Abstract
We have identified a new Dictyostelium p21-activated protein kinase, PAKc, that we demonstrate to be required for proper chemotaxis. PAKc contains a Rac-GTPase binding (CRIB) and autoinhibitory domain, a PAK-related kinase domain, an N-terminal phosphatidylinositol binding domain, and a C-terminal extension related to the Gβγ binding domain of Saccharomyces cerevisiae Ste20, the latter two domains being required for PAKc transient localization to the plasma membrane. In response to chemoattractant stimulation, PAKc kinase activity is rapidly and transiently activated, with activity levels peaking at ∼10 s. pakc null cells exhibit a loss of polarity and produce multiple lateral pseudopodia when placed in a chemoattractant gradient. PAKc preferentially binds the Dictyostelium Rac protein RacB, and point mutations in the conserved CRIB that abrogate this binding result in misregulated kinase activation and chemotaxis defects. We also demonstrate that a null mutation lacking the PAK family member myosin I heavy chain kinase (MIHCK) shows mild chemotaxis defects, including the formation of lateral pseudopodia. A null strain lacking both PAKc and the PAK family member MIHCK exhibits severe loss of cell movement, suggesting that PAKc and MIHCK may cooperate to regulate a common chemotaxis pathway.
INTRODUCTION
PAKs, p21-activated protein kinases, are involved in a wide range of signaling pathways in eukaryotic cells, many of which control the cytoskeleton. In yeast, the PAKs Ste20 and Cla4 regulate the pheromone-induced mitogen-activated protein (MAP) kinase pathway controlling mating and cell polarization, respectively. In mammalian cells, two subfamilies of PAKs have been identified (Jaffer and Chernoff, 2002; Bokoch, 2003). PAK1, a member of the canonical form of mammalian PAK, was initially identified as a Rac-GTP binding protein (Manser et al., 1995). PAK1 has a wide range of cellular functions, including the regulation of the actin/myosin cytoskeleton (Bokoch, 2003). PAK1 controls cell movement and F-actin polymerization in neutrophils and other mammalian cell types through the negative regulation of LIM kinase and has a coordinating ability to control leading edge function (Daniels et al., 1999; Edwards et al., 1999; Li et al., 2003). PAK1 localizes to the leading edge of chemotaxing neutrophils where it also acts as an adaptor protein binding the Cdc42 guanine nucleotide exchange factor (GEF) PIXα that is required to induce Cdc42 activation in response to chemoattractant stimulation in neutrophils (Dharmawardhane et al., 1999; Li et al., 2003). Abrogation of PAK1 function in these cells causes a loss of cell polarity and abnormal subcellular localization of other signaling components required for leading edge function (Li et al., 2003). PAK1 also phosphorylates and regulates the myosin light chain kinase and directly controls myosin heavy chain assembly (van Leeuwen et al., 1999; Bokoch, 2000). PAK1 is a regulator of Rac1 and MEK1 in mammalian MAP kinase pathways, is an activator of the nuclear factor-κB (NF-κB) pathway, and can act as a negative regulator of apoptosis (Bokoch, 2000). In Saccharomyces cerevisiae, the PAK family members Cla4 and Ste20 also control the dynamics of the actin cytoskeleton (Eby et al., 1998).
Dictyostelium has two previously identified PAKs that play regulatory roles in controlling the cytoskeleton. PAKa is required for myosin II assembly during both cytokinesis and chemotaxis and the phenotypes of paka null cells are highly similar to those of myosin II (myoII) null cells (Chung and Firtel, 1999). paka null cells are unable to undergo cytokinesis when grown in suspension, forming large multinucleate cells. During chemotaxis, PAKa is required for myosin II assembly in the posterior of the cells. paka null cells form numerous lateral pseudopodia and are unable to effectively retract the cell's posterior. PAKa colocalizes with assembled myosin II at the posterior of chemotaxing cells and the contractile ring of cells undergoing cytokinesis. During chemotaxis, PAKa is positively activated by direct phosphorylation by Akt/PKB (Chung et al., 2001). Recently, mammalian PAK1 was shown to be phosphorylated by Akt/PKB, which mediates interaction with Nck and cell migration (Zhou et al., 2003). The second previously identified PAK is a myosin I kinase (Lee et al., 1996). Myosin I has been linked to the Arp2/3 complex and is important for controlling chemotaxis (de la Roche and Cote, 2001; Falk et al., 2003). Myosin heavy chain kinase null cells lack a strong phenotype (Lee et al., 1996; de la Roche and Cote, 2001), although phosphorylation of myosin I is required for its function and point mutations in the myosin I phosphorylation site result in strong chemotaxis phenotypes (Wessels et al., 1996; Gliksman et al., 2001; Falk et al., 2003). This apparent contradiction suggests that the identified myosin I kinase is probably one of a number of kinases able to phosphorylate and regulate myosin I function.
The hallmark of canonical PAKs is a Cdc42 and Rac interacting binding (CRIB) domain and a highly conserved Ser/Thr protein kinase domain (Jaffer and Chernoff, 2002; Bokoch, 2003). PAKs are activated through the binding of GTP-bound Cdc42 and Rac to the CRIB domain that results in a release of autoinhibition, which is mediated by interaction of the autoinhibitory domain with the kinase domain (Brown et al., 1996; Sells et al., 1997; Zhao et al., 1998; Tu and Wigler, 1999; Zenke et al., 1999; Lei et al., 2000). In addition, the N termini of PAKs contain a variety of regulatory and protein–protein interaction domains. Yeast Cla4 has an N-terminal pleckstrin homology (PH) domain, whereas Ste20 has a C-terminal Gβγ binding domain that is required for Ste20 function (Leeuw et al., 1998). Mammalian PAK1 contains a domain that is homologous to Ste20 at the C terminus; PAK1 binds Gβγ, although this C-terminal domain is insufficient for this binding (Li et al., 2003). PAK1 contains a polyproline domain that interacts with the Src homology (SH)3 domains of the adaptor proteins Nck and Grb2, an acidic domain of unknown function, as well as the domain required for interaction of the Cdc42/Rac GEF PIXα (Bokoch, 2003). Dictyostelium PAKa has an N-terminal regulatory domain that is necessary and sufficient for its proper subcellular localization (Chung and Firtel, 1999). In addition to the CRIB domain, the domain contains an Akt/PKB phosphorylation site, polyproline domains that can bind the SH3 domain of mammalian Nck, and an acidic domain.
We have identified a new Dictyostelium PAK, PAKc, which we demonstrate is required for proper chemotaxis. pakc null cells are less polarized than wild-type cells and produce multiple, lateral pseudopodia. A null mutation of a second PAK, MIHCK, exhibits phenotypes similar to those of pakc null cells. A double knockout strain moves extremely poorly, suggesting that PAKc is essential for chemotaxis in a mihck null background and that both PAK proteins potentially function on the same pathway. PAKc has an N-terminal PH domain, a CRIB domain, a protein kinase domain, and a C-terminal extension that is related to the Ste20 Gβγ binding domain. All four domains are required for proper PAKc function. We found that in response to global stimulation, PAKc is transiently activated and translocates from the cytosol to the plasma membrane. Analysis of the phenotypes of point mutations of PAKc indicates that it plays an important role in controlling chemotaxis.
MATERIALS AND METHODS
Creation of Knockout Strains
A pakC knockout construct was made by inserting the blasticidin resistance (Sutoh, 1993) cassette into a BamHI site created at base 560 of the PAKC cDNA. A 5′-fragment was amplified from cDNA by PCR by using the primers GTTTTTACTAGTTCACCAGATAAAGAGGGTG and GTTTTTAGATCTATTCATCAGGAAGTGCAG and subsequently digested with the enzymes SpeI and BglII. A 3′ fragment was amplified using the primers GTTTTTAGATCTCAAATTTAACTCTTAGTATG and GTTTTTCTCGAGAAAAAATTAAACATTTCTTGCGGC and subsequently digested with the enzymes BglII and XhoI. The vector was digested with SpeI and XhoI and the DNA was transformed into Dictyostelium cells. The positive knockout clones were confirmed by Southern blot analysis. Two independent clones were picked and examined. Both showed the same chemotaxis defects described in the Results.
To create a MIHCK knockout, a fragment of MIHCK was amplified by polymerase chain reaction (PCR) by using primers GTTTTTCTCGAGCCTGTTAGCAACGTTGGTAATAAAC (at nucleotide position 1070) and AATATCTCTATGAATTCTATGTTG (at nucleotide 2057) and subsequently digested with XhoI and EcoRI. A mihck knockout construct was made by inserting the blasticidin resistance cassette into an internal BamHI site at nucleotide position 1686. The vector was digested with XhoI and EcoRI and the DNA was transformed to KAx-3. Positive knockout clones were confirmed by Southern blot analysis.
To create a mihck/pakc double knockout strain, a mihck knockout construct was made by inserting the hygromycin resistance cassette into an internal BamHI site instead of the blasticidin resistance cassette as described above.
Plasmids
A full-length of PAKc cDNA clone was obtained by screening a 12–16 h developmental λZAP library (Schnitzler et al., 1994) with a probe amplified from genomic DNA containing PH and CRIB domains by PCR.
PAKc was tagged at the NH2 terminus with the myc epitope by using the primer 5′-GTTTTTGAATTCAAAATGGAACAAAAATTAATTTCAGAAGAAGATTTAAATATTTCATTAGAAGATCTATC-3′. For expression experiments, wild-type PAKc and PAKc mutants were subcloned into the DIP-J expression vector.
cAMP Stimulation of Dictyostelium Cells
To produce aggregation-competent cells, log-phase vegetative cells were washed twice with 12 mM Na+/K+ phosphate buffer and resuspended at a density of 5 × 106 cells/ml in Na+/K+ phosphate buffer (Insall et al., 1994; Meili et al., 1999). Cells were pulsed with 30 nM cAMP at 6 min intervals for 5 h. We spun down cells and resuspended them to 2 × 107 cells/ml in Na+/K+ phosphate buffer, pH 6.2, and bubbled them for 10 min. We stimulated cells with 1 μM cAMP and took time points as indicated.
PAKc Kinase Activity Assay
The PAKc kinase activity assay is a modification of that used for Akt/PKB (Meili et al., 1999). Cells were lysed with an equal volume of 2× NP-40 lysis buffer (2× phosphate-buffered saline [PBS], 100 mM NaF, 2% NP-40, 4 mM EDTA, 2 mM pyrophosphate, 2 mM Na3VO4, and protease inhibitors leupeptin and aprotinin) on ice for 10 min. The lysate was precleared by centrifugation for 10 min. To immunoprecipitate myc-PAKc, 1 μl of anti-myc monoclonal antibody (mAb) from Invitrogen (1 mg/ml) and 30 μl of a 50% slurry of protein G-Sepharose were added to the supernatant. The beads were washed twice with lysis buffer and twice with kinase buffer (25 mM MOPS, pH 7.4, 25 mM β-glycerophosphate, 20 mM MgCl2, and 1 mM dithiothreitol [DTT]). We incubated beads with 75 μl of kinase buffer containing 5 μM cold ATP, 10 μCi of [γ-32P]ATP, and 5 μg of H2B as substrate and incubated at room temperature for 15 min. The reaction was stopped by the addition of 25 μl of 5× sample buffer (250 mM Tris, 500 mM DTT, 10% SDS, 50% glycerol, and 0.5% bromphenol blue) and boiled for 2 min. Samples were separated by 10% SDS-PAGE, blotted onto a polyvinylidene difluoride (PVDF) membrane, and exposed to film. Experiments were repeated at least three independent times, always including a wild-type control for comparison.
Chemotaxis Assay
Chemotaxis analysis was performed as described previously (Funamoto et al., 2001). Briefly, log-phase vegetative cells were washed twice and resuspended at a density of 5 × 106 cells/ml with 12 mM Na+/K+ phosphate buffer and pulsed with 30 nM cAMP for 5 h at 6-min intervals. Pulsed cells were plated in Na+/K+ phosphate buffer at a density of 6 × 104 cells/cm2 onto a plate with a hole covered by a 0.17-mm glass coverslip. An Eppendorf Patchman micromanipulator with a glass capillary needle filled with 150 μM cAMP solution was brought into the field of view of an inverted microscope. The response of the cells was recorded by time-lapse video. Computer analysis was performed using DIAS software (Soll and Voss, 1998; Wessels et al., 1998). At least five cells were analyzed, and the chemotaxis assays were performed at least three times on separate days.
Immunofluorescence Staining
Pulsed cells were spotted on a coverslip and sat for 15 min. Cells were fixed with -20°C methanol for 10 min, washed, and incubated with anti-myc antibody (1/500 dilution) in PBS containing 0.5% bovine serum albumin (BSA) and 0.05% Tween 20 for 1 h. After washing, cells were incubated with fluorescein isothiocyanate-labeled anti-mouse antibodies for 1 h. After washing, cells were observed with a 100× oil immersion lens on a Leica microscope.
F-Actin Polymerization and Myosin II Assembly
F-actin polymerization and myosin II assembly were assayed as described previously (Chung et al., 2000) with modifications as described by M. Mendoza, F. Du, U. Tang, H. Ma, and R.A. Firtel (unpublished data).
Two-Hybrid Assays
We performed two types of yeast two-hybrid assays. In both assays, a DNA fragment encoding the CRIB domain of Dictyostelium PAKc (SAVSQPFNLKHEVHVDFNSATGFSGLPKEWEVILKSSN) was amplified by PCR and cloned in the appropriate vector. The first assay followed the protocol described previously (Lee et al., 1997).
For the second assay, the DNA fragment encoding the CRIB domain of Dictyostelium PAKc was cloned into the yeast two-hybrid vector pACT2 (BD Biosciences Clontech, Palo Alto, CA). DNA fragments carrying the RacB mutations were generated from wild-type cDNA by PCR-based, site-directed mutagenesis. We followed the protocols for the Matchmaker two-hybrid system from BD Biosciences Clontech for the two-hybrid assays. After co-transformation of yeast strain Y190, interactions were assessed semiquantitatively by colony-lift β-galactosidase filter assay and quantitatively by liquid culture β-galactosidase assay by using 2-nitrophenyl-β-d-galactopyranoside as a substrate. The results were similar for both assays.
Phospholipid Binding Assay
The PH domains of GRP1 and PAKc were subcloned to a glutathtione S-transferase (GST) vector carrying a protein kinase A (PKA) phosphorylation site (SRRASV) between the GST and PH domains. Recombinant protein was purified from a subsaturating culture of Escherichia coli that had been induced to express the fusion proteins with 0.1 mM isopropyl β-d-thiogalactoside at room temperature for 1 h. Washed cells were suspended in PBS containing leupeptin and aprotinin and sonicated. Cell debris was removed by centrifugation and Triton X-100 was added to the supernatant to 1%. Glutathione beads were added and the suspension was rocked in the cold room (4°C) for 30 min. The beads were pelleted, washed three times with PBS containing 1% Triton, and washed once with PBS.
The GST fusion protein was then labeled with 32PO4. GST fusion protein on the beads was washed once with PKA buffer (50 mM PO4, pH 7.2, 10 mM MgCl2, 5 mM NaF, 4.5 mM DTT, 6.25 μg/ml aprotinin, and 6.25 μg/ml leupeptin) and then resuspended in PKA buffer with 1.25 mCi/ml [32PγATP] and 112 μg/ml PKA catalytic domain protein and incubated at room temperature for 30 min. The beads were washed 4 times in PBS containing 5 mM DTT. The GST fusion protein was eluted with 15 mM reduced glutathione in 50 mM Tris, pH 8.0, overnight.
Lipids were dotted onto PVDF membrane and dried overnight. The membrane was blocked with 0.1% BSA (fatty acid free, CAS 9048-46-8; Sigma-Aldrich, St. Louis, MO) in Tris-buffered saline/Tween 20 (TBST) for 1 h. 32P-Labeled GST fusion protein was added to the blocked membrane and incubated for 1 h at room temperature. The membrane was washed three times with TSBT and then exposed to film.
Gβγ Binding
Wild-type KAx-3 and gβ null cells (Lilly et al., 1993) were cotransformed with myc-tagged PAKc and Gβ-yellow fluorescent protein (YFP) (Jin et al., 2000; Janetopoulos et al., 2001). The cells were selected for the expression of Gβ-YFP by immunofluorescence and myc-PAKC by Western blot. Because of the possibility that the membrane association of Gβγ through the prenylation of the Gγ subunit might interfere with coimmunoprecipitation, a second set of cells were cotransformed with myc-PAKc, Gβγ-YFP, and Gγ with a point mutation in the CAAX box that prevents prenylation and thus membrane association (Zhang et al., 2001). Cells were then selected for YFP expression and myc-PAKc expression by Western blot. To determine whether the cells also expressed the mutant Gγ protein, we compared membrane and cytosolic localization of YFP. In cells cotransformed only with myc-PAKc and Gβ-YFP, all of the YFP fluorescence was on the plasma membrane. In the second set of transformants, we selected cells in which a significant fraction of the Gβ-YFP was cytosolic, as would be expected if the cells were expressing the mutant Gγ subunit, consistent with previous reports (Zhang et al., 2001).
To examine the possible coimmunoprecipitation of myc-PAKC with Gβγ (cotransformed with myc-PAKc and Gβ-YFP or cotransformed with myc-PAKc and Gβ-YFP and mutant Gγ), cells were pulsed for 5 h with 30 nM cAMP as used for the PAKc activation and chemotaxis assays. Cells were then activated with the addition of cAMP to 1 μM and time points were taken at 0 (before addition of cAMP), 10, and 30 s. Cells were lysed with 2 volumes of lysis buffer and immunoprecipitated with the anti-myc antibody. Immunoprecipitates were analyzed by SDS-PAGE and Western blots by using both anti-myc and anti-green fluorescent protein (GFP) antibodies, which effectively immunoprecipitates the Gβγ-YFP. In these experiments, two different lysis buffers we tested: lysis buffer 1 containing PBS, 50 mM NaF, 1% NP-40, 2 mM EDTA, pH 7.2, 1 mM Na pyrophosphate, 1 mM orthovanadate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin; lysis buffer 2 containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 15 mM NaF, 2 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 20 mM β-glycerophosphate, 1 mM orthovanadate, 1 mM Na pyrophosphate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin.
In a second set of experiments, cells were cotransformed with myc-PAKC in hemagglutinin (HA)-tagged Gβγ. Experiments were performed as described above.
RESULTS
Structure and Expression Pattern of PAKc
PAKc was identified in a bioinformatics search for proteins in the Dictyostelium genome that contain a CRIB domain and are therefore potential downstream effectors of Rac family GTPases. At the time PAKc was identified, the full sequence of the gene was unavailable in the databases. A clone carrying the full PAKc open reading frame (ORF) was identified and sequenced and used for subsequent studies (see Materials and Methods). The sequence agreed with the predicted ORF from the sequence obtained from the Dictyostelium genomic sequencing project.
Figure 1, A and B, show the sequence and domain organization of PAKc. PAKc contains a PH domain very close to its N terminus, which is immediately followed by a CRIB domain. Sequence analysis of the CRIB domain and adjacent sequences (Figure 1D) suggests that, as in the PAK1 family of PAKs, there is an overlapping autoinhibitory domain (Brown et al., 1996; Sells et al., 1997; Zhao et al., 1998; Tu and Wigler, 1999; Zenke et al., 1999; Lei et al., 2000).
Figure 1.
The main structure and expression of PAKc. (A) The derived amino acid sequence of PAKa is shown. The four domains, the PH domain, the CRIB/autoinhibitory domain, the kinase domain, and the putative Gβγ binding domain, are boxed. (B) Domain structure of PAKc. (C) Developmental pattern of expression of PAKc. Total cell RNA was isolated from developing cells plated on nonnutrient agar at the indicated times. 0 is vegetatively growing cells; 4 h represents the time of early/midaggregation. Aggregates have formed by 8 h and tipped aggregates by 12 h; 24 h represents the mature fruiting body. Casein kinase II, which is expressed equally throughout growth and development, is used as a loading control. (D) Sequence comparisons of the PAKc CRIB/autoinhibitory domain compared with human PAK1 (AAC24716), S. pombe PAK1 (NP-596626), Dictyostelium MIHCK (AAC71063), Dictyostelium PAKa (AAD28470), and mouse PAK3 (AAB35358). The residues that were mutated in order to examine the function of the CRIB/autoinhibitory domain are indicated. (E) The C-terminal extension of PAKc is shown compared with that of mouse PAK3 (AAB35358), rat PAK1 (2201417A), S. cerevisiae Ste20 (Q03497), and S. cerevisiae Cla4 (CAA57879). The residues that are mutated in PAKc are indicated. (F) Sequence comparison of the PAKc PH domain to other PH domains: mouse GRP1 (NP-035311), Dictyostelium Akt/PKB (P54644), Dictyostelium CRAC (P35401), Dictyostelium PhdA (AAC29444), S. cerevisiae Cla4 (CAA57879), and mouse PLC3 (Q8R3B1).
To confirm that the PAKc CRIB domain interacts with GTP-bound forms of Dictyostelium Rac GTPases, we reproduced the two-hybrid assay with constitutively active and dominant negative forms of Dictyostelium RacB. These experiments are essentially the same as those from a separate study in which we identified Dictyostelium RacB-GTP as the Dictyostelium Rac showing the strongest interaction with the PAKc CRIB/autoinhibitory domain (CRIB/AI: linked Rac binding, autoinhibitory domain; Park et al., 2004). As shown in Table 1A and in Park et al. (2004), the PAKc CRIB/AI domain exhibits a strong interaction with constitutively active forms of RacB (RacBQ61L and RacBG12V) but not the dominant negative form (RacBS17N). (These experiments are reproduced here to allow comparison of the binding of the mutant forms of the RacB CRIB.)
Table 1.
Yeast two-hybrid analysis of interactions of the PAKc CRIB/AI domain with RacB
| A.
|
|
|---|---|
| RacB
|
PAKc
|
| CRIB/AI | |
| RacB G12V | +++++ |
| RacB Q61L | +++++ |
| RacB N17S | - |
| B.
|
|
|---|---|
| CRIB/AI
|
Interaction
|
| RacB Q61L | |
| Wild-type | +++++ |
| L144F | +++ |
| H121,124L | - |
A. Relative interactions between constitutively active (RacBG12V and RacBQ61L) and dominant negative (RacBN17S) forms of RacB and the CRIB and autoinhibitory domain (CRIB/AI).
B. Relative interactions between RacBQ61L and wild-type CRIB/AI and mutant forms of the CRIB/AI domain.
The PAKc C terminus has a protein kinase domain related to that of PAKs followed by a C-terminal extension that is homologous to the Gβγ binding domain found in Ste20 and the C-terminal domain of mammalian PAK1 subfamily members (Figure 1E). The presence of a putative PAK kinase and a CRIB domain suggested that PAKc is a member of the p21-activated protein kinase family. Sequence comparison of the putative Gβγ binding domain revealed strong conservation of the C-terminal domains in S. cerevisiae Ste20 and mammalian PAK1. The PAKc PH domain indicates conservation in a wide range of Dictyostelium, mammalian, and yeast PH domains (Figure 1F). The highly conserved arginine 34 is required for inositol phosphate binding (Fukuda et al., 1996; Salim et al., 1996). In contrast to Dictyostelium PAKa and the mammalian PAK1–3 subfamily, which have long N-terminal domains, PAKc is more compact and has no other identifiable domains.
To examine the developmental expression of the gene encoding PAKc (pak3), we performed RNA blot analysis with RNA isolated from growing cells at different stages of development. The results in Figure 1C show that PAKc is preferentially expressed during vegetative growth and in the earliest stages of development. RNA levels decrease significantly by 8 h in development, because there is a shift from aggregation to cell type differentiation and morphogenesis of the multicellular organism. The gene is expressed, but at significantly lower levels during later stages of development.
Regulation of PAKc Kinase Activity
Because PAKs often control the actin/myosin cytoskeleton, we investigated whether PAKc kinase activity is responsive to chemotactic stimulation. For this purpose, we used an immunoprecipitate kinase assay similar to that which we previously used to examine the chemoattractant-mediated kinase activity of Akt/PKB, PAKa, and RCK1 (see Materials and Methods; Meili et al., 1999; Chung et al., 2001; Sun and Firtel, 2003). As shown in Figure 2, PAKc has a very low basal activity in unstimulated (0-s time point), aggregation-competent cells (cells fully responsive to the chemoattractant cAMP). There is a peak of activity 10 s after cAMP stimulation and then activity decreases rapidly to a plateau at about the basal level by 20 s for the time frame assayed. To confirm that the kinase activity observed is dependent on the PAKc kinase domain, we created a Lys→ Ala mutation at residue 234, the Lys expected to be required for ATP binding and kinase activity. As depicted (Figure 2), myc-PAKcK234A exhibits no kinase activity. These results demonstrate that PAKc is rapidly and transiently activated by chemoattractant stimulation.
Figure 2.
Kinase activity of wild-type and mutant PAKc proteins expressed in wild-type (A), pakc null (B), and pi3k1/2 null cell (C) backgrounds. Myc-tagged wild-type PAKc and mutant PAKcs were expressed in either wild-type cells or pakc null cells. Aggregation-competent cells were stimulated with cAMP and kinase activity was measured on immunoprecipitated PAKc isolated from cell extracts at the time points indicated. 0 represents unstimulated cells. Bottom, Western blot against anti-myc antibody and indication of the amount of PAKc in each immunoprecipitate. In all experiments, cells were compared with wild-type PAKc. All panels have the same level of expression and gels were run in parallel. The exception is the panel of PAKcR34C expressed in pakc null cells. The autoradiogram was overexposed to demonstrate the lack of PAKcR34C inducible kinase activity. In this case, a 10-s wild-type time point was run in parallel. The intensity of this band should be compared with the 10-s time point of wild-type PAKc in the left-hand panel.
The kinase activities of mammalian PAK1 and Schizosaccharamyces pombe PAK1 are regulated through the CRIB/AI domain (Brown et al., 1996; Sells et al., 1997; Zhao et al., 1998; Tu and Wigler, 1999; Zenke et al., 1999; Lei et al., 2000). To investigate the regulation of PAKc, we created a set of mutated PAKc proteins that, based on mutant studies in mammalian and S. pombe PAK1s, should either disrupt the ability of the PAKc putative CRIB domain to bind activated Rac (PAKcH121,124L, equivalent to hPAK1H83,86L) and/or disrupt the function of the autoinhibitory domain (PAKcL144F, equivalent to hPAK1L107F). We examined the effects of these mutations on PAKc kinase activity. In these experiments, wild-type PAKc was assayed in parallel on the same gels, enabling us to make quantitative comparisons. Western blot analyses of the immunoprecipitated myc-PAKc were performed to provide an indication of the amount of PAKc in the kinase reaction and to act as a control for sample loading in the kinase assay.
PAKcH121,124L, which is expected to lead to the loss of Rac/Cdc42-GTP binding in PAK1, exhibits a high basal activity, whether it is expressed in wild-type cells or in pakc null cells (Figure 2, A and B, respectively). The activity does not change after chemoattractant stimulation. Mammalian PAK1 carrying these mutations also exhibits an elevated basal activity (Sells et al., 1997; Daniels et al., 1999). The increase in basal kinase activity suggests that the H121,124L mutation, in addition to disrupting interaction with Rac-GTP, either lies within and thus disrupts the autoinhibitory domain or changes the conformation of the adjacent autoinhibitory domain and thereby disrupts its function. When the conserved Leu within the autoinhibitory domain was mutated to Phe (PAKcL144F), we observed an even higher basal kinase activity (Figure 2), similar to that which was observed for mammalian PAK1 (Zenke et al., 1999). However, in contrast to the activity of PAKcH121,124L, PAKcL144F activity was significantly induced in response to chemoattractant stimulation, and the activity returned to the elevated basal activity with kinetics somewhat slower than those observed in wild-type cells. Similar results were obtained when PAKcL144F was expressed in either a wild-type or a pakc null strain (created by homologous recombination; see Materials and Methods).
To confirm that the CRIB/AI domains carrying the H121,124L and L144F mutations exhibited the expected Rac-GTP binding properties, we used a yeast two-hybrid assay. The results summarized in Table 1B show that the H121,124L mutation abrogated RacB-GTP binding, whereas the L144F mutation in the CRIB/AI bound Rac-GTP with a slightly reduced affinity compared with the wild-type CRIB/AI, consistent with results from studies in other systems. We suggest that the PAKcL144F disrupts the autoinhibitory domain leading to higher basal activity, and chemoattractant stimulation results in further PAKc kinase activity. From the analysis of the PAKcH121,124L mutation, we presume the induced activity takes place through RacB-GTP or another Rac-GTP by interacting with the CRIB domain.
We also observed that the level of expression of PAKcL144F is significantly lower than the expression of the wild-type or other mutant forms of PAKc when expressed in either the wild-type or a pakc null background. The lower level of expression of this mutant PAKc was observed in multiple clonal isolates from multiple transformations (data not shown). We expect that a high level of expression of this mutant protein leads to growth defects that prevent us from isolating high expression clones. The specific activity of PAKcL144F is therefore extremely high compared with wild-type PAKc or with PAKcH121,124L.
To understand the possible requirements of the PH domain and C-terminal extension, we examined the kinase activity of PAKc carrying mutations in these domains. PAKcR34C carries an Arg→Cys amino acid substitution in the highly conserved arginine of the PAKc PH domain. This amino acid substitution was initially identified in the PH domain-containing kinase Btk (Bruton's tyrosine kinase) and blocked its ability to be activated (Li et al., 1995; Fukuda et al., 1996). Furthermore, studies on a variety of PH domain-containing proteins, including Akt/PKB from several organisms and PhdA from Dictyostelium, have demonstrated that this amino acid substitution results in the loss of inositol phosphate binding (Salim et al., 1996; R. Meili et al., 1999, 2000; Dowler et al., 2000; Funamoto et al., 2001; Meili, S. Funamoto, and R.A. Firtel, unpublished observation). As can be seen in Figure 2B, PAKcR34C exhibits no chemoattractant-mediated increase in kinase activity. We also examined PAKc carrying mutations in two conserved residues (PAKcS466A,P470A) in the C-terminal extension that is homologous to the Gβγ binding domain of Ste20. PAKcS466A,P470A has an extremely low basal activity, even lower than that which is observed for PAKcR34C. PAKcS466A,P470A kinase activity is not induced upon chemoattractant stimulation. These studies suggest that both of these domains are required for in vivo PAKc activation (see Discussion). In the case of PAKcS466A,P470A (Figure 2A), which exhibits little if any basal kinase activity, we cannot exclude the possibility that the C-terminal extension/putative Gβγ binding domain is essential for the normal conformation of PAKc and disruption of this domain may abrogate all PAKc kinase activity.
Because PAKc has a PH domain, we examined whether PAKc activation is dependant on PI3K by examining PAKc's activation in a strain lacking the two class I PI3Ks PI3K1 and PI3K2 (pi3k1/2 null cells, Funamoto et al., 2001). As depicted in Figure 2C, PAKc shows the same activation in wild-type and pi3k1/2 null cells.
PAKc Is Required for Proper Chemotaxis
Our finding that PAKc kinase activity is rapidly and transiently stimulated upon chemoattractant stimulation suggested that PAKc may be involved in controlling chemotaxis. To examine this, we compared the ability of the pakc null and wild-type cells to chemotax to a micropipette containing the chemoattractant cAMP (see Materials and Methods). DIAS computer software was used to calculate chemotaxis parameters (Soll and Voss, 1998; Wessels et al., 1998). This analysis is presented in Table 2.
Table 2.
DIAS analysis of chemotaxis
| Vector | PAKc | PAKc | PAKc K234A Kinase dead | PAKc R34C | PAKc S466A, P470A Gβγ “binding site” | ||
|---|---|---|---|---|---|---|---|
| Strain background | KAx-3 | PAKc null | KAx-3 | PAKc null | KAx-3 | KAx-3 | KAx-3 |
| Speed (μm/min) | 9.96 ± 0.70 | 8.92 ± 0.55 | 5.58 ± 0.77 | 6.94 ± 0.83 | 3.70 ± 0.37 | 11.3 ± 0.01 | 9.18 ± 2.04 |
| Dir ch (deg) | 24.1 ± 4.0 | 29.3 ± 1.3 | 55.8 ± 1.65 | 55.8 ± 11.2 | 38.6 ± 4.21 | 19.6 ± 1.71 | 25.1 ± 7.39 |
| Roundness | 50.4 ± 3.0 | 68.2 ± 5.0 | 44.5 ± 2.04 | 59.9 ± 3.66 | 57.7 ± 2.92 | 59.2 ± 2.00 | 63.7 ± 5.46 |
| Directionality | 0.78 ± 0.01 | 0.73 ± 0.03 | 0.43 ± 0.05 | 0.48 ± 0.11 | 0.59 ± 0.04 | 0.81 ± 0.01 | 0.74 ± 0.09 |
| Vector | PAKc ΔPH domain | PAKc H121, 124L | PAKc L144F | PAKc ΔPH domain | PAKc H121, 124L | PAKc L144F | ||
|---|---|---|---|---|---|---|---|---|
| Strain background | KAx3 | KAx-3 | KAx-3 | PAKc null | PAKc null | PAKc null | MIHCK null | MIHCK/PAKc null |
| Speed (μm/min) | 7.98 ± 0.79 | 6.36 ± 0.51 | 11.5 ± 0.48 | 9.45 ± 0.75 | 8.13 ± 0.35 | 11.1 ± 0.68 | 9.55 ± 0.70 | 3.49 ± 0.07 |
| Dir ch (deg) | 37.0 ± 11.4 | 34.5 ± 1.17 | 28.2 ± 1.71 | 28.1 ± 8.28 | 51.2 ± 2.15 | 43.6 ± 11.4 | 23.6 ± 2.94 | 62.2 ± 5.02 |
| Roundness | 48.6 ± 4.81 | 58.4 ± 3.07 | 72.1 ± 2.28 | 58.9 ± 4.06 | 48.2 ± 0.65 | 47.7 ± 6.17 | 60.0 ± 0.36 | 64.0 ± 5.01 |
| Directionality | 0.67 ± 0.09 | 0.71 ± 0.01 | 0.75 ± 0.01 | 0.79 ± 0.04 | 0.49 ± 0.01 | 0.57 ± 0.06 | 0.81 ± 0.10 | 0.33 ± 0.07 |
DIAS analysis of chemotaxis. Numbers are mean values ± standard deviation. Dir ch, direction of change of direction. Speed indicates speed of cell's centroid movement along the total path. Direction change is a relative measure of the number and frequency of turns the cell makes. Larger numbers indicate more turns and less efficient chemotaxis. Directionality is a measure of the linearity of the pathway. Cells moving in a straight line to the needle have a directionality of 1.00. Roundness is an indication of the polarity of the cells. Larger numbers indicate the cells are more round (less polarized).
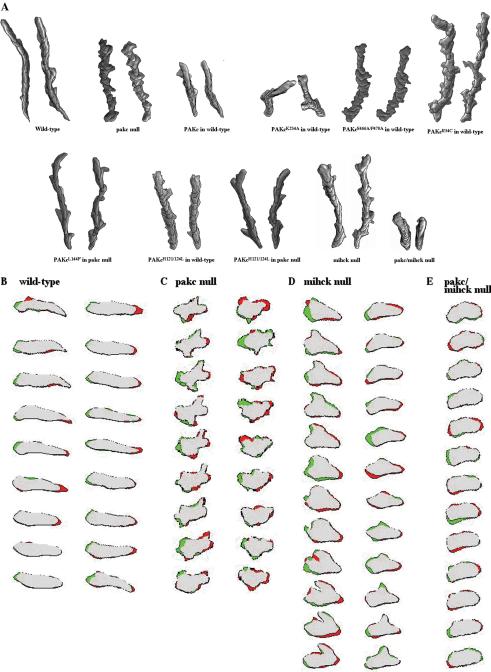
As shown in Figure 3A and Table 2, wild-type cells are highly polarized, primarily protrude a single pseudopod in the direction of the chemoattractant source, and move directionally to the chemoattractant source with a speed of ∼10 μm/s. In contrast, pakc null cells are more rounded and amoeboid-shaped than wild-type cells and produce multiple pseudopodia from the lateral sides and back of the cells as well as the front of cells, but move with only a slightly reduced speed (∼9 μm/s). The inability of the pakc null strain to inhibit the formation of lateral pseudopodia is strikingly illustrated in the analysis in Figure 3, B and C. In this DIAS analysis, new membrane protrusions are indicated in green and areas of membrane contraction are in red. In wild-type cells, membrane protrusions occur primarily at the side of the cell facing the chemoattractant source, whereas membrane retractions occur primarily on the side opposite the chemoattractant source, leading to a highly polarized cell and persistence in the direction of cell movement (Figure 3B). In contrast, pakc null cells protrude pseudopodia along most of the perimeter of the cells with pseudopodia sometimes forming at the side of the plasma membrane facing away from the chemoattractant source (Figure 3C). Further, one often observes areas of membrane retraction at the side of the membrane facing the chemoattractant source. When we examine the images more closely, we observe that in pakc null cells, the amount of membrane protrusion/retraction, as visualized by the amount of red and green domains, seems greater than in wild-type cells. The analysis suggests that pakc null cells have more active membrane protrusion than wild-type cells. The speed of the cells (as measured by the speed of movement of the centroid) is only slightly reduced, although the distance traveled toward the micropipette shows a greater reduction, as might be expected if the cells form pseudopodia along the cell's perimeter. These results demonstrate that pakc null cells are unable to properly regulate the direction of pseudopod formation and inhibit lateral pseudopod formation.
Figure 3.
DIAS analysis of wild-type cells and cells expressing wild-type and mutant PAKc. (A) The figure depicts the overlapping DIAS image analysis of chemotaxing cells. Overlapping images were captured at 1-min intervals. Cells for analysis were randomly chosen and the paths examined represent a 10-min interval taken from the middle of the chemotaxis movie. The images illustrate the shape, total distance moved, and the directionality of movement. (B–E) Changes in membrane protrusions (green) and retractions (red) in wild-type (KAx-3), pakc null, mihck null, and pakc/mihck null cells. The first image in the series is on top and sequential images, taken every 6 s, are shown below. Two different cells are shown for the wild-type and pakc null strains. For the mihck null strain, one cell is shown with the first series of images on the left. These cells exhibit extremes of cell shape with a very nonpolar phenotype and large lateral extensions and retractions, whereas at other times, the phenotype is less severe.
Expression in wild-type cells of PAKcR34C or PAKcS466A,P470A, which are not activated in response to chemoattractant stimulation, also exhibits strong phenotypes, including reduced polarity and production of multiple lateral pseudopodia, similar to the phenotypes exhibited by pakc null cells (Figure 3A and Table 2). Although PAKcS466A,P470A-expressing cells show significant changes in direction, the overall movement of the cell (the efficiency by which it moves up the chemoattractant gradient) remains high. Expression of a kinase dead PAKc (PAKcK234A) in wild-type cells results in a severe loss of cell movement, suggesting that PAKcK234A functions as a dominant negative protein, possibly by interacting with and inhibiting other components in the PAKc pathway (see below).
Overexpression of wild-type PAKc causes a reduction of speed and loss of directionality. This was observed whether PAKc was overexpressed in wild-type or pakc null cells (the level of PAKc expression was higher than in wild-type cells as judged by RNA levels; our unpublished data). pakc null and wild-type cells expressing the misregulated PAKc proteins PAKcH121,124L and PAKcL144F exhibited mild phenotypes (Table 2 and Figure 3A; our unpublished data).
The ability to respond to the chemoattractant cAMP is a developmentally regulated process, which is induced by oscillatory pulses of cAMP that induce the aggregation response. To test whether pakc null cells are cAMP responsive, we examined the induction of two cAMP-induced genes: cAR1, which encodes the major cAMP receptor required for aggregation; and csA (gp80), a cell adhesion molecule involved in aggregation. As shown in Figure 4, the kinetics and level of expression of the cAR1 and csA mRNAs in response to exogenous cAMP pulsing are the same in wild-type and pakc null cells.
Figure 4.
RNA blot analysis. Total cell RNA was isolated from vegetative cells of wild-type, pakc null, mihck null, and mihck/pakc null cells. After electrophoresis on formaldehyde-containing agarose gels, the RNA was blotted onto a Magnagraph 0.22-μm nylon membrane and probed with a nick-translated fragment from either cAR1 or csA.
Control of F-Actin Polymerization and Myosin II Assembly
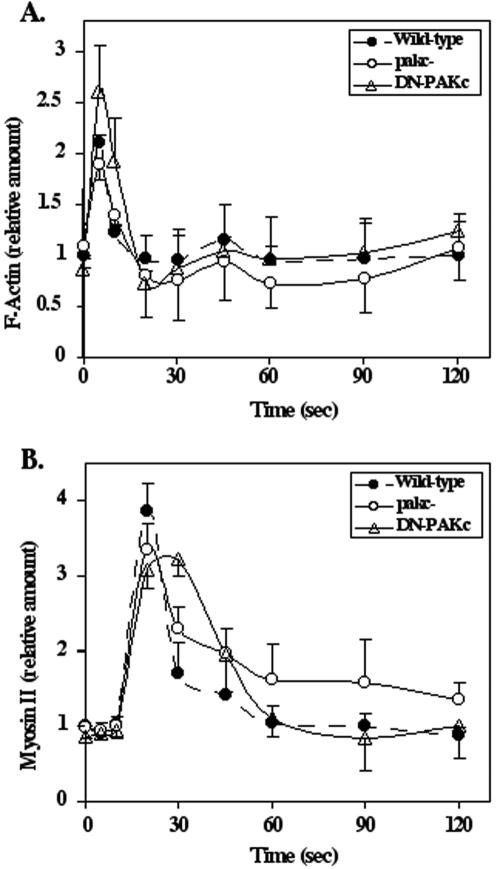
Myosin II null cells or mutants that abrogate the ability to assemble myosin II (e.g., paka null strains) also form lateral pseudopodia, similar to the behavior of pakc null cells. To determine whether the pakc null phenotype results from altered kinetics or the level of myosin II assembly and F-actin polymerization, we quantified F-actin polymerization and myosin II assembly in the Triton-insoluble, cytoskeletal fraction in response to chemoattractant stimulation in wild-type, pakc null, and PAKc kinase dead (PAKcK234A) cells. In wild-type cells, F-actin assembly in response to chemoattractant stimulation is biphasic (Condeelis et al., 1990). The first peak, which is associated with the cringe response, is very transient, rising rapidly and peaking at 5 s. After decreasing to almost basal levels by 20–30 s, there is a second lower and broader peak, which is associated with pseudopod extension. As shown in Figure 5A, the kinetics and level of F-actin assembly are slightly lower in the first and second peaks of F-actin polymerization, but within statistical variation, in pakc null cells compared with wild-type cells. In PAKcK234A cells, the first peak is elevated, but there is no difference observed in the second peak. For myosin II assembly, the kinetics of assembly show an extended shoulder on the myosin II assembly profile for pakc null cells that is statistically higher than that of wild-type cells (Figure 5B). Cells expressing PAKcK234A have a significantly broader peak of activation compared with wild-type cells. Whereas some effect is observed on myosin II and F-actin assembly and polymerization, the effects are not large.
Figure 5.
Kinetics of F-actin polymerization and myosin II assembly in the Triton-insoluble, cytoskeletal fraction. The kinetics of F-actin polymerization (A) and myosin II assembly (B) in response to chemoattractant stimulation are shown for wild-type, pakc null, and PAKc kinase dead (PAKcK234A) cells. See Materials and Methods for details.
Possible Interactions between PAKc and Other PAK Proteins
As described above, expression of the kinase dead PAKc (PAKcK234A) in wild-type cells results in a decrease in cell polarity and a significant loss in speed and directionality. One explanation to account for our observation that the PAKcK234A phenotype is more severe than that of pakc null cells is that PAKcK234A may complex with one or more proteins required for chemotaxis, blocking their function, possibly including other PAKs.
To test these possibilities, we wanted to examine the genetic interactions between PAKc and other Dictyostelium PAKs. Dictyostelium has two PAK proteins in addition to PAKc that have been described in the literature: PAKa and MIHCK, both of which have CRIB/AI domains (Lee et al., 1996; Chung and Firtel, 1999). PAKa is required for myosin II assembly; paka null cells exhibit defects in cytokinesis and chemotaxis. Because pakc null cells do not exhibit defects in cytokinesis (our unpublished data) or reduced myosin II assembly (Figure 5B), it is unlikely that PAKc and PAKa function in the same biochemical pathway. Myosin I proteins are regulated by phosphorylation (de la Roche and Cote, 2001; Gliksman et al., 2001; Falk et al., 2003) and MIHCK is regulated by Rac-GTP (Lee et al., 1996, 1998). MIHCK, like PAKc, has a CRIB domain, but lacks the putative Gβγ-binding C-terminal extension. An initial examination of mihck null cells suggested that the strain did not exhibit any gross developmental or growth defects or defects in phagocytosis or pinocytosis (de la Roche and Cote, 2001). To examine MIHCK function in more detail, we made a MIHCK gene knockout in our wild-type KAx-3 strain. As illustrated in Figure 3, A and D, and Table 2, mihck null cells exhibit mild chemotaxis defects and, like pakc null cells, mihck null cells show a loss of cell polarity and the formation of multiple lateral pseudopodia. We postulated that if PAKc and MIHCK had redundant functions, we may uncover a more severe phenotype if we created a double knockout of the genes encoding MIHCK and PAKc. The results in Figure 3, A and E, and Table 2 indicate that mihck/pakc double knockout cells exhibit strong chemotaxis defects with some loss of polarity and a very significant loss of speed. The results demonstrate that in a genetic background lacking MIHCK, PAKc is required for chemotaxis.
To confirm that the mihck null and the mihck/pakc double knockout cells are developmentally competent, the expression of cAR1 and csA mRNAs was assayed in response to cAMP pulses. As shown in Figure 4, mihck null and mihck/pakc double knockout cells show the same kinetics and level of cAR1 and csA mRNA induction as wild-type and pakc null cells.
Because of the homology between the C-terminal extension of PAKc and the Gβγ, we attempted to determine whether a complex formed in vivo between PAKc and Gβγ. Such a complex was identified in the case of mammalian PAK1 (Li et al., 2003). Wild-type and gβ null cells (Lilly et al., 1993) were cotransformed with myc-PAKc and Gβ-YFP (Jin et al., 2000; Janetopoulos et al., 2001). Cells also were cotransformed with myc-PAKc and HA-tagged Gβ (Devreotes, personal communication). Stable transformants were then screened for the expression of both proteins (see Materials and Methods). To examine possible interaction between PAKc and Gβγ, the strains were pulsed with cAMP for 5 h, the same conditions used for the PAKc kinase and chemotaxis assays. Cells were lysed before addition of cAMP or 10 and 30 s after stimulation and then potential complexes were immunoprecipitated using an anti-myc, GFP, or HA antibody, as described in Materials and Methods. The immunoprecipitates were then analyzed by SDS-PAGE/Western blot for the presence of the other protein. In the assays in which HA- or YFP-Gβ was immunoprecipitated, no myc-PAKc was observed by Western blotting. Similarly, when myc-PAKc was immunoprecipitated, no HA- or YFP-Gβ was observed (our unpublished data). Because complexes might not be solubilized from the plasma membrane, we also did a triple cotransformation with myc-PAKc and Gβ-YFP and Gγ that carries a C→ S mutation in the prenylation site that would block lipid modification of Dictyostelium Gγ and thus its membrane localization (Zhang et al., 2001). Cells coexpressing all three proteins were identified (see Materials and Methods). There was no detectable coimmunoprecipitation between myc-PAKc and tagged Gβγ (our unpublished data).
Subcellular Localization of PAKc
Many signaling proteins that are involved in mediating chemotaxis exhibit a change in subcellular localization upon chemoattractant stimulation (Iijima et al., 2002; Devreotes and Janetopoulos, 2003; Merlot and Firtel, 2003). For example, the PH domain-containing proteins CRAC, Akt/PKB, and PhdA rapidly and transiently localize to the plasma membrane in response to chemoattractant stimulation in a phosphatidylinositol-3-kinase–dependent manner (Parent et al., 1998; Meili et al., 1999; Funamoto et al., 2001, 2002). The presence of a PH domain in PAKc suggests that PAKc may interact with membranes to properly regulate its activity. To examine PAKc's subcellular localization, we expressed PAKc tagged with either the myc epitope or GFP. Although GFP fusions make it easier to examine the dynamics of subcellular localization, we have found for some proteins that GFP fusions do not effectively complement the null mutation and some do not exhibit the same subcellular localization as either the endogenous protein or the small epitope-tagged version of the protein (Sobko et al., 2002). We found this also to be the case with GFP-PAKc (our unpublished observation) and therefore performed our studies by using myc-PAKc.
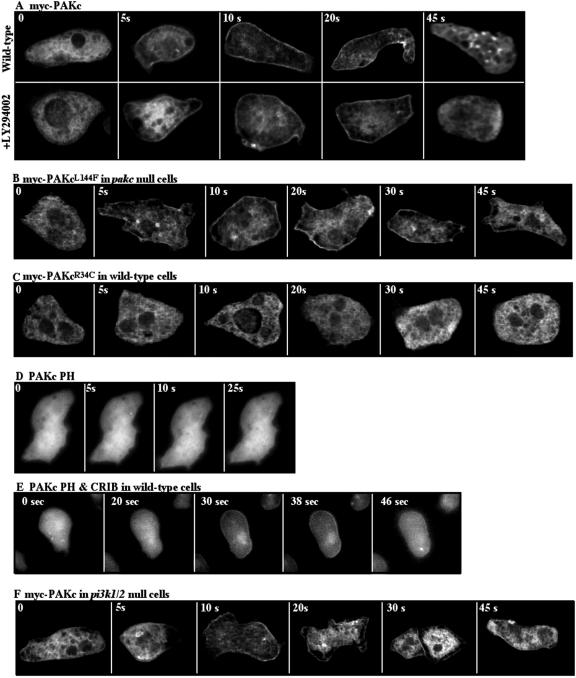
As illustrated in Figure 6A, myc-PAKc is cytosolic in unstimulated cells and localizes to the plasma membrane in response to a global chemoattractant stimulation (cells are rapidly bathed in a high concentration of chemoattractant so that all receptors are maximally occupied) with kinetics consistent with those of PAKc activation. PAKcL144F, which has a mutation in the putative autoinhibitory domain, also translocates to the cortex but remains associated with the cortex for an extended time in either wild-type or pakc null cells (Figure 6B; our unpublished data). This observation parallels our finding that the kinase activity of PAKcL144F remains elevated for an extended time compared with wild-type PAKc and suggests that the inactivation of PAKc kinase activity may control cortical delocalization of PAKc. PAKcR34C, which carries a point mutation in the PH domain, remains cytosolic in either wild-type or pakc null cells, indicating that a functional PH domain is required for PAKc cortical localization (Figure 6C; our unpublished data). Similarly, PAKcS466A,P470A, which contains amino acid substitutions in the putative Gβγ binding domain, does not localize to the cortex upon chemoattractant stimulation (our unpublished data). Neither of these mutant PAKc proteins exhibit chemoattractant-mediated kinase activity, providing a correlation between the function of these domains, membrane localization, and kinase activation. Many of the proteins that translocate to the plasma membrane in response to chemoattractant stimulation also are localized at the leading edge in chemotaxing cells. However, when chemotaxing cells were examined, we observed no enhancement of PAKc at the leading edge (our unpublished data).
Figure 6.
Subcellular localization of PAKc. Myc-tagged wild-type and mutant PAKc proteins (A–C) were expressed. The subcellular localization of PAKc before (0 s) or at various times after cAMP stimulation was examined by indirect immunofluorescence by using deconvolution microscopy (see Materials and Methods). (D and E) Subcellular localization of GFP-tagged PH domain alone and GFP-tagged PH-CRIB domains, respectively. (F) Subcellular localization of myc-PAKc in pi3k1/2 null cells.
We also examined the subcellular localization of the PH domain alone and the PH domain in association with the CRIB domain. The PH domain alone was cytosolic and did not exhibit any membrane or cortical localization in response to chemoattractant stimulation (Figure 6D). The PH domain plus the CRIB domain exhibited a weak membrane localization in response to chemoattractant stimulation (Figure 6E), the kinetics of which were significantly slower than those of the whole protein. The explanation for this is unknown, but it may involve the requirement of a higher level of accumulation of appropriate lipid products in the membrane for this domain alone. Because wild-type PAKc localizes to the cortex upon chemoattractant stimulation but PAKc carrying a mutation in either the PH or C-terminal domain does not localize, we suggest that cooperative interactions mediated by all three domains are required and that the PH domain, CRIB domain, and C-terminal extension all participate in PAKc's membrane localization.
Phosphatidylinositol Lipid Binding Profile of the PAKc PH Domain
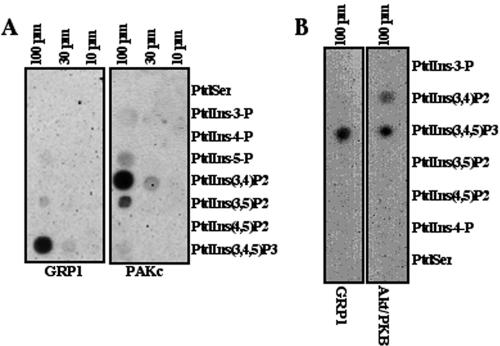
Because of the essential role of the PAKc PH domain in controlling PAKc subcellular localization and activation, we examined the potential lipid binding of this domain. A subset of PH domains bind phosphatidylinositols and are essential to localize proteins to membrane compartments (Dowler et al., 1999, 2000). To investigate whether the PAKc PH domain binds phosphatidylinositols, a GST fusion of the PAKc PH domain was expressed in E. coli, purified by affinity chromatography, and assayed for the ability to bind a panel of phospholipids by using a dot blot assay (Dowler et al., 2000). As illustrated in Figure 7A, the PAKc PH domain shows the strongest interaction with PI(3,4)P2, a product of class I PI3Ks, and weaker interaction with PI(3)P and PI(5)P. Unexpectedly, no interaction above the background level was observed with PI(3,4,5)P3. The PH domain of GRP1, which is specific for PI(3,4,5)P3 (Dowler et al., 2000), was used as a control for PI(3,4,5)P3 binding. For comparison, the PH domain of Dictyostelium Akt/PKB also was compared using an internal control of GRP1. As previously demonstrated for the PH domain of human Aktα (Dowler et al., 1999), the Dictyostelium Akt/PKB PH domain shows specificity for PI(3,4,5)P2 and PI(3,4)P2. The overall binding profile of the PAKc PH domain is significantly different from that of the PH domain of Dictyostelium or mammalian Akt/PKB, which has a preference for binding to PI(3,4,5)P3 and PI(3,4)P2 (Dowler et al., 1999).
Figure 7.
Binding of the PAKc and GRP1 PH domains (A) or Dictyostelium Akt/PKB and GRP1 PH domains (B) to different phospholipids on a lipid dot blot is shown. S, phosphatidyl serine; 3, PI(3)P; 4, PI(4)P; 5, PI(5)P; 3,4, PI(3,4)P2; 3,5, PI(3,5)P2; 4,5, PI(4,5)P2; and 3,4,5, PI(3,4,5)P3. See Materials and Methods for details.
Because of the PAKc exhibits preferential binding that is specific for PI(3,4)P2, we examined whether PAKc cortical translocation is dependent on the PI3K pathway. This was examined by pretreating wild-type cells for 10 min with 30 μM LY294002, which is sufficient to inhibit >95% of chemoattractant-mediated Akt/PKB activation (Meili et al., 1999, 2000) and examining myc-PAKc translocation in parallel with PAKc translocation in untreated wild-type cells. As can be seen in Figure 6A, LY294002 did not block PAKc translocation. In addition, we examined PAKc translocation in pi3k1/2 null cells. As expected from our observation that PAKc is activated normally in pi3k1/2 null cells, myc-PAKc also translocated to the cortex in this strain (Figure 6F), although the translocation may be slightly weaker than in wild-type cells.
DISCUSSION
Phenotype and Regulation of PAKc
We have identified a new member of the Dictyostelium PAK family of p21-activated protein kinases. PAKc contains a phosphatidylinositol lipid-binding PH domain, a CRIB or Rac family member GTPase binding domain, a conserved PAK-family kinase domain, and a C-terminal domain that is related to the Gβγ binding, C-terminal domain in the S. cerevisiae PAK STE20. PAKc is rapidly and transiently activated in response to chemoattractant stimulation and localizes to the plasma membrane with activity peaking at 10 s after stimulation. We demonstrate that functional PH, CRIB, and C-terminal domains, along with a functional kinase domain, are all required for chemoattractant-mediated PAKc activation. Our observation that mutations in the PH or C-terminal domain abrogate cortical translocation suggests that this translocation is required for PAKc activation. We also found that point mutations in the CRIB domain abrogate binding to RacB-GTP, which is expected to be associated with the plasma membrane. The inability of PAKc to localize to the cortex, the presumed site of Rac activation, would be expected to preclude PAKc's activation. Although we could not detect a complex, because of the conserved putative Gβγ-binding C-terminal domain, we believe it is possible that PAKc, like PAK1 (Li et al., 2003), also may interact with the Gβγ subunit and this interaction may be required for PAKc's activation downstream from chemoattractant receptors. Because we do not detect even a basal kinase activity with the PAKc carrying the mutation in the C-terminal domain, we cannot exclude the possibility that this domain is required intrinsically for PAKc kinase activity.
pakc null cells exhibit significant defects in cell polarization and are unable to properly inhibit the formation of lateral pseudopodia when placed in a chemoattractant gradient. In contrast to wild-type cells, pakc null cells produce pseudopod extensions and membrane retractions on all regions of the plasma membrane independent of the direction of the chemoattractant source. The extent of these protrusions and retractions seems to be high compared with wild-type cells as judged by the DIAS computer analysis, with the caveat that we are only able to look at the cell in two dimensions (we cannot examine extensions/retractions above our plane of focus). Thus, PAKc is required for inhibiting lateral pseudopodia and helping restrict pseudopod formation to the side of the plasma membrane facing the chemoattractant source. Interestingly, the speed of pakc null cell (centroid) movement is similar to that of wild-type cells and the cells are able to move up the chemoattractant gradient. Wild-type cells expressing PAKcS466A,P470A or PAKcR34C exhibit phenotypes similar to those of pakc null cells. We interpret this observation to suggest that the mutant PAKcs interact with and presumably interfere with other components of the signaling pathway, leading to a pakc null phenotype.
Our analysis of PAKc mutants suggests that, like mammalian PAK1, Dictyostelium PAKc has overlapping GTPase binding and autoinhibitory domains and we demonstrate that the Rac binding/autoinhibitory domains are important in controlling PAKc activity. Point mutations in two conserved histidine residues that abrogate binding of PAK1 to cdc42 and affect GTPase binding in S. pombe PAK lead to a PAKc that exhibits a higher basal level of kinase activity but cannot be activated upon chemoattractant stimulation (Sells et al., 1997, 1998; Tu and Wigler, 1999; Zenke et al., 1999; Bokoch, 2003). The PAKc CRIB domain preferentially binds the GTP-bound form of Dictyostelium RacB (Parks et al., 2004). Consistent with the results in mammalian PAK1 and S. pombe PAK (Sells et al., 1997, 1998), we find that mutations in two conserved histidines of PAKc, which abrogate binding to RacB-GTP, cause elevated basal activity that cannot be further activated in response to chemoattractant stimulation. Mutation of the conserved phenylalanine in the autoinhibitory domain results in a PAKc that has extremely high basal activity that can be further activated in response to chemoattractant stimulation. The increased, chemoattractant-mediated activity of this PAK remains elevated for significantly longer than wild-type PAK and remains associated with the plasma membrane for a longer period of time. We find that the transient membrane localization of PAKc in response to chemoattractant stimulation is dependent on the PH domain, as a point mutation in the domain abrogates membrane localization, but the PH domain is insufficient for PAKc localization. Constructs carrying the CRIB and PH domains or the PH domain alone do not effectively localize. Interestingly, the PH domain exhibits a strong preferential binding to PI(3,4)P2, a PI3K product. However, PAKc cortical localization occurs in pi3k1/2 null cells or cells treated with LY294002, suggesting that the PH domain may bind other phospholipids in vivo in the context of the whole protein or that PI(3,4)P2 may be produced via a pathway that does not depend on class I PI3Ks. In contrast to other signaling components that translocate to the plasma membrane in response to global stimulation, PAKc shows no observable cortical localization when cells are placed in a chemoattractant gradient. Although it is possible that PAKc does not localize under the conditions of a chemoattractant gradient, we suspect that the level of PAKc on the plasma membrane, whether uniformly localized or preferentially localized at the front or the back of cells, may be too low to be detected by our methods.
The pakc/mihck Double Null Strain Is Unable to Effectively Move in a Chemoattractant Gradient
We observe that cells expressing kinase dead PAKc (PAKcK234A) exhibit very strong chemotaxis defects, being unable to effectively move (significantly reduced speed). We hypothesized that PAKcK234A may have a dominant negative effect on the PAKc pathway by inhibiting the function of components of the pathway, possibly another PAK. Two other Dictyostelium PAKs have been identified and characterized. paka null cells, like pakc null cells, are defective in lateral pseudopod inhibition, although the cell morphologies of the two strains are different (Chung and Firtel, 1999). Furthermore, PAKa and PAKc seem to regulate distinct pathways; PAKa, but not PAKc, is required for myosin II assembly.
Myosin I proteins are regulated by phosphorylation and the PAK MIHCK was identified as a myosin I kinase and found to be activated by Rac-GTP (Lee et al., 1996, 1998; Gliksman et al., 2001; Falk et al., 2003). Published studies suggested that mihck null cells did not exhibit overt developmental phenotypes (de la Roche and Cote, 2001); our examination of a mihck null strain demonstrates that it exhibits chemotaxis defects similar to those of pakc null cells, although not as severe. A double knockout strain (pakc/mihck null cells) has a severe chemotaxis defect similar to that of PAKcK234A-expressing cells, exhibiting a very reduced speed. These results suggest the possibility that PAKc and MIHCK provide partially redundant functions in a common pathway. However, there are other explanations, including the possibility that mutations on different pathways could synergize to produce a more severe phenotype than that of either of the two mutant strains. A single knockout of the Dictyostelium myosin I MyoIF leads to phenotypes that include the formation of multiple pseudopodia and reduced cell polarity, and a double knockout of myoIF and myoIB exhibits a severe loss of speed, similar to the phenotype of pakc/mihck null cells. However, we do not know whether the similarities of the MyoIB/F and PAKc/MIHCK phenotypes result from PAKc/MIHCK controlling myosin I function or if they are coincidental. We attempted to assay activated PAKc for activity on myosin I, but we were unable to demonstrate phosphorylation (S. Lee and R. Firtel, unpublished observation), and we did not have an appropriate positive control.
Acknowledgments
We are indebted to P. Devreotes for the expression vectors for Gβ-YFP and Gγ carrying a mutant in the prenylation site. We thank J. Feramisco and the University of California, San Diego, Cancer Center Imaging Core for assistance in the deconvolution microscopy. We thank S. Taylor at University of California, San Diego, for the PKA catalytic subunit. This work was supported by grants from the Deutsche Forschungsgemeinschaft (RI 1034/2) and the Köln Fortune program to F.R. and U.S. Public Health Service grants to R.A.F.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–04–0323. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–04–0323.
References
- Bokoch, G.M. (2000). Regulation of cell function by Rho family GTPases. Immunol. Res. 21, 139-148. [DOI] [PubMed] [Google Scholar]
- Bokoch, G.M. (2003). Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743-781. [DOI] [PubMed] [Google Scholar]
- Brown, J.L., Stowers, L., Baer, M., Trejo, J., Coughlin, S., and Chant, J. (1996). Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr. Biol. 6, 598-605. [DOI] [PubMed] [Google Scholar]
- Chung, C.Y., and Firtel, R.A. (1999). PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 147, 559-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C.Y., Lee, S., Briscoe, C., Ellsworth, C., and Firtel, R.A. (2000). Role of Rac in controlling the actin cytoskeleton and chemotaxis in motile cells. Proc. Natl. Acad. Sci. USA 97, 5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C.Y., Potikyan, G., and Firtel, R.A. (2001). Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7, 937-947. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., Bresnick, A., Demma, M., Dharmawardhane, S., Eddy, R., Hall, A.L., Sauterer, R., and Warren, V. (1990). Mechanisms of amoeboid chemotaxis: an evaluation of the cortical expansion model. Dev. Genet. 11, 333-340. [DOI] [PubMed] [Google Scholar]
- Daniels, R.H., Zenke, F.T., and Bokoch, G.M. (1999). alphaPix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J. Biol. Chem. 274, 6047-6050. [DOI] [PubMed] [Google Scholar]
- de la Roche, M.A., and Cote, G.P. (2001). Regulation of Dictyostelium myosin I and II. Biochim. Biophys. Acta 1525, 245-261. [DOI] [PubMed] [Google Scholar]
- Devreotes, P., and Janetopoulos, C. (2003). Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 278, 20445-20448. [DOI] [PubMed] [Google Scholar]
- Dharmawardhane, S., Brownson, D., Lennartz, M., and Bokoch, G.M. (1999). Localization of p21-activated kinase 1 (PAK1) to pseudopodia, membrane ruffles, and phagocytic cups in activated human neutrophils. J. Leukoc. Biol. 66, 521-527. [DOI] [PubMed] [Google Scholar]
- Dowler, S., Currie, R.A., Downes, C.P., and Alessi, D.R. (1999). DAPP 1, a dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem. J. 342, 7-12. [PMC free article] [PubMed] [Google Scholar]
- Dowler, S., Currie, R.A., Campbell, D.G., Deak, M., Kular, G., Downes, C.P., and Alessi, D.R. (2000). Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eby, J.J., Holly, S.P., van Drogen, F., Grishin, A.V., Peter, M., Drubin, D.G., and Blumer, K.J. (1998). Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr. Biol. 8, 967-970. [DOI] [PubMed] [Google Scholar]
- Edwards, D.C., Sanders, L.C., Bokoch, G.M., and Gill, G.N. (1999). Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253-259. [DOI] [PubMed] [Google Scholar]
- Falk, D.L., Wessels, D., Jenkins, L., Pham, T., Kuhl, S., Titus, M.A., and Soll, D.R. (2003). Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J. Cell Sci. 116, 3985-3999. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Kojima, T., Kabayama, H., and Mikoshiba, K. (1996). Mutation of the pleckstrin homology domain of Bruton's tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J. Biol. Chem. 271, 30303-30306. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., Meili, R., Lee, S., Parry, L., and Firtel, R.A. (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611-623. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., Milan, K., Meili, R., and Firtel, R. (2001). Role of phosphatidylinositol 3′ kinase and a downstream plekstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153, 795-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliksman, N.R., Santoyo, G., Novak, K.D., and Titus, M.A. (2001). Myosin I phosphorylation is increased by chemotactic stimulation. J. Biol. Chem. 276, 5235-5239. [DOI] [PubMed] [Google Scholar]
- Iijima, M., Huang, Y.E., and Devreotes, P. (2002). Temporal and spatial regulation of chemotaxis. Dev. Cell 3, 469-478. [DOI] [PubMed] [Google Scholar]
- Insall, R.H., Soede, R.D.M., Schaap, P., and Devreotes, P.N. (1994). Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol. Biol. Cell 5, 703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffer, Z.M., and Chernoff, J. (2002). p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell. Biol. 34, 713-717. [DOI] [PubMed] [Google Scholar]
- Janetopoulos, C., Jin, T., and Devreotes, P. (2001). Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science 291, 2408-2411. [DOI] [PubMed] [Google Scholar]
- Jin, T., Zhang, N., Long, Y., Parent, C.A., and Devreotes, P.N. (2000). Localization of the G protein betagamma complex in living cells during chemotaxis. Science 287, 1034-1036. [DOI] [PubMed] [Google Scholar]
- Lee, S., Escalante, R., and Firtel, R.A. (1997). A Ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium. Development 124, 983-996. [DOI] [PubMed] [Google Scholar]
- Lee, S.F., Egelhoff, T.T., Mahasneh, A., and Cote, G.P. (1996). Cloning and characterization of a Dictyostelium myosin I heavy chain kinase activated by Cdc42 and Rac. J. Biol. Chem. 271, 27044-27048. [DOI] [PubMed] [Google Scholar]
- Lee, S.F., Mahasneh, A., de la Roche, M., and Cote, G.P. (1998). Regulation of the p21-activated kinase-related Dictyostelium myosin I heavy chain kinase by autophosphorylation, acidic phospholipids, and Ca2+-calmodulin. J. Biol. Chem. 273, 27911-27917. [DOI] [PubMed] [Google Scholar]
- Leeuw, T., Wu, C., Schrag, J.D., Whiteway, M., Thomas, D.Y., and Leberer, E. (1998). Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391, 191-195. [DOI] [PubMed] [Google Scholar]
- Lei, M., Lu, W., Meng, W., Parrini, M.C., Eck, M.J., Mayer, B.J., and Harrison, S.C. (2000). Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102, 387-397. [DOI] [PubMed] [Google Scholar]
- Li, T., Tsukada, S., Satterthwaite, A., Havlik, M.H., Park, H., Takatsu, K., and Witte, O.N. (1995). Activation of Bruton's tyrosine kinase (BTK) by a point mutation in its pleckstrin homology (PH) domain. Immunity 2, 451-460. [DOI] [PubMed] [Google Scholar]
- Li, Z., Hannigan, M., Mo, Z., Liu, B., Lu, W., Wu, Y., Smrcka, A.V., Wu, G., Li, L., Liu, M., Huang, C.K., and Wu, D. (2003). Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell 114, 215-227. [DOI] [PubMed] [Google Scholar]
- Lilly, P., Wu, L.J., Welker, D.L., and Devreotes, P.N. (1993). A G-protein beta-subunit is essential for Dictyostelium development. Genes Dev. 7, 986-995. [DOI] [PubMed] [Google Scholar]
- Manser, E., Chong, C., Zhao, Z.S., Leung, T., Michael, G., Hall, C., and Lim, L. (1995). Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J. Biol. Chem. 270, 25070-25078. [DOI] [PubMed] [Google Scholar]
- Meili, R., Ellsworth, C., and Firtel, R.A. (2000). A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol. 10, 708-717. [DOI] [PubMed] [Google Scholar]
- Meili, R., Ellsworth, C., Lee, S., Reddy, T.B., Ma, H., and Firtel, R.A. (1999). Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 18, 2092-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., and Firtel, R.A. (2003). Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 116, 3471-3478. [DOI] [PubMed] [Google Scholar]
- Parent, C.A., Blacklock, B.J., Froehlich, W.M., Murphy, D.B., and Devreotes, P.N. (1998). G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95, 81-91. [DOI] [PubMed] [Google Scholar]
- Park, K.C., Rivero, F., Meili, R., Lee, S., Apone, F., and Firtel, R.A. (2004). Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Salim, K., et al. (1996). Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 15, 6241-6250. [PMC free article] [PubMed] [Google Scholar]
- Schnitzler, G.R., Fischer, W.H., and Firtel, R.A. (1994). Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Dev. 8, 502-514. [DOI] [PubMed] [Google Scholar]
- Sells, M.A., Barratt, J.T., Caviston, J., Ottilie, S., Leberer, E., and Chernoff, J. (1998). Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J. Biol. Chem. 273, 18490-18498. [DOI] [PubMed] [Google Scholar]
- Sells, M.A., Knaus, U.G., Bagrodia, S., Ambrose, D.M., Bokoch, G.M., and Chernoff, J. (1997). Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7, 202-210. [DOI] [PubMed] [Google Scholar]
- Sobko, A., Ma, H., and Firtel, R.A. (2002). Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2, 745-756. [DOI] [PubMed] [Google Scholar]
- Soll, D.R., and Voss, E. (1998). Two- and three-dimensional computer systems for analyzing how animal cells crawl. In: Motion Analysis of Living Cells, eds. D.R. Soll and D. Wessels, New York: Wiley-Liss, 25-52.
- Sun, B., and Firtel, R.A. (2003). A regulator of G protein signaling-containing kinase is important for chemotaxis and multicellular development in Dictyostelium. Mol. Biol. Cell 14, 1727-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh, K. (1993). A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid 30, 150-154. [DOI] [PubMed] [Google Scholar]
- Tu, H., and Wigler, M. (1999). Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19, 602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, F.N., van Delft, S., Kain, H.E., van der Kammen, R.A., and Collard, J.G. (1999). Rac regulates phosphorylation of the myosin-II heavy chain, actinomyosin disassembly and cell spreading. Nat. Cell Biol. 1, 242-248. [DOI] [PubMed] [Google Scholar]
- Wessels, D., Titus, M., and Soll, D.R. (1996). A Dictyostelium myosin I plays a crucial role in regulating the frequency of pseudopods formed on the substratum. Cell Motil. Cytoskeleton 33, 64-79. [DOI] [PubMed] [Google Scholar]
- Wessels, D., Voss, E., Von Bergen, N., Burns, R., Stites, J., and Soll, D.R. (1998). A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskeleton 41, 225-246. [DOI] [PubMed] [Google Scholar]
- Zhang, N., Long, Y., and Devreotes, P.N. (2001). Ggamma in Dictyostelium: its role in localization of Gbetagamma to the membrane is required for chemotaxis in shallow gradients. Mol. Biol. Cell 12, 3204-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke, F.T., King, C.C., Bohl, B.P., and Bokoch, G.M. (1999). Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J. Biol. Chem. 274, 32565-32573. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.S., Manser, E., Chen, X.Q., Chong, C., Leung, T., and Lim, L. (1998). A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18, 2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G.L., Zhuo, Y., King, C.C., Fryer, B.H., Bokoch, G.M., and Field, J. (2003). Akt phosphorylation of serine 21 on Pak1 modulates Nck binding and cell migration. Mol. Cell. Biol. 23, 8058-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]