Abstract
Increased phosphorylation of dynein IC IC138 correlates with decreases in flagellar microtubule sliding and phototaxis defects. To test the hypothesis that regulation of IC138 phosphorylation controls flagellar bending, we cloned the IC138 gene. IC138 encodes a novel protein with a calculated mass of 111 kDa and is predicted to form seven WD-repeats at the C terminus. IC138 maps near the BOP5 locus, and bop5-1 contains a point mutation resulting in a truncated IC138 lacking the C terminus, including the seventh WD-repeat. bop5-1 cells display wild-type flagellar beat frequency but swim slower than wild-type cells, suggesting that bop5-1 is altered in its ability to control flagellar waveform. Swimming speed is rescued in bop5-1 transformants containing the wild-type IC138, confirming that BOP5 encodes IC138. With the exception of the roadblock-related light chain, LC7b, all the other known components of the I1 complex, including the truncated IC138, are assembled in bop5-1 axonemes. Thus, the bop5-1 motility phenotype reveals a role for IC138 and LC7b in the control of flagellar bending. IC138 is hyperphosphorylated in paralyzed flagellar mutants lacking radial spoke and central pair components, further indicating a role for the radial spokes and central pair apparatus in control of IC138 phosphorylation and regulation of flagellar waveform.
INTRODUCTION
Our goal is to determine the mechanisms that regulate ciliary and eukaryotic flagellar bending. Based on informative mutations in Chlamydomonas, and effective in vitro functional studies, a surprisingly complex array of different dynein motors is required for generation and control of normal ciliary and flagellar bending (Mitchell, 1994; Gibbons, 1995; Porter, 1996; Porter and Sale, 2000; DiBella and King, 2001; Kamiya, 2002). For example, the outer arm dyneins are homogeneous structures responsible for control of beat frequency and power required for movement (Satir et al., 1993; Brokaw, 1994; DiBella and King, 2001; Kamiya, 2002). The inner arm dyneins, however, are more complex, composed of at least seven different dynein subspecies precisely organized in a 96-nm repeat pattern along each doublet microtubule of the axoneme (Porter, 1996; Porter and Sale, 2000). Diverse data indicate the inner arm dyneins control the size and shape of the flagellar bend (Brokaw and Kamiya, 1987; Brokaw, 1994; Kamiya, 2002). The mechanism for control of flagellar waveform involves additional structures (e.g., radial spokes, central pair apparatus, and the dynein regulatory complex) and control of dynein phosphorylation (Porter and Sale, 2000; DiBella and King, 2001; Kamiya, 2002; Smith and Yang, 2004).
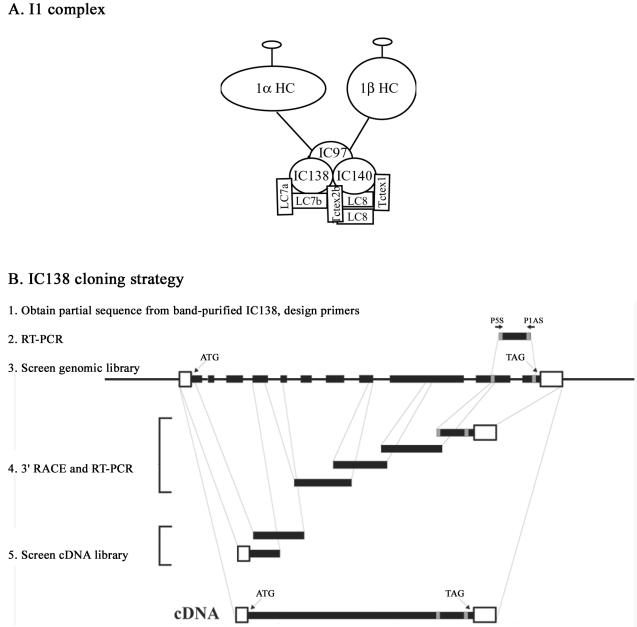
The present study is focused on a single inner arm dynein, the I1 complex, also called the f-dynein (Goodenough et al., 1987; Piperno et al., 1990; Kagami and Kamiya, 1992; Porter et al., 1992). The I1 complex is a tripartite structure, or triad, located near the base of the S1 radial spoke, at the proximal end of the axonemal 96-nm repeat (Goodenough and Heuser, 1985; Piperno et al., 1990; Mastronarde et al., 1992; Smith and Sale, 1992b). This large 18S, two-headed complex (Figure 1A) is composed of two heavy chains, 1α and 1β; three intermediate chains, IC140, IC138, and IC97 (also referred to as IC110); and several light chains, including LC7a, LC7b, LC8, Tctex1, and Tctex2b (Goodenough and Heuser, 1985; Piperno et al., 1990; Smith and Sale, 1991, 1992b; Porter et al., 1992; King and Patel-King, 1995; Harrison et al., 1998; Perrone et al., 1998; Yang and Sale, 1998; Myster et al., 1999; Perrone et al., 2000; DiBella et al., 2004a,b).
Figure 1.
Inner arm dynein I1 and IC138 cloning strategy. (A) Illustration of the inner arm dynein I1. The I1 dynein complex is composed of two heavy chains (1α Dhc and 1β Dhc), three intermediate chains (IC140, IC138, and IC97—also called IC110), and several light chains (LC8, LC7a, LC7b, Tctex1, and Tctex2b). (B) Primers (P5S and P1AS) were designed from sequence obtained from purified IC138 and used for RT-PCR. Sequences obtained by RT-PCR were then used to design additional primers for screening Chlamydomonas genomic libraries. The sequences obtained from those screens were then used to screen cDNA libraries to generate the full-length IC138 cDNA. The IC138 gene contains 11 exons (solid bars). The IC138 sequence can be obtained from GenBank under the accession no. AY743342.
Various data indicate that the I1 complex is an unusual dynein motor and plays a key regulatory role in the axoneme. Unlike other inner arm dyneins, the isolated I1 complex does not efficiently translocate microtubules in in vitro motility assays (Smith and Sale, 1991; Kagami and Kamiya, 1992). Mutations in I1 result in failure of control of normal ciliary and flagellar waveform and phototaxis in Chlamydomonas, indicating a regulatory role in these processes (Brokaw and Kamiya, 1987; King and Dutcher, 1997). Additionally, mutations in I1 subunits suppress paralysis in a central pair mutant, indicating a functional link between I1 activity and the central pair apparatus mechanism for control of microtubule sliding (Porter et al., 1992). Consistent with this idea, in vitro functional assays, by using isolated axonemes or reconstituted axonemes, have revealed that the I1 complex plays a key role in the regulation of microtubule sliding and that the mechanism involves changes in I1 phosphorylation (Smith and Sale, 1992a,b; Howard et al., 1994; Habermacher and Sale, 1996, 1997; Yang and Sale, 2000; Smith, 2002). In these in vitro experiments, IC138 was the only I1 subunit that was phosphorylated, and increased phosphorylation of IC138 correlated with decreased dynein-driven microtubule sliding (Habermacher and Sale, 1997; King and Dutcher, 1997; Yang and Sale, 2000). Furthermore, abnormal phosphorylation of IC138 also results in a failure in Chlamydomonas phototaxis (King and Dutcher, 1997). Together, the data indicate changes in IC138 phosphorylation regulate I1 activity and microtubule sliding.
To further test the hypothesis that IC138 is a regulatory phosphoprotein, we cloned the gene and began characterization of mutant strains defective in IC138. We determined that like several other dynein intermediate chains, IC138 is a WD-repeat protein. The IC138 gene maps near the BOP5 locus (Dutcher et al., 1988), and the bop5-1 mutant displays a slow swimming phenotype that is rescued by the wild-type IC138 gene. The bop5-1 mutation results in the truncation of IC138 just before the last WD-repeat. Surprisingly, in bop5-1 axonemes, the truncated IC138 assembles with all of the other known I1 subunits with the exception of LC7b. Consistent with a recent report (DiBella et al., 2004b), these results indicate that IC138 anchors LC7b in the I1 complex and that LC7b is required for normal control of bending. Furthermore, IC138 is hyperphosphorylated in axonemes from paralyzed flagella mutants lacking radial spokes and central pair apparatus. These data are consistent with the hypothesis that I1 is part of a regulatory network, including the radial spokes and central pair apparatus, which controls flagellar waveform.
MATERIALS AND METHODS
Cell Strains and Growth Conditions
The following Chlamydomonas strains were obtained from the Chlamydomonas Genetic Center (Dr. E. H. Harris, Department of Biology, Duke University, Durham, NC): CC-125 (wild-type, 137c mt+), CC-124 (wild-type, 137c mt-), CC-1036 (pf18 mt+), CC-1035 (pf17 mt+), CC-899 (pf10 mt+), CC-1877 (pf28 mt-), CC-2244 (oda9 mt+), and CC-3921 (ida7-1 [5b10]). The bop5-1 strain was obtained from S. K. Dutcher (Washington University School of Medicine, St. Louis, MO) (Dutcher et al., 1988), and the 5A strain was described previously (Perrone et al., 1998). Double mutant strains bop5oda7, bop5pf17, bop5pf18 were isolated from nonparental ditype tetrads. Cells were grown in either Trisacetate-phosphate medium or in modified Sager Granick minimal medium (Sager and Granick, 1953) with aeration on a 14:10-h light/dark cycle.
Isolation of Axonemes, Dynein Purification, and Biochemical Analyses
Flagella were isolated by the dibucaine method and demembranated using Nonidet (NP-40; Calbiochem, San Diego, CA) as described previously (Witman, 1986). Axonemes were resuspended in 10 mM HEPES, 5 mM MgSO4, 1 mM dithiothreitol (DTT), 0.5 mM EDTA, 30 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 0.6 trypsin inhibitor units aprotinin (HMDE-Na). Dynein extraction and sucrose gradient fractionation were performed as described previously (Smith and Sale, 1992b), and ion exchange by fast-performance liquid chromatography was performed using a Mono-Q column (Amersham Biosciences, Piscataway, NJ) as described previously (Goodenough et al., 1987; Kagami and Kamiya, 1992; Porter et al., 1992).
SDS-PAGE was performed as described previously (Smith and Sale, 1991), but by using Precision Plus protein standards (Bio-Rad, Hercules, CA) to estimate protein size. To visualize dynein heavy chains, samples were separated on 2.5–5.0% acrylamide and 3–8 M urea gels (Smith and Sale, 1991) and silver stained. Protein cross-linking was carried out as described previously (King et al., 1991). Briefly, axonemes (1–2 mg/ml) were treated with 0.0–5.0 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDC; Pierce Chemical, Rockford, IL) for 1 h at room temperature in HMDE-Na. The reaction was terminated by the addition of β-mercaptoethanol to a final concentration of 20× the EDC concentration. Samples were immediately fixed for gel electrophoresis. Phosphatase treatment was performed by resuspending axonemes at 2 mg/ml in HMDE-Na and then incubating with 1.5 U of calf intestinal alkaline phosphatase (CIP; Promega, Madison, WI) per 100 μg of axonemal protein at room temperature for 30 min (King and Dutcher, 1997). The reaction was terminated by the addition of 5× sample gel buffer. Samples were resolved by 5% SDS-PAGE and analyzed by Western blotting.
Molecular Approaches
For cloning IC138 our strategy was to obtain partial amino acid sequence from band-purified protein and design primers for reverse transcription-polymerase chain reaction (RT-PCR) as described previously (Yang and Sale, 1998; Yang et al., 2004) and outlined in Figure 1B. Briefly, axonemes were isolated from pf28 cells, and the I1 complex was further purified by zonal centrifugation through a 5–20% sucrose gradient. Fractions (0.5 ml) were collected and resolved by 7% SDS-PAGE. The protein band corresponding to IC138 was excised from the gel and microsequenced (performed by John Leszyk, University of Massachusetts, Worcester, MA). Peptides obtained are listed in Table 1. The peptide sequences were used to design degenerate primers that were used for RT-PCR on total RNA purified from wild-type cells 45 min after deflagellation. A degenerate primer set “P1AS1” (ATGY(C,T)TCCTCN(A,C,G,T)AGN(A,C,G,T)GTR(A,G)TCR(A,G)TA) and “P5S1” (ATAY(C,T) AGY(C,T)GAR(A,G)CAR(A,G)TAY(C,T)CTN(A,C,G,T)GA) yielded a 450-base pair fragment, which was used to design additional primers for screening a Chlamydomonas λfixII genomic library provided by E. F. Smith (Dartmouth College, Hanover, NH) (Figure 1B). Partial sequences obtained from the genomic screen were used to design additional primers that were used for 3′ rapid amplification of cDNA ends and an additional round of RT-PCR. The resulting sequences were used to screen the λgt10 cDNA library obtained from G. Pazour (University of Massachusetts, Amherst, MA).
Table 1.
Amino acid sequences of peptides obtained by direct microsequencing of band purified IC138
| Peptide 1 | AYRLYNVSHEYDTLEEO......... (P1AS) |
| Peptide 2 | ANPDLLAVGYGSYAFGSGTPGAGAAGDPL |
| Peptide 3 | GGAGDTTTPNSE |
| Peptide 4 | TPKPLLSLNPTVLK |
| Peptide 5 | CSTSYSEOYLESYR.................. (P5S) |
| Peptide 6 | LEIWDFALSTVKPVMHQ |
| Peptide 7* | ATGVQATAWDISDTFR |
| Peptide 8* | AAUPEQQDAFISR |
| Peptide 9* | GAVLPSISQLAGGVA |
| Peptide 10* | PSAYHGQSMAFGAQPSYM |
| Peptide 11* | DMFISS |
The sequences used for design of primers P5S and P1AS are underlined, and the asterisk (*) indicates peptides identified by subsequent mass spectrometry analysis of purified IC138.
Restriction fragment length polymorphism (RFLP) mapping of the IC138 gene was performed as described previously (Porter et al., 1996; Perrone et al., 2000). The IC138 gene was used as a probe on genomic Southern blots to identify an EcoRI/XhoI RFLP between two polymorphic Chlamydomonas strains. The IC138 gene was then hybridized to mapping filters containing DNA isolated from tetrad progeny between multiply marked Chlamydomonas strains and cosegregation of the RFLP was analyzed with respect to >50 genetic and molecular markers. Linkage to the genetic mutation pf9 (∼29 cM) and the two molecular markers CK1 (∼30 cM) and CNC63 (∼43 cM) placed the IC138 gene on the left arm of linkage group XII/XIII. The position of the IC138 gene on linkage group XII/XIII was more precisely determined using sequence from the 3′ untranslated region and single nucleotide polymorphisms (Kathir et al., 2003).
The ability of the IC138 gene to rescue the bop5-1 motility defect was tested by cotransformation as described previously (Perrone et al., 1998). Briefly, bop5-1 was crossed into an arginine requiring background (arg7) then bop5-1arg7 cells were cotransformed using a selectable marker, pARG7.8, and a full-length genomic clone of IC138, pHx. Arg-positive transformants were picked into liquid medium and scored under a dissecting scope for possible rescue of the bop5-1 swimming phenotype. Cells with apparent wild-type motility were further analyzed by phase contrast microscopy to measure swimming speed, as described below.
Antibody Production and Western Analyses
An insert including nucleotides 1–677 of IC138 (cDNA clone B4) was subcloned into the pET-28(a)+ expression vector (Novagen, Madison, WI). The His-tagged B4 peptide was expressed in inclusion bodies of Escherichia coli (BL21). The inclusion bodies were solubilized in 8 M urea, and the His-tagged fusion protein was purified on a Ni+ column following the manufacturer's instructions (Novagen). The purified fusion protein remained soluble following dialysis in phosphate-buffered saline (PBS) and was used to generate polyclonal antibodies in rabbits (Spring Valley Laboratories, Woodbine, MD).
Western blotting was performed on polyvinylidene diflurodie. The IC138 antibody was used at a dilution of 1:20,000. The following rabbit polyclonal antibodies were used at the indicated dilutions: α-IC140 (1:10,000), α-1α Dhc (1:10), α-LC8 (1:100), α-Tctex1 (1:50), α-Tctex2b (1:50), α-LC7b (1:50), and α-LC7a (1:50) (King and Patel-King, 1995; King et al., 1996; Myster et al., 1997; Yang and Sale, 1998; Bowman et al., 1999; DiBella et al., 2004a,b). The horseradish peroxidase-conjugated goat anti-rabbit secondary was used and immunoreactivity detected by enhanced chemiluminescence (Amersham Biosciences).
Analysis of Motility and Microtubule Sliding Assay
The forward swimming velocity of freely swimming cells was measured as described previously using phase contrast microcopy and video tape analysis (Porter et al., 1992; Myster et al., 1997, 1999). The average swimming speed for each strain was calculated from a minimum of 20 individual cells. Beat frequency was measured stroboscopically using dark field microscopy and synchronization of strobe frequency with the flagellar beat (Mitchell and Kang, 1991).
Microtubule sliding velocity was measured as described previously (Howard et al., 1994; Habermacher and Sale, 1996, 1997). Briefly, isolated flagella were first resuspended in a buffer containing 10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 50 mM potassium acetate (HMDEK), with no protease inhibitors. An aliquot of flagella was demembranated with 0.5% NP-40 in HMDEK, and 8–10 μl of axonemes was added to a glass perfusion chamber constructed with a coverslip mounted on a slide by using double stick tape. The axonemes were then washed with 50 μl of HMDEK containing 1 mM ATP. Microtubule sliding was initiated by the addition of HMDEK + ATP supplemented with 1–2 μg/ml protease (Nagarse; Sigma-Aldrich, St. Louis, MO). Microtubule sliding was visualized using dark field microscopy through a Ziess Axiovert 35 and recorded by a silicon intensified camera (VE-1000; Dage-MTI, Michigan City, IN) and a VCR equipped with a jog/shuttle device as described previously (Habermacher and Sale, 1996). The average microtubule sliding velocity was calculated from a minimum of six experiments with a sample size of at least 60 independent axonemes.
RESULTS
IC138 Sequence Analysis
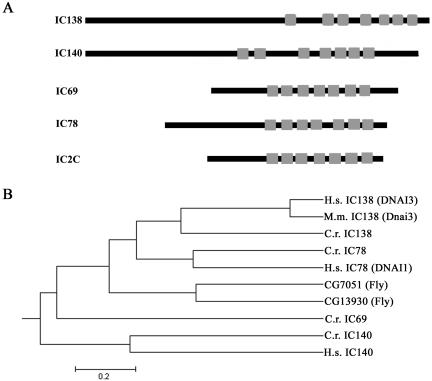
The initial strategy for cloning IC138 was to obtain partial peptide sequence (Table 1), design oligonucleotide primers for RT-PCR, and use the PCR product for screening libraries, and additional PCR (Figure 1B). Sense and antisense degenerate primers P1AS1 and P5S1, listed in Materials and Methods, were designed based on the underlined amino acids indicated in Table 1. We obtained the gene and cDNA sequences as outlined in the legend of Figure 1. The Chlamydomonas IC138 is found in six BAC clones (17c8, 23k17, 4p7, 9d3, 29i21, and 32d15) and is located in scaffold 16, contig 37 (Chlamydomonas reinhardtii genome version 2; http://genome.jgi-psf.org/chlre2/chlre2.home.html). IC138 also found in two expressed sequence tags (BQ818669.1 and AV643468.1). The full-length genomic IC138 clone (GenBank accession no. AY743342) is novel and contains 11 exons and 10 introns (Figure 1B). The predicted protein is novel and composed of 1057 amino acids with a predicted mass of 111.1 kDa, a calculated pI of 5.73, and contains the peptides listed in Table 1 and underlined in Figure 2. Consistent with the other members of the dynein IC family (Wilkerson et al., 1995; Yang and Sale, 1998), the sequence predicted a series of seven WD-repeats in the C-terminal portion of the protein (bold text, Figure 2). Alignment of IC138 with known Chlamydomonas dynein IC proteins indicates that IC138 contains a similar arrangement of seven WD-repeats (Figure 3 and that IC138 is more similar to the outer dynein arm IC78 compared with IC69 (Figure 3B).
Figure 2.
The IC138 gene encodes a WD-repeat containing protein. The IC138 cDNA predicts a protein of 1057 amino acids and ∼111 kDa with a calculated pI of 5.73. Peptide sequences obtained from microsequencing purified IC138 are underlined; the N-terminal sequence that was used to create the His-tagged fusion protein for the purpose of producing the IC138 polyclonal antibody is italicized. IC138, like other IC dyneins, is predicted to contain seven WD-repeats, which are indicated in bold. The residues enclosed in the shaded box are missing in bop5-1.
Figure 3.
IC138 structural domains and homologues. (A) Schematic illustration of the dynein intermediate chains comparing the positions of the seven WD/β-sheet repeats among several axonemal dynein intermediate chains. Alignment of these sequences indicates that the WD/β-sheet repeats are all clustered within the C-terminal portion of the proteins. (B) The MegAlign Program (DNASTAR) was used to generate a phylogenetic tree of dynein intermediate chains. (C) Alignment of the Chlamydomonas IC138 (C.r. IC138) and its homologues human IC138 (DNAI3), mouse IC138 (Dnai3).
Database searches for sequences similar to Chlamydomonas IC138 revealed a human IC138 homologue (GenBank accession no. NM_024763) and mouse IC138 homologue (GenBank accession no. XM_143950). Following the standard nomenclature, we propose that the human IC138 be referred to as DNAI3 and the mouse IC138 as Dnai3. DNAI3 has been mapped to position 1p32.1 in the human genome, whereas Dnai3 has been mapped to chromosome 4 in the mouse genome. IC138 and DNAI3 share 34% identity and 49% similarity, whereas there is 30% identity and 56% similarity between IC138 and Dnai3 (Figure 3C). There is 58% identity and 68% similarity between DNAI3 and Dnai3. Motif prediction programs indicate both DNAI3 and Dnai3 are WD-repeat proteins, similar in organization to other dynein IC proteins. DNAI3 has five canonical WD-repeats, whereas Dnai3 is predicted to have six WD-repeats, all of which are clustered in the C-terminal region of both proteins. The existence of human and mouse homologues of IC138 suggests that these proteins also may have evolutionarily conserved roles. These genes become candidates for loci involved in primary cilia dyskinesia (El Zein et al., 2003).
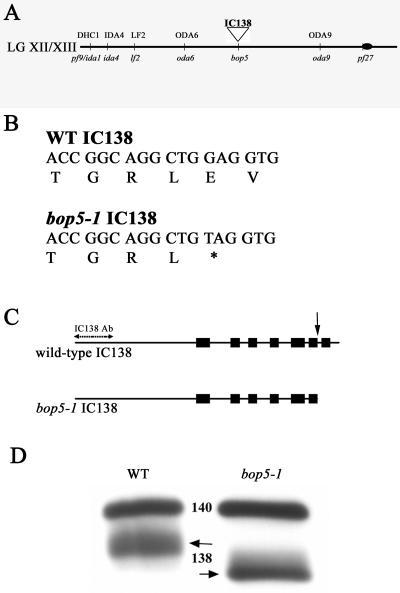
The IC138 Gene Maps Near the BOP5 Locus and IC138 Is Truncated in bop5-1 Axonemes
Molecular mapping procedures placed the IC138 gene on the left arm of linkage group XII/XIII, between two outer arm dynein loci, ODA6 and ODA9 (Figure 4A; Kathir et al., 2003). This map position places the IC138 gene in proximity to a novel motility mutation, bop5-1, whose mutant gene product had not previously been identified. bop5-1 was first isolated as a suppressor of the Chlamydomonas mutant pf10, which exhibits abnormal swimming speed and ineffective flagellar beat (Dutcher et al., 1988). The bop5-1 mutation partially suppresses the pf10 motility defect; however, neither the bop5-1pf10 double mutant nor the bop5-1 suppressor alone is wild-type (Dutcher et al., 1988). For example, the average swimming speed of bop5-1 cells is significantly slower than wild-type cells (see below). To determine whether the IC138 gene is defective in bop5-1, we sequenced the entire IC138 gene in bop5-1 and found that it contained a single point mutation at nucleotide 4725 that changes a guanine to a thymine. The conceptual translation suggests that the mutation converts codon 974 from glutamic acid to a premature stop codon (Figure 4B) that would result in the premature termination of the seventh WD-repeat (Figure 4C). Western blot analysis by using a polyclonal antibody to the first 148 N-terminal amino acids of IC138 indicates that IC138 is present and truncated in bop5-1 axonemes (Figure 4D). Thus, despite the truncation of a WD-repeat structure, IC138 assembles in bop5-1 axonemes (see Discussion).
Figure 4.
IC138 is truncated in bop5-1. (A) Abbreviated map of Chlamydomonas linkage group XII/XIII. The IC138 gene was mapped to the left arm of linkage group XII/XIII based on linkage to the genetic marker pf9 (∼29 cM) and a series of molecular markers as described in Materials and Methods. (B) Sequence analysis of IC138 from the bop5-1 allele revealed a point mutation at nucleotide 4725 that results in a premature stop codon, 974E → STOP. (C) Positions of the seven WD/β-sheet repeats are indicated by boxes. The bop5-1 IC138 nucleotide sequence predicts a truncated protein that lacks the last 84 amino acids, including the last WD-repeat (arrow), producing a truncated protein. (D) Axonemes from wild-type and bop5-1 cells probed with a polyclonal antibody generated against a fusion protein containing the first N-terminal 148 amino acids of IC138 (Figure 2, italicized sequence), which specifically recognizes a protein at ∼118 kDa corresponding to IC138, and a polyclonal IC140 antibody (Yang and Sale, 1998).
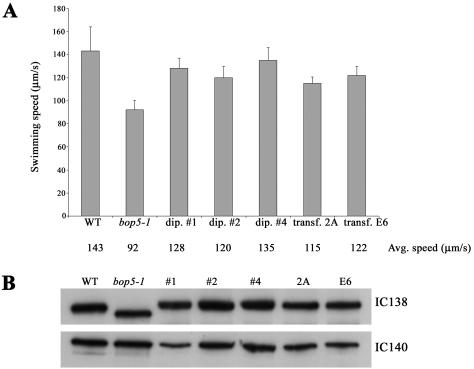
To verify that the motility defects seen in bop5-1 cells are caused by the mutation in IC138, bop5-1 cells were transformed with the wild-type IC138 gene and assayed for rescue of the motility phenotype. As shown in Figure 5A, the swimming speeds of the transformants (2A, E6) are significantly faster than the bop5-1 mutant cells and only slightly slower than typical wild-type, haploid cells. Similarly, swimming speed was rescued in heterozygous diploids (Figure 5A, dip# 1, 2, and 4). Other experiments confirmed that the swimming speeds of the heterozygous, diploid cells were indistinguishable from homozygous, wild-type diploid cells. Consistent with these observations, Western blot analysis revealed that only the full-length, wild-type IC138 polypeptide was found in the isolated axonemes prepared from the transformants and diploid cells (Figure 5B). Identical results were observed in four independent experiments. Collectively, these findings demonstrate that the bop5-1 phenotype is due to the mutation in IC138. We conclude that the BOP5 locus encodes IC138. Furthermore, in diploids and bop5-1 transformed with wild-type IC138, the full-length IC138 is preferentially assembled in axonemes.
Figure 5.
IC138 rescues motility in bop5-1. bop5-1 cells that had been transformed with full-length IC138 and stable diploids containing both the wild-type IC138 gene and the bop5-1 IC138 gene were analyzed. (A) Swimming speeds of freely swimming cells were measured. The decreased swimming speed seen in bop5-1 is rescued by wild-type IC138. (B) By Western analysis, only the full-length IC138 is found in the axonemes of the stable diploids (#1, #2, and #4) and the bop5-1 cells transformed with the wild-type IC138 gene (2A, E6).
The LC7b Subunit of I1 Fails to Assemble in bop5-1
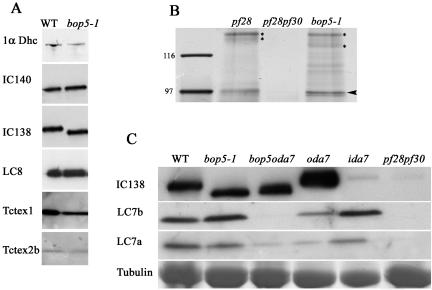
We postulated that truncation of IC138 in bop5-1 would result in a failure to assemble a complete I1 complex. We used Western blot analysis and protein stains to determine whether the I1 complex is accurately assembled in bop5-1. Wild-type and bop5-1 axonemes contain similar amounts of I1 proteins, including the 1α Dhc, IC140, IC138, LC8, Tctex1, and Tctex2b (Figure 6A). These results were verified in three independent experiments. Furthermore, based on silver stained gels, bop5-1 axonemes also contain 1β Dhc (our unpublished data) and IC97 (Figure 6B).
Figure 6.
LC7b fails to assemble in I1 from bop5-1. (A) Western analyses were performed on axonemes isolated from wild-type and bop5-1 cells by using antibodies to the 1α dynein heavy chain (1α Dhc), two intermediate chains (IC138 and IC140), and the light chains (LC8, Tctex1, and Tctex2b). (B) Silver-stained SDS-PAGE of I1 fractions indicating that IC97 (arrowhead) is assembled in axonemes from bop5-1 cells in an amount comparable with the wild-type positive control derived from pf28 axonemes and lacking in the negative control derived from pf28pf30 axonemes. IC140, IC138, and truncated IC138 are indicated with asterisks. (C) Further analysis of bop5-1 and the double mutant bop5-1oda7 reveals that LC7b, but not LC7a fails to assemble when IC138 is truncated. The tubulin region of the gel was excised and Coomassie stained to demonstrate protein loads.
The dynein light chains LC7a and LC7b are located in the outer dynein arm as well as the inner dynein arm I1 complex (Piperno and Luck, 1979; Pfister et al., 1982; Harrison et al., 1998; DiBella et al., 2004b). Therefore, to determine whether LC7a and LC7b are present in the I1 complex it was necessary to perform the Western blot on axonemes from the double mutant bop5-1oda7, which contains the truncated IC138 and is missing the outer arm dynein. LC7a is reduced in oda7 axonemes, as expected, as well as in axonemes from the bop5-1oda7 double mutant. However, although it is present in oda7 axonemes, LC7b is missing in axonemes isolated from the bop5-1oda7 (Figure 6C), consistent with its failure to assemble in the I1 complex in bop5-1 axonemes. This result was repeated in three independent experiments and indicates that the C terminus, including the last WD-repeat, is required for LC7b assembly into I1. We were unable to analyze LC8 in a similar manner because LC8 is also found in the radial spoke, in addition to the inner arm dynein I1 and the outer arm dynein (Piperno and Luck, 1979; Pfister et al., 1982; Harrison et al., 1998; Yang et al., 2001).
We also examined whether the truncation in IC138 in bop5-1 alters the stability of the I1 complex after salt extraction and purification by MonoQ chromatography or zonal centrifugation on sucrose gradients. As described before for the I1 complex in wild-type axonemes, IC138 and IC140 from bop5-1 copurify in the f-fraction (our unpublished data; Kagami and Kamiya, 1992), as well as in the 18S I1 complex peak fractions on sucrose gradients (our unpublished data; Smith and Sale, 1991; Porter et al., 1992). Thus, despite the C-terminal truncation in IC138 and the failure to assemble LC7b, the I1 dynein still forms a discrete 18S complex in bop5-1 axonemes.
EDC Cross-Linking of IC138 to a ∼97-kDa Axonemal Protein
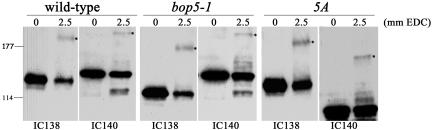
Previous studies have shown that IC140 forms a ∼250-kDa cross-linked product when using a zero length cross-linker EDC (Yang and Sale, 1998). To determine whether the ∼250-kDa cross-linked product is a result of a direct interaction between IC138 and IC140, we performed EDC cross-linking on isolated axonemes and analyzed the results by Western blots by using antibodies to IC138 and IC140. In wild-type axonemes, an ∼250-kDa cross-linked product is observed using both IC138 and IC140 antibodies (Figure 7). This result could be explained by three possibilities: 1) IC138 and IC140 cross-linked to each other, 2) both proteins independently cross-linked to a third protein, or 3) both IC138 and IC140 are cross-linked to different proteins of similar sizes.
Figure 7.
EDC cross-links IC138 to an ∼97-kDa axonemal protein. Axonemal proteins were isolated from wild-type (WT), bop5-1 and 5A (truncated IC140) cells and cross-linked using the zero-length cross-linker EDC. The samples were then separated on a 3–6% gradient gel and transferred to nitrocellulose. The blots were probed with α-IC140 or α-IC138. The IC138 and IC140 EDC cross-linked products (indicated by the asterisk) are coincidentally at the same position for WT axonemes (∼250 kDa) but has shifted in the bop5-1 IC138 blot as well as the 5A IC140 blot by ∼97 kDa in each case, indicating that IC138 and IC140 both interact with an axonemal protein of ∼97 kDa.
To distinguish these possibilities, we took advantage of both the bop5-1 mutant and an ida7 allele that contains a truncated IC140, “5A” (Perrone et al., 1998). The prediction is that if IC138 and IC140 are cross-linking to each other, then in the presence of either a truncated IC138, as in the case of bop5-1, or a truncated IC140, as in the case of 5A, the sizes of the IC138 and IC140 cross-linked product should be identical on Western blots. However, in the bop5-1 axonemes the IC138 cross-linked product is smaller than the IC140 cross-linked product and, in 5A axonemes, the IC140 cross-linked product is smaller than the IC138 cross-linked product (Figure 7). Moreover, in every case the cross-linked product is ∼97 kDa larger than the parent band. These results indicate that IC138 and IC140 are not cross-linked to each other using EDC. Instead, both proteins independently interact with another protein, possibly the third IC protein in I1, IC97. Further testing of this model will require antibodies to IC97.
IC138 Is Hyperphosphorylated in Paralyzed Flagellar Mutants
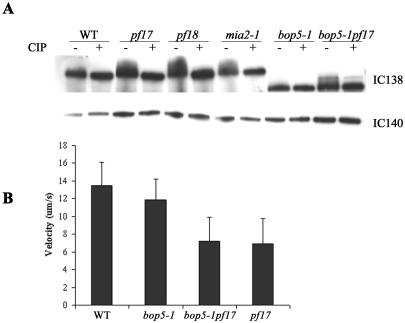
As reviewed in the Introduction, the basis for our focus on IC138 is that changes in IC138 phosphorylation correlate with changes in the control of dynein-driven microtubule sliding and the regulation of flagellar motility (Habermacher and Sale, 1997; King and Dutcher, 1997; Yang and Sale, 2000). For example, inhibition of microtubule sliding, which is characteristic of paralyzed flagella mutants, correlates with increased phosphorylation of IC138. Furthermore, mutations that result in hyperphosphorylation of IC138 also result in decreased axonemal microtubule sliding velocity and failure to phototax (King and Dutcher, 1997). To further test the idea that IC138 is abnormally phosphorylated in paralyzed flagellar mutants, we developed a Western blot based assay of IC138 phosphorylation. The strategy was to perform Western blots on control axonemes, or axonemes first treated with alkaline phosphatase (CIP), before preparation for SDS-PAGE, and then examine changes in IC138 mobility in acrylamide gels (King and Dutcher, 1997).
This assay revealed that the phosphorylation state of IC138 is altered in paralyzed flagellar mutants (Figure 8). In wild-type axonemes, IC138 runs as a relatively discrete band on 5% gels irrespective of CIP treatment (Figure 8A, compare lanes 1 and 2). In mia2-1 cells, which are known to contain hyperphosphorylated IC138 (King and Dutcher, 1997), IC138 migrates as a smeared band. After alkaline phosphatase treatment, the mia2-1 IC138 migrates as a discrete band (Figure 8A, lane 8), similar to IC138 from wild-type axonemes. As predicted from previous work (Habermacher and Sale, 1997), IC138 in axonemes from paralyzed flagellar mutants pf17 (lacking radial spoke head) or pf18 (lacking the central pair apparatus) is not a discrete band (Figure 8A, lanes 3 and 5). Rather, IC138 seems to run as a more slowly migrating and smeared band, consistent with a more highly phosphorylated IC138. Consistent with this interpretation, alkaline phosphatase treatment resulted in a discrete IC138 band, similar to the IC138 migration pattern in wild-type axonemes (Figure 8A, lanes 4 and 6). Similar results were obtained for axonemes isolated from pf14 cells (our unpublished data).
Figure 8.
IC138 is hyperphosphorylated in radial spoke and central pair mutants. To determine whether of the phosphorylation state of IC138 is altered in paralyzed flagellar mutants, axonemes isolated from wild-type, pf17, pf18, mia2-1, bop5-1, and bop5-1pf17 were treated with CIP. (A) Based on a comparison of CIP-treated (-) versus control, untreated (+) axonemes, IC138 seems to be hyperphosphorylated in mia2-1 and the paralyzed mutants pf17, pf18, and bop5-1pf17. In contrast, IC138 from wild type and bop5-1 is not hyperphosphorylated. (B) Flagella were isolated and demembranated immediately preceding the addition of ATP and protease. Microtubule sliding velocity is expressed as micrometers per second.
To determine whether the bop5-1 mutation might affect the phosphorylation state of IC138, we also analyzed bop5-1 in a radial spoke or central pair mutant background. Interestingly, bop5-1 does not suppress paralysis in the radial spoke mutant pf17, or in the central pair mutant pf18 (our unpublished data). In addition, the truncated IC138 in axonemes from bop5-1 migrates as a discrete band, indicating the truncation does not result in any obvious changes in the phosphorylation of IC138 (Figure 8A, lane 9). However, IC138 in the double paralyzed mutant, bop5-1pf17, slowly migrates as a smeared band, characteristic of pf mutants (Figure 8A, lane 11). A similar migration pattern was observed for IC138 in axonemes isolated from a bop5-1pf18 double mutant (our unpublished data). Thus, the bop5-1 IC138 subunit seems similar to the wild-type IC138 subunit with respect to its pattern of phosphorylation.
To further assess how the bop5-1 IC138 might alter motility, we also measured microtubule sliding velocity in axonemes by using a videomicroscopy assay. The microtubule sliding velocities of wild-type and bop5-1 axonemes were essentially identical (Figure 8B). The velocity of microtubule sliding in the double mutant bop5-1pf17 was identical to that of pf17 axonemes (Figure 8B). Moreover, addition of kinase inhibitors (Habermacher and Sale, 1997; Yang and Sale, 2000) restored microtubule sliding to near wild-type velocity for both pf17 and bop5-1pf17. The truncation of IC138 in bop5-1 therefore does not interfere with the rescue of microtubule sliding induced by protein kinase inhibitors.
DISCUSSION
Here, we further analyze flagellar dynein IC IC138, a regulatory subunit of inner arm dynein I1. Key conclusions from this study include that IC138 is a WD-repeat protein and that in vivo the WD-repeat structure is important for assembly of the roadblock-like light chain, LC7b, a newly identified component of the I1 complex (DiBella et al., 2004b). The basis for these conclusions is that the IC138 gene rescues the bop5-1 motility defects; that the bop5-1 allele contains a mutation that results in the deletion of the seventh WD-repeat of IC138; and although the truncated IC138 and the other I1 proteins assemble in bop5-1 axonemes, LC7b fails to assemble in the complex. Significantly, whereas bop5-1 cells display wild-type beat frequency, the cells swim slowly. Therefore, a relatively modest truncation in IC138 structure reveals the importance of the WD-repeat structure for roadblock light chain assembly, inner arm I1 complex function and control of flagellar waveform.
Sequence and secondary structural predictions have shown that most dynein intermediate chains studied are members of the WD-repeat protein family (Ogawa et al., 1995; Wilkerson et al., 1995; Yang and Sale, 1998). The exceptions to date include dynein IC1 from sea urchin sperm tail axonemes and dynein IC3 from Ciona sperm tail axonemes, neither of which contains WD-repeats (Ogawa et al., 1996; Padma et al., 2001). Using the same criteria established in Yang and Sale (1998), including both a classical definition of a WD-repeat (Neer et al., 1994; Smith et al., 1999) and predicted β-sheet “propeller” structure, not strictly conforming to the traditional WD definitions (Faber et al., 1995; Neer and Smith, 1996), IC138 is another member of the dynein IC WD/β-sheet family, predicted to contain seven repeats located near the C terminus. As proposed previously, one simple model is that each WD-repeat dynein IC contains seven WD/β-sheet repeats (Yang and Sale, 1998) and that the repeats are likely organized into a β-sheet propeller structure (Wall et al., 1995; Lambright et al., 1996; Sondek et al., 1996) to mediate protein–protein interactions required for dynein assembly and anchoring to cargo. Given this model, it may be surprising that IC138 lacking the seventh WD/β-sheet repeat can still assemble and support much of the assembly of the I1 complex. Thus, this observation is one of the most informative features of bop5-1. An additional informative feature of the bop5-1 phenotype, as discussed below, is that the WD/β-sheet repeat may mediate dynein light chain anchoring in the I1 complex.
The WD/β-sheet repeat class of dynein intermediate chains is thought to be located at the base of the dynein structure (Figure 1A), in a position to mediate assembly and anchoring to cargo (King and Witman, 1990; Paschal et al., 1992; Ogawa et al., 1995; Perrone et al., 1998; Yang and Sale, 1998). Evidence includes direct electron microscopic (EM) immuno-localization of IC78 in outer arm dynein (King and Witman, 1990), and IC74 in cytoplasmic dynein (Steffan et al., 1996), and EM analysis of inner arm dynein I1 mutants lacking either of the motor domains (Myster et al., 1999; Perrone et al., 2000; Porter and Sale, 2000). In Chlamydomonas and humans, mutant alleles of IC genes result in failure of outer dynein assembly, demonstrating their essential role (Mitchell and Kang, 1991; Wilkerson et al., 1995; Pennarun et al., 1999; DiBella and King, 2001; Kamiya, 2002). Furthermore, cross-linking studies revealed that IC78 and IC69 physically interact, indicating they are both located at the base of the outer arm dynein (King et al., 1991). Sequence comparison between intermediate chains of the outer and the inner arm dyneins suggest that IC138 is most similar to IC78 (Ogawa et al., 1995; Wilkerson et al., 1995). This conclusion is consistent with the previous proposal that IC140 in the inner arm dynein I1 is most similar to IC69 in outer arm dynein (Yang and Sale, 1998). Thus, based on EM analysis of I1 mutants and analogy to the outer arm dynein intermediate chains, both IC140 and IC138 are likely located at the base of the I1 complex mediating assembly and function of inner arm dynein as illustrated (Figure 1A).
The bop5-1 mutant, originally recovered as a suppressor of the pf10 motility phenotype (Dutcher et al., 1988), has proven to be informative with respect to IC138 structure and function and the role of I1 in control of flagellar bending. The phenotype of the bop5-1 mutant is distinct from other mutants that fail to assemble the complete I1 complex. I1 mutants swim forward with a significantly reduced velocity (Perrone et al., 1998, 2000) and altered waveform (Brokaw and Kamiya, 1987). The I1 mutants also show phototaxis defects (King and Dutcher, 1997). In contrast, the bop5-1 mutant does not show a phototaxis defect, and its swimming speed is only slightly reduced compared with mutants that completely lack I1. Thus, bop5-1 axonemes seem to retain some I1 function. Similar results also have been observed in other I1 mutants that assemble truncated Dhc subunits (Myster et al., 1999; Perrone et al., 2000). However, the specific changes in waveform in bop5-1 will require direct, high-resolution analysis of flagellar bending.
We also have repeated Dutcher's original observations that in a bop5-1pf10 double mutant, bop5-1 partially suppresses the motility phenotype in pf10, resulting in more effective forward swimming (Dutcher et al., 1988). The pf10 gene product is likely required for assembly of an unknown axonemal component involved in regulation of flagellar waveform. Thus far, study of pf10 axonemes has not revealed alterations in the central pair, radial spokes and dynein regulatory complex components. However, in preliminary studies, structural defects in the inner arm region have been observed in pf10 axonemes (Porter and O'Toole, unpublished data), consistent with defective regulation in waveform.
Direct sequence analysis revealed a point mutation in bop5-1 that results in a premature stop codon predicted to truncate a portion of the C terminus, including the last WD-repeat. Notably, Western blot analysis of bop5-1 axonemes, by using antiserum to the N terminus fusion protein, confirmed the IC138 is indeed truncated and migrates ∼10% faster on SDS-PAGE. Therefore, the truncated IC138 assembles in axonemes from bop5-1, and based on a combination of Western blots and protein stains, most of the other known I1 subunits also assemble and can be salt extracted as a relatively stable and intact 18S I1 complex (see Results). The one exception is that LC7b (DiBella et al., 2004b) fails to assemble in the I1 complex in bop5-1 axonemes, suggesting that IC138 plays a role in anchoring this dynein light chain.
The failure of LC7b assembly in the I1 complex of bop5-1 axonemes is one of the most revealing outcomes of this study. LC7b is a newly identified member of the roadblock class of dynein light chains (DiBella et al., 2004b). Like other dynein light chains, including LC8 (King and Patel-King, 1995; Pazour et al., 1998), LC7a (Bowman et al., 1999) and Tctex1 (King et al., 1996; Harrison et al., 1998), LC7b is a subunit of more than one dynein isoform (DiBella et al., 2004b). Our analysis of bop5-1 and the double mutant bop5-1oda7 indicate that IC138, and in particular the WD-repeat domain, directly anchors LC7b in the I1 complex. This conclusion is consistent with cross-linking studies by using EDC, indicating a direct interaction between IC138 and LC7b (for detailed discussion, see DiBella et al., 2004b). Our result also suggests that the LC7b–IC138 interaction either occurs in the WD-repeat structure of IC138 or that the intact WD-repeat structure is required for LC7b assembly. The observation that LC7b is missing from the I1 complex in bop5-1 may reveal a general role of the WD-repeat structure in dynein intermediate chains for localization of the roadblock family of dynein light chains. Given the cyclic barrel shaped structure predicted for the seven WD/β-sheet repeat structure (Wall et al., 1995), the first and seventh WD-repeats are likely to structurally interact. Thus, the structure formed by the initial and terminal WD-repeats may form a common, conserved platform for anchoring the roadblock class of dynein light chains. Consistent with this model, the roadblock binding domain maps near the initial WD-repeat in IC74 (Susalka et al., 2002) and LC7b fails to assemble into I1 when IC138 is lacking the terminal WD-repeat. In contrast, other light chains, including LC8 and Tctex1, seem to be anchored in non-WD-repeat domains (Lo et al., 2001; Mok et al., 2001; Makokha et al., 2002).
As discussed in the Introduction, diverse evidence indicates that changes in phosphorylation of IC138 correlate with control of dynein-driven microtubule sliding and regulation of flagellar bending (Habermacher and Sale, 1997; King and Dutcher, 1997; Yang and Sale, 2000). These studies further indicated that increased phosphorylation of IC138 — “hyperphosphorylation”— correlated with inhibition of dynein activity and altered flagellar waveform (King and Dutcher, 1997). In the current studies, we postulated that IC138 in paralyzed flagellar mutants also would be hyperphosphorylated. Consistent with this hypothesis, our Western blot assay revealed that IC138 is hyperphosphorylated in axonemes from paralyzed flagellar axonemes defective in radial spoke or central pair assembly. This result supports a model in which a network of structures including the central pair apparatus, radial spokes, inner arm dynein I1, and IC138, and possibly the dynein regulatory complex, participate in control of flagellar motility (Porter and Sale, 2000; Smith and Yang, 2004).
Additionally, this result provides further support for the idea that, in the absence of the radial spoke or central pair components, dyneins are “globally” inactivated (Huang et al., 1982). The phosphorylation of IC138 is apparently diagnostic of inhibition of dynein. Furthermore, bop5-1 does not suppress paralysis in the radial spoke and central pair mutants examined, and IC138 remains hyperphosphorylated. However, the global phosphorylation of IC138 is evidently not sufficient to cause paralysis. For example, mutant cells totally lacking the I1 complex (pf9, pf30, ida1, ida2, or ida7) or defective in IC138 phosphorylation (mia2-1) are motile (Brokaw and Kamiya, 1987; Kamiya et al., 1991; Porter et al., 1992; King and Dutcher, 1997; Perrone et al., 1998). However, it is important to note that each of these mutants is defective in flagellar waveform and phototaxis (King and Dutcher, 1997), consistent with a role for the I1 complex in control of flagellar bending.
The bop5-1 motility phenotype further indicates that IC138 plays a key role in control of inner arm dynein I1 and regulation of flagellar waveform. Here, we also show that the bop5-1 mutant has a slow swimming phenotype but displays wild-type beat frequency. The simplest interpretation is that the bop5-1 mutation, truncation of IC138, alters flagellar waveform. In accordance with this interpretation, transformation of bop5-1 with the wild-type IC138 gene restores swimming speed. The transformed bop5-1 cells also assemble the full-length, wild-type IC138 in the axoneme, indicating that rescue involves replacement of the truncated IC138. The bop5-1 motility defect is therefore likely caused by the IC138 C-terminal truncation and is likely to involve the failure of LC7b assembly in the I1 complex. Because LC7b is also closely associated with several outer arm dynein subunits (DiBella et al., 2004b), one hypothesis is that both LC7b and IC138 may be important for the efficient coordination of inner arm and outer arm dynein activity. Further studies will be needed to address this interesting possibility.
Acknowledgments
We are grateful to Drs. Steve King and Linda DiBella (University of Connecticut, Storr, CT) for antibodies to the dynein light chains and sharing data on roadblock light chains LC7a and LC7b before publication, Dr. Carolyn Silflow (University of Minnesota, St. Paul, MN) for fine resolution mapping of IC138, and Drs. Pete Lefebvre (University of Minnesota), Elizabeth Smith (Dartmouth College), and Greg Pazour (University of Massachusetts) for DNA libraries. We thank Dr. Maureen Wirschell and Laura A. Fox for support and comments on the manuscript. T.W.H. was supported by a National Institutes of Health postdoctoral training award GM-00680, and this work was supported by grants from the National Institutes of Health (GM-68101 to P.Y, GM-55667 to M.E.P, and GM-51173 to W.S.S.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–08–0694. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0694.
References
- Bowman, A.B., Patel-King, R.S., Benashski, S.E., McCaffery, J.M., Goldstein, L.S., and King, S.M. (1999). Drosophila roadblock and Chlamydomonas LC 7, a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146, 165-180. [PMC free article] [PubMed] [Google Scholar]
- Brokaw, C.J. (1994). Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil. Cytoskeleton 28, 199-204. [DOI] [PubMed] [Google Scholar]
- Brokaw, C.J., and Kamiya, R. (1987). Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton 8, 68-75. [DOI] [PubMed] [Google Scholar]
- DiBella, L.M., and King, S.M. (2001). Dynein motors of the Chlamydomonas flagellum. Int. Rev. Cytol. 210, 227-268. [DOI] [PubMed] [Google Scholar]
- DiBella, L.M., Skatato, M., Patel-King, R.S., Pazour, G.J., and King, S.M. (2004a). The light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell 15, 4633-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella, L.M., Smith, E.F., Patel-King, R.S., Wakabayashi, K.I., and King, S.M. (2004b). A novel Tctex2-related light chain is required for stability of Inner dynein arm I1 and motor function in the Chlamydomonas flagellum. J. Biol. Chem. 279, 21666-21676. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K., Gibbons, W., and Inwood, W.B. (1988). A genetic analysis of suppressors of the PF10 mutation in Chlamydomonas reinhardtii. Genetics 120, 965-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zein, L., Omran, H., and Bouvagnet, P. (2003). Lateralization defects and ciliary dyskinesia: lessons from algae. Trends Genet. 19, 162-167. [DOI] [PubMed] [Google Scholar]
- Faber, H.R., Groom, C.R., Baker, H.M., Morgan, W.T., Smith, A., and Baker, E.N. (1995). 1.8 Å crystal structure of the C-terminal domain of rabbit serum haemopexin. Structure 3, 551-559. [DOI] [PubMed] [Google Scholar]
- Gibbons, I.R. (1995). Dynein family of motor proteins: present status and future questions. Cell Motil. Cytoskeleton 32, 136-144. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W., Gebhart, B., Mermall, V., Mitchell, D.R., and Heuser, J.E. (1987). High-pressure liquid chromatography fractionation of Chlamydomonas dynein extracts and characterization of inner-arm dynein subunits. J. Mol. Biol. 194, 481-494. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1985). Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100, 2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher, G., and Sale, W.S. (1996). Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J. Cell Sci. 109, 1899-1907. [DOI] [PubMed] [Google Scholar]
- Habermacher, G., and Sale, W.S. (1997). Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 136, 167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, A., Olds-Clarke, P., and King, S.M. (1998). Identification of the t complex-encoded cytoplasmic dynein light chain tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J. Cell Biol. 140, 1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, D.R., Habermacher, G., Glass, D.B., Smith, E.F., and Sale, W.S. (1994). Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J. Cell Biol. 127, 1683-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Ramanis, Z., and Luck, D.J. (1982). Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28, 115-124. [DOI] [PubMed] [Google Scholar]
- Kagami, O., and Kamiya, R. (1992). Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci. 103, 653-664. [Google Scholar]
- Kamiya, R. (2002). Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219, 115-155. [DOI] [PubMed] [Google Scholar]
- Kamiya, R., Kurimoto, E., and Muto, E. (1991). Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 112, 441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir, P., LaVoie, M., Brazelton, W.J., Haas, N.A., Lefebvre, P.A., and Silflow, C.D. (2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2, 362-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.J., and Dutcher, S.K. (1997). Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J. Cell Biol. 136, 177-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., Dillman, J.F., 3rd, Benashski, S.E., Lye, R.J., Patel-King, R.S., and Pfister, K.K. (1996). The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J. Biol. Chem. 271, 32281-32287. [DOI] [PubMed] [Google Scholar]
- King, S.M., and Patel-King, R.S. (1995). The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270, 11445-11452. [DOI] [PubMed] [Google Scholar]
- King, S.M., Wilkerson, C.G., and Witman, G.B. (1991). The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J. Biol. Chem. 266, 8401-8407. [PubMed] [Google Scholar]
- King, S.M., and Witman, G.B. (1990). Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265, 19807-19811. [PubMed] [Google Scholar]
- Lambright, D.G., Sondek, J., Bohm, A., Skiba, N.P., Hamm, H.E., and Sigler, P.B. (1996). The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379, 311-319. [DOI] [PubMed] [Google Scholar]
- Lo, K.W., Naisbitt, S., Fan, J.S., Sheng, M., and Zhang, M. (2001). The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J. Biol. Chem. 276, 14059-14066. [DOI] [PubMed] [Google Scholar]
- Makokha, M., Hare, M., Li, M., Hays, T., and Barbar, E. (2002). Interactions of cytoplasmic dynein light chains Tctex-1 and LC8 with the intermediate chain IC74. Biochemistry 41, 4302-4311. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., O'Toole, E.T., McDonald, K.L., McIntosh, J.R., and Porter, M.E. (1992). Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas J. Cell Biol. 118, 1145-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.R. (1994). Cell and molecular biology of flagellar dyneins. Int. Rev. Cytol. 155, 141-180. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and Kang, Y. (1991). Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 113, 835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, Y.K., Lo, K.W., and Zhang, M. (2001). Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J. Biol. Chem. 276, 14067-14074. [DOI] [PubMed] [Google Scholar]
- Myster, S.H., Knott, J.A., O'Toole, E., and Porter, M.E. (1997). The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol. Biol. Cell 8, 607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster, S.H., Knott, J.A., Wysocki, K.M., O'Toole, E., and Porter, M.E. (1999). Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas. J. Cell Biol. 146, 801-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297-300. [DOI] [PubMed] [Google Scholar]
- Neer, E.J., and Smith, T.F. (1996). G protein heterodimers: new structures propel new questions. Cell 84, 175-178. [DOI] [PubMed] [Google Scholar]
- Ogawa, K., Kamiya, R., Wilkerson, C.G., and Witman, G.B. (1995). Interspecies conservation of outer arm dynein intermediate chain sequences defines two intermediate chain subclasses. Mol. Biol. Cell 6, 685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, K., Takai, H., Ogiwara, A., Yokota, E., Shimizu, T., Inaba, K., and Mohri, H. (1996). Is outer arm dynein intermediate chain 1 multifunctional? Mol. Biol. Cell 7, 1895-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma, P., Hozumi, A., Ogawa, K., and Inaba, K. (2001). Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275, 177-183. [DOI] [PubMed] [Google Scholar]
- Paschal, B.M., Mikami, A., Pfister, K.K., and Vallee, R.B. (1992). Homology of the 74-kD cytoplasmic dynein subunit with a flagellar dynein polypeptide suggests an intracellular targeting function. J. Cell Biol. 118, 1133-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Wilkerson, C.G., and Witman, G.B. (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun, G., Escudier, E., Chapelin, C., Bridoux, A.M., Cacheux, V., Roger, G., Clement, A., Goossens, M., Amselem, S., and Duriez, B. (1999). Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 65, 1508-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone, C.A., Myster, S.H., Bower, R., O'Toole, E.T., and Porter, M.E. (2000). Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1beta dynein heavy chain. Mol. Biol. Cell 11, 2297-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone, C.A., Yang, P., O'Toole, E., Sale, W.S., and Porter, M.E. (1998). The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol. Biol. Cell 9, 3351-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, K.K., Fay, R.B., and Witman, G.B. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. Cytoskeleton 2, 525-547. [DOI] [PubMed] [Google Scholar]
- Piperno, G., and Luck, D.J. (1979). Axonemal adenosine triphosphatases from flagella of Chlamydomonas reinhardtii. Purification of two dyneins. J. Biol. Chem. 254, 3084-3090. [PubMed] [Google Scholar]
- Piperno, G., Ramanis, Z., Smith, E.F., and Sale, W.S. (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110, 379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E. (1996). Axonemal dyneins: assembly, organization, and regulation. Curr. Opin. Cell Biol. 8, 10-17. [DOI] [PubMed] [Google Scholar]
- Porter, M.E., Knott, J.A., Myster, S.H., and Farlow, S.J. (1996). The dynein gene family in Chlamydomonas reinhardtii. Genetics 144, 569-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., Power, J., and Dutcher, S.K. (1992). Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J. Cell Biol. 118, 1163-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., and Sale, W.S. (2000). The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151, F37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R., and Granick, S. (1953). Nutritional studies with Chlamydomonas reinhardtii. N.Y. Acad. Sci. 56, 831-838. [DOI] [PubMed] [Google Scholar]
- Satir, P., Barkalow, K., and Hamasaki, T. (1993). The control of ciliary beat frequency. Trends Cell Biol. 3, 409-412. [DOI] [PubMed] [Google Scholar]
- Smith, E.F. (2002). Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskeleton 52, 33-42. [DOI] [PubMed] [Google Scholar]
- Smith, E.F., and Sale, W.S. (1991). Microtubule binding and translocation by inner dynein arm subtype I1. Cell Motil. Cytoskeleton 18, 258-268. [DOI] [PubMed] [Google Scholar]
- Smith, E.F., and Sale, W.S. (1992a). Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science 257, 1557-1559. [DOI] [PubMed] [Google Scholar]
- Smith, E.F., and Sale, W.S. (1992b). Structural and functional reconstitution of inner dynein arms in Chlamydomonas flagellar axonemes. J. Cell Biol. 117, 573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E.F., and Yang, P. (2004). The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton 57, 8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. (1999). The WD-repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181-185. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., and Sigler, P.B. (1996). Crystal structure of a G-protein beta gamma dimer at 2.1Å resolution. Nature 379, 369-374. [DOI] [PubMed] [Google Scholar]
- Steffan, W., Hodgkinson, J.L., and Wiche, G. (1996). Immunogold localisation of the intermediate chain within the protein complex of cytoplasmic dynein. J. Struct. Biol. 117, 227-235. [DOI] [PubMed] [Google Scholar]
- Susalka, S.J., Nikulina, K., Salata, M.W., Vaughanm, P.S., King, S.M., Vaughan, K.T. Pfister, K.E. (2002). The roadblock light chain binds a novel region of the cytoplasmic dtynein intermediate chain. J. Biol. Chem. 277, 32939-32946. [DOI] [PubMed] [Google Scholar]
- Wall, M.A., Coleman, D.E., Lee, E., Iniguez-Lluhi, J.A., Posner, B.A., Gilman, A.G., and Sprang, S.R. (1995). The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83, 1047-1058. [DOI] [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., Koutoulis, A., Pazour, G.J., and Witman, G.B. (1995). The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129, 169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. [DOI] [PubMed] [Google Scholar]
- Yang, P., Diener, D.R., Rosenbum, J.L., and Sale, W.S. (2001). Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153, 1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., and Sale, W.S. (1998). The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9, 3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., and Sale, W.S. (2000). Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J. Biol. Chem. 275, 18905-18912. [DOI] [PubMed] [Google Scholar]
- Yang, P., Yang, C., and Sale, W.S. (2004). Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]