Abstract
TIA-1 is an RNA binding protein that promotes the assembly of stress granules (SGs), discrete cytoplasmic inclusions into which stalled translation initiation complexes are dynamically recruited in cells subjected to environmental stress. The RNA recognition motifs of TIA-1 are linked to a glutamine-rich prion-related domain (PRD). Truncation mutants lacking the PRD domain do not induce spontaneous SGs and are not recruited to arsenite-induced SGs, whereas the PRD forms aggregates that are recruited to SGs in low-level–expressing cells but prevent SG assembly in high-level–expressing cells. The PRD of TIA-1 exhibits many characteristics of prions: concentration-dependent aggregation that is inhibited by the molecular chaperone heat shock protein (HSP)70; resistance to protease digestion; sequestration of HSP27, HSP40, and HSP70; and induction of HSP70, a feedback regulator of PRD disaggregation. Substitution of the PRD with the aggregation domain of a yeast prion, SUP35-NM, reconstitutes SG assembly, confirming that a prion domain can mediate the assembly of SGs. Mouse embryomic fibroblasts (MEFs) lacking TIA-1 exhibit impaired ability to form SGs, although they exhibit normal phosphorylation of eukaryotic initiation factor (eIF)2α in response to arsenite. Our results reveal that prion-like aggregation of TIA-1 regulates SG formation downstream of eIF2α phosphorylation in response to stress.
INTRODUCTION
TIA-1 and TIAR are related RNA binding proteins that promote general translational arrest in environmentally stressed cells (Anderson and Kedersha, 2002a). Stress-induced translational arrest is initiated by the activation of PKR, PERK, HRI, and/or GCN2, serine/threonine kinases that phosphorylate the translation initiation factor eIF2α (Srivastava et al., 1998; Kaufman, 1999; Harding et al., 2000a,b; Kimball et al., 2001; Zhan et al., 2002; Jefferson and Kimball, 2003). Phosphorylation of eIF2α reduces the availability of the eIF2-GTP-tRNA Meti ternary complex that loads the initiator tRNA onto the 40S ribosomal subunit, resulting in ternary complex deficient 48S preinitiation complexes that cannot recruit the 60S ribosomal subunit to begin protein translation. Such stalled initiation complexes are dynamically routed by TIA-1 and TIAR into discrete cytoplasmic foci known as stress granules (SGs) (Anderson and Kedersha, 2002a), which form rapidly during stress as the equilibrium is shifted between translating and stalled messenger ribonucleoproteins (mRNPs). Thus, SG formation can be induced by several different stimuli: eIF2α phosphorylation, which prevents initiation and thus inhibits polysome assembly; treatment with puromycin, which promotes premature termination and polysome disassembly; or acute energy starvation (Anderson and Kedersha, 2002a). The latter treatment can induce SGs without measurably increasing eIF2α phosphorylation, suggesting that a separate energy-dependent step is involved in SG dynamics (Kedersha et al., 2002). SG assembly also is regulated by the RasGAP-associated endoribonuclease G3BP (Tourriere et al., 2003). In neurons, fragile X mental retardation protein promotes the assembly of neuronal granules that are structurally and functionally similar to SGs (Mazroui et al., 2002; Wickens and Goldstrohm, 2003).
TIA-1 and TIAR are modular proteins composed of three amino-terminal RNA recognition motifs and a carboxy-terminal glutamine-rich motif that is structurally related to prion protein (Tian et al., 1991; Kawakami et al., 1992). Overexpressed TIA-1 induces SG formation and represses reporter gene expression, whereas the isolated prion-related domain (PRD) of TIA-1 forms cytoplasmic microaggregates that sequester endogenous TIA-1 and TIAR and promote expression of cotransfected reporter genes (Kedersha et al., 1999, 2000a,b). These data suggest that the PRD is capable of self-oligomerization and that sequestration of full-length TIA proteins prevents their repressive effects on reporter gene expression. Given that the aggregation of the TIA proteins into SGs is reversible in cells exposed to a sublethal stress, but irreversible in cells exposed to a lethal stress (Kedersha et al., 1999), factors that regulate TIA aggregation may thus influence cell survival.
Prions are infectious proteins that can exist in at least two interchangeable forms that exhibit different physical properties, especially solubility (Riesner, 2003). In animals, prion protein can assume two conformations: the soluble conformer (PrPC) is rich in α-helices, whereas the insoluble scrapie conformer (PrPSc) is rich in β-pleated sheets. PrPSc has the capacity to convert PrPC into PrPSc, a requisite feature of infectious propagation (Chesebro, 2003; Riesner, 2003). In yeast, several proteins possessing prion-related domains serve as protein-based epigenetic elements that confer distinct, hereditable phenotypes. SUP35 is a translation termination factor that possesses a prion-like domain (i.e., the NM domain) at its amino terminus (Serio and Lindquist, 2001), which can adopt two distinct conformations. In (psi–) strains, SUP35 assumes a soluble conformation that mediates normal termination at stop codons. In (PSI+) strains, SUP35 assumes an aggregation-prone conformation that is incapable of affecting normal termination. The structural transition between soluble and aggregation-prone conformations is regulated by molecular chaperones such as heat shock protein (HSP)104, HSP70, and HSP40 (Chernoff et al., 1999; Newnam et al., 1999; Kushnirov et al., 2000; Kryndushkin et al., 2002; Schwimmer and Masison, 2002; Jones and Masison, 2003; Jones et al., 2003), as well as prion-related epigenetic modifiers such as RNQ1 (Sondheimer and Lindquist, 2000).
We have investigated the role of the PRD of TIA-1 in the assembly of SGs. Our results reveal that the PRD of TIA-1 forms a protein aggregate that serves as a scaffold onto which abortive preinitiation complexes are routed to form SGs. The NM domain from the yeast prion SUP35 can replace the PRD and reconstitute this function, supporting the idea that prion-like homotypic aggregation mediates the coalescence of untranslated mRNPs at SGs. Like SUP35, the aggregation of the PRD induces the expression of, and is regulated by, HSP70. We suggest that TIA-1–mediated translational repression is regulated by HSP70-catalyzed structural transitions within its PRD.
MATERIALS AND METHODS
Cell Lines
COS7 cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% fetal bovine serum. TIA-1 KO, TIAR KO, and WT cell lines were generated in our laboratory as described previously (Li et al., 2002). The S51A knock-in mouse embryonic fibroblasts (MEFs) were a kind gift from Donalyn Scheuner and Randall Kaufman (Scheuner et al., 2001).
Antibodies and Reagents
Anti-hemagglutinin (HA) monoclonal antibody was purchased from Covance (Princeton, NJ). Antibodies against eIF3-p116 (N20), TIA-1, HuR, eIF5, p70S6K1, and lamin A/E were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-eIF4G1 was obtained from AbCam (Cambridge, MA). Antibodies against eIF2-α (phospho-specific), HSP90, HSP70, HSP56, HSP40, and HSP27 were purchased from Stressgen Technologies (Victoria, British Colombia, Canada). Monoclonal anti-FLAG antibody, Hoechst dye (33258), and sodium arsenite were purchased from Sigma-Aldrich (St. Louis, MO). Antibody against G3BP was a kind gift from Imed Gallouzi (McGill University, Montreal, Canada). All secondary antibodies for immunofluorescence and Western blotting (Cy2-, Cy3-, Cy5-, and horseradish peroxidase-conjugates) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
COS Cell Transfections
COS7 cells were transfected as described previously (Kedersha et al., 2000a) with modifications as follows. Cells were incubated with DNA-SuperFect complexes for 4–8 h, and then trypsinized and replated to parallel plates for immunofluorescence (24-well plates containing 11-mm coverslips), Western blotting (12-well plate with no coverslips), and Northern blotting (10-cm dish). To induce stress granule formation in selected samples, arsenite (0.5 mM) was added to the media 30 min to 1 h before fixation and/or harvest.
Transmission Electron Microscopy
COS-7 cells were transfected with pMT2-TIA-1 or pMT2-TIA-1-PRD by using DEAE-dextran. After 48–72 h, cells were fixed with 1.25% glutaraldehyde in cacodylate buffer containing 1% CaC2 and postfixed in 1% OsO4. After dehydration and embedding, the samples were sectioned, stained with lead citrate, and examined with a JEM 100 CX II electron microscope.
Immunoelectron Microscopy
COS-7 transfectants were prepared for immunocryoelectron microscopy in the electron microscopy core facility at the Dana-Farber Cancer Institute (Boston, MA) by using methods that have been described previously (Medley et al., 1996) and examined with a JEOL 100CX electron microscope. Isotype control antibodies produced no detectable staining (our unpublished data).
Plasmid Construction
The construction of TIA-1 and TIA-1-PRD (originally referred to as TIA-1ΔRRM) in both pMT2-HA and pSRα-HA-green fluorescent protein (GFP) vectors was described previously (Tian et al., 1991; Kedersha et al., 2000a). Using previously constructed or gifted coding regions as templates, the coding regions used in this study were cloned into the pSRα-GFP-HA, pcDNA3-FLAG, or pEYFP (Invitrogen, Carlsbad, CA) vectors via a polymerase chain reaction (PCR) strategy. PCR products were amplified using primers listed in Table 1 and reaction conditions as listed in Table 2. The PCR products were digested with restriction enzymes as indicated and inserted in frame into the tagged vector, which was similarly cut. The Sup35NM coding region was a kind gift from Dr. Susan Lindquist (Massachusetts Institute of Technology, Cambridge, MA). The coding region of murine HSC70 was a kind gift from Dr. Jean-Claude Mercier (Institut National dela Recherche Agronomique, Jouy-en-Josas Cedex, France). The coding region of murine HSP70 was a kind gift from Dr. Stuart Calderwood (Harvard Medical School, Boston, MA). All plasmid constructs used in this study were verified by sequencing.
Table 1.
Primer sequences
| Plasmid Construction
|
||||||
| Construct | Primer Sequences | Restriction sites | Vector | |||
| pSRα-HA-TIA-1 | 5′-CACAGAATTCATGGAGGACGAGATG-3′ | EcoRI | pSRα-HA | |||
| 5′-TATATAGTCGACTCACTGGGTTTCATAC-3′ | SalI | |||||
| pSRα-HA-RRM | 5′-CACAGAATTCATGGAGGACGAGATG-3′ | EcoRI | pSRα-HA | |||
| 5′-TATATAGTCGACTCATAGTTGTTCTGTTAGC-3′ | SalI | |||||
| pSRα-HA-PRD | 5′-CACAGAATTCATGCGTCAGACTTTTTC-3′ | EcoRI | pSRα-HA | |||
| 5′-TATATAGTCGACTCACTGGGTTTCATAC-3′ | SalI | |||||
| pSRα-HA-Sup35NM | 5′-CACAGAATTCATGTCGGATTCAAACC-3′ | EcoRI | pSRα-HA | |||
| 5′-TATATAGTCGACTCACAAATTGTTATTGTAGTTG-3′ | SalI | |||||
| pSRα-HA-RRM/NM | 5′-CACAGAATTCATGGAGGACGAGATG-3′ | EcoRI | pSRα-HA | |||
| 5′-TATATCTAGATAGTTGTTCTGTTAGC-3′ | XbaI | |||||
| 5′-TATATCTAGAATGTCGGATTCAAACC-3′ | XbaI | |||||
| 5′-TATATAGTCGACTCACAAATTGTTATTGTAGTTG-3′ | blunt | |||||
| pcDNA3-FLAG-HSP70 | 5′-CTCTCGGATCCGCCAAGAACACGGCGATC-3′ | BamHI | pcDNA3-FLAG | |||
| 5′-CACACGAATTCCTAATCCACCTCCTCGAT-3′ | EcoRI | |||||
| pcDNA3-FLAG-HSC70 | 5′-CTCTCGGATCCAAGGGACCTGCAGTTGGC-3′ | BamHI | pcDNA3-FLAG | |||
| 5′-CACACGAATTCTTAATCCACCTCTTCAAT-3′ | EcoRI | |||||
| Digoxygenin-labeled DNA probes
|
||||||
| Probe | Primer sequence | Fragment size | Template | |||
| HSP27 | 5′-GTCAAGACCAAGGATG-3′ | 229 bases | pcDNA3-HSP27 | |||
| 5′-GACTCGAAGGTGACTG-3′ | ||||||
| HSP70 gene 2 UTR | 5′-AAGTGGACTGTTGGGACTCAAGGACTTTG-3′ | 239 bases | Heat shocked | |||
| 5′-CAAACAAACTCGTACAGAAGGTG-3′ | COS7 cDNA | |||||
| RT-PCR analysis
|
||||||
| Primer sequence | ||||||
| HSP70 coding region | 5′-ATAACGGCTAGCCTGAGGA-3′ | |||||
| 5′-GTCCGACTGCACCACCGGG-3′ | ||||||
| HSP70 gene 1 UTR | 5′-GAGCTTCAAGACTTTGCATTTCTTAG-3′ | |||||
| 5′-GGGCATCACTTGAATTTTAAAG-3′ | ||||||
| HSP70 gene 2 UTR | 5′-AAGTGGACTGTTGGGACTCAAGGACTTTG-3′ | |||||
| 5′-CAAACAAACTCGTACAGAAGGTG-3′ | ||||||
| ATF4 | 5′-CTGTGGGTCTGCCCGTCCCAAAC-3′ | |||||
| 5′-TCAACTAGGGGACCCTTTTC-3′ | ||||||
| GADD34 | 5′-GAGCAGCTTGCTCGGGATCGC-3′ | |||||
| 5′-TCAGCCACGCCTCCCACTG-3′ | ||||||
| XBP1 | 5′-CTTTGTGGTTGAGAACCAGG-3′ | |||||
| 5′-GGGAGCTCCTCCAGGCTG-3′ | ||||||
Table 2.
Reaction conditions
| Method | Reaction components | Thermal conditions |
|---|---|---|
| Plasmid construction | 1.25 mM dNTP mix (Qbiogene) | 94°C 45 s/60°C 30 s/68°C 1 min 30 s |
| 2.5 U of Platinum Pfx polymerase (Invitrogen) | 25 cycles | |
| 5 μl of solution Q (Promega, Madison, WI) | ||
| 5 μl of 10× Pfx buffer | ||
| 2.5 mM magnesium | ||
| 100 pmol/primer | ||
| 10 ng of template | ||
| Double-distilled H2O to 50 μl | ||
| Digoxygenin-labeled DNA probes | 10 μl of PCR DIG labeling mix (Roche Diagnostics) | 95°C 1 min/50°C 1 min/72°C 30 s |
| 2.5 U of Taq Polymerase (Fisher Scientific, Pittsburgh, PA) | 30 cycles | |
| 10 μl of 10× Taq buffer A | ||
| 1.5 mM magnesium | ||
| 100 pmol/primer | ||
| 10 ng of template | ||
| Double-distilled H2O to 100 μl | ||
| Reverse transcription | 10 U of AMV-RT (Promega) | 42°C 60 min/75°C 10 min |
| 4 μl of 5× AMV-RT buffer | ||
| 2 pmol of (oligo)dT15 | ||
| 40 U of RNAse inhibitor (Promega) | ||
| 2.5 μg of RNA from heat-shocked COS7 | ||
| 1.25 mM dNTP mix | ||
| Double-distilled H2O to 20 μl |
Western Blot Analysis
Western blot samples were processed as described previously (Kedersha et al., 2000a) with the following modification: samples were acetone precipitated in 60% acetone and resuspended in SDS sample buffer after lysis, boiling, and sonication. Proteins were resolved, and blots were processed as described previously (Kedersha et al., 2000a).
Digoxygenin-labeled DNA Probes
Digoxygenin-labeled DNA probes were created for nonisotopic Northern blot analysis by using PCR. Primers, templates, and probe lengths are listed in Table 1. Reagents used in reverse transcription and PCR reactions are listed in Table 2. To make the HSP70 3′-UTR probe, RNA was extracted from COS7 cells that had been heat-shocked at 42°C for 90 min before harvest, by using the RNAqueous kit (Ambion, Austin, TX) per manufacturer's protocol. A reverse transcription reaction was performed, and then from the resulting cDNA, a PCR reaction was performed under the same cycling conditions as described.
NonIsotopic Northern Blot Analysis
RNA was extracted from COS7 cells by using the RNAqueous kit (Ambion) per the manufacturer's protocol. Ten to 30 μg of RNA per sample was run on a 1.2% agarose-MOPS gel with ∼1.9% formaldehyde. RNA was transferred from the gel to a nylon membrane by using the TurboBlotter rapid downward transfer system (Schliecher & Schuell, Keene, NH) per manufacturer's protocol. After the transfer, the membranes were dried at 80°C for 2 h and then UV cross-linked. The membranes were rehydrated in 2× SSC and then prehybridized for 30 min in Dig-Easy Hyb granule hybridization solution (Roche Diagnostics, Indianapolis, IN). Digoxygenin-labeled DNA probes were denatured at 95°C for 5 min and then diluted in 200 μl of hybridization solution and added to the solution on the prehybridized membrane. Membranes were hybridized for 4–24 h at 50°C. After hybridization, membranes were washed twice for 5 min in 2× SSC + 0.1% SDS at room temperature and then washed twice for 15 min in 0.5× SSC + 0.1% SDS at 65°C. Membranes were blocked in a 1× solution of blocking reagent (Roche Diagnostics) for 30 min and then incubated for 30 min in anti-digoxygenin-AP antibody (Roche Diagnostics) diluted 1:5000 in 1× blocking reagent. Membranes were then washed twice for 15 min in DIG wash buffer (Roche Diagnostics) and once for 5 min in DIG detection buffer (Roche Diagnostics) and then incubated for 5 min in CDP-Star ready-to-use (Roche Diagnostics) to produce a chemiluminescence visualized on BioMax MR film (Eastman Kodak, Rochester, NY).
Immunofluorescence Microscopy and Cell Scoring
Cells were fixed and stained as described previously (Kedersha et al., 2000). Cells were viewed and photographed using a Nikon Eclipse 800 microscope with charge-coupled device-SPOT RT digital camera. The images were compiled using Adobe Photoshop software (version 7.0; Adobe Systems, Mountain View, CA).
For the stress granule formation assay, the pSRα-HA constructs were used. Slides were blinded and cells scored for stress granules. Certain samples in which the transfectants contained stress granules were then rescored to determine the presence or absence of the transfected protein at the stress granule. For the recombinant protein relocalization assay, the pMT2-HA constructs were used because these plasmids yield more protein that enhances the effectiveness of the assay. Slides were blinded and transfected cells were scored based on the localization of the majority (>50%) of the recombinant protein. The cells were divided into three categories: >50% nuclear localization, equal localization between cytoplasm and nucleus, and >50% cytoplasmic localization. Approximately 100 transfected cells were counted for each sample. The graphic data shown is the average of three to four independent experiments.
Reverse Transcription (RT)-PCR Analysis
RNA was extracted from COS7 cells 36–48 h posttransfection, or postheat shock, by using the RNAqueous kit (Ambion) per the manufacturer's protocol. Reverse transcription and PCR reactions were performed as described under “Digoxygenin-labeled DNA Probes” by using 1.25 mM dNTP mix (Qbiogene, Carlsbad, CA) instead of the DIG labeling mix. The primers used are listed in Table 1.
Soluble-Insoluble Fractionation of Lysates
COS7 cells were transfected as described above. At various times posttransfection, cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in ice-cold extraction buffer (Akakura et al., 2001) containing 10 mM Tris, pH 7.4, 1 mM MgCl2, 0.2% Tween 20, 10 mM sodium molybdate, and protease inhibitors. The lysates were scraped from the dishes and collected and then sonicated twice for 2 min in a Branson cup sonicator at setting 8, in ice water. The samples were immediately centrifuged at 14,000 × g for 20 min at 4°C to pellet insoluble material, and then the supernatant was removed and both fractions were boiled in 2% SDS, precipitated with 60% acetone, and resuspended in equal volumes of reducing SDS-PAGE sample buffer before SDS-PAGE and immunoblotting as described.
Protease Digestion
Resistance of the different forms of TIA-1 to protease K digestion was used to assess the state of the PRD in cells. COS7 cells were transfected with HA-TIA-1, HA-PRD, GFP-TIA-1, or GFP-PRD and harvested by scraping the cells into PBS at different time points as indicated. Cells were then pelleted, frozen, and thawed into the same buffer as used in the fractionation experiments (10 mM Tris, pH 7.4, 10 mM MgCl2, 0.2% Tween 20, and 10 mM sodium molybdate). Lysates were extensively sonicated to disrupt protein aggregates and then digested with protease K before SDS-PAGE-immunoblot analysis. Reactions were stopped by the addition of 2% SDS, 2% dithiothreitol and boiling for 10 min. Blots were probed with antibodies reactive with HA, TIA-1 (a polyclonal antisera reactive with the extreme carboxy terminus of TIA-1), or HSP40.
RESULTS
The TIA-1-PRD Is Required for the Assembly of SGs
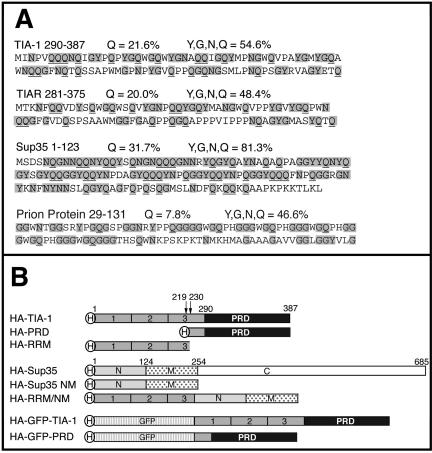
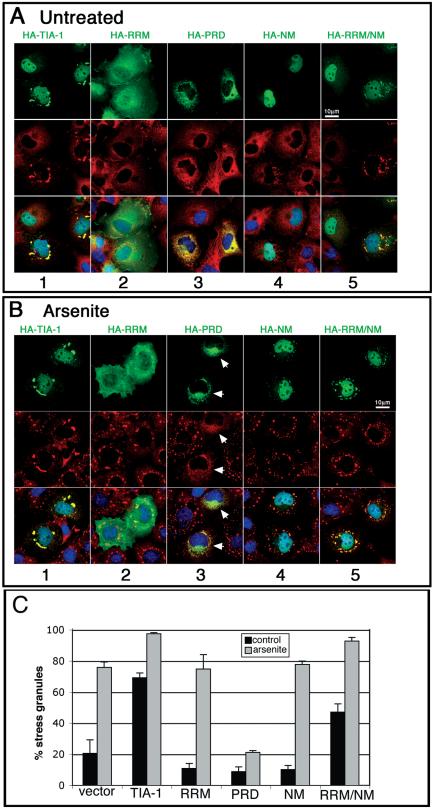
The carboxyl termini of TIA-1 and TIAR have an amino acid composition (∼20% Q; ∼50% QNYG) that is similar to the aggregation domains of mammalian and yeast prion proteins (Figure 1A). Intramolecular interactions between polar amino acids within these domains promote the assembly of homotypic or heterotypic oligomers (Perutz, 1994). To determine whether the PRD of TIA-1 contributes to the assembly of SGs, we compared the subcellular localization of full-length HA-TIA-1, HA-PRD, and HA-RRM (a truncation mutant composed of most of the TIA-1 RNA binding domains; Figure 1B). COS7 cells were transiently transfected with each construct and then processed for two-color immunofluorescence with anti-HA to visualize recombinant proteins (Figure 2A, top, green), and anti-eIF3, to visualize an endogenous marker of SGs (Figure 2A, middle, red). Like the endogenous protein, recombinant HA-TIA-1 is concentrated in the nucleus where it is excluded from nucleoli, but it is otherwise homogeneously distributed. Overexpression of HA-TIA-1 induces the assembly of eIF3-containing SGs in 70% of transfected cells (quantified in Figure 2C). These spontaneous SGs also contain poly(A+) RNA, PABP, HuR, and other SG markers (our unpublished data), and they are dissolved upon treatment with cycloheximide (Kedersha et al., 2000a), indicating that HA-TIA-1 overexpression induces the assembly of actual SGs rather than merely forming recombinant HA-TIA-1 aggregates due to overexpression. On treatment with arsenite, an inducer of oxidative stress, SGs are observed in nearly 100% of HA-TIA-1 transfectants (Figure 2B, column 1). When the PRD is deleted from TIA-1, the recombinant protein (HA-RRM) no longer induces the assembly of spontaneous SGs (Figure 2A, column 2). Moreover, HA-RRM is not recruited to arsenite-induced SGs, nor does it block SG formation (Figure 2B, column 2; C). Thus, the TIA-1 PRD is required for the spontaneous assembly of SGs and for recruitment of TIA-1 to arsenite-induced SGs. Overexpressed HA-PRD forms cytoplasmic microaggregates that sequester endogenous TIA-1 and TIAR (Kedersha et al., 1999), as well as some eIF3 (Figure 2A, column 3) and prevents the assembly of arsenite-induced SGs (Figure 2B, column 3, arrows).
Figure 1.
Prion-like domains in TIA-1, TIAR, Sup 35, and human prion protein. (A) Amino acid composition of the prion-like domains of TIA-1 and TIAR, the N-terminal prion domain of Sup35, and the aggregation domain of human prion protein. Y, G, N, and Q residues are shaded, and glutamine residues (Q) are underlined. (B) Schematic representation of TIA-1 and Sup35 constructs and full-length proteins. The PRD constructs in pMT2-HA and pSRα-HA-GFP vectors begin at methionine 230. The pSRα-HA-PRD initiates at methionine 219. H indicates the N-terminal HA tag.
Figure 2.
Replacement of the PRD of TIA-1 with the NM domain of yeast Sup35 restores normal function to the RRM truncation mutant. (A and B) Immunofluorescent micrographs of transiently expressed HA-tagged TIA-1, RRM, PRD, SUP35-NM, and RRM/NM in pSRα vector expressed in COS7 cells. Cells were untreated (A) or treated with 0.5 mM arsenite for 45 min (B) to induce stress granule formation before fixation and staining for HA (green), eIF3 (red), and DNA (blue). Bars, 10 μm. (C) Quantification of the percent of transfectants containing stress granules, with and without arsenite treatment. At least 100 transfectants were scored for each construct, and results averaged from three independent experiments.
Sup35-NM Can Substitute for TIA-1-PRD in Promoting the Assembly of SGs
To determine whether prion-like aggregation can mediate the assembly of SGs, we made constructs encoding SUP35-NM, the aggregation domain of the yeast prion SUP35, as well as chimeric constructs linking HA-RRM to SUP35-NM (Figure 1B, HA-RRM/NM). In the absence of arsenite, HA-SUP35-NM is predominantly nuclear in COS cells (Figure 2A, column 4) and is rarely observed in cytoplasmic aggregates (Figure 2A, column 4; C for quantification). Unlike HA-PRD, HA-SUP35-NM does not inhibit the arsenite-induced assembly of SGs (Figure 2B, column 4). However, HA-SUP35-NM is weakly recruited to arsenite-induced SGs, suggesting that it may assemble heterotypic aggregates with some SG component(s) (Figure 2B, column 4). When Sup35-NM is fused to the RNA recognition motifs (RRMs) of TIA-1 in lieu of the PRD, chimeric recombinant HA-RRM/NM induces the assembly of spontaneous SGs (Figure 2A, column 5) in 50% of transfected cells (Figure 2C), and it is efficiently recruited to arsenite-induced SGs (Figure 2B, column 5). We conclude that the prion-like NM domain from SUP35 can functionally substitute for the TIA-1 PRD and reconstitute its ability to assemble SGs.
Aggregated TIA-1 Associates with Ribosomes; Aggregated PRD Does Not
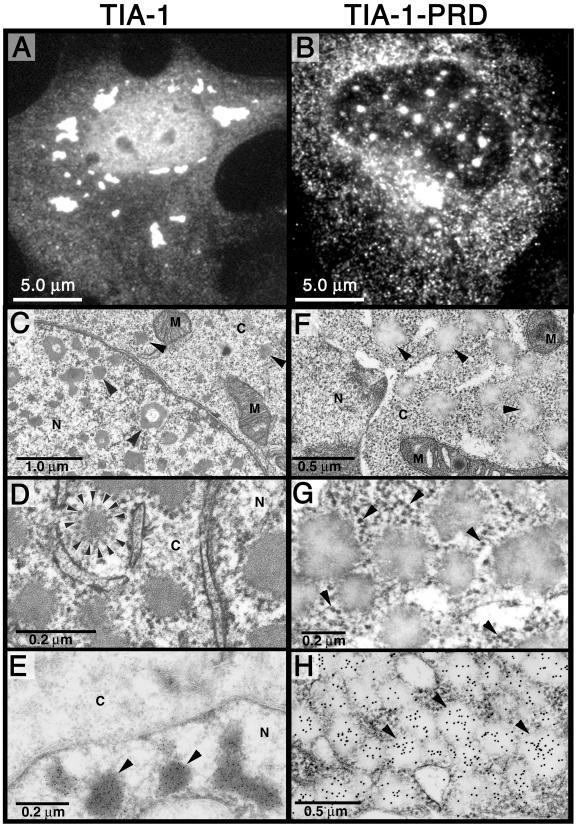
To examine the aggregates induced by overexpression of TIA-1 or the PRD at higher magnification, COS transfectants expressing untagged recombinant TIA-1 (Figure 3A) or PRD (Figure 3B) also were examined using transmission electron microscopy. Consistent with the results obtained using light microscopy (Figure 3, A and B), the TIA-1 transfectants display cytoplasmic and nuclear aggregates that average 0.1–0.2 μm in diameter and strongly stain with lead citrate (Figure 3, C and D). Nuclear aggregates are typically irregular and often donut shaped. Cytoplasmic aggregates are rimmed by a corona of particles that resemble ribosomes or ribosomal subunits (Figure 3D, arrowheads); this result is striking because small ribosomal subunits are among the core components of SGs (Anderson and Kedersha, 2002a; Kedersha et al., 2002). Immunogold staining using an antibody specific for the C terminus of TIA-1 confirms that these aggregates contain recombinant TIA-1 protein (Figure 3E). Cells transfected with PRD (Figure 3, F–H) contain more regular globular aggregates of similar size (0.1–0.2 μm) that stain poorly with lead citrate and are almost entirely cytoplasmic (Figure 3F, arrowheads). Unlike the TIA-1 aggregates, the PRD aggregates are not associated with ribosomelike particles (Figure 3G, arrowheads indicate ribosome-like particles), but they do stain with anti-TIA-1 antibody (Figure 3H), indicating that they contain recombinant PRD. Because lead citrate has a high affinity for RNA, these data suggest that the TIA-1 aggregates, but not the PRD aggregates, contain RNA. This conclusion is consistent with the finding that full-length TIA-1 induces spontaneous SGs containing eIF3 (Figure 2) and polyA(+) RNA (our unpublished data) and is also consistent with the inability of the PRD to bind RNA in vitro (Dember et al., 1996). The ribosome-associated cytoplasmic aggregates assembled by full-length TIA-1 are likely to be subcomponents of SGs. Interestingly, electron micrographs of plant SGs reveal that they are also composed of small aggregates that coalesce to form granules that are visible by light microscopy (Nover et al., 1989a,b). It is likely that the 1.0- to 2.0-μm individual SGs revealed by light microscopy are composed of considerably smaller aggregates that contain a protein core generated by self-assembling PRD-PRD interactions, to which abortive initiation complexes are recruited by the RRMs of TIA-1. In the absence of the RRMs, the PRD forms similar microaggregates (hereafter referred to as PRD microaggregates) that cannot bind RNA and thus do not become functional SGs.
Figure 3.
Aggregated forms of TIA-1 and PRD in COS7 cells. (A and B) Light micrographs of COS7 cells expressing recombinant HA-TIA-1 (A) or HA-PRD (B) stained for HA. (C–H) Transmission electron micrographs of COS7 TIA-1 (untagged) transfectants (C–E) or untagged PRD transfectants (F–H). (C) Arrowheads indicate nuclear and cytoplasmic inclusions of TIA-1 not observed in vector transfectants. N, nucleus; C, cytoplasm; M, mitochondria. (D) Enlarged view of cytoplasmic inclusions of TIA-1. Arrowheads indicate ribosome-like particles surrounding the cytoplasmic aggregates. (E) Immunoelectron micrograph of TIA-1 transfectant, labeled with anti-TIA-1 antibody and visualized using a gold-conjugated anti-mouse Ig. Arrowheads indicate nuclear inclusions of TIA-1. (F) Transmission electron micrograph of PRD transfectant. Arrowheads indicate cytoplasmic aggregates of recombinant PRD. (G) Enlarged view of cytoplasmic aggregates. Arrowheads indicate ribosome-like particles that are not associated with the PRD aggregates. (H) Immunoelectron micrograph of PRD transfectant stained for TIA-1 by using immunogold. Arrowheads indicate cytoplasmic aggregates of PRD, which stain less intensely with lead citrate than do TIA-1 aggregates but are strongly stained with anti-TIA-1 immunogold.
Concentration-dependent Aggregation of PRD Correlates with SG Assembly
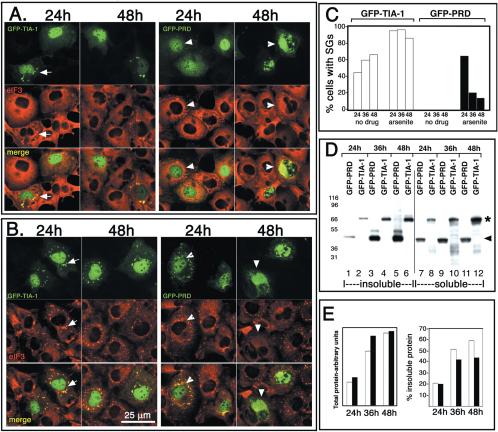
We confirmed that TIA-1 promotes, and PRD inhibits, the assembly of SGs by using GFP-TIA-1 and GFP-PRD (Figure 4). GFP-TIA-1 (Figure 4A, top; GFP fluorescence, green) induces the spontaneous assembly of SGs when expressed in COS cells (Figure 4A, arrows) and is also quantitatively recruited to arsenite-induced SGs (Figure 4B, arrows). The percentage of GFP-TIA-1 transfectants exhibiting spontaneous SGs increases with time after transfection (Figure 4C, white bars), suggesting that such SG induction is concentration dependent, because the amount of GFP-TIA-1 more than doubles between the 24- and the 36-h time point (Figure 4E, white bars).
Figure 4.
GFP-PRD progressively forms insoluble aggregates that prevent SG assembly. (A) COS7 cells expressing GFP-tagged versions of TIA-1 or PRD shown at 24 and 48 h after transfection. GFP-TIA-1 (green) expression induces spontaneous SG assembly (arrows, confirmed by eIF3 staining, red), whereas GFP-PRD does not induce SGs (green, arrowheads) and becomes predominantly cytoplasmic at later times (48 h). (B) COS7 transfectants as in A, treated with 0.5 mM sodium arsenite for 45 min Bar, 25 um. (C) Quantification of the percentage of GFP-TIA-1 (white bars) or GFP-PRD (black bars) transfectants that exhibit SGs. Transfected cells were cultured for 24, 36, or 48 h and either untreated or exposed to arsenite (0.5 μM) for 45 min. Data are expressed as the percentage of transfected cells with eIF3+ SGs. Data shown are typical of three independent experiments. (D) Western blot of GFP-PRD and GFP-TIA-1 protein in detergent-soluble and detergent-insoluble fractions prepared from cells harvested at the indicated times. Asterisk indicates the mobility of GFP-TIA-1; arrowhead indicates mobility of GFP-PRD. (E) Densitometric quantification of the data shown in D. White bars, GFP-TIA-1; black bars, GFP-PRD.
GFP-PRD behaves somewhat like endogenous TIA-1 at 24 h after transfection: the GFP-PRD is predominantly nuclear (Figure 4A), and it is recruited to arsenite-induced SGs (Figure 4B, green, arrowheads); however, no spontaneous SGs are induced. In cells transfected for longer than 32 h, GFP-PRD becomes predominantly cytoplasmic (Figure 4A, 48 h); and at higher magnification, it occurs in microaggregates similar to the PRD microaggregates formed by HA-PRD (Figure 3B) or untagged PRD (our unpublished data). In cells where the recombinant GFP-PRD is predominantly cytoplasmic and microaggregated, SGs are not formed in response to arsenite (Figure 4B, 48 h, arrowheads; quantified in C). Thus, GFP seems to increase the solubility and thereby delay the microaggregation of the PRD and the inhibition of SG assembly. Between 24 and 36 h, the amount of GFP-PRD present in these transfectants more than doubles (Figure 4E, white bars), suggesting that a critical threshold is reached, whereupon GFP-PRD irreversibly forms aggregates in vivo. Comparing serial dilutions of overexpressed GFP-PRD with undiluted nontransfected controls on immunoblots (using an antipeptide antibody against the PRD and correcting for dilution factor and transfection efficiency), we estimate that GFP-PRD is expressed at ∼100-fold molar excess to endogenous TIA-1 after 48 h (our unpublished data).
To investigate the correlation between apparent aggregation (as determined by immunofluorescence) and solubility, transfected cells harvested at different time points were sonicated in detergent-containing buffer and then fractionated into detergent-soluble and -insoluble fractions by high-speed centrifugation as described in Materials and Methods. Under these fractionation conditions, endogenous SG components such as TIA-1, HuR, and eIF3 are equally soluble in arsenite-treated cells as in control cells (see Supplemental Figure 1, lanes 1 and 2). The expression of GFP-TIA-1 and GFP-PRD was quantified using immunoblotting analysis and an antipeptide antibody recognizing the C terminus of TIA-1. At 24 h, when both GFP-TIA-1 (Figure 4A, 24 h, green, arrows) and GFP-PRD (Figure 4A, 24 h, green, arrowheads) are predominantly nuclear and when GFP-PRD is recruited to arsenite-induced SGs (Figure 4B, green, arrowheads; C), the solubility of both proteins is similar (Figure 4D, quantified in E). With increasing time and expression levels, the solubility of each protein is reduced. Notably, at the 36- and 48-h time points, GFP-TIA-1 displays a range of low-molecular-weight degradation products that are exclusively soluble (Figure 4D, lanes 10 and 12). In contrast, GFP-PRD does not seem to be degraded but instead displays a range of higher molecular weight species that are exclusively insoluble (Figure 4D, lanes 3 and 5; quantified in E). This suggests that cells are able to degrade SG-associated, soluble GFP-TIA-1, whereas microaggregated, insoluble GFP-PRD becomes cross-linked. Detergent insolubility, protease resistance, and formation of higher molecular weight covalent adducts are characteristic of prion-like proteins in their aggregated state (Aguzzi and Polymenidou, 2004). Together, the data indicate that the aggregation of the PRD is concentration dependent, and its assembly into insoluble microaggregates is required for it to inhibit SG assembly.
PRD Aggregates Are Resistant to Proteolytic Digestion
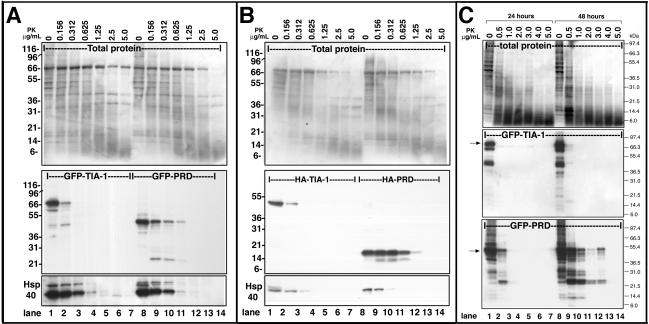
One of the hallmarks of prion-like proteins is the resistance of aggregation-prone conformers to protease treatment. To compare the sensitivity of full-length TIA-1 and PRD to protease digestion, we expressed GFP- or HA-tagged forms of each protein in COS7 cells. Detergent lysates were prepared and were extensively sonicated (twice for 2 min by using a Branson sonicator) to completely disrupt cellular structure before treatment with protease K. Cell lysates were treated with different concentrations of protease K and then subjected to SDS-PAGE and blotted using a commercial antibody against the extreme C terminus of TIA-1 (Figure 5, A and B, middle). Both HA-tagged (Figure 5B) and GFP-tagged forms of PRD exhibit markedly more resistance to protease K digestion over time than similarly tagged forms of the full-length protein (Figure 5A). Whereas GFP-TIA-1 is completely digested by 0.312 μg/ml protease K (Figure 5A, middle, lane 3), GFP-PRD exhibits partial resistance when treated with twice as much protease K (Figure 1A, lane 11). Ponceau red staining of total protein (top) as well as the digestion of endogenous HSP40 on the same blot (bottom) serves as loading controls and also confirms that the protease was equally active in both sets of digestions.
Figure 5.
Protease resistance of GFP-PRD. (A) GFP-PRD is more resistant to proteolysis that GFP-TIA-1. COS7 cell lysates containing GFP-TIA-1 or GFP-PRD were prepared from 48-h transfectants and then incubated with the indicated concentrations of protease K for the times indicated at room temperature. Top, ponceau red staining of total protein. Middle, blot probed with anti-TIA-1 antibody against the C-terminal region. Bottom, blot probed for endogenous HSP40. (B) HA-TIA-1 and HA-PRD lysates, treated as in A. (C) Protease resistance of GFP-PRD increases with time. COS7 cells expressing GFP-TIA-1 or GFP-PRD were cultured for 24 or 48 h, lysed, and sonicated, and then treated with the indicated concentrations of protease K for 30 min at room temperature. Top, total protein. Middle, digestion of GFP-TIA-1. Bottom, digestion of GFP-PRD; blots probed with the antibody against the TIA-1 C terminus.
To determine whether protease resistance was related to aggregation and insolubility of the PRD, we next examined lysates obtained from COS cells transfected for 24 versus 48 h (Figure 4). Whereas GFP-TIA-1 is fully sensitive to protease K digestion at both early and late time points (Figure 5C, middle), the protease resistance of GFP-PRD is increased between 24 and 48 h (Figure 5C, bottom). These results suggest that the PRD acquires protease K resistance concurrent with its cytoplasmic aggregation in vivo, its ability to prevent SG assembly, and its detergent insolubility.
The PRD of TIA-1 Induces the Expression of HSP70
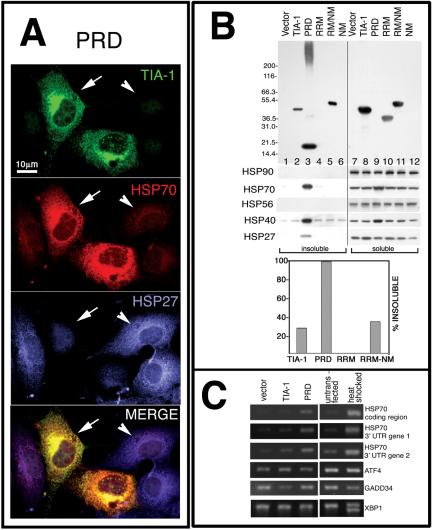
Prion-like proteins can assume either aggregation-prone or soluble conformations (Serio and Lindquist, 2001). The regulated transition between these structural states is facilitated by molecular chaperones (e.g., HSP70, HSP40, HSP90, and HSP104) (Serio and Lindquist, 2001). Moreover, aggregates of the yeast prion SUP35 trigger a stress response that induces the expression of molecular chaperones (Schwimmer and Masison, 2002). We therefore examined the expression levels of various chaperones in cells transfected with HA-TIA-1, HA-PRD, or HA-RRM. Immunofluorescent microscopy reveals that cells containing HA-PRD microaggregates (Figure 6A, green, arrows) express more HSP70 (Figure 6A, red) than nontransfected cells (Figure 6A, arrowheads). The HSP70 colocalizes with HA-PRD in both cytoplasmic and nuclear aggregates, looking yellow in the merged view (Figure 6A). In the PRD-transfected cells, the signal for HSP27 (Figure 6A, blue, compare transfected cells [arrows] to nontransfected cell [arrowheads]) looks diminished. Cells overexpressing full-length HA-TIA-1 or HA-RRM exhibit no alterations in HSP70 or HSP27 (our unpublished data). The effect of HA-PRD seems specific for certain chaperones, because its overexpression does not result in increased expression of HSP90, HSC70, HSP56, or p23 as determined by immunofluorescence (our unpublished data).
Figure 6.
PRD aggregation induces HSP70 expression and sequesters HSP27. (A) Immunofluorescence micrographs of HA-PRD transfectants at 46 h, stained for TIA-1 (green), endogenous HSP70 (red), and endogenous HSP27 (blue). (B) Solubility of different HA-tagged proteins. COS7 transfectants were fractionated into insoluble and soluble fractions as described, resolved by PAGE, and probed with antibodies against HA, HSP90, HSP70, and HSP27. Top, blot probed with anti-HA; bottom, blot probed for HSP70, HSP40, and HSP27 as indicated. Graph shows quantification of percentage of insoluble recombinant protein. The HA-NM construct was not detected by Western blot, and its solubility could not be determined. (C) RT-PCR analysis of RNA from pMT2 vector and TIA-1 and PRD transfectants. Activation of the unfolded protein response causes the splicing of XBP1 mRNA to its smaller, active form. Primers used for this PCR reaction spanned the spliced region, creating a smaller band upon activation of the unfolded protein response.
The exact colocalization of HSP70 with HA-PRD suggests that these proteins may directly or indirectly interact. We therefore used detergent fractionation and immunoblotting to determine whether HSP70 and HSP27 remain soluble or associate with insoluble HA-PRD. As shown in Figure 6B, HA-PRD is almost entirely insoluble (Figure 6B, compare lanes 3 and 9), whereas HA-RRM is almost entirely soluble (Figure 6B, compare lanes 4 and 10). HSP70 is entirely soluble except in cells transfected with HA-PRD, where much of it occurs in the insoluble fraction (Figure 6B, lane 3). In addition, an increased amount of HSP70 is found in the soluble fraction in HA-PRD transfectants (Figure 6B, lane 9), consistent with its increased expression revealed by immunofluorescence (Figure 6A). HSP27 is similarly rendered insoluble by HA-PRD expression, but no changes in total amount of HSP27 protein are observed, nor does its mRNA increase as determined by Northern blotting analysis (our unpublished data). These results suggest that HSP27 is present in the PRD microaggregates, but it is inaccessible to the anti-HSP27 antibody in situ. Given that HSP70 requires protein cofactors for its regulated chaperone activities, we also blotted for several other HSP70 cochaperones. Like HSP70, HSP40 exhibits insolubility and increased expression in response to HA-PRD expression (Figure 6B); it also colocalizes with HA-PRD by immunofluorescence (our unpublished data). In contrast, the expression and solubility of HSP56 and HSP90 are unaffected by HA-PRD expression (Figure 6B). Together, the data indicate that insoluble HA-PRD microaggregates contain HSP27, HSP40, and HSP70 and that HSP70 and HSP40 expression are strongly induced by microaggregated PRD.
HSP70 is encoded by two separate genes that have identical 5′ untranslated and coding regions, but different 3′ untranslated regions (3′UTR) (Huang et al., 2001). To determine which of the HSP70 transcripts are induced by HA-PRD, we selected primers specific for the 3′ UTR of each gene to amplify transcripts by RT-PCR. This analysis reveals that the expression of both HSP70 transcripts is increased in response to HA-PRD expression, while the expression of HSP70 mRNA is not increased in cells transfected with vector control or HA-TIA-1 (Figure 6C). We also analyzed components of the endoplasmic-reticulum stress-induced pathway (Harding et al., 2002; Liu and Kaufman, 2003) and find that ATF4 and GADD34 mRNAs are not induced by HA-PRD, nor is processing of XBP-1 mRNA detected. Thus, the expression of the PRD does not trigger a generalized stress response, but selectively induces the transcription of HSP70 mRNA.
Recombinant HSP70 Inhibits the Aggregation of PRD and Prevents the Induction of Endogenous HSP70
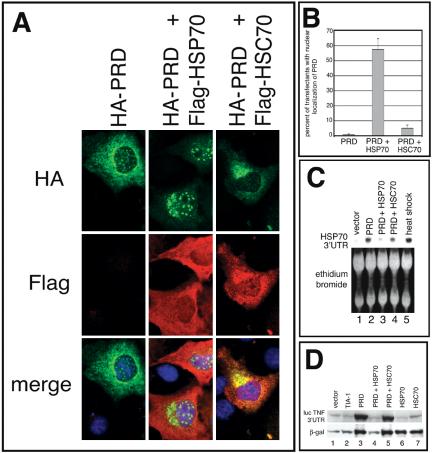
To determine whether the cytoplasmic aggregation of PRD is regulated by HSP70, we compared the subcellular localization of HA-PRD in COS-7 cells cotransfected with FLAG-HSP70 or FLAG-HSC70. In the absence of enforced expression of HSP70, HA-PRD accumulates in cytoplasmic microaggregates and at discrete nuclear foci (Figure 7A, left; also see Figure 3B). In cells cotransfected with FLAG-HSP70, the cytoplasmic aggregates are greatly reduced or disappear, whereas the nuclear foci become more prominent (Figure 7A, middle). These changes are not observed in cells cotransfected with HSC70 (Figure 7A, right). The average percentage of transfectants (n = 3) in which HA-PRD is relocalized to the nucleus in the presence of HSP70 and HSC70 is quantified in Figure 7B. These results suggest that HSP70, but not HSC70, can alter the conformation of HA-PRD so as to prevent its cytosolic aggregation and restore its nuclear localization.
Figure 7.
Overexpression of exogenous HSP70, but not HSC70, reverses the effects of the PRD on reporter gene expression. (A) Immunofluorescence micrographs of transfectants expressing the HA-PRD truncation mutant cotransfected with pcDNA3 vector, FLAG-tagged HSP70, or FLAG-tagged HSC70, stained for HA (green), FLAG (red), or DNA (blue). (B) Quantification of effects of vector, HSP70, or HSC70 on nuclear localization of HA-PRD in COS7. Cells were stained with anti-HA and anti-FLAG antibodies and manually scored for nuclear localization of HA-PRD. The results are the average of four independent experiments. (C) Northern blot analysis of COS7 transfectants coexpressing HA-PRD with pcDNA3 vector, HSP70, or HSC70. The HSP70 3′UTR probe recognized only the endogenous levels of HSP70 mRNA in all samples, not the exogenous HSP70 mRNA. RNA extracted from untransfected, heat shocked COS7 cells (42°C for 90 min) was used as a positive control. (D) Immunoblot showing effects of different constructs on reporter gene expression. Equal amounts of protein were loaded, and expression of the indicated constructs was verified by reprobing the same blot (our unpublished data).
If the cytoplasmic aggregation of HA-PRD triggers the induction of HSP70, enforced expression of recombinant HSP70, which inhibits the formation of cytoplasmic aggregates, should inhibit the PRD-induced transcription of endogenous HSP70. We used Northern blotting analysis to quantify the expression of induced HSP70 mRNA (using a probe for the 3′untranslated region (UTR), which recognizes gene 2, but not the recombinant construct) in cells transfected with HA-PRD, which were cotransfected with either HSP70 or HSC70, or a vector control. As shown in Figure 7C (lane 2), HA-PRD induces the expression of endogenous HSP70 mRNA. Cotransfection with recombinant HSP70 (lane 3), but not HSC70 (lane 4), prevents the induction of endogenous HSP70 mRNA. Heat shock was included as a positive control for the induction of HSP70 mRNA (lane 5).
We have previously reported that the PRD of TIA-1 enhances reporter gene expression (Kedersha et al., 2000a), an effect attributed to its ability to sequester endogenous TIA-1 in cytoplasmic aggregates and thus inactivate the function of TIA-1 as a translational silencer. If PRD aggregation is required for this effect, then coexpression of HSP70 (which inhibits cytosolic aggregation) but not HSC70 (which does not) should reverse the enhanced expression of reporter genes induced by PRD coexpression. To assess the effects of PRD aggregation on protein expression, reporter constructs encoding either β-galactosidase or luciferase containing the 3′UTR of tumor necrosis factor-α (which contains the TIA binding sites and is translationally repressed by TIA; Kedersha et al., 2000a) were cotransfected with either vector, full-length HA-TIA-1, HA-PRD alone, HA-PRD with HSP70, HA-PRD with HSC70, HSP70 alone, or HSC70 alone. After 48 h, total protein lysates were examined by immunoblotting to determine the levels of reporter gene expression. As shown in Figure 7D and as reported previously (Kedersha et al., 2000a), expression of HA-PRD (lane 3) enhances expression of both reporters, whereas vector (lane 1) and TIA-1 (lane 2) do not. Coexpression of HSP70 with HA-PRD, which drives the HA-PRD to the nucleus (Figure 7A, middle), reduces the expression of reporter genes to baseline levels (lane 4). In contrast, coexpression of HSC70 with HA-TIA-1 does not alter TIA-1-PRD localization (Figure 7A, right) nor repress reporter gene expression (Figure 7D, lane 5). The amount of recombinant HA-PRD is not affected by HSP70 or HSC70 coexpression as determined by Western blot with anti-HA (our unpublished data). Together, the data indicate that the subcellular localization and aggregation state of TIA-1 regulate its effects on gene expression.
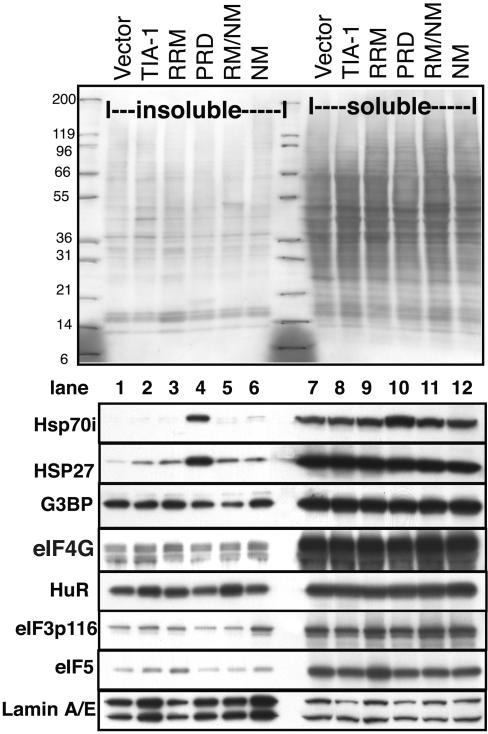
Solubility of Other SG Markers Is Not Altered by PRD Expression
The ability of overexpressed PRD to inhibit SG assembly could be directly due to immobilization of endogenous TIA-1/TIAR via protein–protein interactions, or indirectly by trapping other factors necessary for SG assembly in the aggregates. Candidate SG assembly factors include chaperones such as HSP70/HSP27/HSP40, or other SG components such as G3BP or eIF3. The data presented thus far demonstrate a PRD–HSP interaction and support a mechanism whereby HSP70-catalyzed conformational changes drive PRD aggregation, but they do not rule out the possibility that other SG components are involved. We therefore examined the soluble and insoluble fractions containing the different TIA constructs (as shown in Figure 6B), including the PRD for the presence of other SG markers. Figure 8 shows that the solubility of SG components G3BP, eIF4G, HuR, and eIF3 are not altered by overexpression of the PRD (Figure 8, lane 4). Similarly, PRD overexpression does not alter the sedimentation of other non-SG translation factors such as eIF5, nor lamin A/E (included as a loading control). We conclude that TIA-1 and molecular chaperones are key regulators of SG assembly/disassembly.
Figure 8.
Solubility of SG components in cells expressing mutant forms of TIA-1. COS transfectants were fractionated into soluble and insoluble fractions as described in Figure 6B and probed for additional SG markers. Top, ponceau red staining showing total protein. Numbers indicate mobility of molecular weight markers in kilodaltons. Blots were probed for HSP70, HSP27, G3BP, eIF4G, HuR, eIF3p116, eIF5, and lamin A/E (as a loading control), as indicated.
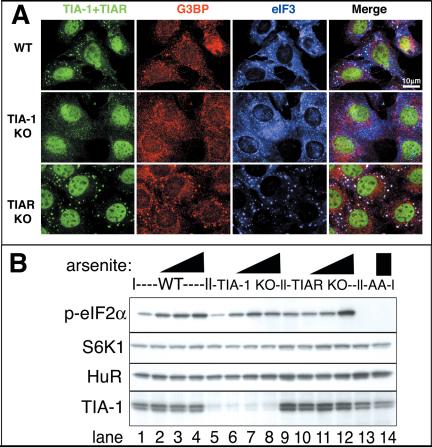
TIA-1 KO MEFs Exhibit Impaired Ability to Form SGs
To examine the importance of TIA-1 in the regulation of SG assembly, we subjected MEFs derived from wild-type, TIA-1 KO, or TIAR KO mouse embryos to arsenite, heat shock, and other stresses. Whereas wild-type and TIAR KO MEFs readily assembled SGs in response to arsenite treatment, TIA-1 KO MEFs seem deficient in their ability to assemble morphologically discrete SGs, as determined using antibodies against three independent markers of SGs: TIA (green), G3BP (red), and eIF3 (blue) (Figure 9A). Although a minority of TIA-1 KO cells were able to assemble normal SGs, the majority of the TIA-1 KO MEFs exhibited only marginal ability to assemble distinct SGs (Figure 9A, middle row). Similar results were obtained using other stresses such as heat shock (our unpublished data), as were seen using other markers of SGs such as HuR, eIF4E, and eIF4G (our unpublished data). Surprisingly, the TIAR KO MEFs exhibited an enhanced ability to form SGs compared with WT MEFs (compare Figure 9A, bottom row, with A, top row). To determine whether these differences in SG assembly were mediated by loss of TIA-1, or instead indicative of impaired ability to phosphorylate eIF2α in response to stress, we treated MEFs with different concentrations of arsenite and analyzed protein extracts for the presence of phospho-eIF2α by using a phospho-specific antibody (Figure 9B). Both wild-type (lanes 1–4) and TIA-1/TIAR KO MEFs (lanes 5–8 and 9–12) exhibited a similar increase in phospho-eIF2α upon arsenite treatment. As a control for the specificity of the phospho-specific antibody, lysates from MEFs derived from mice homozygous for a mutant form of eIF2α that cannot be phosphorylated (S51A mutant; Scheuner et al., 2001) exhibited no detectable signal (lanes 13 and 14).
Figure 9.
TIA-1 KO MEFs exhibit impaired SG assembly. (A) MEFs from wild-type, TIA-1 –/– or TIAR –/– MEFs were exposed to 0.5 mM arsenite for 30 min and then fixed and stained for TIA (green, using a mixture of anti-TIA-1 and TIAR antibodies), G3BP (red), and eIF3 (blue). Bar, 10 μm. (B) MEFs from wild-type, TIA-1 –/–, TIAR –/–, or S51A mutant cells (as a nonphosphorylated control) were untreated (lanes 1, 5, 9, and 13) or treated for 30 min with 0.125 mM arsenite (lanes 2, 6, and 10), 0.5 mM arsenite (lanes 3, 7, and 11), or 0.25 mM arsenite (lanes 4, 8, 12, and 14). Blots were probed for phospho-eIF2α, p70S6 kinase (as a loading control), HuR, and TIA-1.
The enhanced ability of TIAR KO MEFs to form SGs was unexpected but reproducible in several trials and with different stimuli such as heat shock (our unpublished data). These cells seem to exhibit somewhat enhanced phospho-eIF2α in response to the highest concentration of arsenite (lane 12), but this does not explain their enhanced ability to form SGs in response to heat shock by using conditions in which no increase in phospho-eIF2α relative to the other cells was observed, despite enhanced SG assembly (our unpublished data). Exclusive of eIF2α phosphorylation, an additional factor that might explain this difference is that TIAR KO cells seem to compensate for loss of TIAR by up-regulation of TIA-1 compared with wild-type MEFs (compare lanes 9–12 with 1–4 and 13 and 14 in the bottom panels of Figure 9B), which has been reported previously [see Figure 7 of Li et al. (2002)]. This is not an adaptive response in tissue culture, because similar compensatory changes are seen in tissues from the knockout (KO) animals (N.K., unpublished data). Therefore, the relative ability of the different MEFs to assemble SGs correlates with the amount of TIA-1 protein they express. To determine whether the partial ability to assemble SGs in the TIA-1 KO MEFs was due to TIAR, we attempted to knock down TIAR in the TIA-1 KO MEFs by using RNA interference. However, this resulted in extreme vacuolation of the cells and subsequent cell death, consistent with previous reports indicating that either TIA-1 or TIAR is required for cell viability (Le Guiner et al., 2003). Whether SG assembly is required for viability, or whether other TIA-mediated functions such as splicing are critical for viability remains to be determined.
DISCUSSION
Previous work from our laboratory and others has established that SGs are dynamic aggregates of abortive translation initiation complexes that form in response to stress-induced phosphorylation of eIF2α (Kedersha et al., 1999, 2000a,b, 2002; Kimball et al., 2003). TIA-1 and TIAR are RNA binding proteins that link the phosphorylation of eIF2α to the assembly of SGs. The TIA proteins are composed of an RNA binding domain and a glutamine-rich PRD, which has been proposed to self-aggregate and thereby drive the assembly of SGs (Anderson and Kedersha, 2002b). Here, we show that both the RNA binding domains and the PRD are required for SG assembly: the RRMs are required to recruit RNA to the SG, whereas the PRD is required to create the cytoplasmic aggregates to which the SG components are recruited. We present evidence that the HSP70-regulated aggregation of the PRD is mechanistically similar to the aggregation of mammalian and yeast prion proteins and that its aggregation regulates TIA-1 localization and function.
The aggregation domain of TIA-1 is rich in polar amino acids (Figure 1A) that promote homotypic oligomerization (Perutz et al., 1994). When overexpressed in COS cells, recombinant PRD spontaneously assembles microaggregates that coalesce to form larger cytoplasmic inclusions (Figure 3B and F–H), demonstrating its propensity to aggregate at high concentrations. These cytoplasmic PRD aggregates quantitatively recruit endogenous TIA-1 and TIAR and sequester them in the cytoplasm, thereby preventing their normal shuttling (N.K. and P.A., unpublished observation). Similar induced aggregation occurs in yeast cells, wherein overexpressed SUP35 forms aggregates that sequester endogenous SUP35, by inducing a conformational change in endogenous SUP35 that renders it aggregation prone (Serio et al., 2000). In mammalian cells, mutant aggregation-prone huntingtin promotes the aggregation of wild-type huntingtin both in vitro and in vivo (Busch et al., 2003). In GFP-PRD transfectants, the concentration-dependent appearance of detergent-insoluble aggregates correlates with the inhibition of SG assembly and the appearance of higher molecular weight species, the molecular nature of which remains to be determined (Figure 4D). This suggests that TIA-1 displays a similar conformation-dependent change of function, the soluble form being a translational silencer and the aggregated form promoting stress-induced translational arrest via SG assembly. However, it remains to be determined whether PRD microaggregates can act as molecular seeds to directly catalyze the ordered aggregation of soluble TIA-1.
The aggregated form of mammalian prion protein is strongly resistant to protease digestion (Riesner, 2003), as are the aggregation-prone conformers of yeast prion-like proteins (Masison and Wickner, 1995; King et al., 1997; Komar et al., 1997). We show here that the assembly of detergent-insoluble GFP-PRD aggregates correlates with the acquisition of GFP-PRD protease resistance, suggesting that the PRD assumes a protease-resistant conformation only upon aggregation. We have not detected protease-resistant species of endogenous TIA-1 in lysates from arsenite-treated cells, but because photobleaching studies indicate that TIA-1 very rapidly shuttles in and out of SGs with a residence time of seconds (Kedersha et al., 2000a), this result is not surprising. It is likely that conformational changes in TIA-1 that drive SG assembly in vivo are very rapid and continually reversed by the action of chaperones in an ATP-dependent manner (see below). The aggregated GFP-PRD seems to represent a more stable form of aggregate, possibly similar to the metastable protease resistant forms of prion protein or huntingtin seen in various human pathologies. Indeed, TIA-1 has been reported to be a component of huntingtin inclusions (Waelter et al., 2001).
The aggregation of the PRD is specifically regulated by HSP70 (Figure 7), because overexpressed HSP70, but not HSC70, prevents the cytoplasmic aggregation of PRD. Thus, HSP70 either dissolves PRD aggregates or prevents PRD aggregates from forming in vivo. This specific requirement for HSP70 suggests that levels of available HSP70 may regulate SG disassembly during recovery from stress. Because the aggregation of the yeast prion SUP35 is highly dependent on specific molecular chaperones such as HSP104, HSP70, and HSP40 (Newnam et al., 1999), the specific requirement for HSP70 in the regulation of PRD aggregation further underscores the prion-like nature of TIA-1 aggregation.
Yeast strains in which SUP35 assumes its aggregationprone conformation express increased amounts of HSP70 (Schwimmer and Masison, 2002). PRD aggregates induce the transcription of HSP70 mRNA, but they do not induce transcription of ATF4 or GADD34 mRNA, or processing of XBP-1 mRNA. Thus, PRD aggregates do not induce a general stress response but rather selectively induce the transcription of HSP70 mRNA. The cosedimentation of HSP70 protein with HA-PRD (Figure 6B) and the precise colocalization of PRD and HSP70 (Figure 6A) suggest that HSP70 interacts, directly or indirectly, with aggregated PRD, consistent with the finding that HSP70 prevents cytosolic PRD aggregation. Interactions between HSP70 and the PRD also may determine how the PRD induces the expression of HSP70 transcripts. Stress-induced transcription of HSP70 mRNA is induced by the nuclear translocation of the transcription heat shock factor (HSF)1. In unstressed cells, HSP70 maintains HSF1 in a conformation that masks a nuclear import signal (Cotto and Morimoto, 1999). On stress, the accumulation of unfolded proteins displaces HSP70 from HSF1, allowing HSF1 to move to the nucleus and induce the expression of HSP70 mRNA. In a similar manner, PRD aggregation could divert HSP70 away from HSF1, allowing its nuclear import and transcription of HSP70 mRNA. By this mechanism, the PRD may constitute a conformational switch that regulates SG assembly in response to HSP70 activity. It is interesting to note that HSP70 mRNA is excluded from SGs during heat shock (Kedersha and Anderson, 2002), conditions when its translation is permitted, while that of most housekeeping mRNAs is prevented. Although the mechanism whereby HSP70 mRNA is excluded from SGs is not known, its exclusion from SGs likely contributes to its preferential translation during stress.
The behavior of the RRM/NM chimeric protein provides further evidence that prion-like aggregation drives the assembly of SGs, because its expression promotes the assembly of spontaneous SGs that are indistinguishable from those induced by wild-type TIA-1 (Figure 2A, compare column 5 and 1). Moreover, RRM/NM is quantitatively recruited to arsenite-induced SGs (Figure 2C), indicating that SUP35-NM can functionally replace the PRD of TIA-1. Surprisingly, the isolated NM domain of SUP35 does not form spontaneous aggregates at concentrations achieved in COS transfectants, nor does it inhibit the assembly of arsenite-induced SGs. It is, however, weakly recruited to arsenite-induced SGs, suggesting that it can interact with some SG components, possibly the PRDs of TIA-1 or TIAR.
In yeast, the de novo appearance of the aggregation-prone conformation of SUP35 requires the presence of other prion proteins (e.g., RNQ1, URE2, and NEW1) (Derkatch et al., 2001), indicating that the prion-like domain from one protein can influence the aggregation of the prion-like domain of another protein. Although SUP35-NM does not spontaneously aggregate in COS cells, it is recruited to arsenite-induced SGs. This result suggests that some component of SGs can convert SUP35-NM to an aggregation-prone conformation. Similarly, HA-RRM/NM–induced SGs efficiently recruit endogenous TIA-1 (our unpublished data). The ability of SGs containing aggregated TIA-1 to recruit SUP35-NM and vice versa suggests that the aggregation-prone domains of these proteins interact to influence the transition between soluble and aggregation-prone conformations. Such an interaction might be direct or indirect (i.e., by competition for chaperones required to maintain both proteins in soluble form).
Previous studies indicated that the phosphorylation of serine 51 on eIF2α is necessary and sufficient to induce SGs (Kedersha et al., 1999). However, the ability of metabolic inhibitors such as mitochondrial poisons or glucose deprivation to induce SGs without any measurable increase in the phosphorylation of eIF2α was unexplained (Kedersha et al., 2002). eIF2α phosphorylation inhibits protein translation by depleting the eIF2-GTP-tRNA Meti ternary complex. We previously speculated that metabolic inhibitors might similarly deplete the levels of ternary complex by reducing the availability of GTP (Anderson and Kedersha, 2002a,b), but present data suggest an alternative or additional mechanism by which energy starvation might promote SG assembly: HSP70-induced conformational changes are ATP dependent (Young et al., 2003). Consequently, energy starvation could prevent HSP70-induced solubilization of the TIA-1 PRD, and the resulting aggregation of the PRD could promote the assembly of SGs. By this mechanism, the driving force for SG formation in response to metabolic inhibitors may be a reduced rate of SG disassembly, rather than an increased rate of SG assembly. Recent data indicate that basal phosphorylation of eIF2α is absolutely required for the assembly of SGs, because cells homozygous for a nonphosphorylatable form of eIF2α (S51A) are unable to form SGs in response to a broad range of stress stimuli, including energy starvation (N.K. and P.A., unpublished data). Together, these results indicate that basal, but not hyperphosphorylation of eIF2α is required for the assembly of SGs. The nature of the essential role for phospho-eIF2α in the assembly of SGs remains to be determined.
Interestingly, PRD is found in nuclear speckles where it colocalizes with HSP70 (Figure 6A). These speckles become more prominent when HSP70 is coexpressed (Figure 7A) and also contain splicing factors such as SC35 (N.K. and P.A., unpublished observations). In addition to its role in the assembly of cytoplasmic SGs, TIA-1 promotes the alternative splicing of selected heterogeneous nuclear RNAs (Del Gatto-Konczak et al., 2000; Forch et al., 2000; Zhu et al., 2003). By regulating the aggregation of the TIA-1 PRD, HSP70 may affect TIA-1–induced splicing events in the nucleus.
The ability of protein aggregates to promote human disease has been well described. The large number of proteins that are prone to pathological aggregation implies that the propensity to aggregate is essential for normal cellular functions (Kopito and Ron, 2003). This is certainly true in yeast, where controlled aggregation regulates the functions of a handful of proteins (Serio and Lindquist, 2001). In Aplysia, prion-like aggregation of CPEB (Si et al., 2003) may play a role in the maintenance of long-term synaptic changes associated with memory storage. Our results suggest that prion-like aggregation of TIA-1 is another example of this type of regulatory control. Given that aggregation-prone and prion-related domains from different proteins have a propensity to interact with one another and/or compete for the same chaperones, it will be important to determine whether interactions between TIA-1, TIAR, CPEB, huntingtin, and other aggregation-prone proteins contribute to normal or abnormal cellular function.
Supplementary Material
Acknowledgments
We thank Dr. Steven Wax for making the pCDNA3-FL-HSP70 and pCDNA3-FL-HSC70 constructs, Dr. Wei Li for constructing the pSRα-HA-GFP-PRD, Dr. Imed Gallouzi for the G3BP antibody, and Drs. Donalyn Scheuner and Randall Kaufman for the AA MEFs. We thank the members of the Anderson laboratory for helpful discussions and advice. This work was supported by National Institutes of Health grants AI33600 and AI50167 and by a biomedical science award from the Arthritis Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–08–0715. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0715.
Abbreviations used: eIF, eukaryotic initiation factor, HSF, heat shock factor; HSP, heat shock protein; PRD, prion-related domain; RRM, RNA recognition motif; SG, stress granules; UTR, untranslated region.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aguzzi, A., and Polymenidou, M. (2004). Mammalian prion biology: one century of evolving concepts. Cell 116, 313–327. [DOI] [PubMed] [Google Scholar]
- Akakura, S., Yoshida, M., Yoneda, Y., and Horinouchi, S. (2001). A role for Hsc70 in regulating nucleocytoplasmic transport of a temperature-sensitive p53 (p53Val-135). J. Biol. Chem. 276, 14649–14657. [DOI] [PubMed] [Google Scholar]
- Anderson, P., and Kedersha, N. (2002a). Stressful initiations. J. Cell Sci. 115, 3227–3234. [DOI] [PubMed] [Google Scholar]
- Anderson, P., and Kedersha, N. (2002b). Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A., Engemann, S., Lurz, R., Okazawa, H., Lehrach, H., and Wanker, E.E. (2003). Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J. Biol. Chem. 278, 41452–41461. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y.O., Newnam, G.P., Kumar, J., Allen, K., and Zink, A.D. (1999). Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19, 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro, B. (2003). Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull 66, 1–20. [DOI] [PubMed] [Google Scholar]
- Cotto, J.J., and Morimoto, R.I. (1999). Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem. Soc. Symp. 64, 105–118. [PubMed] [Google Scholar]
- Del Gatto-Konczak, F., Bourgeois, C.F., Le Guiner, C., Kister, L., Gesnel, M.C., Stevenin, J., and Breathnach, R. (2000). The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20, 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember, L.M., Kim, N.D., Liu, K.Q., and Anderson, P. (1996). Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 271, 2783–2788. [DOI] [PubMed] [Google Scholar]
- Derkatch, I.L., Bradley, M.E., Hong, J.Y., and Liebman, S.W. (2001). Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106, 171–182. [DOI] [PubMed] [Google Scholar]
- Forch, P., Puig, O., Kedersha, N., Martinez, C., Granneman, S., Seraphin, B., Anderson, P., and Valcarcel, J. (2000). The apoptosis promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Calfon, M., Urano, F., Novoa, I., and Ron, D. (2002). Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell. Dev. Biol. 18, 575–599. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000a). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000b). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904. [DOI] [PubMed] [Google Scholar]
- Huang, L., Mivechi, N.F., and Moskophidis, D. (2001). Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol. Cell. Biol. 21, 8575–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, L.S., and Kimball, S.R. (2003). Amino acids as regulators of gene expression at the level of mRNA translation. J. Nutr. 133, 2046S–2051S. [DOI] [PubMed] [Google Scholar]
- Jones, G.W., and Masison, D.C. (2003). Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G.W., Song, Y., and Masison, D.C. (2003). Deletion of the Hsp70 chaperone gene SSB causes hypersensitivity to guanidine toxicity and curing of the [PSI+] prion by increasing guanidine uptake in yeast. Mol. Genet. Genomics 269, 304–311. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- Kawakami, A., Tian, Q., Duan, X., Streuli, M., Schlossman, S.F., and Anderson, P. (1992). Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89, 8681–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., and Anderson, P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I.J., Stahl, J., and Anderson, P. (2002). Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Cho, M.R., Li, W., Yacono, P.W., Chen, S., Gilks, N., Golan, D.E., and Anderson, P. (2000a). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Cho, M., Li, W., Yacono, P., Chen, S., Golan, D., and Anderson, P. (2000b). Mammalian stress granules: highly dynamic sites of mRNA triage during stress induced translational arrest. Cold Spring Harbor Press: Cold Spring Harbor.
- Kedersha, N.L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball, S.R., Clemens, M.J., Tilleray, V.J., Wek, R.C., Horetsky, R.L., and Jefferson, L.S. (2001). The double-stranded RNA-activated protein kinase PKR is dispensable for regulation of translation initiation in response to either calcium mobilization from the endoplasmic reticulum or essential amino acid starvation. Biochem. Biophys. Res. Commun. 280, 293–300. [DOI] [PubMed] [Google Scholar]
- Kimball, S.R., Horetsky, R.L., Ron, D., Jefferson, L.S., and Harding, H.P. (2003). Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. 284, C273–C284. [DOI] [PubMed] [Google Scholar]
- King, C.Y., Tittmann, P., Gross, H., Gebert, R., Aebi, M., and Wuthrich, K. (1997). Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar, A.A., Lesnik, T., Cullin, C., Guillemet, E., Ehrlich, R., and Reiss, C. (1997). Differential resistance to proteinase K digestion of the yeast prion-like (Ure2p) protein synthesized in vitro in wheat germ extract and rabbit reticulocyte lysate cell-free translation systems. FEBS Lett. 415, 6–10. [DOI] [PubMed] [Google Scholar]
- Kopito, R., and Ron, D. (2003). Conformational disease. Nat. Cell Biol. 2, E207–E209. [DOI] [PubMed] [Google Scholar]
- Kryndushkin, D.S., Smirnov, V.N., Ter-Avanesyan, M.D., and Kushnirov, V.V. (2002). Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277, 23702–23708. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V.V., Kryndushkin, D.S., Boguta, M., Smirnov, V.N., and Ter-Avanesyan, M.D. (2000). Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10, 1443–1446. [DOI] [PubMed] [Google Scholar]
- Le Guiner, C., Gesnel, M.C., and Breathnach, R. (2003). TIA-1 or TIAR is required for DT40 cell viability. J. Biol. Chem. 278, 10465–10476. [DOI] [PubMed] [Google Scholar]
- Li, W., Li, Y., Kedersha, N., Anderson, P., Emara, M., Swiderek, K.M., Moreno, G.T., and Brinton, M.A. (2002). Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76, 11989–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.Y., and Kaufman, R.J. (2003). The unfolded protein response. J. Cell Sci. 116, 1861–1862. [DOI] [PubMed] [Google Scholar]
- Masison, D.C., and Wickner, R.B. (1995). Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Mazroui, R., Huot, M.E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E.W. (2002). Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007–3017. [DOI] [PubMed] [Google Scholar]
- Medley, Q.G., Kedersha, N., O'Brien, S., Tian, Q., Schlossman, S.F., Streuli, M., and Anderson, P. (1996). Characterization of GMP-17, a granule membrane protein that moves to the plasma membrane of natural killer cells following target cell recognition. Proc. Natl. Acad. Sci. USA 93, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam, G.P., Wegrzyn, R.D., Lindquist, S.L., and Chernoff, Y.O. (1999). Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L., Scharf, K., and Neumann, D. (1989a). Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell Biol. 9, 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L., Scharf, K.D., and Neumann, D. (1989b). Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9, 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz, M.F., Johnson, T., Suzuki, M., and Finch, J.T. (1994). Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 91, 5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner, D. (2003). Biochemistry and structure of PrP(C) and PrP(Sc). Br. Med. Bull 66, 21–33. [DOI] [PubMed] [Google Scholar]
- Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R.J. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176. [DOI] [PubMed] [Google Scholar]
- Schwimmer, C., and Masison, D.C. (2002). Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22, 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio, T.R., Cashikar, A.G., Kowal, A.S., Sawicki, G.J., Moslehi, J.J., Serpell, L., Arnsdorf, M.F., and Lindquist, S.L. (2000). Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Serio, T.R., and Lindquist, S.L. (2001). [PSI+], SUP35, and chaperones. Adv. Protein Chem. 57, 335–366. [DOI] [PubMed] [Google Scholar]
- Si, K., Lindquist, S., and Kandel, E.R. (2003). A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 115, 879–891. [DOI] [PubMed] [Google Scholar]
- Sondheimer, N., and Lindquist, S. (2000). Rnq 1, an epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172. [DOI] [PubMed] [Google Scholar]
- Srivastava, S.P., Kumar, K.U., and Kaufman, R.J. (1998). Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273, 2416–2423. [DOI] [PubMed] [Google Scholar]
- Tian, Q., Streuli, M., Saito, H., Schlossman, S.F., and Anderson, P. (1991). A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67, 629–639. [DOI] [PubMed] [Google Scholar]
- Tourriere, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J.M., Bertrand, E., and Tazi, J. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H., and Wanker, E.E. (2001). Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12, 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens, M., and Goldstrohm, A. (2003). Molecular biology. A place to die, a place to sleep. Science 300, 735–755. [DOI] [PubMed] [Google Scholar]
- Young, J.C., Barral, J.M., and Ulrich Hartl, F. (2003). More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28, 541–547. [DOI] [PubMed] [Google Scholar]
- Zhan, K., Vattem, K.M., Bauer, B.N., Dever, T.E., Chen, J.J., and Wek, R.C. (2002). Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stresses. Mol. Cell. Biol. 22, 7134–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Hasman, R.A., Young, K.M., Kedersha, N.L., and Lou, H. (2003). U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 23, 5959–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.