-phase genes were identified by clustering, including those found using membrane arrays (Werner-Washburne et al., 2002) or by data-mining (see Materials and Methods). Although there are likely to be >127 genes induced in cells in stationary-phase cultures, this list represents genes whose mRNA levels were reproducibly detectable in cells of stationary-phase cultures. Transcripts from all of the 127 stationary-phase genes also were reported as induced in stationary phase from experiments examining entry into stationary phase (Gasch et al., 2000).
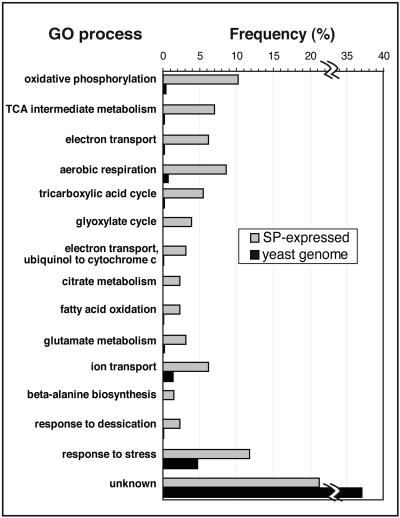
Of the 14 most significant annotations determined by GO analysis (p ≤ 0.001) for the 127 stationary-phase genes shown (Figure 5), eight are associated with mitochondrial function, i.e., aerobic respiration, tricarboxylic acid cycle, other electron transport, and citrate metabolism. This underscores the importance of mitochondrial function, especially ATP synthesis for cells in stationary-phase cultures. Stationary-phase genes also are involved in ion transport, including ATP1, ATP2, ATP3, and ATP18, which are required for ATP synthesis, and responses to stress (15 genes), including desiccation (HSP12, SIP18, and GRE1). Although the largest number of genes, 27/127 (21%), encoded proteins with unknown functions, this is less than the percentage of genes in the entire yeast genome encoding proteins with unknown functions (38%) (SGD).
Figure 5.
Gene ontology-defined processes for SP-expressed genes. The 15 most significant annotations (see Materials and Methods and Web Supplement) for members of the SP-expressed genes (n = 127) are listed in order of decreasing significance. The percentage of genes identified with a given process is shown for SP-expressed genes (gray bars) and for the annotated yeast genome (black bars). Genes can be present in multiple categories.
Phenotypic Analysis of Strains Carrying Deletions in SP-expressed Genes
To determine whether any of the SP-expressed genes might be essential for stationary-phase, we selected 107 genes that were identified by clustering, and examined strains carrying deletions in these genes (Winzeler et al., 1999). Mutants in 12 of the 107 genes could not be examined because either the genes were essential for exponential growth or the strains could not be grown from the deletion library (see Web Supplement).
The remaining 95 deletion strains were examined for survival during the postdiauxic and stationary-phases at 30 and 37°C. Incubation at 37°C was used to test the stringency of a gene's requirement for viability in stationary-phase (see Materials and Methods). The genes identified in this screen are termed stationary-phase-essential genes (SP-essential genes).
Strains with deletions in any of seven genes, GCR1, SDH2, CIT1, MDH1, COX7, ATP3, or KGD1 exhibited a 10- to 105-fold decrease in viability after 16 d of growth at 30°C (Table 2). Twenty-eight mutants reproducibly lost viability (Table 2) after 16 d when shifted to 37°C at day 2. Four strains exhibited variable loss of viability after 16 d (14 at 37°C). Based on the day at which loss of viability was detected, mutants were divided into five groups: those that lost viability at 37°C by 2, 6, 9, or 16 d after inoculation (Table 2) and a final group that exhibited some variability in loss of viability at 37°C by 16 d (see Web Supplement).
Table 2.
Loss of viability over time of mutants in SP-essential genes
| Fold reductiona in colony-forming units relative to Parentalb
|
||||||
|---|---|---|---|---|---|---|
| Days at 37°Cc
|
Days at 30°C
|
|||||
| Gene | Strain | 2 | 6 | 9 | 16 | 16 |
| parentalb | +d | + | + | + | + | |
| ATP1 | Δybl099w | 10-2 | -d | - | - | +e |
| ATP2 | Δyjr121w | 10-1 | - | - | - | +e |
| QCR7 | Δydr529c | + | 10-3 | - | - | 10-1 |
| SDH2 | Δyll041c | + | 10-3 | - | - | 10-3 |
| RIP1 | Δyel024w | + | 10-2 | - | - | +e |
| CIT1 | Δynr001c | + | 10-2 | - | - | 10-1 |
| COX6 | Δyhr051w | + | 10-2 | - | - | +e |
| SDH4 | Δydr178w | + | 10-2 | - | - | +e |
| MDH1 | Δykl085w | + | 10-2 | - | - | 10-5 |
| COX7 | Δymr256c | + | + | 10-4 | - | 10-2 |
| ATP3 | Δybr039w | + | 10-1 | 10-2 | - | 10-1 |
| KGD1 | Δyil125w | + | 10-1 | 10-2 | - | 10-1 |
| ETR1 | Δybr026c | + | + | 10-2 | - | +e |
| SOD2 | Δyhr008c | + | + | 10-2 | - | + |
| ADY2 | Δycr010c | + | + | 10-2 | - | + |
| MOH1 | Δybl049w | + | + | 10-1 | - | + |
| HBT1 | Δydl223c | + | + | 10-1 | - | + |
| GTT1 | Δyir038c | + | + | 10-2 | 10-3 | + |
| SPG5 | Δymr191w | + | + | + | 10-3 | + |
| CTA1 | Δydr256c | + | + | + | 10-3 | + |
| POR1 | Δynl055c | + | + | + | 10-3 | + |
| OM45 | Δyil136w | + | + | + | 10-2 | + |
| FMP45 | Δydl222c | + | + | + | 10-2 | + |
| FAA1 | Δyor317w | + | + | 10-1 | 10-2 | + |
| SPG4 | Δymr107w | + | + | 10-1 | 10-1 | + |
| PST2 | Δydr032c | + | + | + | 10-1 | + |
| GLK1 | Δycl040w | + | + | + | 10-1 | + |
| PNC1 | Δygl037c | + | + | + | 10-1 | + |
Average from two or more experiments unless noted.
MATa BY4742 or MATa BY4741.
Cultures were shifted to 37°C after 2 d of growth at 30°C.
No reduction in viability (+), 10-5 or more reduction in viability (-).
Data from one experiment.
Strains with deletions in ATP1 and ATP2, genes involved in ATP synthesis, lost viability after 2 d (Table 2), which is considered the postdiauxic phase of growth. The severity of this phenotype demonstrates the necessity for ATP as cells make the transition to growth on nonfermentable carbon sources.
A second group of mutants lost viability after 6 d in culture (4 d after being transferred to 37°C) (Table 2). All of these genes encode products required for mitochondrial functions (SGD and YPD Bioknowledge Library), including dehydrogenases (SDH2, SDH4, and MDH1) and components of the oxidative phosphorylation electron-transport chain (QCR7, COX6).
A third group of mutants lost viability after 9 d in culture (7 d at 37°C). Genes in this group also encode proteins with a variety of mitochondrial functions, including the electron transport chain (COX7), dehydrogenases (ETR1, KGD1) (Repetto and Tzagoloff, 1989), and superoxide dismutase (SOD2). Nonmitochondrial genes in this group encode proteins involved in cell wall morphogenesis (HBT1) (Dittmar et al., 2002; Young et al., 2002), a glutathione transferase (GTT1) (Lazarow and Kunau, 1997; Choi et al., 1998) and a putative ammonia transport protein that is essential for survival of yeast cells in colonies (ADY2) (Paiva et al., 2004).
A fourth group of genes were found to be essential much later in stationary-phase (14 d at 37°C) and included FMP45 that encodes a mitochondrial protein thought to be involved in sphingolipid metabolism (Runner and Brewster, 2003); PNC1, a gene in the NAD(+) salvage pathway (Ghislain et al., 2002); and GLK1, which is involved in glucokinase activity required for trehalose degradation (Bisson and Fraenkel, 1983). FAA1 is involved in N-myristoylation (Johnson et al., 1994). CTA1, encoding peroxisomal catalase, is one of two genes identified that is required for protection from reactive oxygen species (Mihara and Sato, 1985; Yaffe et al., 1989). These varied functions suggest that long-term survival in stationary-phase depends on many biochemical functions, including posttranslational protein modification, transport, and specific enzyme activity.
A fifth group of mutants lost viability in at least two out of four trials (see Web Supplement). These included NGR1, encoding a growth regulator that is highly conserved among eukaryotes (Lee and Moss, 1993), and ALD3, an aldehyde dehydrogenase induced in response to stress (Boucherie, 1985). Two genes, SPG1 and SPG3, were identified in this group and encode proteins with unknown function.
Evolutionary Distribution of Stationary Phase-expressed and SP-Essential Genes
We had previously hypothesized that because quiescence is likely to have been the state of the first cell and almost all cells retain the ability to become quiescent, it is likely that genes essential for this state would be conserved in all three major domains of life (Braun et al., 1996; Woese et al. 1990). To determine whether the phylogenetic distribution of SP-expressed and SP-essential genes differed from the distribution of all yeast genes, we evaluated all yeast genes and SP-expressed and SP-essential genes by using the Clusters of Orthologous Groups databases (COG/KOG/TWOG), which includes 63 prokaryotic and seven eukaryotic genomes (http://www.ncbi.nlm.nih.gov/COG/new/). These databases have been used previously for phylogenetic analyses, including an analysis of the differences in phylogenetic conservation between essential and all genes in yeast and Caenorhabditis elegans (Koonin et al., 2004).
The phylogenetic distribution of both SP-expressed and SP-essential genes differs significantly from the distribution of all yeast genes (Figure 6). Significantly more SP-expressed and SP-essential genes are highly conserved across all three phylogenetic domains (Euk/Prok/Arch), and more SP-essential genes are found in the eukaryote/prokaryote domains than would be expected based on the distribution of all yeast genes. This indicates that both SP-expressed and SP-essential genes are more highly conserved than yeast genes in general. Fewer genes than expected in both the SP-expressed and SP-essential gene sets are found only in eukaryotic organisms, in addition to S. cerevisiae. The results of this analysis are consistent with the hypothesis that yeast genes expressed during, and essential for survival in stationary-phase, are more broadly conserved than would be predicted based on the phylogenetic distribution of yeast genes as a whole.
DISCUSSION
This study examines, at the genomic level, changes in gene expression in quiescent yeast cells during the transition to growth in glucose-rich medium. Using microarray and qRT-PCR analysis, we identified four distinct and highly reproducible patterns of gene expression during this process. There is a rapid decrease in stationary-phase mRNAs and an extremely rapid induction of the machinery to synthesize and assemble ribosomes. Interestingly, the most substantial change in expression of a number of genes occurred within 5 min after refeeding.
Because quiescent cells have very low rates of macromolecular synthesis (Gray et al., 2004), it is reasonable to ask how these cells could mount such a rapid and massive response? The level of cellular organization at which this response occurs, e.g., through de novo transcription or via another method for rapidly producing full-length transcripts, is not known. Preliminary data suggest mRNA may be present but sequestered (Aragon, unpublished data). More frequent sampling during the time course as well as other analyses will be required to more thoroughly understand this process.
Our analysis identified the binding sites for Rap1 and BUF (also known as the heterotrimeric replication-protein A, RPA) in front of genes whose transcripts increase rapidly with refeeding. Because Rap1 and BUF (RPA) function both as activators and repressors of gene expression (Kurtz and Shore, 1991; Singh and Samson, 1995; Iftode et al., 1999), their exact roles here are unknown. However, both Rap1p and the RPA are known to function in chromatin remodeling (Luche et al., 1993; Iftode et al., 1999; Yu and Morse, 1999). RPA, a single-stranded DNA binding protein complex, is required for DNA denaturation, replication, repair, stability, as well as in transcriptional regulation. Rap1p has been shown to open chromatin and this assists subsequent activator binding (Yu and Morse, 1999). Thus, chromatin remodeling mediated by Rap1p and RPA may be critical to achieving the rapid increase in expression levels during exit from stationary-phase, perhaps during the transition from condensed G0 chromosomes in quiescent cells (Piñon, 1978) to interphase chromosomes in cells reentering the cell cycle.
The promoter motif GATGAG, the reverse complement of the RPA-binding motif, also has been identified in genomic searches for regulatory elements (Tavazoie et al., 1999; Hughes et al., 2000; Bussemaker et al., 2001; Sudarsanam et al., 2002). Interestingly, this sequence is similar to the PAC box consensus sequence [TG(A/C)GATGAG] that is found in promoters of genes encoding subunits for RNA polymerase I and III (Dequard-Chablat et al., 1991). The PAC box also has been found to be associated with coregulated mRNAs in other microarray datasets (Sudarsanam et al., 2002). If the GATGAG functions as a PAC box, this would indicate a shared cis-regulatory element between rRNA and ribosomal protein genes.
This study has added a significant number to the list of genes known to be essential for survival in stationary-phase. The combined condition of stationary-phase and 37°C we used to detect SP-essential genes might have identified mutants with phenotypes not specifically associated with stationary-phase, especially because a large number of our SP-essential mutants (23 of 32) are annotated as having growth defects on nonfermentable carbon sources. We believe that this assay did identify genes that are essential in stationary-phase at 37°C for several reasons. All of the mutant strains survived for 16 d at 30°C, several days past the postdiauxic phase where cell metabolism changes from fermentation to use of nonfermentable carbon sources. Thirty-nine of the 95 strains that did not exhibit growth defects in our screen also are annotated as having growth defects on nonfermentable carbon sources. Thus, this screen did not simply select for strains with growth defects on nonfermentable carbon sources. In addition, we also did not simply identify strains that lost viability at 37°C, because only one of the 32 mutants has been shown to exhibit growth defects after 20 generations of exponential growth at 37°C (Nislow, personal communication).
An important finding of our work is that there is a real relationship between mitochondrial function and survival in stationary-phase. The requirement for OM45 and POR1 underscores the importance of this activity in quiescent cells. Just as the phrase “died on plates” was a catchall for stationary-phase mutants before the process of entry into stationary-phase was well studied, “does not grow on nonfermentable carbon sources” is also a category that will become more clearly defined as we learn more about mitochondrial function during the yeast life cycle.
Especially interesting to us were the nine SP-essential genes that are not required for growth on nonfermentable carbon sources: MOH1, PST2, FAA1, HBT1, SPG1, SPG3, SPG4, SPG5, and FMP45. Moh1p is a myristolated protein of unknown function whose sequence is related to Snf7p (SGD); Fmp45p (YDL222C) is a protein involved in sphingolipid metabolism that localizes to the mitochondria as well as to the cell cortex (Young et al., 2002), and Pst2p is a protein secreted by regenerating protoplasts (SGD). Faa1p is a protein with long-chain fatty acid ligase activity involved in myristolation (SGD); and Hbt1p is a protein implicated in cell wall morphogenesis that is a substrate of Hub1p, a ubiquitin-like protein (SGD). Thus, posttranslational modifications and the activity of many proteins not involved in macromolecular synthesis seem to be increasingly important for survival as cells persist in stationary-phase at 37°C.
SP-essential genes identified in other studies include ARD1 and NAT1, encoding proteins required for N-terminal acetylation (Mullen et al., 1989); UBI4, encoding ubiquitin (Finley et al., 1987); MOH1 (Ashrafi et al., 1998) and SSA1 and SSA2 (as a double mutant), which encode cytoplasmic chaperones (Craig and Jacobsen, 1984). In addition, a bcy1 mutant strain containing defective regulatory subunits of protein kinase A (PKA) was used to demonstrate the importance of down-regulation of PKA for survival in stationary-phase (Peck et al., 1997). These results are consistent with the hypothesis that posttranslational modifications are critical for stationary-phase survival.
Finally, because of the broad conservation of SP-essential genes, it was of interest to identify their functional significance in other organisms. Twenty-three of the 32 essential genes identified here have human homologues and, of these, at least 13 are involved in apoptosis or are associated with human diseases, including cancer (see Web Supplement). The human homologues of MOH1 and NGR1 are known to induce apoptosis (YPD Bioknowledge Library) and the NGR1/RBP1 homologue inhibits growth when overexpressed in yeast (Akada et al., 1997). Of the five homologues associated with various cancers, four (HBT1, ALD3, CTA1, and SDH2) are mutated or down-regulated in tumor cells (Foury, 1997), whereas FAA1 is induced in colon cancer and RIP1 deficiency may be associated with mitochondrial myopathy. (YPD Bioknowledge Library). QCR7 does not have a human homologue but encodes a protein homologous to C. elegans T02H6, a mitochondrial protein thought to regulate cellular respiration and life span length (Hodges et al., 1999).
In conclusion, these studies indicate that examination of the quiescent state in yeast can provide valuable insights into such diverse biological processes as evolution and human disease. In addition, the evolutionary conservation of important human genes and yeast orthologues involved in the processes of entry into, survival in, and exit from the quiescent state makes comparative genomic studies of the quiescent state very compelling.
Acknowledgments
We thank Jose Weber for technical support with printing arrays, and Ed Thomas and David Haaland for help with the experimental design. We are grateful to George Davidson, Kevin Boyack, and Brian Wylie for help with VxInsight, and we thank Matt Crawford for help with YPD Bioknowledge Library (Incyte Systems, Palo Alto, CA). We also are grateful to Mordechai Choder, Michael Gilchrist, Monica Manginell, Ed Braun, and members of our laboratory for helpful discussions. This work was supported by grants from the National Science Foundation (MCB-0092364) and National Institutes of Health (HG-02262) to M.W.W. and National Institutes of Health (GM-67593) to M.W.W., G.A.Q., and S.S.A.-A.; National Science Foundation (DBI-0314435) to M.J.M; and National Institutes of Health (IMSD GM-60201) and U.S. Department of Agriculture (99-38422-8034) to A.D.A.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–11–0856. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–11–0856.
References
- Akada, R., Yamamoto, J., and Yamashita, I. (1997). Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254, 267–274. [DOI] [PubMed] [Google Scholar]
- Alexander, B.D., and Perfect, J.R. (1997). Antifungal resistance trends towards the year 2000 - implications for therapy and new approaches. Drugs 54, 657–678. [DOI] [PubMed] [Google Scholar]
- Ashrafi, K., Farazi, T.A., and Gordon, J.I. (1998). A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary-phase. J. Biol. Chem. 273, 25864–25864. [DOI] [PubMed] [Google Scholar]
- Ashrafi, K., Sinclair, D., Gordon, J.I., and Guarente, L. (1999). Passage through stationary-phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96, 9100–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson, L.F., and Fraenkel, D.G. (1983). Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherie, H. (1985). Protein synthesis during transition and stationary-phases under glucose limitation in Saccharomyces cerevisiae. J. Bacteriol. 161, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, E.L., Fuge, E.K., Padilla, P.A., and Werner-Washburne, M. (1996). A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J. Bacteriol. 178, 6865–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker, H.J., Li, H., and Siggia, E.D. (2001). Regulatory element detection using correlation with expression. Nat. Genet. 27, 167–171. [DOI] [PubMed] [Google Scholar]
- Choder, M. (1991). A general topoisomerase I-dependent transcriptional repression in the stationary-phase of yeast. Genes Dev. 5, 2315–2326. [DOI] [PubMed] [Google Scholar]
- Choi, J., Lou, W., and Vancura, A. (1998). A novel membrane-bound glutathione S-transferase functions in the stationary-phase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 29915–29922. [DOI] [PubMed] [Google Scholar]
- Conlon, E.M., Liu, X.S., Lieb, J.D., and Liu, J.S. (2003). Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl. Acad. Sci. USA 100, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, E.A., and Jacobsen, K. (1984). Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell 38, 841–849. [DOI] [PubMed] [Google Scholar]
- Crauwels, M., Donaton, M.C.V., Pernambuco, M.B., Winderickx, J., deWinde, J.H., and Thevelein, J.M. (1997). The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (fgm) pathway. Microbiology 143, 2627–2637. [DOI] [PubMed] [Google Scholar]
- Davidson, G.S., Wylie, B.N., and Boyack, K. (2001). Cluster stability and the use of noise in interpretation of clustering. Proc. IEEE Info. Vis. 2001, 23–30. [Google Scholar]
- Dequard-Chablat, M., Riva, M., Carles, C., and Sentenac, A. (1991). RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J. Biol. Chem. 266, 15300–15307. [PubMed] [Google Scholar]
- Dittmar, G.A., Wilkinson, C.R., Jedrzejewski, P.T., and Finley, D. (2002). Role of an ubiquitin-like modification in polarized morphogenesis. Science 295, 2442–2446. [DOI] [PubMed] [Google Scholar]
- Duntze, W., Neumann, D., Gancedo, J.M., Atzpodien, W., and Holzer, H. (1969). Studies on the regulation and localization of the glyoxylate cycle enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 10, 83–89. [DOI] [PubMed] [Google Scholar]
- Finley, D., Özaynak, E., and Varshavsky, A. (1987). The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Foury, F. (1997). Human genetic diseases: a cross-talk between man and yeast. Gene 195, 1–10. [DOI] [PubMed] [Google Scholar]
- Fuge, E.K., Braun, E.L., and Werner-Washburne, M. (1994). Protein-synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 176, 5802–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain, M., Talla, E., and Francois, J.M. (2002). Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19, 215–224. [DOI] [PubMed] [Google Scholar]
- Granot, D., and Snyder, M. (1993). Carbon source induces growth of stationary-phase yeast cells, independent of carbon source metabolism. Yeast 9, 465–479. [DOI] [PubMed] [Google Scholar]
- Gray, J.V., Petsko, G.A., Johnston, G.C., Ringe, D., Singer, R.A., and Werner-Washburne, M. (2004). “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P.K. (2002). Stationary-phase in yeast. Curr. Opin. Microbiol. 5, 602–607. [DOI] [PubMed] [Google Scholar]
- Hodges, P.E., McKee, A.H.Z., Davis, B.P., Payne, and W.E., Garrels, J.I. (1999). Yeast proteome database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 27, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.D., Estep, P.W., Tavazoie, S., and Church, G.M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Iftode, C., Daniely, Y., and Borowiec, J.A. (1999). Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180. [DOI] [PubMed] [Google Scholar]
- Johnson, D.R., Knoll, L.J., Levin, D.E., and Gordon, J.I. (1994). Saccharomyces cerevisiae contains 4 fatty-acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid-metabolism. J. Cell Biol. 127, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jona, G., Choder, M., and Gileadi, O. (2000). Glucose starvation induces a drastic reduction in the rates of both transcription and degradation of mRNA in yeast. Biochim. Biophys. Acta 1491, 37–48. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., et al. (2004). A comprehensive evolutionary classification of proteins encoded incomplete eukaryotic genomes. Genome Biol. 5, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, S., and Shore, D. (1991). Rap1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5, 616–628. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and Kunau, W. (1997). Peroxisomes in the molecular and cellular biology of the yeast Saccharomyces, in The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Cell Cycle and Cell Biology, Cold Spring Harbor, NY: Cold Spring Harbor Press, 547–606.
- Lee, F.J., and Moss, J. (1993). An RNA-binding protein gene (RBP1) of Saccharomyces cerevisiae encodes a putative glucose-repressible protein containing two RNA recognition motifs. J. Biol. Chem. 268, 15080–15087. [PubMed] [Google Scholar]
- Lewis, D.L., and Gattie, D.K. (1991). The ecology of quiescent microbes. ASM News 57, 27–32. [Google Scholar]
- Lewis, K. (2000). Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64, 503–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luche, R.M., Smart, W.C., Marion, T., Tillman, M., Sumrada, R.A., and Cooper, T.G. (1993). Saccharomyces cerevisiae BUF protein binds to sequences participating in DNA replication in addition to those mediating transcriptional repression (URS1) and activation. Mol. Cell. Biol. 9, 5749–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, M.J., Aragon, A.D., Rodriguez, A.L., Weber, J.M., Timlin, J.A., Sinclair, M.B., Haaland, D.M., and Werner-Washburne, M. (2003). Identification and removal of contaminating fluorescence from commercial and inhouse printed DNA microarrays. Nucleic Acids Res. 13, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara, K., and Sato, R. (1985). Molecular cloning and sequencing of cDNA for yeast porin, an outer mitochondrial membrane protein: a search for targeting signal in the primary structure. EMBO J. 4, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta, K., Tsujii, R., Warner, J.R., and Nishiyama, M. (1998). The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res. 26, 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, J.R., Kayne, P.S., Moerschell, R.P., Tsunasawa, S., Gribskov, M., Colavito-Shepanski, M., Grunstein, M., Sherman, F., and Sternglanz, R. (1989). Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva, S., Devaux, F., Barbosa, S., Jacq, C., and Casal, M. (2004). Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast 21, 201–210. [DOI] [PubMed] [Google Scholar]
- Parrish, N.M., Dick, J.D., and Bishai, W.R. (1998). Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6, 107–112. [DOI] [PubMed] [Google Scholar]
- Peck, V.M., Fuge, E.K., Padilla, P.A., Gomez, M.A., and WernerWashburne, M. (1997). Yeast bcy1 mutants with stationary-phase-specific defects. Curr. Genet. 32, 83–92. [DOI] [PubMed] [Google Scholar]
- Piñon, R. (1978). Folded chromosomes in non-cycling yeast cells. Evidence for a characteristic G0 form. Chromosoma 67, 263–274. [DOI] [PubMed] [Google Scholar]
- Repetto, B., and Tzagoloff, A. (1989). Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol. Cell. Biol. 9, 2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runner, V.M., and Brewster, J.L. (2003). A genetic screen for yeast genes induced by sustained osmotic stress. Yeast 20, 913–920. [DOI] [PubMed] [Google Scholar]
- Singh, K.K., and Samson, L. (1995). Replication protein A binds to regulatory elements in yeast DNA repair and DNA metabolism genes. Proc. Natl. Acad. Sci. USA 92, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam, P., Pilpel, Y., and Church, G.M. (2002). Genome-wide co-occurrence of promoter elements reveals a cis-regulatory cassette of rRNA transcription motifs in Saccharomyces cerevisiae. Genome Res. 12, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie, S., Hughes, J.D., Cambell, M.J., Cho, R.J., and Church, G.M. (1999). Systematic determination of genetic network architecture. Nat. Genet. 22, 281–285. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. (2001). Creating the gene ontology resource: design and implementation. Genome Res. 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, M.P., and Garrett, S. (1994). Suppression of a yeast cyclic-AMP-dependent protein-kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally-regulated mouse gene. Mol. Cell. Biol. 14, 5619–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne, M., Braun, E., Johnston, G.C., and Singer, R.A. (1993). Stationary-phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57, 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne, M., Wylie, B., Boyack, K., Fuge, E., Galbraith, J., Weber, J., and Davidson, G. (2002). Comparative analysis of multiple genome-scale data sets. Genome Res. 12, 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., and Wheelis, M.L. (1990). Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc. Natl. Acad. Sci. USA 87, 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Zhang, N.S., Hayes, A., Butler, P., and Oliver, S. (2001). Comprehensive analysis of the yeast stationary-phase transcriptome. Yeast 18, S101–S101. [Google Scholar]
- Yaffe, M.P., Jensen, R.E., and Guido, E.C. (1989). The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J. Biol. Chem. 264, 21091–21096. [PubMed] [Google Scholar]
- Young, M.E., Karpova, T.S., Brügger, B., Moschenross, D.M., Wang, G.K., Schneiter, R., Wieland, F.T., and Cooper, J.A. (2002). The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.N., and Morse, R.H. (1999). Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]