Abstract
Ligand binding to cell surface receptors initiates both signal transduction and endocytosis. Although signaling may continue within the endocytic compartment, down-regulation is the major mechanism that controls the concentration of cell surface receptors, their ability to receive environmental signals, and the ultimate strength of biological signaling. Internalization, recycling, and trafficking of receptor tyrosine kinases (RTKs) within the endosome compartment are each regulated to control the overall process of down-regulation. We have identified the Na+/H+ exchanger regulatory factor (NHERF) as an important molecular component that stabilizes epidermal growth factor receptors (EGFRs) at the cell surface to restrict receptor down-regulation. The NH2-terminal PDZ domain (PDZ 1) of NHERF specifically binds to an internal peptide motif located within the COOH-terminal regulatory domain of EGFR. Expression of NHERF slows the rate of EGF-induced receptor degradation. A point mutation that abolishes the PDZ 1 recognition sequence of EGFR enhances the rate of ligand-induced endocytosis and down-regulation of EGFR. Similarly, expression of a dominant negative mutant of NHERF enhances EGF-induced receptor down-regulation. In contrast to β-adrenergic receptors where NHERF enhances recycling of internalized receptors, NHERF stabilizes EGFR at the cell surface and slows the rate of endocytosis without affecting recycling. Although the mechanisms differ, for both RTKs and G protein-coupled receptors, the overall effect of NHERF is to enhance the fraction of receptors present at the cell surface.
INTRODUCTION
Ligand-induced down-regulation of receptor tyrosine kinases (RTKs) is the major cellular mechanism that controls the concentration of cell surface receptors and thus biological responses to extracellular ligands (Sorkin and von Zastrow, 2002). Mechanisms providing for down-regulation can be overcome when RTKs such as the epidermal growth factor receptor (EGFR) and erbB2 are amplified and overexpressed, contributing to excessive tumor cell growth (Yarden and Sliwkowski, 2001). The known general features of EGFR down-regulation include 1) a ligand-induced increase in the rate of endocytosis of ∼10-fold (Wiley et al., 1991). This is the rate-limiting step in the process of down-regulation and most endocytosis occurs via clathrin-coated pits (Carpenter et al., 1982). Sequences located in the regulatory COOH terminus of EGFR are essential for this process (Chang et al., 1993). 2) Migration through progressively acidified endosomes where pH-sensitive dissociation of ligand from receptor occurs (French et al., 1995). 3) Transit of endosomes containing EGFR to the vicinity of the endoplasmic reticulum where the tyrosine phosphatase PTP1B resides (Haj et al., 2002). Dephosphorylation of EGFR by PTP1B releases SH2 and PTB domain signaling proteins associated with activated, self-phosphorylated EGFR so these can be reused. 4) Trafficking to the lysosome where receptor and ligand are degraded (Futter et al., 1996).
Short peptide motifs (sequence codes) within the cytoplasmic domains of RTKs are necessary for both endocytosis and trafficking to lysosomes. The COOH terminus of EGFR contains aYXXθ endocytic motif that binds the μ subunit of adaptor protein 2 and NPXY type endocytic motifs whose molecular target is not clearly defined (Sorkin and Carpenter, 1993; Nesterov et al., 1995). Lysosomal targeting sequences (YLVI) are also present (Opresko et al., 1995; Kornilova et al., 1996; Jones et al., 2002). Additionally, monoubiquitination catalyzed by cbl for EGFR provides cargo information that is recognized by the cellular sorting machinery (Levkowitz et al., 1998; Mosesson et al., 2003).
Additional mechanisms are important in controlling the overall process of receptor down-regulation. In the present studies, we identify the Na+/H+ exchanger regulatory factor (NHERF) as an important molecular component that stabilizes EGFR at the cell surface and retards receptor down-regulation. The NH2-terminal PDZ domain (PDZ 1) of NHERF specifically binds to an internal peptide motif within the 164-aa COOH-terminal regulatory domain of EGFR. A point mutation within this motif that abolishes PDZ 1 binding enhances the rate of ligand-induced EGFR down-regulation. A mutant form of NHERF that does not bind EGFR also enhances EGFR down-regulation, supporting the conclusion that NHERF functions to retain EGFR at the cell surface. NHERF, which is important for ligand-induced trafficking of β2-adrenergic receptors (Cao et al., 1999), is also an important regulator of ligand-induced EGFR down-regulation.
MATERIALS AND METHODS
Reagents
A polyclonal anti-hEGFR antiserum was made by immunizing rabbits with a KLH-coupled peptide corresponding to the extreme COOH terminus of hEGFR, amino acids SEFIGA (Zhong et al., 2002). The SEFIGA antibody was purified by absorption to an immobilized peptide affinity column. The 528 mouse monoclonal anti-EGFR was used as described previously (Gill et al., 1984). 125I-EGF was obtained from PerkinElmer Life and Analytical Sciences (Boston, MA). The EGFR tyrosine kinase inhibitor tyrphostin 1478 was obtained from Sigma-Aldrich (St. Louis, MO). Monoclonal mouse anti-FLAG IgG H+L, anti-phosphotyrosine antibody PY99 and anti-phospho-extracellular signal-regulated kinase (ERK) antibody E4 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and Texas Red EGF was from Molecular Probes (Eugene, OR). Goat anti-NHERF was obtained from Santa Cruz Biotechnology.
Isolation of NHERF
The COOH terminus of hEGFR, residues 1022–1186, was cloned in frame as an EcoRI/BamHI fragment into pGEX-2TK (Pharmacia, Peapack, NY). GST-TK-EGFR 1022–1186 was expressed in BL21 DE3pLysS cells and purified on glutathione-agarose. The probe was labeled at the protein-kinase A phosphorylation site by using [γ32P]ATP and the purified catalytic subunit of A-kinase provided by Dr. Susan Taylor. A protein–protein interaction overlay screen of an e16 mouse embryo λ Ex Lox expression library (Novagen, San Diego, CA) was performed as described previously (Jurata et al., 1996). Positive clones were plaque purified, subcloned from the phage, and inserts were identified by DNA sequencing.
Mutations and Expression Vectors
Mutations in hEGFR (L1043F and L1063F) were introduced using QuikChange (QIAGEN, Valencia, CA) according to the manufacturer's instructions. The same mutations were introduced into GST-EGFR 1022–1186. The consensus sequence GYGF was mutated to GYAA in the PDZ 1 (G25A/F26A) and PDZ 2 (G163A/F164A) domains of NHERF also by using the QuikChange procedure. All mutations were verified by DNA sequencing. Primer sequences are available upon request.
Wild-type (WT) and mutant EGFR were cloned into the PX vector for preparation of stable B82 cell lines (Lin et al., 1986) and into the pCEP4 vector (Invitrogen, Carlsbad, CA) for expression in human embryonic kidney (HEK) 293 cells. WT and L1043F EGFR cell lines were matched for EGFR number as assessed by Western blotting. NHERF was cloned into a modified pcDNA vector that contains an NH2-terminal luciferase leader and FLAG epitope tag (Jurata and Gill, 1997).
Glutathione S-Transferase (GST) Pulldown Assays
NHERF PDZ 1 (aa 12–97) and PDZ 2 (aa 151–236) were cloned in frame into pGEX-5 × -3 as BamHI/XhoI fragments by using polymerase chain reaction (PCR). GST-PDZ domains and GST-EGFR 1022–1186 (WT and mutant) were expressed in BL21 cells and purified on glutathione-agarose affinity columns. FLAG-tagged NHERF or EGFR were prepared by transfecting HEK 293 cells with the expression plasmids by using the Effectene Reagent (QIAGEN). Cells were lysed in buffer containing 25 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1% Triton X-100, 200 μM leupeptin, 400 mM benzamidine, 2 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride; insoluble material was removed by centrifugation. The supernatant was incubated with 20 μg of immobilized GST-fusion proteins at 4°C for 3 h, washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween 20, boiled in Laemmli sample buffer, and separated on 4–12% bis-Tris PAGE (NuPage; Invitrogen). Proteins were transferred to nitrocellulose membranes overnight; blots were blocked with PBS/0.1% Tween 20/2% bovine serum albumin, and proteins were detected using anti-FLAG or SEFIGA antibodies. Blots were developed using enhanced chemiluminescence (ECL) and scanned with an LKB Ultrascan XL enhanced laser densitometer.
Preparation of Stable Expressor B82 Cell Lines
Mouse B82 L cells that do not express endogenous EGFR were transfected with WT or L1043F EGFR in the PX vector that contains a mutant dihydrofolate reductase gene (Chen et al., 1987) and with WT, mutant PDZ 1, or mutant PDZ 2 NHERF in pcDNA3 FLAG that carries a neoR gene. Clonal lines were selected using 1 μM methotrexate and 500 μg/ml G418. Expression in clonal lines was verified using Western blotting.
Measurement of Internalization and Recycling Rates
The specific internalization rates of WT, L1043F EGFR, and WT EGFR in cells expressing NHERF mutant in its PDZ 1 domain were determined by measuring the initial rates of endocytosis of 125I-EGF (Chang et al., 1993). Data are plotted as described by Wiley and Cunningham (1982)), and endocytic rate constants were calculated from the slope of the line. κe defines the probability of a ligand-occupied receptor being internalized in 1 min at 37°C under initial rate conditions. The rate of recycling (κx) of internalized 125I-EGF was measured using a modification of the method described by French et al. (1995). B82 cells expressing WT hEGFR, L1043F mutant EGFR, and WT EGFR with WT NHERF or PDZ 1 mutant NHERF were characterized using this steadystate sorting assay that measures the fraction of 125I EGF ligand sorted to recycling versus degradation as a function of the number of intracellular ligand molecules. B82 cells were incubated for 2–3 h at 37°C with the following concentrations of 125I-EGF: 0.1, 0.3, 1.0, 3.0, 10.0, and 30.0 ng/ml. Cells were then washed with room temperature PBS and incubated on ice for 2 min with a mild acid strip containing no urea. The acid strip was aspirated to remove any surface bound ligand, and the cells were washed two times with room temperature PBS. Cells were then returned to 37°C with excess unlabeled EGF (1 μg/ml). After incubating for 10 min, media were collected and centrifuged at 10,000 rpm to separate recycled and degraded 125I-EGF by using a centrifugal filter unit with a 5000 molecular weight cut-off (Millipore, Billerica, MA). Cells were then places back on ice, washed five times with ice-cold WHIPS buffer (Wiley et al., 1991), and incubated for 8 min with 2 M urea acid strip. The acid strip was removed from the cells, and radioactivity was counted to determine the amount of surface bound ligand. Cells were solubilized in 1 N NaOH, and the radioactivity was used to determine the amount of intracellular ligand. From the media samples, the radioactive counts were used to calculate the values of recycled and degraded ligand at each concentration.
Immunofluoresence Microscopy
COS 7 cells were transfected with pcDNA-FLAG-NHERF by using Effectine, grown on coverslips for 16 h, placed on ice, rinsed, and incubated with 500 ng/ml Texas Red-labeled epidermal growth factor (EGF) for 60 min. Medium was removed and cells were washed to remove unbound EGF. Cells were transferred to 37°C for the indicated times and then fixed with 2% paraformaldehyde/PBS. Cells were blocked with 2.5% fetal bovine serum and permeabilized with 0.1% Triton X-100. Cells were stained with anti-FLAG antibody (1:5000), washed and incubated with goat anti-mouse IgG H+L chains conjugated to Alexa Fluor 488. Omission of primary antibody was used as a negative control. Cells were viewed using a 63×/1.4 numerical aperture Zeiss oil immersion objective on a Zeiss Axioskop fluorescence microscope equipped with a 640 × 480 pixel COHU Interline Transfer charge-coupled device camera (Coher, San Diego, CA).
RESULTS
Interaction of NHERF with Sequences within the COOH Terminus of EGFR
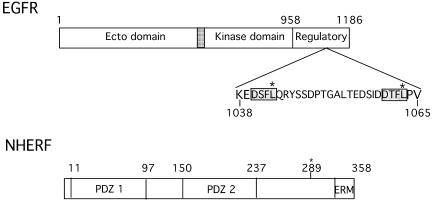
The COOH terminus of EGFR distal to the tyrosine kinase core (residues 958-1186) is a regulatory domain that contains both the principal sites of autophosphorylation and sequence codes necessary for trafficking within the endocytic system (Figure 1). To identify proteins that interact with regulatory sequences in this region, we screened an e16 mouse embryo expression library with [32P]GST EGFR 1022–1186. From a screen of 1.2 × 106 colonies, we isolated four clones, all of which corresponded to full-length NHERF. NHERF, which was initially identified as a cofactor for cAMP-mediated inhibition of the renal Na+/H+ exchanger isoform 3 (NHE3) (Weinman et al., 1993; Yun et al., 1997), has subsequently been implicated in a number of processes, including trafficking of G protein-coupled receptors (Cao et al., 1999), function of the cystic fibrosis transmembrane conductance regulator (CFTR) (Hall et al., 1998a) and signaling via the parathyroid hormone and platelet-derived growth factor (PDGF) receptors (Maudsley et al., 2000; Voltz et al., 2001; Mahon et al., 2002). As shown in Figure 1, NHERF contains two PDZ domains and a COOH-terminal region that binds Ezrin, Radixin, Merlin, and Moesin (ERM domain) (Bretscher et al., 2000). The EGFR-interacting protein isolated corresponds to NHERF 1; a second closely related isoform, NHERF 2 (E3KARP), exists (Yun et al., 1997).
Figure 1.
Schematic representation of EGFR and NHERF. Domain boundaries of EGFR and NHERF are shown. Potential PDZ domain recognition sites within the COOH terminus of EGFR (residues 1022–1186) are shaded and boxed. Sites of mutation are marked (*). The identified serine phosphorylation site at residue 289 in NHERF is marked (*).
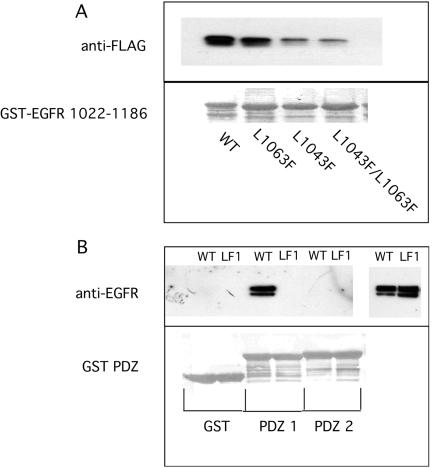
Erb B2 and erb B4 contain COOH-terminal PDZ domain binding motifs, whereas EGFR (erb B1) and erb B3 do not. The COOH terminus of Erb B2 binds both the PDZ domains of erbin and of Lin 7, and these interactions have been implicated in the membrane distribution and surface retention of erb B2 (Borg et al., 2000; Huang et al., 2001; Shelly et al., 2003). The COOH terminus of EGFR does not bind the PDZ domains of either erbin or Lin 7. Because EGFR lacks a COOH-terminal PDZ binding motif, we sought an internal sequence corresponding to those present at the COOH terminus of well documented NHERF targets such as the β2-adrenergic receptor, the purinergic receptor P2Y1, and CFTR (amino acids DSLL, DTSL, and DTRL, respectively) (Hall et al., 1998a,b; Cao et al., 1999). Two related sequences in EGFR 1022–1186 were identified: DSFL and DTFL (Figure 1). Point mutations were made in each of these two sequences, singly or together, and interactions with NHERF were investigated using GST pulldown assays. As shown in Figure 2A, FLAG-epitope–tagged NHERF interacts strongly with GST-EGFR 1022–1186. Mutation of site 1 (L1043F) markedly reduced the interaction (87% decrease), whereas mutation of site 2 (L1063F) had lesser effects (46% reduction). Mutation of both sites (L1043F/L1063F) largely abolished NHERF binding (90% reduction). These results identify the DSFL sequence at residues 1040–1043 of EGFR as the principal site where NHERF binds.
Figure 2.
Identification of the NHERF binding site in the COOH terminus of EGFR. (A) FLAG-tagged NHERF was expressed in HEK 293 cells, and equal aliquots of cell extract were incubated with GST-EGFR 1022–1186 without or with the indicated point mutations. After washing, bound NHERF was identified by Western blotting using anti-FLAG antibody. Binding was quantitated by densitometry. The lower panel shows Poinceau S staining of the indicated GST-fusion proteins used. (B) WT or L1043F (LF1) EGFR were expressed in HEK 293 cells, and equal aliquots of cell extract were incubated with GST-PDZ 1, GST-PDZ 2 of NHERF, or GST. After washing, bound EGFR was identified by Western blotting by using the SE-FIGA antibody. Five percent load is shown on the right. The lower panel shows Ponceau S staining of the GST-fusion proteins used.
Specificity of the NHERF PDZ 1 Domain for the COOH Terminus of EGFR
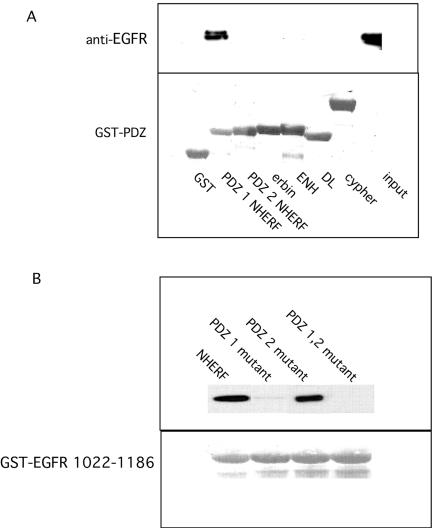
To identify the PDZ domain of NHERF that recognized the site in the COOH terminus of EGFR, PDZ 1 and PDZ 2 domains of NHERF were cloned into pGEX-5 × -3, and the resulting GST fusion proteins were incubated with HEK 293 extracts from cells expressing EGFR. As shown in Figure 3A, PDZ 1 of NHERF specifically recognized EGFR. EGFR was not recognized by PDZ 2 of NHERF nor by PDZ domains of erbin (Borg et al., 2000), cypher (Zhou et al., 1999), enigma homolog (Kuroda et al., 1996), or disks large (Woods and Bryant, 1989).
Figure 3.
Identification of NHERF PDZ 1 as the domain that recognizes EGFR. (A) GST-fusions with the indicated PDZ domains were incubated with extracts from HEK 293 cells expressing EGFR. After washing extensively, bound EGFR was detected by Western blotting by using the SEFIGA anti-EGFR antibody. ENH, enigma homolog; DL, disks large. The lower panel shows Poinceau S staining of the GST-fusion proteins used. (B) FLAG-tagged NHERFs without or with the indicated PDZ domain mutations were expressed in HEK 293 cells and incubated with GST-EGFR 1022–1186. Bound NHERF was identified by Western blotting by using anti-FLAG antibody. The lower panel shows Ponceau S staining of the GST-fusion proteins used.
The specificity of PDZ 1 for recognition of EGFR was confirmed by analysis of the interaction of WT and mutant G25A/F26A PDZ 1 or mutant G163A/F164A PDZ 2 or NHERF containing both mutations with GST-EGFR1022–1186. As shown in Figure 3B, WT and the PDZ 2 NHERF mutant (G163A/F164A) bound to GST-EGFR1022–1186. However, no binding was observed with NHERF that contained a mutation in the PDZ 1 domain (G25A/F26A NHERF) or with NHERF that contained mutations in both PDZ domains.
The specificity of PDZ 1 of NHERF for binding to the COOH terminus of EGFR was assessed by creating the same point mutation studied in GST-EGFR 1022–1186 in holo EGFR. WT EGFR or L1043F EGFR was expressed in HEK 293 cells, and solubilized receptors were incubated with GST-PDZ 1, GST-PDZ 2 of NHERF, or GST alone. WT EGFR specifically bound to GST-PDZ 1 but did not bind to GST-PDZ 2 or to GST alone (Figure 2B). The L1043F mutant EGFR failed to bind to GST-PDZ 1, confirming that PDZ 1 of NHERF specifically recognizes the DSFL1043 sequence in EGFR.
Independence of NHERF Interaction from the Activation State of EGFR
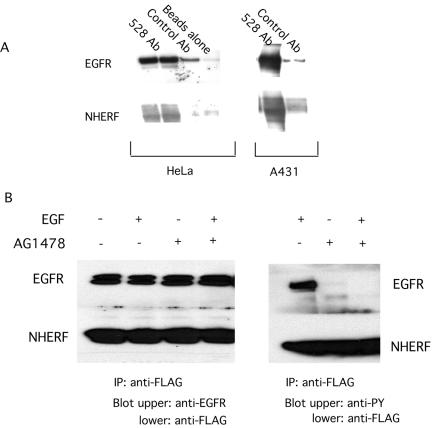
To confirm the interaction between EGFR and NHERF, we immunoprecipitated endogenous EGFR from HeLa and A431 cells and probed for endogenous NHERF. As shown in Figure 4A, NHERF coimmunoprecipitates with EGFR, indicating that endogenous EGFR and NHERF associate in vivo.
Figure 4.
Interaction of EGFR with NHERF in the absence or presence of the tyrosine kinase inhibitor tyrphostin 1478. (A) Coimmunoprecipitation of endogenous EGFR and NHERF. HeLa and A431 cells were lysed and EGFRs were immunoprecipitated using the mouse monoclonal antibody 528. Immunoprecipitates were dissolved and analyzed by Western blotting by using the anti-EGFR antibody SEFIGA and the anti-NHERF antibody at 1:1000 dilution. Only minor amounts of EGFR and NHERF were detected using control IgG precipitates. (B) Direct interaction of NHERF with unmodified EGFR sequences. EGFR and FLAG-tagged NHERF were expressed in HEK 293 cell. Cells were treated with 0.5 μM tyrphostin 1478 for 2 min. EGF (100 ng/ml) was then added where indicated for 5 min. Cells were solubilized in the lysis buffer containing 1% Triton X-100 used for GST pulldown assays and immunoprecipitated with anti-FLAG antibody. Western blotting was carried out using SEFIGA anti-EGFR or anti-phosphotyrosine antibodies. The presence of NHERF in immunoprecipitates was confirmed by separately analyzing the lower half of the gel by using anti-FLAG antibodies. Left, equal amounts of EGFR in immune complexes in cells treated without or with EGF and tyrphostin 1478. Right, confirms that tyrphostin 1478 inhibited autophosphorylation of EGFR. No proteins were detected using control IgG as an immunoprecipitant (our unpublished data).
NHERF was initially identified based on interaction with a nonphosphorylated COOH-terminal fragment of EGFR, suggesting the interaction was independent of the ligand-activated, autophosphorylated state of EGFR. Erbin preferentially interacts with nonactivated erb B2 (Borg et al., 2000), whereas NHERF preferentially interacts with ligand bound but not kinase-active platelet-derived growth factor receptor (PDGFR) (Demoulin et al., 2003). To investigate possible effects of EGFR activation on interactions with NHERF, we used immunoprecipitation of proteins that were coexpressed in HEK 293 cells. As shown in Figure 4B, the interaction of NHERF with EGFR was independent of the addition of EGF. To evaluate a possible role of autophosphorylation, the EGFR tyrosine kinase inhibitor tyrphostin 1478 was added to HEK 293 cells before the addition of EGF. Although the inhibitor effectively blocked EGF-induced receptor autophosphorylation, it did not affect interactions of NHERF with EGFR. The interaction of the two proteins thus seems independent of ligand binding and autophosphorylation of EGFR.
Effect of Expression of NHERF on Down-Regulation of EGFR
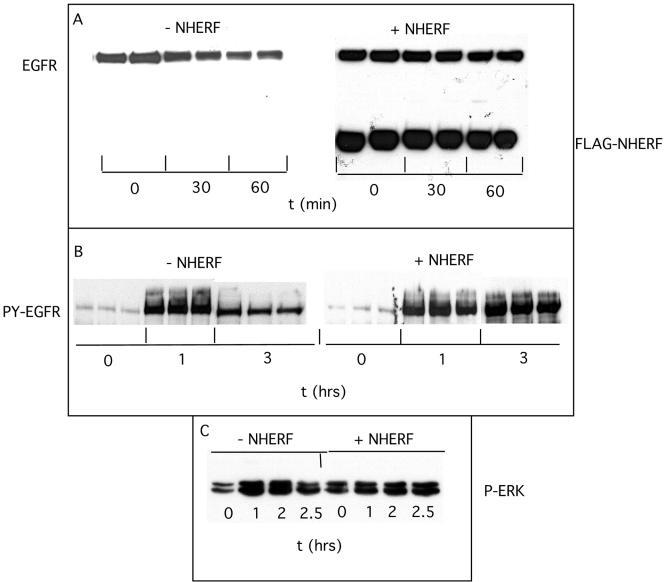
To investigate the effects of NHERF on EGFR, FLAG-tagged NHERF was transfected into HeLa cells, and the time course of ligand-induced endogenous EGFR down-regulation was measured. As shown in Figure 5A, NHERF reduced the rate of EGF-induced EGFR down-regulation as measured by loss of EGFR mass. This effect was confirmed in multiple experiments indicating that NHERF prolonged the half-life of ligand-activated EGFR in HeLa cells approximately twofold (t1/2-NHERF 55 min vs. t1/2 + NHERF 140 min). NHERF did not affect the levels of EGFR in the absence of EGF (our unpublished data). Loss of EGFR mass approximates kdeg because the rate of new receptor synthesis compared with receptor turnover is <10% (Lauffen-burger, 1993). Western blotting with an anti-phosphotyrosine antibody indicated that NHERF prolonged activation of EGFR as assessed by self-phosphorylation (Figure 5B). When triplicate assays were performed, EGF increased tyrosine phosphorylation of EGFR 21-fold at 60 min. By 180 min, this had decreased by 70% in the absence of NHERF, but there was no decrease in the presence of NHERF. Thus, NHERF decreased ligand-induced loss of EGFR mass and prolonged the activated state of EGFR. The effects of NHERF on EGFR were accompanied by prolonged activation of the downstream signaling protein ERK (Figure 5C). In the absence of NHERF, EGF increased phospho-ERK fivefold at 60 min, and this activation decreased by 60% at 150 min. In the presence of NHERF, ERK remained fully active at 150 min.
Figure 5.
Effects of NHERF on down-regulation of EGFR. FLAG-tagged NHERF expression constructs were transfected into HeLa cells. Sixteen hours later, cells were treated without or with 100 nM EGF for the indicated times. (A) Cells were solubilized with hot Laemmeli sample buffer and EGFR mass was detected by Western blotting by using the SEFIGA antibody. (B) Tyrosine phosphorylated EGFR were detected using PY99 anti-phospho-tyrosine antibody. (C) Phospho-ERK was detected using the E4 antibody. Blots were developed using ECL and scanned with an LKB ultrascan XL enhanced lazer densitometer.
Effects of Mutation of the NHERF Binding Site in the COOH Terminus of EGFR and of Expression of Mutant NHERF on Ligand-Induced Receptor Down-Regulation
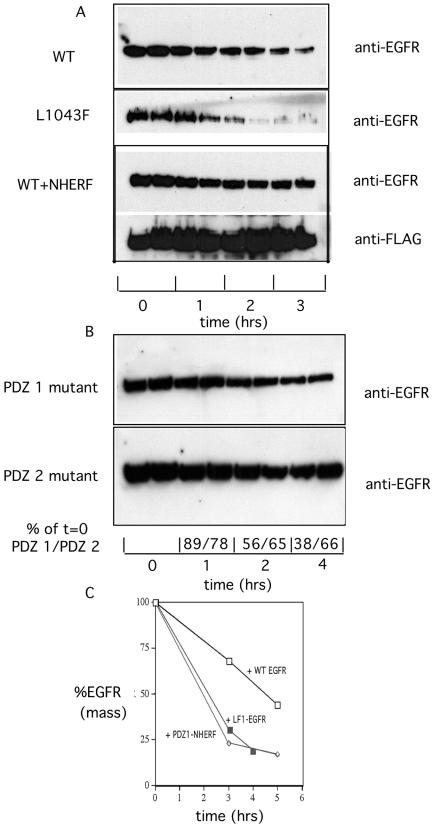
To further assess the functional importance of the EGFR–N-HERF interaction, EGFR containing a point mutation in the principal binding site for PDZ 1 of NHERF (L1043F EGFR) was stably expressed in B82 cells that lack endogenous EGFR (Lin et al., 1986). WT or NHERF mutant in PDZ 1 that abolishes binding to EGFR (G25A/F26A NHERF) or in PDZ 2 that does not (G163A/F164A NHERF) was introduced into B82 cells expressing WT EGFR, and clonal expressor lines also were selected. After selection of stable expressor cell lines, the kinetics of ligand-induced EGFR down-regulation were measured. At the indicated times after addition of 100 nM EGF, whole cell EGFR mass was determined by Western blotting and densitometry. The time course of the decrease in EGFR mass is shown in Figure 6A. As observed in transient transfections in HeLa cells, expression of NHERF slowed the rate of EGF-induced receptor degradation in B82 cells. In contrast, the rate of ligand-induced degradation of L1043F EGFR was increased compared with that of WT EGFR. The cell lines contained the same number of WT or L1043F mutant EGFR per cell (our unpublished data). From multiple experiments, the calculated t1/2 for ligand-treated WT EGFR was 3.7 ± 0.76 h compared with 1.97 ± 0.46 h for L1043F EGFR in B82 cells. Mutating the site of NHERF interaction thus significantly accelerated the rate of ligand-induced EGFR down-regulation. This result supports the observation that NHERF retards ligand-induced receptor down-regulation. To further test this hypothesis, we mutated PDZ 1 of NHERF (G25A/F26A) and PDZ 2 of NHERF (G163A/F164A) and stably expressed these forms of NHERF in B82 cells containing WT EGFR. Because NHERF forms specific dimers (Lau and Hall, 2001), the PDZ 1 NHERF mutant, which failed to bind the COOH terminus of EGFR, was predicted to act as a dominant negative inhibitor of endogenous NHERF. As shown in Figure 6B, the mutant G25A/F26A NHERF significantly enhanced the rate of EGF-induced WT EGFR down-regulation (from multple experiments t1/2 = 2.17 ± 0.29 h). In contrast, the G163A/F164A PDZ 2 mutant NHERF, which did not affect EGFR binding, acted like WT NHERF in stabilizing EGFR. Densitometry analysis indicated that 4 h after addition of EGF cells expressing the PDZ 1 mutant NHERF had lost 62% of EGFR compared with cells expressing the PDZ 2 mutant that had lost only 32% of EGFR. Together, these results from multiple experiments summarized in Figure 6C indicate that NHERF slows the rate of ligand-induced EGFR down-regulation.
Figure 6.
Effects of abolishing NHERF interactions on the rate of ligand-induced EGFR down-regulation. Clonal B82 cell lines stably expressing WT EGFR, L1043F EGFR, or WT EGFR plus G25A/F26A or G163A/F164A NHERF were treated with 100 nM EGF for the indicated times. Total EGFR were extracted with hot Laemmli sample buffer, and EGFR mass was determined by Western blotting by using the anti-EGFR SEFIGA antibody. Quantitation of triplicate samples at each time was done by densitometry. Similar results were obtained in six experiments of similar design. (A) Time course of down-regulation of WT EGFR, L1043F EGFR, and WT EGFR plus NHERF. (B) Time course of down-regulation of WT EGFR in B82 cells expressing NHERF mutant in PDZ 1 or in PDZ 2. A comparison of average densitometry values, expressed as percentage of t = 0, is shown below the Western blots. (C) Summary of rates of down-regulation of WT EGFR (□), L1043F EGFR (▵), and WT EGFR plus mutant PDZ 1 NHERF (⋄).
Retention of EGFR at the Cell Surface
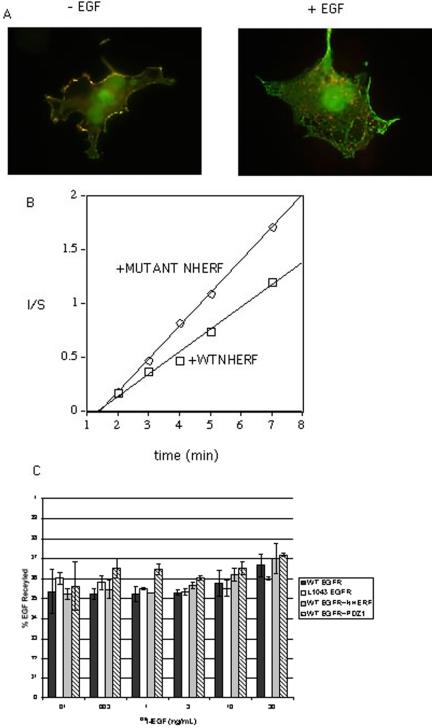
NHERF could slow the rate of ligand-induced receptor down-regulation by retaining EGFR at the cell surface or by enhancing the rate of recycling of endocytosed EGFR or by a combination of both. We first examined the colocalization of EGFR and NHERF. As shown in Figure 7A, both receptor-bound EGF and NHERF are colocalized at the surface of COS7 cells. After 10 min at 37°C, EGF internalized via receptor-mediated endocytosis into early endosomes. NHERF, however, remained at the cell surface and failed to cointernalize with EGF. These results suggest that the principal interaction between EGFR and NHERF occurs at the cell surface.
Figure 7.
Colocalization of EGFR and NHERF at the cell surface. (A) COS 7 cells expressing WT EGFR and FLAG-tagged NHERF were incubated on ice with 500 ng/ml Texas Red-labeled EGF for 1 h. Cells were washed to remove unbound EGF. Cells were shifted to 37°C for 15 min, fixed, and stained with Alexa fluor 488-labeled anti-FLAG antibody. EGF (red), NHERF (green), and overlay (yellow). (B) In/Sur plot of the initial rate of endocytosis of 125I-EGF in B82 cells expressing WT EGFR without (□) or with mutant PDZ 1 NHERF (○). In/Sur plots of the initial rates of endocytosis of WT EGFR in cells expressing wild-type and PDZ 1 mutant NHERF are shown. (C) Recycling of 125I-EGF in B82 cells expressing WT EGFR, L1043F EGFR, WT EGFR with WT NHERF, or with mutant PDZ 1 NHERF. Bar graphs show the percentage of recycled EGF for each cell type with SD (n = 3). Corresponding recycling rate constants (min-1) were WT EGFR = 0.0338 ± 0.0080, L1043F EGFR = 0.0275 ± 0.0097, WT EGFR plus WT NHERF = 0.0267 ± 0.0078, and WT EGFR plus PDZ1 mutant NHERF = 0.0242 ± 0.0050 (n = 3).
To evaluate the relative effects of NHERF on endocytosis and on recycling of EGFR, we compared the rates of endocytosis and recycling of 125I-EGF in B82 cells expressing WT EGFR without or with PDZ 1 mutant NHERF. The rate of endocytosis of 125I-EGF was measured under initial rate conditions and plotted as the relative amount of surface-bound and internalized ligand as a function of time (the In/Sur plot of Wiley and Cunningham, 1982). As shown in Figure 7B, in cells expressing PDZ1 mutant NHERF, ligand-occupied EGFRs have an endocytic rate constant 1.5-fold higher than that in cells expressing WT NHERF (κe = 0.23 ± 0.03 for WT vs. 0.35 ± 0.07 for PDZ 1 mutant NHERF (n = 5; ±SEM). In replicate experiments the κe for L1043F EGFR was directly compared with that of WT EGFR and was found to be 1.5-fold greater (ke = 0.31 vs. 0.21.
The rates of recycling of endocytosed 125I-EGF were compared in cells expressing WT EGFR, L1043F EGFR, or WT EGFR in the presence of WT NHERF or in the presence of the PDZ 1 mutant NHERF (Figure 7C). Recycling was assessed by a steady-state sorting assay (French et al., 1995). The recycling rate constants were between 0.024 and 0.033 min-1, and the percentage recycled was between 50 and 60% for all four cell types. The rates of recycling of ligand and the percentage recycled were thus identical in these four cell types that exhibit significantly different ligand-induced rates of degradation.
DISCUSSION
NHERF is an adaptor protein that plays a larger biological role than its originally identified function to mediate cAMP inhibition of renal Na+/H+ exchanger 3 (Voltz et al., 2001). NHERF contains two PDZ domains and a COOH-terminal domain that binds the ERM family of proteins, which link membrane proteins to the actin cytoskeleton. Although PDZ domains classically bind to short specific sequence motifs located at the extreme COOH terminus of membrane proteins by forming an antiparallel β sheet whose pentultimate hydrophobic amino acid fits into a hydrophobic pocket in the PDZ domain (Doyle et al., 1996), binding to nonterminal β hairpin structures in proteins also occurs and uses the same PDZ domain binding site (Oschkinat, 1999).
The interactions between EGFR and NHERF involve the NH2-terminal PDZ domain (PDZ 1) of NHERF and an internal sequence in the COOH-terminal regulatory domain of EGFR. This EGFR sequence, DSFL, is homologous to the classical COOH-terminal sequences of the β-adrenergic receptor, the purinergic P2Y1 receptor, and CFTR that also bind PDZ 1 of NHERF (Hall et al., 1998a; Cao et al., 1999). The interaction between EGFR and PDZ 1 of NHERF seems specific as NHERF PDZ 2, and a variety of PDZ domains from other proteins failed to recognize EGFR.
NHERF and EGFR are colocalized at the cell membrane. The major effect of the interaction between the two proteins is to stabilize EGFR at this location. Mutation of a single amino acid in the COOH terminus of EGFR that abolishes binding of PDZ 1 of NHERF approximately doubles the rate of ligand-induced EGFR down-regulation. This results from an enhanced rate of endocytosis of ligand-occupied EGFR. Comparison of the rates of recycling of internalized ligand in B82 cells expressing WT EGFR, L1043F EGFR, or WT EGFR with WT NHERF or PDZ 1 mutant NHERF indicated that NHERF does not affect the rate of recycling. The observations that abolishing NHERF interactions enhance ligand-induced down-regulation indicate that NHERF functions to limit the rate of EGFR down-regulation. Interaction with NHERF may tether EGFR at the cell surface; binding of NHERF also may interfer with binding of Grb 2 to nearby sites (aa 1068 and 1086). Because Grb 2 binding to activated EGFR is necessary for ligand-induced internalization of receptors (Jiang et al., 2003), this would decrease the rate of endocytosis.
The effects of NHERF on the rate of endocytosis of EGFR seem sufficient to account for the overall effects on ligand-induced down-regulation of EGFR as predicted from kinetic analysis. The basic process can be expressed as d Ci/dt = keCs - kxCi - kdegCi, where Ci is the amount of intracellular ligand–receptor complex, Cs is the amount of cell surface ligand–receptor complex, kdeg is the degradation rate constant, ke is the endocytic rate constant, and kx the recycling rate constant. Assuming Ci is at roughly steady state due to complexes being internalized, recycled, and degraded: kdeg = ke Cs/[1 + (kx/kdeg)]. Thus, the degradation rate will be diminished twofold when ke/[1 + kx/kdeg] is diminished ∼twofold if the value of Cs remains relatively unchanged.
In the limiting case where most internalized ligand is degraded rather than being recycled (kx/kdeg <<1), the rate of degradation will be diminished twofold when the value of ke is diminished twofold. This is compatible with the fact that endocytic internalization is rate-limiting for degradation. In the opposite extreme where most of the internalized ligand is recycled rather than degraded (kx/kdeg>1), the degradation rate will diminish twofold when the value of ke/kx is diminished twofold. The measured effects of NHERF on ke thus accounts for its principal effects on kdeg. Reduction of ke and/or increase of kx will serve to increase Cs, at least moderately so the factor by which the degradation rate is diminished will be somewhat less than that by which ke or ke/kx is reduced.
NHERF binds to the extreme COOH terminus of several G protein-coupled receptors including these for β2-adrenergic, κ-opioid, and purinergic P2Y1 agonists (Hall et al., 1998a; Li et al., 2002). β-Adrenergic agonists promote NHERF association with the receptor (Hall et al., 1998a); similarly, an agonist for the κ-opioid receptor promotes NHERF association (Li et al., 2002). Binding of NHERF to these ligand-activated GPCR decreased ligand-induced down-regulation (Cao et al., 1999; Li et al., 2002). β-Adrenergic receptors that lack the COOH-terminal NHERF binding site fail to recycle from endosomes to the plasma membrane efficiently and are sorted to lysosomes (Cao et al., 1999). Transplantation of the COOH-terminal DSLL motif that binds NHERF from the β-adrenergic receptor to the δ-opioid receptor rerouted the latter from a degradative to a recycling pathway (Gage et al., 2001). Similarly NHERF binding to the κ-opioid receptor increased the rate of recycling of internalized receptors (Li et al., 2002). Recycling of β2-adrenergic receptors is negatively controlled by phosphorylation (Cao et al., 1999). GPCR-kinase 5 phosphorylates Ser 411 in the DSLL recognition sequence of β2-adrenergic receptors, disrupting both NHERF interaction and recycling, resulting in increased ligand-induced receptor degradation.
Like these GPCR, binding of NHERF to EGFR enhances the fraction of receptors at the cell surface. In contrast to β2-adrenergic receptors that efficiently recycle to the cell surface in the process of resensitization, EGFR more efficiently traffic to lysosomes where they are degraded. Although NHERF and EGFR are colocalized at the cell surface, these dissociate upon receptor-mediated endocytosis with NHERF remaining at the cell surface. The principal effect of NHERF is to retain EGFR at the cell surface. Regulated recycling of EGFR by processes such as cbl-mediated ubiquitination (Levkowitz et al., 1998) and association with the protein CAML (Tran et al., 2003) seem distinct from the effects of NHERF. Decreased rates of down-regulation enhance signaling by EGFR as assessed by prolonged autophosphorylation and activation of ERK. NHERF binds to the extreme COOH terminus of PDGFR and potentiates PDGF signaling in a process dependent on oligomerization of NHERF (Maudsley et al., 2000).
PDZ domain-containing proteins affect the cell surface distribution of other erb B family members. Erbin specifically interacts with the extreme COOH terminus of erbB2 and functions to localize erbB2 to the basolateral membrane of human intestinal epithelial cells (Borg et al., 2000) and likely to postsynaptic membranes at the neuromuscular junction (Huang et al., 2001). Expression of erbin increases erb B2 surface expression. Lin 7, initially identified in Caenorhabditis elegans as a PDZ domain protein that recognized the extreme COOH terminus of Let23, the EGFR ortholog, localizes it at the basolateral surface of vulval precursor cells (Simske et al., 1996; Kaech et al., 1998). The PDZ domain of hLin7 binds to the extreme COOH terminus of erb B2 and stabilizes erbB2 at the cell surface (Shelly et al., 2003). Stabilization at the cell surface seemed to result from both decreased endocytosis and increased recycling of erbB2. Interestingly an NH2-terminal fragment of hLin7 that interacts with the kinase domain of all erbB family members (the KID domain) seems necessary for receptor maturation and basolateral targeting (Shelly et al., 2003).
PDZ domains in a variety of proteins thus specifically recognize receptors and function in retaining these at the cell surface. NHERF acts to retain EGFR at the cell surface principally through inhibiting ligand-induced endocytosis. This effect enhances EGFR signaling.
Acknowledgments
We thank Dr. Marilyn Farquhar for helpful discussions and Dr. Qi Zhong for expert advice and help with mutant constructions. This study was supported by National Institutes of Health grants CA-58689 (to G.N.G.) and CA-96504 (to D.A.L.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0239. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0239.
Abbreviations used: CFTR, cycstic fibrosis transmembrane conductance regulator; EGFR, epidermal growth factor receptor; ERM, Ezrin, Radizin, Moesin binding domain; GPCR, G protein-coupled receptor; NHERF, Na+/H+ exchanger regulatory factor; PDZ, domain initially identified in PSD 95, discs large and Z0-1 proteins; RTK, receptor tyrosine kinase.
References
- Borg, J.-P., Marchetto, S., Le Bivic, A., Ollendorff, V., Jaulin-Bastard, F., Saito, H. Fournier, E., Adélaïde, J., Margolis, B., and Birnbaum, D. (2000). ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat. Cell Biol. 2, 407-414. [DOI] [PubMed] [Google Scholar]
- Bretscher, A., Chambers, D., Nguyen, R., and Reczek, D. (2000). ERM-Merlin and EPP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 16, 113-143. [DOI] [PubMed] [Google Scholar]
- Cao, T.T., Deacon, H.W., Reczek, D., Bretscher, A., and von Zastrow, M. (1999). A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β 2-adrenergic receptor. Nature 401, 286-290. [DOI] [PubMed] [Google Scholar]
- Carpenter, J.-L., et al. (1982). Co-localization of 125I-epidermal growth factor and ferritin-low density lipoprotein in coated pits: a quantitative electron microscopic study in normal and mutant human fibroblasts. J. Cell Biol. 95, 73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-P., et al. (1993). Ligand-induced internalization of the EGF receptor is mediated by multiple endocytic codes analogous to the tyrosine motif found in constitutively internalized receptors. J. Biol. Chem. 268, 19312-19320. [PubMed] [Google Scholar]
- Chen, W.C., Lazar, C.S., Poenie, M., Tsien, R.Y., Gill, G.N., and Rosenfeld, M.G. (1987). Requirement for instrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature 328, 820-823. [DOI] [PubMed] [Google Scholar]
- Demoulin, J.-B., Seo, J.K., Ekman, S., Grapengiesser, E., Hellman, U., Rönnstrand, L., and Heldin, C.-H. (2003). Ligand-induced recruitment of Na+/H+ exchanger regulatory factor to the platelet-derived growth factor (PDGF) receptor regulates actin cytoskeleton reorganization by PDGF. Biochem. J. 376, 505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D.A., Lee, A., Lewis, J., Kim, E., Sheng, M., and MacKinnon, R. (1996). Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85, 1067-1076. [DOI] [PubMed] [Google Scholar]
- French, A.R., Tadaki, D.K., Niyogi, S.K., and Lauffenburger, D.A. (1995). Intracellular trafficking of epidermal growth factor family ligands is directly influenced by the pH sensitivity of the receptor/ligand interaction. J. Biol. Chem. 270, 4334-4340. [DOI] [PubMed] [Google Scholar]
- Futter, C.E., Pearse, A., Hewlett, L.J., and Hopkins, C.R. (1996). Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132, 1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, R.M., Kim, H-A., Cao, T.T., and von Zastrow, M. (2001). A transplantable sorting signal that is sufficient to mediate rapid recycling of G protein-coupled receptors. J. Biol. Chem. 276, 44712-44720. [DOI] [PubMed] [Google Scholar]
- Gill, G.N., Kawamoto, T., Cochet, C., Le., A., Sato, J.D., Masui, H., McCloud, C., and Mendelsohn, J. (1984). Momoclonal anti-epidermal growth factor receptor antibodies which are inhibitors of epidermal growth factor binding and antagonists of epidermal growth factor-stimulated tyrosine kinase activity. J. Biol. Chem. 259, 7761-7766. [PubMed] [Google Scholar]
- Haj, F.G., Verveer, P.J., Squire, A., Neel, B.G., and Bastiaens, P.I.H. (2002). Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708-1711. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Ostedgaard, L.W., Premont, R.T., Blitzer, J.T., Rahman, N., Welsh, M.J., and Lefkowitz, R.J. (1998a). A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc. Natl. Acad. Sci. USA 95, 8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, R.A., et al. (1998b). The β2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature 392, 626-630. [DOI] [PubMed] [Google Scholar]
- Huang, Y.Z., Wang, Q., Xiong, W.C., and Mei, L. (2001). Erbin is a protein concentrated at postsynaptic membranes that interacts with PSD-95. J. Biol. Chem. 276, 19318-19326. [DOI] [PubMed] [Google Scholar]
- Jiang, X., Huang, F., Marusyk, A., and Sorkin, A. (2003). Grb 2 regulates internalization of EGF receptors through clathrin-coated pits. Mol. Biol. Cell 14, 858-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S.M., Foreman, S.K., and Shank, B.B. (2002). EGF receptor down regulation depends on a trafficking motif in the distal tyrosine kinase domain. Am. J. Physiol. 282, C420-C433. [DOI] [PubMed] [Google Scholar]
- Jurata, L.W., Kenny, D.A., and Gill, G.N. (1996). Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl. Acad. Sci. USA 93, 11693-11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata, L.W., and Gill, G.N. (1997). Functional analysis of the nuclear LIM domain interactor NLI. Mol. Cell. Biol. 17, 5688-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., Whitfield, C.W., and Kim, S.K. (1998). The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94, 761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornilova, E., Sorkina, T., Beguinot, L., and Sorkin, A. (1996). Lysosomal targeting of epidermal growth factor receptors via a kinase-dependent pathway is mediated by the receptor carboxyl-terminal residues 1022–1123. J. Biol. Chem. 271, 30340-30346. [DOI] [PubMed] [Google Scholar]
- Kuroda, S., Tokunaga, C., Kiyohara, Y., Higuchi, O., Konishi, H., Mizuno, K., Gill, G.N., and Kikkawa, U. (1996). Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 271, 31029-31032. [DOI] [PubMed] [Google Scholar]
- Lau, A.G., and Hall, R.A. (2001). Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry 40, 8312-8320. [DOI] [PubMed] [Google Scholar]
- Lauffenburger, D. (1993). Receptors: models for binding, trafficking and signaling, ch. 3. Oxford University Press, New York.
- Levkowitz, G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W.Y., Beguinot, L., Geiger, B., and Yarden, Y. (1998). c-Cbl/sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.R., Chen, W.S., Lazar, C.S., Carpenter, C.D., Gill, G.N., Evans, R.M., and Rosenfeld, M.G. (1986). Protein kinase C phosphorlation at Thr654 of the unoccupied EGF receptor and EGF binding regulate functional receptor loss by independent mechanisms. Cell 44, 839-848. [DOI] [PubMed] [Google Scholar]
- Li, J.-G., Chen, C., and Liu-Chen, L.-Y. (2002). Ezrin-Radixin-Moesin-binding Phosphoprotein-50/Na+/H+ exchanger regulatory factor (EBP50/NHERF) blocks U50,488H-induced down-regulation of the human κ opioid receptor by enhancing its recycling rate. J. Biol. Chem. 277, 27545-27552. [DOI] [PubMed] [Google Scholar]
- Mahon, M.J., Donowitz, M., Yun, C.C., and Segre, G.V. (2002). Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signaling. Nature. 417, 858-861. [DOI] [PubMed] [Google Scholar]
- Maudsley, S., Zamah, A.M., Rahman, N., Blitzer, J.T., Luttrell, L.M., Lefkowitz, R.J., and Hall, R.A. (2000). Platelet-derived growth factor receptor association with Na+/H+ exchanger regulatory factor potentiates receptor activity. Mol. Cell. Biol. 20, 8352-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson, Y., Shtiegman, K., Katz, M., Zwang, Y., Vereb, G., Szollosi, J., and Yarden, Y. (2003). Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 278, 21323-21326. [DOI] [PubMed] [Google Scholar]
- Nesterov, A., Wiley, H.S., and Gill, G.N. (1995). Ligand-induced endocytosis of epidermal growth factor receptors that are defective in binding clathrin adaptors. Proc. Natl. Acad. Sci. USA 92, 8719-8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko, L.K., Chang, C.P., Will, B.H., Burke, P.M., Gill, G.N., and Wiley, H.S. (1995). Endocytosis and lysosomal targeting of epidermal growth factor receptors are mediated by distinct sequences independent of the tyrosine kinase domain. J. Biol. Chem. 270, 4325-4333. [DOI] [PubMed] [Google Scholar]
- Oschkinat, H. (1999). A new type of PDZ domain recognition. Nat. Struct. Biol. 7, 408-410. [DOI] [PubMed] [Google Scholar]
- Shelly, M., Mosesson, Y., Citri, A., Lavi, S., Zwang, Y., Melamed-Book, N., Aroeti, B., and Yarden, Y. (2003). Polar expression of ErbB-2/HER2 in epithelia: bimodal regulation by Lin-7. Dev. Cell 5, 475-486. [DOI] [PubMed] [Google Scholar]
- Simske, J.S., Kaech, S.M., Harp, S.A., and Kim, S.K. (1996). LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85, 195-204. [DOI] [PubMed] [Google Scholar]
- Sorkin, A., and Carpenter, G. (1993). Interaction of activated EGF receptor with coated pit adaptins. Science 261, 612-615. [DOI] [PubMed] [Google Scholar]
- Sorkin, A., and von Zastrow, M. (2002). Signal transduction and endocytosis: close encounters of many kinds. Nature 3, 600-614. [DOI] [PubMed] [Google Scholar]
- Tran, D.D., Russell, H.R., Sutor, S.L., van Deursen, J., and Bram, R.J. (2003). CAML is required for efficient EGF receptor recycling. Dev. Cell 5, 245-254. [DOI] [PubMed] [Google Scholar]
- Voltz, J.W., Weinman, E.J., and Shenolikar, S. (2001). Expanding the role of NHERF, a PDZ-domain containing protein adapter, to growth regulation. Oncogene 20, 6309-6314. [DOI] [PubMed] [Google Scholar]
- Weinman, E.J., Steplock, D., and Shenolikar, S. (1993). CAMP-mediated inhibition of the renal brush border membrane Na+-H+ exchanger requires a dissociable phosphoprotein cofactor. J. Clin. Investig. 92, 1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, H.S., and Cunningham, D.D. (1982). The endocytotic rate constant a cellular parameter for quantitating receptor-mediated endocytosis. J. Biol. Chem. 257, 4222-4229. [PubMed] [Google Scholar]
- Wiley, H.S., Herbst, J.J., Walsh, B.J., Lauffenberger, D.A., Rosenfeld, M.G., and Gill, G.N. (1991). Role of tyrosine kinase activity in endocytosis, compartmentation and down regulation of the EGF receptor. J. Biol. Chem. 266, 11083-11094. [PubMed] [Google Scholar]
- Woods, D.F., and Bryant, P.J. (1989). Molecular cloning of the lethal (1) discs large-1 oncogene of Drosophila. Dev. Biol. 134, 222-235. [DOI] [PubMed] [Google Scholar]
- Yarden, Y., and Sliwkowski, M.X. (2001). Untangling the erbB signaling network. Mol. Cell. Biol. 127-137. [DOI] [PubMed]
- Yun, C.H.C., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C.-M., Weinman, E.J., and Donowitz, M. (1997). cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. USA 94, 3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Q., Lazar, C.S., Tronchere, H., Sato, T., Meerloo, T., Yeo, M., Songyang, Z., Emr, S.D., and Gill, G.N. (2002). Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. USA 99, 6767-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Ruiz-Lozano, P., Martone, M.E., and Chen, J. (1999). Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem. 274, 19807-19813. [DOI] [PubMed] [Google Scholar]