Abstract
Septins are filament-forming proteins that function in cytokinesis in a wide variety of organisms. In budding yeast, the small GTPase Cdc42p triggers the recruitment of septins to the incipient budding site and the assembly of septins into a ring. We herein report that Bni1p and Cla4p, effectors of Cdc42p, are required for the assembly of the septin ring during the initiation of budding but not for its maintenance after the ring converts to a septin collar. In bni1Δ cla4-75-td mutant, septins were recruited to the incipient budding site. However, the septin ring was not assembled, and septins remained at the polarized growing sites. Bni1p, a formin family protein, is a member of the polarisome complex with Spa2p, Bud6p, and Pea2p. All spa2Δ cla4-75-td, bud6Δ cla4-75-td, and pea2Δ cla4-75-td mutants showed defects in septin ring assembly. Bni1p stimulates actin polymerization for the formation of actin cables. Point mutants of BNI1 that are specifically defective in actin cable formation also exhibited septin ring assembly defects in the absence of Cla4p. Consistently, treatment of cla4Δ mutant with the actin inhibitor latrunculin A inhibited septin ring assembly. Our results suggest that polarisome components and Cla4p are required for the initial assembly of the septin ring and that the actin cytoskeleton is involved in this process.
INTRODUCTION
The development of both unicellular and multicellular organisms requires cells to respond to intracellular and extracellular cues that direct growth and division. These signals regulate polarized cell growth, maintenance of cell shape, cell motility, and cytokinesis (Drubin and Nelson, 1996). Polarized cell growth is a complex process that requires the directed organization of the actin cytoskeleton and the coordinated function of many polarity proteins and signal transduction cascades. Cells of the yeast Saccharomyces cerevisiae grow by budding, a process in which a rigid cell wall is locally expanded as a result of polarized secretion (Pruyne and Bretscher, 2000a). Before bud emergence, cells polarize the actin cytoskeleton toward the future bud site and assemble a septin ring at that site.
The septins are a conserved family of filament-forming proteins that play important roles in a variety of cell functions in fungal and animal cells (Longtine et al., 1996; Trimble, 1999; Gladfelter et al., 2001; Longtine and Bi, 2003). Typical septins have a variable N-terminal region, a conserved core that includes the element of a GTP-binding site, and a variable C-terminal region. Septins were first identified as temperature-sensitive cdc mutants in S. cerevisiae (Hartwell, 1971). Septins are assembled into a ring before bud formation and remain as a collar subjacent to the plasma membrane at the mother-bud neck for most of the cell cycle. The septin ring is assembled by the copolymerization of the septins Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p/Sep7p into filaments (Frazier et al., 1998; Field and Kellogg, 1999). Purified septins from yeast form filaments of 7–9-nm diameter and of various lengths (Frazier et al., 1998).
One major role for the septins in various organisms is in cytokinesis (Hartwell, 1971; Neufeld and Rubin, 1994; Kinoshita et al., 1997; Bi et al., 1998; Lippincott and Li, 1998). The septins localize to the division site (cleavage furrow) during cytokinesis (Longtine et al., 1996; Trimble, 1999; Nguyen et al., 2000; Westfall and Momany, 2002). The septins recruit a variety of other proteins, whose correct localization to the neck is critical for the performance of their various functions, including chitin deposition (DeMarini et al., 1997), bud site selection (Chant et al., 1995; Sanders and Herskowitz, 1996), and cell cycle control (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). Thus, the septins have been proposed to function as a scaffold at the bud neck for protein anchoring and organization (Longtine et al., 1996, 2000; Field and Kellogg, 1999; Gladfelter et al., 2001). Additionally, the septins function as a diffusion barrier at the mother-bud junction to prevent membrane-associated proteins from moving freely between the mother and bud compartments (Barral et al., 2000; Takizawa et al., 2000). In spite of our plentiful knowledge of the functions of septins, little is known as to how the septin ring is assembled during budding initiation.
Reorganization of both actin and septins requires the Rho type small GTPase Cdc42p (Pringle et al., 1995). Conditional cdc42 mutant cells are defective in assembly of the septin ring and polarized reorganization of the actin cytoskeleton. Therefore, these cells are defective in bud emergence and localized cell surface growth, and they become arrested as large, multinucleate, unbudded cells at the restrictive growth temperature. Isolation of cdc42 mutants, which specifically display defects in septin ring assembly, and analysis of the effect of Cdc42p GAPs on septin ring assembly suggest that cycles of GTP loading and hydrolysis by Cdc42p play an important role in septin ring assembly (Gladfelter et al., 2002; Smith et al., 2002; Caviston et al., 2003). Although significant effort has been dedicated to deciphering the Cdc42p effector pathways important for actin polarization, little is known about how Cdc42p mediates septin ring assembly. Several effectors of Cdc42p have been identified, including the p21-activated kinase (PAK)-like kinases Ste20p, Cla4p, and Skm1p, a formin family protein Bni1p, and Gic1p and Gic2p (Pruyne and Bretscher, 2000a). Among these effectors, Cla4p is required for normal septin function (Cvrckova et al., 1995; Weiss et al., 2000; Schmidt et al., 2003). However, cla4Δ mutant can still assemble a septin ring. Bni1p and its related protein Bnr1p assemble actin cables, which play a pivotal role in the polarized transport of secretory vesicles as a track for type V myosin, Myo2p (Evangelista et al., 2002; Sagot et al., 2002a; Schott et al., 2002). Bni1p can stimulate polymerization of actin in vitro independent of the Arp2/3 complex (Pruyne et al., 2002; Sagot et al., 2002b). Bni1p, together with Spa2p, Bud6p, and Pea2p, constitutes a large complex termed the 12S polarisome (Sheu et al., 1998; Pruyne and Bretscher, 2000b). Polarisome components are required for apical growth (Sheu et al., 2000); in their absence, vegetative buds grow as spheres rather than ellipsoids. In the course of our study on the genetic interactions between effectors of CDC42, we found that a bni1Δ mutation shows a synthetic lethal interaction with the cla4Δ mutation. The bni1Δ cla4-75-td mutant showed abnormal morphology, which was caused by a defect in the assembly of the septin ring during budding initiation. Mutations in other polarisome components also showed a similar synthetic defect with the cla4Δ mutation in septin ring assembly. Interestingly, actin cables were suggested to be involved in the septin ring assembly, downstream of Bni1p.
MATERIALS AND METHODS
Media and Genetic Techniques
Unless otherwise specified, strains were grown in rich medium YPDA (1% yeast extract [Difco Laboratories, Detroit, MI], 2% bacto-peptone [Difco], 2% glucose, and 0.01% adenine). Strains carrying plasmids were selected in synthetic medium (SD) containing the required nutritional supplements (Sherman, 1991). For induction of the GAL1 promoter, 3% galactose and 0.2% sucrose were used as carbon sources, instead of glucose (YPGA). Sporulation was performed as previously described (Mochida et al., 2002). Standard genetic manipulations of yeast were performed as described (Sherman and Hicks, 1991). Yeast transformations were performed by the lithium acetate method (Elble, 1992; Gietz and Woods, 2002). Escherichia coli strains DH5α and XL1-Blue were used for construction and amplification of plasmids.
Strains and Plasmids
Yeast strains used in this study are listed in Table 1. The complete deletion of the ORFs of BNI1, BUD6, SPA2, PEA2, CLA4, and BNR1 was performed with the use of a PCR-based procedure as described (Longtine et al., 1998b; Goldstein and McCusker, 1999). The ts-degron–tagged cla4-75 (cla4-75-td) construct was integrated at the URA3 locus as previously described (Holly and Blumer, 1999). The enhanced green fluorescent protein-tagged CDC12 (CDC12-EGFP) construct was integrated at the LEU2 locus. To overexpress a green fluorescent protein (GFP)-tagged Cdc42p or Cdc42G12Vp, the PGAL1-GFP-CDC42 or PGAL1-GFP-CDC42G12V construct was integrated at the URA3 locus. The tpm1-2 tpm2Δ, myo2-20, and arp2-2 mutants were backcrossed three times to YEF473 background strains. Plasmids used in this study are listed in Table 2. Schemes for the construction of plasmids and nucleotide sequences of PCR primers are available upon request.
Table 1.
S. cerevisiae strains used in this study
| Straina | Genotype | Reference or source |
|---|---|---|
| ABY530 | MATα lys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2-3,112 ade2-101 myo2-20::HIS3 | Schott et al. (1999) |
| ABY944 | MATalys2-801 ura3-52 his3Δ-200 trp1-1 leu2-3,112 tpm1-2::LEU2 tpm2Δ::HIS3 | Pruyne et al. (1998) |
| K1993 | MATaura3 his3-11,15 trp1-1 leu2-3,112 ade2-1 can1-100 ssd1-Δ2 cdc15-2 | A gift from K. Nasmyth |
| YMW221U | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 ade2-101 arp2-2(G19D)::URA3 | Madania et al. (1999) |
| YEF473 | MATa/α lys2-801/lys2-801 ura3-52/ura3-52 his3Δ-200/his3Δ-200 trp1Δ-63/trp1Δ-63 leu2Δ-1/leu2Δ-1 | Bi and Pringle (1996) |
| YEF2182 | MATabni1Δ::HIS3 bnr1Δ::HIS3 [YEp352-BNI1] | This study |
| YEF2632 | MATabni1Δ::TRP1 bnr1Δ::HIS3 [pRS316-BNR1] | This study |
| YEF2669 | MATabni1-116 bnr1Δ::HIS3 | This study |
| YKT38 | MATalys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 | Mochida et al. (2002) |
| YKT91 | MATamyo3Δ::TRP1 myo5-1::KanMX6 | Toi et al. (2003) |
| YKT382 | MATabni1Δ::HIS3MX6 | This study |
| YKT440 | MATabnr1Δ::HphMX4 HIS3::PGAL1-3HA-BNI1 | This study |
| YKT483 | MATacla4Δ::KanMX6 HIS3::PGAL1-3HA-BNI1 | This study |
| YKT541 | MATabni1Δ::HIS3MX6 cla4Δ::KanMX6 URA3::cla4-75-td | This study |
| YKT618 | MATacla4Δ::KanMX6 URA3::cla4-75-td | This study |
| YKT637 | MATaLEU2::CDC12-EGFP | This study |
| YKT638 | MATabni1-116 cla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT649 | MATabni1Δ::HIS3MX6 cla4Δ::KanMX6 URA3::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT664 | MATabud6Δ::HphMX4 cla4Δ::KanMX6 URA3::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT666 | MATaspa2Δ::HphMX4 cla4Δ::KanMX6 URA3::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT668 | MATapea2Δ::HphMX4 cla4Δ::KanMX6 URA3::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT676 | MATabni1Δ::HphMX4 LEU2::CDC12-EGFP | This study |
| YKT678 | MATacla4Δ::KanMX6 URA3::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT717 | MATacla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT743 | MATabni1-116 bnr1Δ::HphMX4 LEU2::CDC12-EGFP | This study |
| YKT744 | MATabni1-116 bnr1Δ::HphMX4 cla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT811 | MATabni1Δ::HIS3MX6 cla4Δ::KanMX6 TRP1::cla4-75-td LEU2::CDC12-EGFP | This study |
| YKT812 | MATatpm1-2::LEU2 tpm2Δ::HIS3 LEU2::CDC12-EGFP | This study |
| YKT813 | MATatpm1-2::LEU2 tpm2Δ::HIS3 cla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT814 | MATamyo2-20::HIS3 LEU2::CDC12-EGFP | This study |
| YKT815 | MATamyo2-20::HIS3 cla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT816 | MATaarp2-2(G19D)::URA3 LEU2::CDC12-EGFP | This study |
| YKT817 | MATaarp2-2(G19D)::URA3 cla4Δ::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT818 | MATamyo3Δ::TRP1 myo5-1::KanMX6 LEU2::CDC12-EGFP | This study |
| YKT819 | MATamyo3Δ::TRP1 myo5-1::KanMX6 cla4Δ::CgHIS3 LEU2::CDC12-EGFP | This study |
| YKT891 | MATaLEU2::CDC12-EGFP URA3::PGAL1-GFP-CDC42 | This study |
| YKT892 | MATaLEU2::CDC12-EGFP URA3::PGAL1-GFP-CDC42G12V | This study |
| YKT893 | MATabni1Δ::HphMX4 LEU2::CDC12-EGFP URA3::PGAL1-GFP-CDC42 | This study |
| YKT894 | MATabni1Δ::HphMX4 LEU2::CDC12-EGFP URA3::PGAL1-GFP-CDC42G12V | This study |
| YKT939b | MATacdc15-2 LEU2::CDC12-EGFP | This study |
| YKT940b | MATabni1Δ::KanMX6 cdc15-2 LEU2::CDC12-EGFP | This study |
YEF and YKT strains, except for YKT939 and YKT940, are isogenic derivatives of YEF473. For YEF and YKT strains, only relevant genotypes are described.
These strains are derivatives of K1993.
Table 2.
Plasmids used in this study
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pFA6a-TRP1 | TRP1 | Longtine et al. (1998b) |
| pFA6a-KanMX6 | PTEF-KanMX6-TTEF | Longtine et al. (1998b) |
| pAG32 | PTEF-HphMX4-TTEF | Goldstein and McCusker (1999) |
| pcla4-75-degron | cla4-75-ts degron URA3 | Weiss et al. (2000) |
| YCp50-LEU2-BNI1 | BNI1 LEU2 CEN4 | A gift from J. Pringle |
| YEp352-BNI1 | BNI1 URA3 2 μm | A gift from J. Pringle |
| YEp24 | URA3 2 μm | Rose and Broach (1990) |
| pRS316 | URA3 CEN6 | Sikorski and Hieter (1989) |
| pRS316-BNR1 | BNR1 URA3 CEN6 | This study |
| pKT1178 [pRS316-CDC12G] | CDC12-EGFP URA3 CEN6 | Iwase and Toh-e (2001) |
| pKT1254 [pRS316-CLA4] | CLA4 URA3 CEN6 | This study |
| pKT1364 [YEp24-BEM3(Δ1-114)] | BEM3 (Δ1-114) URA3 2 μm | This study |
| YEplac195 | URA3 2 μm | Gietz and Sugino (1988) |
| pKT1365 [YEplac195-RGA1(Δ1-632)] | RGA1 (Δ1-632) URA3 2 μm | This study |
| pRS314 | TRP1 CEN6 | Sikorski and Hieter (1989) |
| pKT1226 [pRS314-BNI1] | BNI1 TRP1 CEN6 | This study |
| pKT1420 [pRS314-BNI1(Δ6-808)] | BNI1 (Δ6-808) TRP1 CEN6 | This study |
| pKT1421 [pRS314-BNI1(Δ826-986)] | BNI1 (Δ826-986) TRP1 CEN6 | This study |
| pKT1422 [pRS314-BNI1(1-1750)] | BNI1 (1-1750) TRP1 CEN6 | This study |
| pKT1423 [pRS314-BNI1(Δ1228-1414)] | BNI1 (Δ1228-1414) TRP1 CEN6 | This study |
| pKT1424 [pRS314-BNI1(Δ1553-1646)] | BNI1 (Δ1553-1646) TRP1 CEN6 | This study |
| pRS305 | LEU2 | Sikorski and Hieter (1989) |
| pKT1452 [pRS305-CDC12-EGFP] | CDC12-EGFP LEU2 | This study |
| pRS306 | URA3 | Sikorski and Hieter (1989) |
| pKT1474 [pRS306-PGAL1-GFP-CDC42] | PGAL1-GFP-CDC42 URA3 | This study |
| pKT1475 [pRS306-PGAL1-GFP-CDC42G12V] | PGAL1-GFP-CDC42G12V URA3 | This study |
Construction of the bni1-116 bnr1Δ Strain
We randomly mutagenized the entire BNI1 gene by a PCR-based method (Cadwell and Joyce, 1992; Caviston et al., 2002). The mutagenized PCR products were mixed with an equal amount of linearized YCp50-LEU2-BNI1, in which a major portion of the BNI1 gene was removed by a restriction digestion with PvuII (sequences from 2329-base pairs downstream of the BNI1 ATG to 337-base pairs downstream of the BNI1 stop codon were removed). This DNA mixture was transformed into YEF2182 (a bni1Δ::HIS3 bnr1Δ::HIS3) containing YEp352-BNI1. Transformants were selected on SD-Leu plates at 24°C and replicated onto SD-Leu plates containing 5-fluoroorotic acid to select for the loss of the URA3-containing plasmid. These Ura– Leu+ colonies were replicated onto two sets of SD-Leu plates that were incubated at 24 and 37°C, respectively, to allow the identification of the temperature-sensitive (ts) mutants. The plasmid carrying the bni1-ts allele was isolated and the bni1-ts ORF was sequenced by a standard protocol.
To integrate a bni1-ts mutation onto the chromosomal locus, the plasmid carrying the mutant allele was digested with SphI and AflII, which released the entire DNA insert carrying the bni1-ts allele from the plasmid backbone. The digestion mixture was transformed into YEF2632 (a bni1Δ::TRP1 bnr1Δ::HIS3) containing the pRS316-BNR1. The transformation mixture was plated onto YPDA plates at 23°C for 3 d and replicated onto synthetic complete (SC) plates containing 5-fluoroorotic acid to select for the loss of the URA3-containing plasmid. The Ura– Trp– colonies were streaked onto two sets of YPDA plates and incubated at 24 and 37°C, respectively, to confirm temperature sensitivity. The ts phenotype of each bni1-ts bnr1Δ strain was fully complemented by a centromere-based plasmid carrying either BNI1 or BNR1 (unpublished results). In this study, we used bni1-116 bnr1Δ strain, one of them. The characterization of this mutant strain will be described elsewhere.
Microscopic Observations
Cells were observed using a Nikon ECRIPS E800 microscope (Nikon Instec, Tokyo, Japan) equipped with HB-10103AF super high-pressure mercury lamp and 1.4NA 100× Plan Apo oil immersion objective (Nikon Instec) with appropriate fluorescence-filter sets (Nikon Instec) or differential interference contrast (DIC) optics. Images were acquired using a cooled digital CCD camera (C4742–95-12NR; Hamamatsu Photonics K.K., Hamamatsu, Japan) and AQUACOSMOS software (Hamamatsu Photonics K.K.). To observe Cdc12p-GFP, cells were fixed for 5 min at room temperature by direct addition of commercial 37% formaldehyde stock (Wako Pure Chemical Industries, Osaka, Japan) to a final concentration of 3.7% in the medium and washed twice with phosphate-buffered saline before mounting on a glass microscope slide. Fixed cells were observed using a GFP bandpass filter set (excitation, 460–500 nm; dichroic mirror, 505 nm; emission, 510–560 nm).
Time-lapse analyses of cell morphology and septin ring assembly were carried out as follows. Cells were grown to an early logarithmic phase in SC medium at 25°C, harvested by brief centrifugation, washed once with SC, and resuspended in SC. The cell suspension was spotted onto a thin layer of SC medium containing 1% agarose on a glass microscope slide, which was quickly covered with a coverslip. Around the edges of the coverslip, a small amount of vaseline (Wako) was applied for sealing. An image at each time point was acquired as described above. During observation, the sample was kept at 37°C by Thermo plate (Tokai HIT Co., Fujinomiya, Japan).
Initial septin ring assembly was monitored by observing Cdc12p-GFP in cells exiting from cell cycle arrest. Cells were synchronized in the G1 phase of the cell cycle by the addition of α-factor and released from the block by removal thereof. In brief, cells were grown to a logarithmic phase, pelleted, and resuspended in YPDA containing 1 μg/ml α-factor (Sigma Chemical, St. Louis) at 0.3–0.5 × 107 cells/ml. When cells were observed to have shmoos, cells were washed with 10 ml of YP (1% yeast extract, 2% bacto-peptone) three times and released back into fresh YPDA at 25 or 37°C. The septin ring assembly phenotypes were also examined in cells exiting from stationary phase. Stationary phase cells were collected as described by Ayscough et al. (1997) with minor modifications. An overnight culture was inoculated in YP containing 2% raffinose and 0.01% adenine and grown for 2 d at 25°C. Cells were pelleted and resuspended in YP containing 1 M sorbitol. Cells were spun at 500 × g for 1 min. Unbudded cells remained in the supernatant fraction. Cells were repeatedly centrifuged until a uniform population of unbudded cells was obtained. These cells were pelleted, resuspended in YPDA or YPGA at 1.0 × 107 cells/ml, and released from stationary phase at 25 or 37°C. These two experiments gave essentially the same results. Therefore, the results of the α-factor arrest-release experiment were presented, unless otherwise mentioned. cdc15-2 and bni1Δ cdc15-2 mutant cells were synchronized as described (Fitch et al., 1992). When the effect of latrunculin A (LAT-A; Wako) was examined, G1-arrested cells were treated with 100 μM LAT-A (added to the medium from a 20 mM stock in DMSO) as described by Ayscough et al. (1997). As a control, an equal volume of DMSO alone was added. At least 100 cells containing polarized Cdc12p-GFP were observed in each experiment, where >90% of them showed a uniform phenotype, unless otherwise mentioned. A representative image of the cells is shown in each figure.
To measure septin ring diameter, cells were released from G1 arrest for 30 min and fixed before bud emergence. The diameter of the septin ring, which was defined as the maximum distance across the septin ring, was measured from the center between outer and inner edges of the septin ring to the opposite center between the edges using Adobe illustrator version 9.0 (Adobe Systems, San Jose, CA). For each determination of the average diameter of septin rings, 100 cells were chosen randomly and measured.
Isolation of Multicopy Suppressors of the bni1-116 cla4Δ mutant
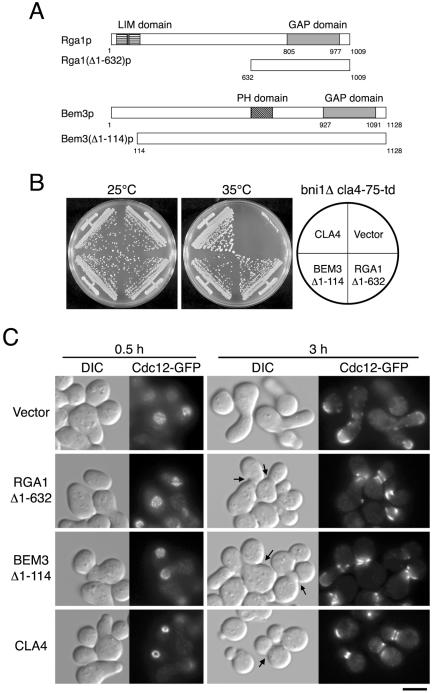
The bni1-116 cla4Δ strain (YKT530) was transformed with a yeast genomic DNA library constructed in the multicopy plasmid YEp24. After transformation, cells were incubated on SD-Ura plates at 25°C for 40 h to allow recovery, replica-plated onto fresh YPDA plates, and then incubated at 35°C for 3 d. About 50,000 transformants were screened, and 12 transformants that reproducibly grew at 35°C were obtained. From each of the transformants, plasmids were recovered for further analysis. PCR revealed that five plasmids contained BNI1 ORF, but no plasmid contained CLA4 ORF. All of the remaining seven plasmids conferred temperature-resistant growth on YKT530. The genes present in the seven plasmids were identified by sequencing both ends of the inserts. The suppressor gene in those was identified by testing individual subcloned fragments for suppressing activity. Two of them were BEM3Δ1-114 and RGA1Δ1-632. The other five genes will be described elsewhere.
RESULTS
Loss of Bni1p Causes Defective Septin Organization in the Absence of Cla4p
The effectors of Cdc42p are involved in cellular polarization through the cell cycle, but the details of their functions are unknown. To further explore these functions, we examined the genetic interactions between effectors of CDC42. Previous reports showed that a cell lacking both Cla4p and Ste20p kinases was inviable (Cvrckova et al., 1995; Holly and Blumer, 1999; Goehring et al., 2003). We examined the genetic interaction of BNI1 with CLA4 or STE20. Interestingly, cla4Δ mutation led to a synthetic lethal interaction with the bni1Δ mutation, whereas there was no genetic interaction for cell growth between ste20Δ and bni1Δ (unpublished results).
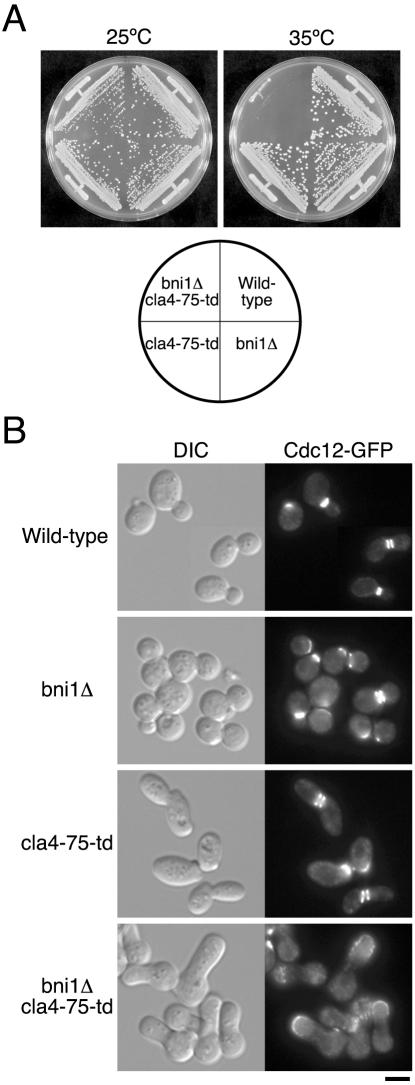
To examine the terminal phenotype of cla4Δ mutant lacking Bni1p, we constructed a series of strains expressing an integrated cla4-75-ts degron construct (cla4-75-td). This version of Cla4p was degraded rapidly after a shift to the restrictive temperature (Holly and Blumer, 1999), and thus the bni1Δ cla4-75-td mutant showed a ts growth (Figure 1A). At the restrictive temperature, bni1Δ cla4-75-td mutant cells exhibited a wide bud neck and abnormally elongated bud morphology (Figure 1B). We examined the organization of septin, a constituent of bud neck filaments, by observing GFP-tagged Cdc12p (Figure 1B). In wild-type and single mutant cells, the septins localized at the bud neck. In contrast, the septins localized as a cap at the polarized growing site in the bni1Δ cla4-75-td mutant, suggesting that Bni1p and Cla4p are redundantly involved in septin organization.
Figure 1.
Loss of BNI1 causes defects in cell growth, cell morphology, and septin organization in the absence of Cla4p. (A) YKT38 (wild-type), YKT382 (bni1Δ), YKT618 (cla4-75-td), and YKT541 (bni1Δ cla4-75-td) were streaked on a YPDA plate, which was incubated at 25 or 35°C for 2 d. (B) YKT637 (CDC12-GFP), YKT676 (bni1Δ CDC12-GFP), YKT678 (cla4-75-td CDC12-GFP), and YKT649 (bni1Δ cla4-75-td CDC12-GFP) were grown in YPDA medium at 25°C and then shifted to 37°C for 2 h. Cells were fixed and observed by DIC and fluorescence microscopy. Bar, 5 μm.
Whereas our study was in progress, Sprague and colleagues reported a similar genetic interaction between BNI1 and CLA4 (Goehring et al., 2003). Our results are consistent with their data, in which they also showed that loss of Bni1p or its interacting proteins (Spa2p, Bud6p, and Pea2p; see below) in a cla4Δ mutant resulted in lethality and caused cells to form elongated buds with mislocalized septin rings. Furthermore, they showed that Bni1p is a Ste20p-dependent phosphoprotein and may be directly regulated by Ste20p. We extended this study and found that, as described below, Bni1p and Cla4p are required for the assembly of the septin ring during budding initiation.
Bni1p and Cla4p Are Required for the Initial Septin Ring Assembly, but Not for the Maintenance of Septin Collar
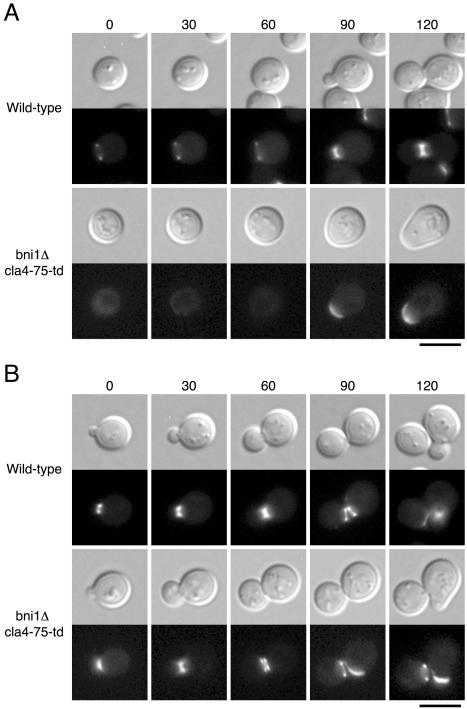
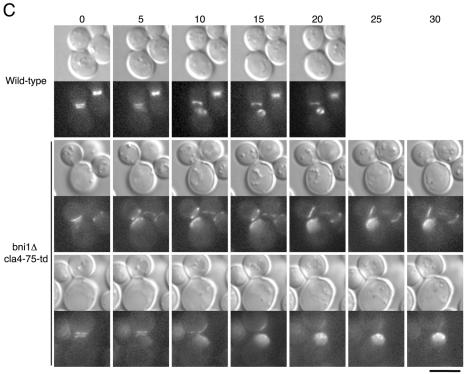
We performed time-lapse observations of a bni1Δ cla4-75-td mutant expressing Cdc12p-GFP. When unbudded bni1Δ cla4-75-td mutant cells that had not yet assembled a septin ring were shifted to 37°C, these cells could not assemble normal septin ring like that of wild-type cells (Figure 2A). Their septins, instead, accumulated as a cap at the incipient budding site and remained at the polarized growing site. In contrast, when budding bni1Δ cla4-75-td mutant cells that had formed a septin collar were shifted to 37°C, these cells could accomplish cytokinesis normally without disorganizing their septin collar (Figure 2B). However, the resulting mother and daughter cells showed the defect in septin ring assembly in the next cell cycle. To observe the initial septin assembly in detail, we performed time-lapse analyses with the 5-min time points. We observed 16 individual bni1Δ cla4-75-td mutant cells and found that none of them could assemble a septin ring at 37°C, suggesting that this mutant cannot assemble a septin ring even with a transient manner (Figure 2C). These results indicate that the bni1 cla4 double mutation causes a defect in the initial assembly of septin ring but not in the maintenance of the formed septin collar.
Figure 2.
bni1Δ cla4-75-td mutant is defective in initial septin ring assembly. Time-lapse microscopy was performed at 37°C on YKT637 (CDC12-GFP) and YKT649 (bni1Δ cla4-75-td CDC12-GFP). (A) Unbudded cells that had not yet assembled a septin ring were shifted to 37°C. (B) Budding cells that had formed a septin collar were shifted to 37°C. (C) Cells in cytokinesis were shifted to 37°C to observe the initial septin assembly. Images were recorded at the indicated time points (minutes). Bars, 5 μm.
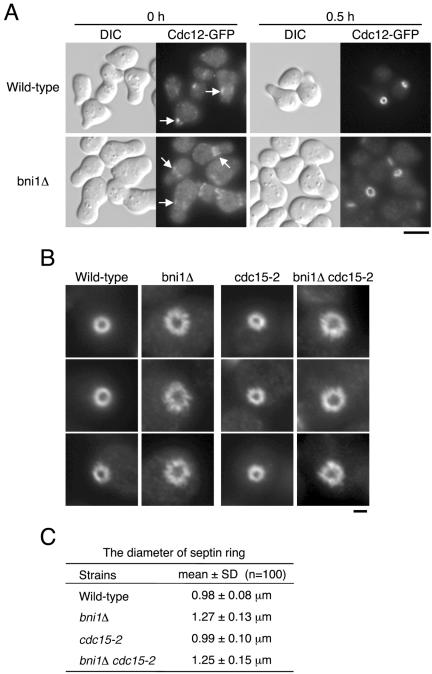
The cla4Δ mutant shows defects in septin functions (Cvrckova et al., 1995; Weiss et al., 2000; Schmidt et al., 2003). Because the septin defects have not been described in the bni1Δ mutant in detail, we carefully observed the septin rings assembled during the initiation of budding in cells exiting from the α-factor–induced G1 arrest. The G1-arrested wild-type cells made a very polarized shmoo, whereas the bni1Δ mutant did not and instead formed a broad blunt projection, as described (Evangelista et al., 1997). Septins formed an array of bars at the base of the projection in G1-arrested cells (Figure 3A), as described (Longtine et al., 1998a). After release from the arrest, we could observe the formation of a septin ring that is clearly bright compared with the shmoo-based septin array (Figure 3A). We found that the septin ring in bni1Δ mutant exiting from G1 arrest displays an irregular and jagged morphology (Figure 3B). This abnormal morphology was found in >95% of bni1Δ mutant cells. We furthermore found that the diameter of the septin ring was greater in the bni1Δ mutant (1.27 ± 0.13 μm) than in wild-type (0.98 ± 0.08 μm) cells (Figure 3C). To confirm the septin ring abnormalities observed in bni1Δ mutant more solidly, we performed other cell synchronization methods. We observed septin rings with a larger diameter in bni1Δ cells released from nutritional arrest, although the jagged morphology was less severe than that seen in the α-factor–arrested cells (unpublished results). However, it was difficult to obtain the statistic data of the initial septin ring morphology because the degree of synchrony, when released from nutritional arrest, was not sufficiently high compared with that achieved with an α-factor block and release. To obtain a better synchrony, we also used a cdc15-2 mutation, which causes a temperature-sensitive arrest in mitotic exit (Fitch et al., 1992). We examined the initial septin ring assembly in cdc15-2 or bni1Δ cdc15-2 mutant exiting from telophase. We found that the initial septin ring in bni1Δ cdc15-2 mutant had a larger diameter (1.25 ± 0.15 μm) than in cdc15-2 mutant (0.99 ± 0.10 μm) and displayed an aberrant morphology as that in bni1Δ mutant exiting from G1 arrest (Figure 3, B and C). These results suggest that the bni1Δ mutation affects the integrity of the septin ring and thus shows a synthetic defect with cla4Δ mutation in septin ring assembly.
Figure 3.
Loss of BNI1 causes defects in the integrity of the septin ring. (A) The septin ring in YKT637 (CDC12-GFP) and YKT676 (bni1Δ CDC12-GFP). Cells were G1-arrested with α-factor and released into fresh medium at 25°C. After 0- and 30-min incubations, cells were fixed and observed by fluorescence microscopy. At 0 min, septins formed an array of bars at the base of the projection (arrows). At 30 min, the initial septin rings were assembled. Bar, 5 μm. (B) The morphology of the septin ring in YKT637 (CDC12-GFP), YKT676 (bni1Δ CDC12-GFP), YKT939 (cdc15-2 CDC12-GFP), and YKT940 (bni1Δ cdc15-2 CDC12-GFP). YKT637 and YKT676 were G1-arrested with α-factor and released for 30 min as in A. YKT939 and YKT940 were arrested at telophase by 2-h incubation at 37°C and then released for 50 and 60 min, respectively, after a shift to 25°C. Cells were fixed and observed by fluorescence microscopy. Bar, 1 μm. (C) The average diameter of the septin ring in YKT637, YKT676, YKT939, and YKT940. To measure the average septin ring diameter, 100 cells of each strain were chosen randomly. The values are means with SD.
The Effects of Bni1p Truncation Mutants on Septin Ring Assembly in the Absence of Cla4p
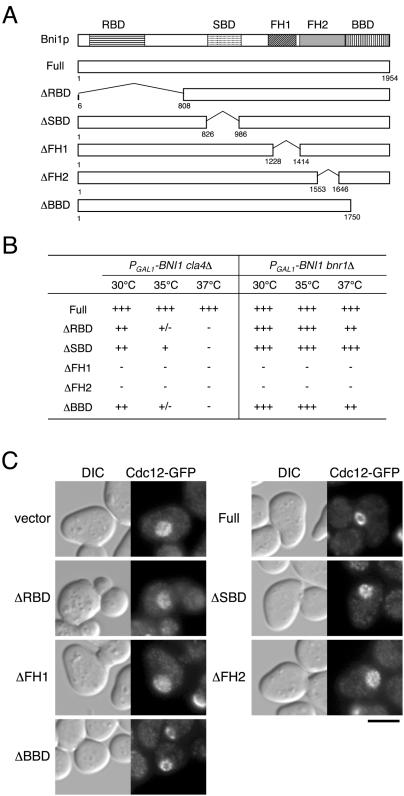
Bni1p is a member of a protein family that is characterized by formin homology (FH) domains, FH1 and FH2. The proline-rich FH1 domain binds to profilin as well as to peptide-recognition modules such as SH3 and WW domains (Wasserman, 1998; Ridley, 1999; Zeller et al., 1999). The FH2 domain is highly conserved, and both FH1 and FH2 are involved in actin filament assembly (Evangelista et al., 2002; Sagot et al., 2002a). Bni1p also has a Rho-binding domain (RBD), which interacts with Rho1p and Cdc42p, in its amino-terminal region (Kohno et al., 1996; Evangelista et al., 1997), a Spa2p-binding domain (SBD) in the middle region (Fujiwara et al., 1998), and a Bud6p-binding domain (BBD) at the carboxyl-terminal region (Evangelista et al., 1997). The bni1Δ mutation shows a synthetic lethal interaction with the bnr1Δ mutation due to defects in actin cable assembly (Evangelista et al., 2002; Sagot et al., 2002a). To examine the effects of Bni1p truncation mutants on cell growth in the absence of Cla4p, we constructed five truncation mutations of BNI1 and tested these for the ability to complement the PGAL1-BNI1 bnr1Δ or PGAL1-BNI1 cla4Δ mutation in glucose medium, in which the activity of GAL1 promoter is repressed (Figure 4A). ΔFH1 and ΔFH2 could not restore the growth of either the PGAL1-BNI1 bnr1Δ or PGAL1-BNI1 cla4Δ mutant at all temperatures tested (Figure 4B). We also examined assembly of the septin ring upon exit from G1 arrest induced by α-factor (Figure 4C). We observed Cdc12p-GFP in bni1Δ cla4-75-td mutants expressing truncated Bni1p after 30- and 60-min incubations at 37°C. Typically, >70% and >90% of bni1Δ cla4-75-td mutant expressing full-length Bni1p assembled a normal septin ring after 30- and 60-min incubations, respectively (Figure 4C, unpublished results). In bni1Δ cla4-75-td mutant expressing either ΔFH1 or ΔFH2, septins formed a cap-like structure at the incipient budding site after a 30-min incubation; this was reminiscent of the phenomena observed in bni1Δ cla4-75-td mutant at 37°C. Less than 5% of cells expressing ΔFH2 could form a septin ring-like structure, which was much fainter and thinner than the septin ring in wild-type cells, but the septins became diffused and formed a cap-like structure on the bud tip by 60 min (unpublished results). Our results suggest that actin cables, which are formed by the action of Bni1p, are required for septin ring assembly in the absence of Cla4p.
Figure 4.
The effects of Bni1p truncation on cell growth and septin ring assembly in the absence of Cla4p. (A) Structure of Bni1p truncation proteins. Each number indicates the amino acid residue. Domains include Rho-binding domain (RBD), Spa2p-binding domain (SBD), formin-homology domain (FH1 and FH2), and Bud6p-binding domain (BBD). Each horizontal bar represents a segment of Bni1p: Full, full-length Bni1p; ΔRBD, Bni1(Δ6–808)p; ΔSBD, Bni1(Δ826-986)p; ΔFH1, Bni1(Δ1228-1414)p; ΔFH2, Bni1(Δ1553-1646)p; and ΔBBD, Bni1(1-1750)p. (B) The growth of either bni1Δ bnr1Δ or bni1Δ cla4Δ mutants expressing a truncated Bni1p. Five truncation mutants of BNI1 were cloned into pRS314, and the resultant plasmids were introduced into YKT440 (PGAL1-BNI1 bnr1Δ) or YKT483 (PGAL1-BNI1 cla4Δ). Growth was examined on YPDA plates at the temperature indicated. Symbols indicate relative growth rate from wild-type (+++) to no growth (–). (C) Initial assembly of septin ring in bni1Δ cla4-75-td mutants expressing truncated Bni1p. The plasmids used in B were introduced into YKT649 (bni1Δ cla4-75-td CDC12-GFP). The transformants were G1-arrested with α-factor at 25°C and released into fresh medium at 37°C. After a 30-min incubation, cells were fixed and observed by DIC and fluorescence microscopy. Bar, 5 μm.
ΔSBD and ΔBBD restored the growth of the PGAL1-BNI1 bnr1Δ mutant at all temperatures tested, suggesting that the interaction of Bni1p with either Spa2p or Bud6p is not required for formation of the actin cables that are sufficient for polarized cell growth. Consistently, a bnr1Δ mutation did not show a synthetic lethal interaction with either a spa2Δ or bud6Δ mutation (unpublished results). Interestingly, neither ΔSBD nor ΔBBD restored the growth of PGAL1-BNI1 cla4Δ mutant in glucose medium at high temperatures, suggesting that the interaction of Bni1p with either Spa2p or Bud6p plays an important role for growth in the absence of Cla4p (Figure 4B). Assembly of the septin ring was examined in the bni1Δ cla4-75-td mutant expressing either ΔSBD or ΔBBD upon exit from G1 arrest at 37°C. When grown for 30 min, >80% of cells expressing ΔBBD and <10% of cells expressing ΔSBD could form a septin ring-like structure at the incipient budding site. This septin ring-like structure appeared fainter and thinner than a normal septin ring in wild-type cells. However, the septins in both mutants became diffused and formed a cap-like structure on the bud tip after a 60-min incubation (unpublished results). Therefore, the interaction of Bni1p with either Spa2p or Bud6p seems to be required for normal septin ring assembly in the absence of Cla4p.
ΔRBD restored the growth of PGAL1-BNI1 bnr1Δ mutant in glucose medium at all temperatures tested. Interestingly, ΔRBD could not restore the growth of PGAL1-BNI1 cla4Δ mutant in glucose medium at high temperatures, suggesting that the interaction between Bni1p and Rho-GTPase is also important for growth in the absence of Cla4p (Figure 4B). The bni1Δ cla4-75-td mutant expressing ΔRBD could not assemble a septin ring upon exit from G1 arrest (Figure 4C). These results suggest that all of the functional domains of Bni1p, which have been identified so far, are required for septin ring assembly during budding initiation, especially at high temperatures in the absence of Cla4p.
Loss of Polarisome Components Causes Defective Septin Ring Assembly in the Absence of Cla4p
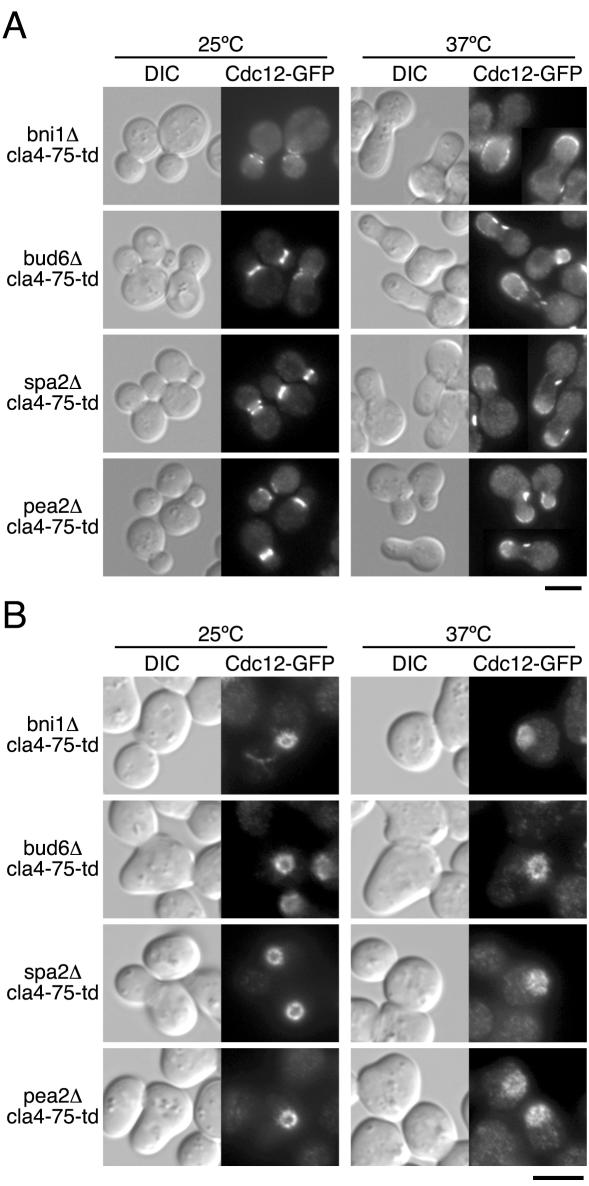
If the interaction between Bni1p and a polarisome component is important for the growth in the absence of Cla4p, polarisome genes would also show the genetic interaction with CLA4. The cla4Δ mutant was crossed with bud6Δ, spa2Δ, or pea2Δ mutant, and the resulting diploid cells were sporulated and dissected for tetrad analysis. The bud6Δ cla4Δ and spa2Δ cla4Δ double mutants exhibited a synthetic lethality at 25°C, and the pea2Δ cla4Δ double mutant exhibited a poor growth phenotype at 25°C (unpublished results). We also examined the terminal phenotype of cla4-75-td mutants lacking a polarisome component. The cell morphological phenotypes of bud6Δ cla4-75-td, spa2Δ cla4-75-td, and pea2Δ cla4-75-td mutants at the restrictive temperature mostly resembled that of the bni1Δ cla4-75-td mutant (Figure 5A). In each strain, Cdc12p-GFP localized as a cap at the polarized growing site or as a patch around the wide bud neck. These results are consistent with the recent report by Sprague and colleagues (Goehring et al., 2003). To further examine whether a polarisome component is required for initial septin ring assembly in the absence of Cla4p, we analyzed the assembly of the septin ring upon exit from G1 arrest at 37°C (Figure 5B). When grown for 30 min, both spa2Δ cla4-75-td and pea2Δ cla4-75-td mutants could not assemble a normal septin ring as well as bni1Δ cla4-75-td mutant. bud6Δ cla4-75-td mutant could form a septin ring-like structure, but an additional 30 min later, this structure became diffused and formed a cap-like structure on the bud tip as well as bni1Δ cla4-75-td mutant carrying ΔBBD (unpublished results). These results suggest that the presence of a polarisome component and its interaction with Bni1p is required for initial septin ring assembly in the absence of Cla4p.
Figure 5.
Defective septin ring assembly in cla4-75-td mutants lacking a polarisome component. (A) YKT649 (bni1Δ cla4-75-td CDC12-GFP), YKT664 (bud6Δ cla4-75-td CDC12-GFP), YKT666 (spa2Δ cla4-75-td CDC12-GFP), and YKT668 (pea2Δ cla4-75-td CDC12-GFP) were grown in YPDA medium at 25°C and then shifted to 37°C for 2 h. Cells were fixed and observed by DIC and fluorescence microscopy. (B) Cells of each strain were G1-arrested with α-factor at 25°C and released into fresh medium at 25°C or 37°C. After a 30-min incubation, cells were fixed and observed by DIC or fluorescence microscopy. Bars, 5 μm.
Actin Polymerization Is Required for Septin Ring Assembly in the Absence of Cla4p
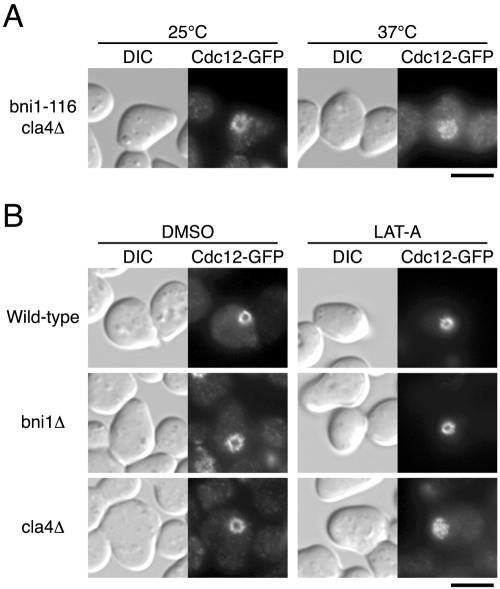
The FH2 domain of Bni1p is required for actin filament assembly in vivo (Evangelista et al., 2002; Sagot et al., 2002a) and can stimulate actin polymerization in vitro (Pruyne et al., 2002; Sagot et al., 2002b). As described above, ΔFH2 could not restore the growth of the bni1Δ cla4Δ double mutant. This result suggests that the actin-nucleating activity of Bni1p may be required for septin ring assembly in the absence of Cla4p. However, it is also possible that the FH2 domain possesses a distinct function in septin ring assembly. To discriminate these possibilities, we isolated a ts-allele of BNI1, bni1-116, which causes amino acid substitutions (V1475A, K1498E, and D1511N) in the FH2 domain. In cells carrying bni1-116 bnr1Δ mutations, actin cables visualized by rhodamine-phalloidin disappear in 2 min after upshift to 37°C (unpublished results). This effect on actin cables was not due to degradation of Bni1-116p, because Bni1-116p-GFP was localized to a bud tip in cells grown at 37°C (unpublished results). The bni1-116 cla4Δ mutant showed growth defects above 35°C and abnormally elongated buds with wide necks, results similar to bni1Δ cla4-75-td mutant cells (unpublished results). We examined assembly of the septin ring upon exit from G1 arrest at 37°C in the bni1-116 cla4Δ mutant. When grown for 30 min, bni1-116 cla4Δ mutant as well as bni1Δ cla4-75-td mutant could not assemble normal septin ring (Figure 6A). bni1-11 (amino acid substitutions: D1511G and K1601R; Evangelista et al., 2002) and bni1-FH2#1 (amino acid substitutions: R1528A and R1530A; Sagot et al., 2002a) are previously characterized mutations in the FH2 domain. Both mutations also cause rapid disassembly of actin cables at 37°C. We confirmed that bni1-11 cla4Δ and bni1-FH2#1 cla4Δ mutants showed the defects in cell growth and septin ring assembly as bni1Δ cla4-75-td mutant (unpublished results). These results suggest that actin polymerization mediated by the FH2 domain of Bni1p is required for the assembly of the septin ring during budding initiation.
Figure 6.
Perturbation of actin polymerization causes defective septin ring assembly in the absence of Cla4p. (A) Point mutant in the FH2 domain of BNI1, characterized as that defective in actin cable assembly, causes the defects in septin ring assembly in the absence of Cla4p. YKT638 (bni1-116 cla4Δ CDC12-GFP) was G1-arrested with α-factor at 25°C and released into fresh medium at 25 or 37°C. After a 30-min incubation, cells were fixed and observed by DIC and fluorescence microscopy. (B) LAT-A inhibits septin ring assembly in cla4Δ mutant. YKT637 (CDC12-GFP), YKT676 (bni1Δ CDC12-GFP), and YKT717 (cla4Δ CDC12-GFP) were G1-arrested with α-factor and released into fresh medium at 25°C in the absence (DMSO only) or presence of LAT-A. After 30-(DMSO) and 60 (LAT-A)-min incubations, cells were fixed and observed by DIC or fluorescence microscopy. Bars, 5 μm.
A previous study suggested that assembly of the septin ring is independent of the actin cytoskeleton (Ayscough et al., 1997). They showed that the actin assembly inhibitor LAT-A did not affect septin ring assembly in wild-type cells. The results described above suggest that LAT-A may inhibit septin ring assembly in the cla4Δ mutant. We analyzed septin ring assembly upon exit from G1 arrest at 25°C in the presence or absence of LAT-A in the wild-type, bni1Δ, and cla4Δ mutants. When grown for 30 min after release from G1 arrest in the absence of LAT-A, each strain assembled a septin ring. An extended diameter and jagged morphology of the septin ring were observed in bni1Δ mutant as shown in Figure 3A, and the periphery of the septin ring in cla4Δ mutant looked fainter compared with wild-type (Figure 6B). After an additional 30 min, each strain formed a small bud and the septin collar localized to the bud neck (unpublished results). The diameter of the septin collar in the bni1Δ mutant was slightly greater than that in wild-type. Less than 10% of the cla4Δ mutant cells localized the septins on the bud tip (unpublished results). When grown for 30 min in the presence of LAT-A, each strain localized the septins at the incipient budding site, but did not assemble a septin ring (unpublished results). An additional 30 min later, >70% of the wild-type and bni1Δ mutant cells assembled a septin ring (Figure 6B), suggesting that LAT-A treatment caused a delay in septin ring assembly. However, the cla4Δ mutant could not assemble a septin ring (Figure 6B), even after a 6-h incubation (unpublished results). Previous study reported that actin perturbation by LAT-A triggers a morphogenesis checkpoint and induces a delay of nuclear division (McMillan et al., 1998). To assess some aspect of cell cycle progression in the LAT-A–treated cells, we monitored the mitotic spindle assembly in the wild-type, bni1Δ, and cla4Δ mutant cells expressing GFP-Tub1p. When grown for 60 min after release from G1 arrest in the presence of LAT-A, the fraction of the cells that assembled a mitotic spindle decreased from 70 to 40% compared with the untreated cells in each strain (unpublished results), suggesting that the LAT-A treatment induces the cell cycle delay. The extent of the cell cycle delay, however, was similar between the wild-type, bni1Δ, and cla4Δ mutants, suggesting that the cla4Δ mutant cannot assemble a septin ring in the presence of LAT-A independent of the cell cycle delay. These results are consistent with our observations that Bni1p-dependent actin polymerization is required for initial septin ring assembly in the absence of Cla4p.
Actin Cable-dependent Transport May Be Required for Septin Ring Assembly in the Absence of Cla4p
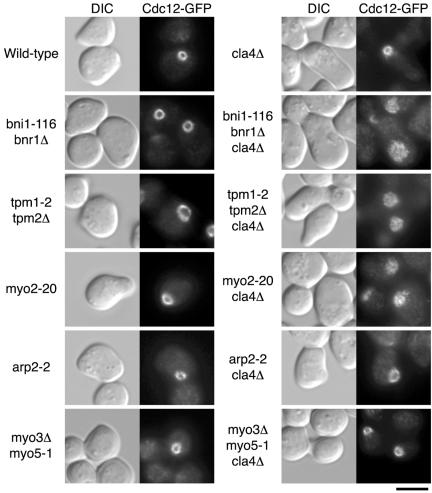
We examined actin cable-related factors for their involvement in septin ring assembly. We constructed cla4Δ mutants combined with actin cable-related ts mutations such as bni1-116 bnr1Δ, tpm1-2 tpm2Δ, and myo2-20 (Pruyne et al., 1998; Schott et al., 1999). Tropomyosins (Tpm1p and Tpm2p) are actin cable-stabilizing proteins. Myo2p, a type V myosin, has a direct role in secretory vesicle targeting using actin cables as tracks for transport. We analyzed septin ring assembly upon exit from G1 arrest at 37°C. When grown for 60 min, >80% of bni1-116 bnr1Δ, tpm1-2 tpm2Δ, and myo2-20 mutants assembled a septin ring (Figure 7). In contrast, bni1-116 bnr1Δ cla4Δ, tpm1-2 tpm2Δ cla4Δ, and myo2-20 cla4Δ mutant cells could not assemble septin rings (Figure 7). Deletion of only BNR1 from cla4Δ mutant affected neither growth nor morphology of the cells (unpublished results). Because loss of Bni1p is known to cause depolarization of cortical actin patches (Kohno et al., 1996; Evangelista et al., 1997), we examined the involvement of an actin patch-related protein in septin ring assembly. We constructed the cla4Δ mutants combined with the arp2-2 and myo3Δ myo5-1 ts mutations (Geli and Riezman, 1996; Madania et al., 1999). Arp2p is a subunit of the Arp2/3 complex, which is the major known contributor to actin nucleation in vivo in yeast (Winter et al., 1999). Myo3/5p, type I myosins, are known as regulators of the Arp2/3 complex (Evangelista et al., 2000; Lechler et al., 2000). Both arp2-2 and myo3Δ myo5-1 mutants showed normal septin ring assembly, and depletion of Cla4p in these mutants did not affect this assembly (Figure 7). These results suggest that actin cable-dependent transport, but not actin patch function, is required for the assembly of a normal septin ring.
Figure 7.
Synthetic effects of cla4Δ mutation and actin-related mutations on septin ring assembly. YKT637 (CDC12-GFP), YKT717 (cla4Δ CDC12-GFP), YKT743 (bni1-116 bnr1Δ CDC12-GFP), YKT744 (bni1-116 bnr1Δ cla4Δ CDC12-GFP), YKT812 (tpm1-2 tpm2Δ CDC12-GFP), YKT813 (tpm1-2 tpm2Δ cla4Δ CDC12-GFP), YKT814 (myo2-20 CDC12-GFP), and YKT815 (myo2-20 cla4Δ CDC12-GFP), YKT816 (arp2-2 CDC12-GFP), YKT817 (arp2-2 cla4Δ CDC12-GFP), YKT818 (myo3Δ myo5-1 CDC12-GFP), and YKT819 (myo3Δ myo5-1 cla4Δ CDC12-GFP) were G1-arrested with α-factor at 25°C and then released into fresh medium at 37°C. After a 60-min incubation, cells were fixed and observed by DIC and fluorescence microscopy. Bar, 5 μm.
Cdc42p and Its GAPs May Be Involved in the Regulation of Septin Ring Assembly by Bni1p and Cla4p
To identify genes involved in septin ring assembly mediated by Bni1p and Cla4p, we isolated multicopy suppressors of the ts growth phenotype of bni1-116 cla4Δ mutant. We isolated truncated fragments of BEM3 and RGA1 that encode GTPase-activating protein (GAP) for Cdc42p (Zheng et al., 1993; Stevenson et al., 1995; Johnson, 1999). Both of the isolated fragments, BEM3Δ1-114 and RGA1Δ1-632, contained a region encoding a GAP domain (Figure 8A). Multicopy BEM3Δ1-114 and RGA1Δ1-632 also suppressed the growth defect of the bni1Δ cla4-75-td mutant (Figure 8B). The full-length BEM3 suppressed the growth defect of the bni1Δ cla4-75-td mutant, but to a lesser extent than that of BEM3Δ1-114, whereas the full-length RGA1 did not, suggesting that an N-terminal region of Rga1p and Bem3p may possess a negative regulatory role for GAP activity (unpublished results). We also examined the assembly of the septin ring upon exit from G1 arrest at 35°C in bni1Δ cla4-75-td mutants carrying multicopy BEM3Δ1-114 or RGA1Δ1-632 (Figure 8C). The cells carrying BEM3Δ1-114 or RGA1Δ1-632 did not assemble septin rings either after 30-min (Figure 8C) or 60-min growth (unpublished results). Surprisingly, after 3 h, they often formed ring-like structures comprised of septins around the bud neck and eventually accomplished cytokinesis (Figure 8C). These results suggest that the defects in septin assembly in bni1Δ cla4-75-td mutant can be alleviated during polarized growth after budding has occurred.
Figure 8.
Suppression of the growth, morphological and septin organization defects of bni1Δ cla4-75-td mutants by multicopy RGA1Δ1-632 or BEM3Δ1-114. (A) Structures of Rga1(Δ1-632)p and Bem3(Δ1-114)p. Each number indicates the amino acid residue. Both of the isolated fragments contained a region encoding a GAP domain. (B) Suppression of the growth defect. YKT811 (bni1Δ cla4-75-td CDC12-GFP) was transformed with plasmid YEp24 (vector), pKT1364 (YEp24-BEM3Δ1-114), pKT1365 (YEplac195-RGA1Δ1-632), or pKT1254 (pRS316-CLA4). The transformants were streaked on a YPDA plate, which was incubated at 25 or 35°C for 2 d. (C) Suppression of the septin assembly defect. Each transformant was G1-arrested with α-factor at 25°C and released into fresh medium at 35°C. After 30-min and 3-h incubations, cells were fixed and observed by DIC and fluorescence microscopy. Arrows indicate the cells in cytokinesis. Bar, 5 μm.
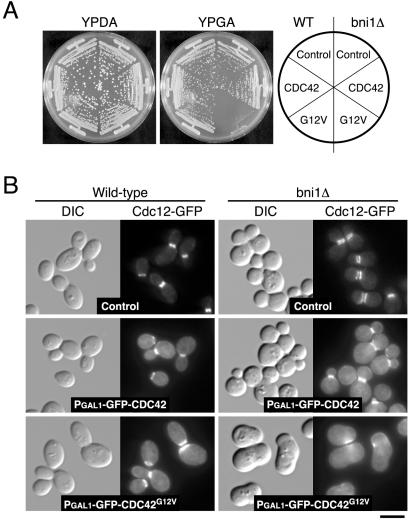
Multicopy BEM3Δ1-114 and RGA1Δ1-632 also suppressed the growth defects of bud6Δ cla4-75-td, spa2Δ cla4-75-td, and pea2Δ cla4-75-td mutants, but not that of bni1-116 bnr1Δ mutant (unpublished results), suggesting that they suppressed the defects caused by the cla4Δ mutation. Because the Cdc42p GAPs seem to down-regulate Cdc42p, expression of a dominant active version of Cdc42p may cause lethality in bni1Δ mutant. However, expression of Cdc42G12Vp caused lethality in wild-type cells (unpublished results). Overexpression of a GFP-tagged dominant active version of Cdc42p, GFP-Cdc42G12Vp, did not inhibit the growth of wild-type cells (Figure 9A), suggesting that its activity is relatively weaker than that of Cdc42G12Vp. GFP-Cdc42G12Vp inhibited the growth of the bni1Δ mutant (Figure 9A), but not the wild-type (Figure 9A) or cla4Δ mutant (unpublished data). This growth inhibition appears to be due to GTP-bound Cdc42p, because GFP-Cdc42p did not inhibit the growth of the bni1Δ mutant. The bni1Δ mutant overexpressing GFP-Cdc42G12Vp also displayed a wide bud neck, although hyperpolarized growth was not remarkable compared with bni1Δ cla4-75-td mutant (Figure 9B). In these cells, the septins formed a broad ring around the bud neck, leading to defects in cytokinesis. These results suggest that the cla4Δ mutation induces effects similar to those caused by accumulation of Cdc42p-GTP and that this results in the synthetic defects in septin ring assembly with the bni1Δ mutation.
Figure 9.
Constitutive activation of Cdc42p is sufficient to cause defective septin organization in the absence of Bni1p. (A) YKT637 (CDC12-GFP), YKT676 (bni1Δ CDC12-GFP), YKT891 (PGAL1-GFP-CDC42 CDC12-GFP), YKT892 (PGAL1-GFP-CDC42G12V CDC12-GFP), YKT893 (PGAL1-GFP-CDC42 bni1Δ CDC12-GFP), and YKT894 (PGAL1-GFP-CDC42G12V bni1Δ CDC12-GFP) were streaked on a YPDA or a YPGA plate, which was incubated at 25°C for 3 d. (B) The strains used in A were released from stationary phase into YPGA medium at 25°C. After a 6-h incubation, cells were fixed and observed by DIC and fluorescence microscopy. Cytosolic and plasma membrane staining in GFP-Cdc42p–expressing cells appear to be due to fluorescence of GFP-Cdc42ps. Bar, 5 μm.
DISCUSSION
The Role of Polarisome Components and Actin Cytoskeleton in Septin Ring Assembly
We showed that the polarisome components (Bni1p, Spa2p, Bud6p, and Pea2p) are required for septin ring assembly in the absence of Cla4p. In double mutants carrying mutations in either of polarisome components and CLA4, Cdc12p-GFP was recruited to the budding site, but a septin ring was not assembled and Cdc12p-GFP was instead localized to the polarized site. Time-lapse analyses indicated that a polarisome component and Cla4p are not required for the stable maintenance of the septin collar, once a septin collar has been formed at the permissive temperature, but that they are specifically required for the initial assembly step of the septin ring. On the contrary, Goehring et al. (2003) reported that the bni1Δ cla4Δ mutant carrying the YCp-cla4-75 plasmid, which contains the original cla4-75 allele, not the cla4-75-ts degron allele, formed a normal initial septin ring, with septin abnormalities only appearing later. We also observed that ∼30% of the bni1Δ cla4Δ mutant cells carrying YCp-cla4-75 initially assembled a septin ring-like structure with deformed morphology, which decayed to a cap as the bud began to grow (unpublished results). The residual activity of Cla4-75p, caused by the lack of rapid degradation and the increased expression from YCp plasmid, may partially promote the initial septin ring assembly in the bni1Δ cla4Δ mutant.
Deletion analysis of Bni1p revealed the importance of Rho-, Spa2p-, and Bud6p-binding domains for septin ring assembly, a clear contrast to the results that these domains are not required for the suppression of growth defects of bni1Δ bnr1Δ mutant. These results suggest that the integrity of the polarisome complex is required for septin ring assembly, in addition to the presence of each component.
We showed that a bni1Δ mutant assembles a septin ring with a wider diameter than that of wild-type cells. The polarisome component mutants display defects in apical growth because they fail to confine the polarized growth site to a small region at the bud tip (Sheu et al., 2000). If polarisome mutants also display this defect during bud site assembly and if the area of bud site assembly determines the width of septin ring, depletion of a polarisome component would result in the assembly of a wider septin ring.
Interestingly, LAT-A treatment restored a septin ring with normal diameter and morphology in bni1Δ mutant. In bni1Δ mutant, Bnr1p assembles actin cables, and thus polarized growth still occurs. The LAT-A treatment completely abolishes polarized growth by inhibiting the actin polymerization. This may allow the bni1Δ mutant to have sufficient time to assemble a septin ring with normal morphology. Consistently, bni1-116 bnr1Δ mutant displayed a septin ring with relatively normal morphology (Figure 7). However, in the absence of CLA4, the bni1-116 bnr1Δ mutant did not assemble a septin ring, suggesting that the septin ring in bni1Δ mutant is altered not only morphologically, but also functionally. This is consistent with previous observations that bni1 and spa2 mutations show synthetic growth defects with cdc12 (Zahner et al., 1996) and cdc10 (Flescher et al., 1993) mutations, respectively.
A Bni1 mutant protein, which is defective in actin cable formation, also shows a defect in septin ring assembly in the absence of Cla4p, suggesting that the actin cytoskeleton is involved in septin ring assembly. Consistently, LAT-A treatment inhibited septin ring assembly in the cla4Δ mutant. Because LAT-A does not inhibit septin ring assembly in wild-type cells, the Cla4p functions in septin assembly appear to be independent of the actin cytoskeleton. Actin cables serve as a track for a type V myosin, Myo2p. Our results that myo2-20 cla4Δ, tpm1-2 tpm2Δ cla4Δ, and bni1-116 bnr1Δ cla4Δ mutants showed a defect in septin ring assembly suggest that Myo2p may be required for transport of a factor specifically required for septin ring assembly to the bud emergence site. Consistent with this, even in the presence of CLA4, the assembly of septin ring was delayed by LAT-A treatment (Figure 6) and in the bni1-116 bnr1Δ, tpm1-2 tpm2Δ, and myo2-20 mutants (unpublished results), although there is another possibility that this delay may be attributed to a cell cycle delay triggered by LAT-A treatment or formin deficiency. However, it is more likely that the Bni1p-catalyzed formation of actin filaments is specifically required for septin ring assembly. In the bni1Δ cla4-75-td mutant, Myo2p is transported to a polarized site and thus polarized growth occurs, because Bnr1p can form actin cables. The Bni1p-specific actin cables may guide polarized growth to a focused region to assemble a septin ring with normal diameter and morphology. Another interesting possibility is that Bni1p provides an actin-based structure, which is different from the actin cables used for polarized transport. Interestingly, in mammalian systems, actin bundles can serve as a template for septin assembly via an actin-binding protein, anillin (Kinoshita et al., 2002). It is an intriguing possibility that the actin cytoskeleton, formed by the action of Bni1p, plays a more direct role in septin assembly in conjunction with polarisome components.
Functional Interactions between Polarisome Components, Cla4p and Cdc42p GAPs for Septin Ring Assembly
Cla4p has also been implicated in the initial assembly of the septin ring (Cvrckova et al., 1995; Weiss et al., 2000). Fluorescence-recovery-after-photobleaching (FRAP) studies have shown that septin ring is labile during budding initiation and mitotic exit and are stable during S, G2, and M phases (Caviston et al., 2003; Dobbelaere et al., 2003). Cla4p is required for this immobilization of the septin ring (Dobbelaere et al., 2003). The defects in septins in the cla4Δ mutant, in conjunction with the spatial defects in polarisome mutants as to where septin rings are assembled, may result in severe defects in the assembly of the septin ring. Deletion of SWE1, which encodes a protein kinase thought to be part of a morphogenesis checkpoint that negatively regulates Clb1, 2p-Cdc28p activity, restores normal bud morphology in cla4 mutant (Longtine et al., 2000; Weiss et al., 2000; Mitchell and Sprague, 2001). Goehring et al. (2003) reported that deletion of SWE1 restores the localization of septins to the mother-bud neck in unsynchronized cultures of bni1Δ cla4-75 mutant. However, we observed septin ring assembly defects in bni1Δ cla4-75-td swe1Δ cells upon release from G1 arrest (unpublished results), suggesting that Cla4p possesses specific functions for septin ring assembly.
We showed that overexpression of BEM3Δ1-114 and RGA1Δ1-632, which encode truncated versions of Cdc42p GAPs, suppressed the growth defects of the bni1Δ cla4-75-td mutant. It was recently reported that a mutant in Cdc42p GAPs (bem3Δ rga1Δ rga2Δ) showed severe defects in the assembly of the septin ring (Caviston et al., 2003). Therefore, it seems that a similar molecular defect underlies the bni1Δ cla4-75-td and bem3Δ rga1Δ rga2Δ mutants. Our results suggest that Cdc42p GAPs are involved in the Cla4p-related pathway for septin ring assembly. Because one of the plausible functions of Cdc42p GAPs is to down-regulate Cdc42p-GTP, Cdc42p-GTP may accumulate in the cla4Δ mutant. Consistently, we showed that a dominant active version of Cdc42p caused a septin ring assembly defect in bni1Δ mutant. Cla4p phosphorylates Cdc24p, resulting in the dissociation of Bem1p from Cdc24p and the subsequent down-regulation of Cdc42p-GTP formation (Gulli et al., 2000), although the other group concluded that the Cla4p-dependent phosphorylation of Cdc24p does not affect the Bem1p-Cdc24p interaction (Bose et al., 2001). Based on the observation that hyper activation of Cdc42p and depletion of Cdc42p GAPs caused defects in septin organization, Gladfelter et al. (2002) proposed that Cdc42p cycling between a GTP-bound “ON” state and GDP-bound “OFF” state is required for normal septin ring assembly. Septin ring assembly defects in the cla4Δ mutant may be partly due to hyper activation of Cdc42p.
We showed that overexpression of Cdc42p GAPs restored septin ring assembly in the bni1Δ cla4-75-td mutant. This suppression appeared to occur during polarized growth, rather than at bud emergence. Similar observations were made in the Cdc42p GAP triple mutant and in cdc42V36G mutant (Caviston et al., 2003). They showed that, by a time-lapse analysis, the septin cap could be converted to a normal ring structure next to bud tip at some point during bud growth and that a bud of normal shape began to form distal to the septin ring. Therefore, a septin collar can be constructed, not only at the initiation of budding, but also during polarized growth. Caviston et al. (2003) proposed that septin ring formation consists of at least two steps: recruitment of septin proteins and their assembly into the septin ring. In this model, both steps depend on Cdc42p, whereas the Cdc42p GAPs and other “assembly factor,” such as Cla4p, Gin4p, and Elm1p, function in the assembly step. How do the truncated Cdc42p GAPs induce the septin collar formation in the bni1Δ cla4-75-td mutant after budding has occurred? The truncated Cdc42p GAPs may partially suppress the defects in the initial septin assembly, to an extent that the effects on the septins cannot be visualized under the microscope, resulting in the formation of a septin collar later on. It is also possible that, due to deregulation by truncation, they could induce septin assembly even during polarized growth. Caviston et al. (2003) proposed that Cdc42p GAPs function as effectors of Cdc42p for septin ring assembly. According to this model, the Cdc42p GAPs-related function of Cla4p may be to interact more directly with septins, rather than to regulate the nucleotide-binding state of Cdc42p. It was recently shown that Cla4p interacts directly with and phosphorylates septins in vitro and in vivo (Versele and Thorner, 2004). Currently, we favor a hypothesis that Cla4p regulates the septin assembly through both direct interactions with septins and regulation of the nucleotide binding state of Cdc42p.
Our observations suggest that polarisome components also function in the assembly step together with Cdc42p GAPs and Cla4p. Further identification of a protein, which directs septin ring assembly through direct interactions with septins under the control of polarisome, Cla4p and Cdc42p GAPs, is an important step toward understanding of the molecular mechanisms of septin ring assembly.
Acknowledgments
We thank Drs. Anthony Bretscher, David Drubin, Kim Nasmyth, John Pringle, Akio Toh-e, and Barbara Winsor for plasmids and yeast strains. We thank Aiko Ishioh, Eriko Itoh, Toshika Matsumoto, Grace Pai, and Kim Tu for excellent technical assistance. This work was supported by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to T.Y. and K.T.), and a National Institutes of Health grant GM59216 and an American Cancer Society grant RSG02039CSM (to E. B.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0254. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0254.
Abbreviations used: BBD, Bud6p-binding domain; DIC, differential interference contrast; FH, formin homology; GAP, GTPase-activating protein; GFP, green fluorescent protein; ts, temperature-sensitive; LAT-A, latrunculin A; RBD, Rho-binding domain; SBD, Spa2p-binding domain.
References
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., Parra, M., Bidlingmaier, S., and Snyder, M. (1999). Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., Mermall, V., Mooseker, M.S., and Snyder, M. (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841–851. [DOI] [PubMed] [Google Scholar]
- Bi, E., and Pringle, J.R. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., Maddox, P., Lew, D.J., Salmon, E.D., McMillan, J.N., Yeh, E., and Pringle, J.R. (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, I., Irazoqui, J.E., Moskow, J.J., Bardes, E.S., Zyla, T.R., and Lew, D.J. (2001). Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 276, 7176–7186. [DOI] [PubMed] [Google Scholar]
- Cadwell, R.C., and Joyce, G.F. (1992). Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 28–33. [DOI] [PubMed] [Google Scholar]
- Caviston, J.P., Tcheperegine, S.E., and Bi, E. (2002). Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc. Natl. Acad. Sci. USA 99, 12185–12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J.P., Longtine, M., Pringle, J.R., and Bi, E. (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., Mischke, M., Mitchell, E., Herskowitz, I., and Pringle, J.R. (1995). Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129, 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., De Virgilio, C., Manser, E., Pringle, J.R., and Nasmyth, K. (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9, 1817–1830. [DOI] [PubMed] [Google Scholar]
- DeMarini, D.J., Adams, A.E., Fares, H., De Virgilio, C., Valle, G., Chuang, J.S., and Pringle, J.R. (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M.S., Hallberg, R.L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345–357. [DOI] [PubMed] [Google Scholar]
- Drubin, D.G., and Nelson, W.J. (1996). Origins of cell polarity. Cell 84, 335–344. [DOI] [PubMed] [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13, 18–20. [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M.S., Chow, C.J., Adames, N., Pringle, J.R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Klebl, B.M., Tong, A.H., Webb, B.A., Leeuw, T., Leberer, E., Whiteway, M., Thomas, D.Y., and Boone, C. (2000). A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 32–41. [DOI] [PubMed] [Google Scholar]
- Field, C.M., and Kellogg, D. (1999). Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 9, 387–394. [DOI] [PubMed] [Google Scholar]
- Fitch, I., Dahmann, C., Surana, U., Amon, A., Nasmyth, K., Goetsch, L., Byers, B., and Futcher, B. (1992). Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher, E.G., Madden, K., and Snyder, M. (1993). Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 122, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier, J.A., Wong, M.L., Longtine, M.S., Pringle, J.R., Mann, M., Mitchison, T.J., and Field, C. (1998). Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, T., Tanaka, K., Mino, A., Kikyo, M., Takahashi, K., Shimizu, K., and Takai, Y. (1998). Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli, M.I., and Riezman, H. (1996). Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272, 533–535. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., Pringle, J.R., and Lew, D.J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681–689. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., Bose, I., Zyla, T.R., Bardes, E.S., and Lew, D.J. (2002). Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, A.S., Mitchell, D.A., Tong, A.H., Keniry, M.E., Boone, C., and Sprague, G.F., Jr. (2003). Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14, 1501–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.L., and McCusker, J.H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gulli, M.P., Jaquenoud, M., Shimada, Y., Niederhauser, G., Wiget, P., and Peter, M. (2000). Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell 6, 1155–1167. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H. (1971). Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69, 265–276. [DOI] [PubMed] [Google Scholar]
- Holly, S.P., and Blumer, K.J. (1999). PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M., and Toh-e, A. (2001). Nis1 encoded by YNL078W: a new neck protein of Saccharomyces cerevisiae. Genes Genet. Syst. 76, 335–343. [DOI] [PubMed] [Google Scholar]
- Johnson, D.I. (1999). Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63, 54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M., Kumar, S., Mizoguchi, A., Ide, C., Kinoshita, A., Haraguchi, T., Hiraoka, Y., and Noda, M. (1997). Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11, 1535–1547. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M., Field, C.M., Coughlin, M.L., Straight, A.F., and Mitchison, T.J. (2002). Self- and actin-templated assembly of mammalian septins. Dev. Cell 3, 791–802. [DOI] [PubMed] [Google Scholar]
- Kohno, H. et al. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Lechler, T., Shevchenko, A., and Li, R. (2000). Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., DeMarini, D.J., Valencik, M.L., Al-Awar, O.S., Fares, H., De Virgilio, C., and Pringle, J.R. (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., Fares, H., and Pringle, J.R. (1998a). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998b). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., Theesfeld, C.L., McMillan, J.N., Weaver, E., Pringle, J.R., and Lew, D.J. (2000). Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., and Bi, E. (2003). Regulation of septin organization and function in yeast. Trends Cell Biol. 13, 403–409. [DOI] [PubMed] [Google Scholar]
- Madania, A., Dumoulin, P., Grava, S., Kitamoto, H., Scharer-Brodbeck, C., Soulard, A., Moreau, V., and Winsor, B. (1999). The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10, 3521–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J.N., Sia, R.A., and Lew, D.J. (1998). A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142, 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.A., and Sprague, G.F., Jr. (2001). The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida, J., Yamamoto, T., Fujimura-Kamada, K., and Tanaka, K. (2002). The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics 160, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T.P., and Rubin, G.M. (1994). The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77, 371–379. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.Q., Sawa, H., Okano, H., and White, J.G. (2000). The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J. Cell Sci. 113, 3825–3837. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., Bi, E., Harkins, H.A., Zahner, J.E., De Virgilio, C., Chant, J., Corrado, K., and Fares, H. (1995). Establishment of cell polarity in yeast. Cold Spring Harbor Symp. Quant. Biol. 60, 729–744. [DOI] [PubMed] [Google Scholar]
- Pruyne, D.W., Schott, D.H., and Bretscher, A. (1998). Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143, 1931–1945. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000a). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113, 365–375. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000b). Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113, 571–585. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J. (1999). Stress fibres take shape. Nat. Cell Biol. 1, E64–E66. [DOI] [PubMed] [Google Scholar]
- Rose, A.B., and Broach, J.R. (1990). Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 185, 234–279. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Klee, S.K., and Pellman, D. (2002a). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42–50. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Rodal, A.A., Moseley, J., Goode, B.L., and Pellman, D. (2002b). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626–631. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., and Herskowitz, I. (1996). The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J. Cell Biol. 134, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M., Varma, A., Drgon, T., Bowers, B., and Cabib, E. (2003). Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell 14, 2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, D., Ho, J., Pruyne, D., and Bretscher, A. (1999). The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147, 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, D.H., Collins, R.N., and Bretscher, A. (2002). Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 156, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sherman, F., and Hicks, J. (1991). Micromanipulation and dissection of asci. Methods Enzymol. 194, 21–37. [DOI] [PubMed] [Google Scholar]
- Sheu, Y.J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., Barral, Y., and Snyder, M. (2000). Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz, M.J., Inouye, C.J., and Thorner, J. (1999). Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G.R., Givan, S.A., Cullen, P., and Sprague, G.F., Jr. (2002). GTPase-activating proteins for Cdc42. Eukaryotic Cell 1, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, B.J., Ferguson, B., De Virgilio, C., Bi, E., Pringle, J.R., Ammerer, G., and Sprague, G.F., Jr. (1995). Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 9, 2949–2963. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., and Vale, R.D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344. [DOI] [PubMed] [Google Scholar]
- Toi, H., Fujimura-Kamada, K., Irie, K., Takai, Y., Todo, S., and Tanaka, K. (2003). She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell 14, 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble, W.S. (1999). Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J. Membr. Biol. 169, 75–81. [DOI] [PubMed] [Google Scholar]
- Versele, M., and Thorner, J. (2004). Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman, S. (1998). FH proteins as cytoskeletal organizers. Trends Cell Biol. 8, 111–115. [DOI] [PubMed] [Google Scholar]
- Weiss, E.L., Bishop, A.C., Shokat, K.M., and Drubin, D.G. (2000). Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2, 677–685. [DOI] [PubMed] [Google Scholar]
- Westfall, P.J., and Momany, M. (2002). Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, D.C., Choe, E.Y., and Li, R. (1999). Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96, 7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner, J.E., Harkins, H.A., and Pringle, J.R. (1996). Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller, R., Haramis, A.G., Zuniga, A., McGuigan, C., Dono, R., Davidson, G., Chabanis, S., and Gibson, T. (1999). Formin defines a large family of morpho-regulatory genes and functions in establishment of the polarising region. Cell Tissue Res. 296, 85–93. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Hart, M.J., Shinjo, K., Evans, T., Bender, A., and Cerione, R.A. (1993). Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J. Biol. Chem. 268, 24629–24634. [PubMed] [Google Scholar]