Abstract
ARVCF, an armadillo-repeat protein of the p120ctn family, associates with classical cadherins and is present in adherens junctions, but its function is poorly understood. Here, we show that ARVCF interacts via a C-terminal PDZ-binding motif with zonula occludens (ZO)-1 and ZO-2. ARVCF and ZO-1 partially colocalize in the vicinity of the apical adhesion complex in polarized epithelial Madin-Darby canine kidney cells. ARVCF, ZO-1, and E-cadherin form a complex and are recruited to sites of initial cell-cell contact in sparse cell cultures. E-cadherin binding and plasma membrane localization of ARVCF require the PDZ-binding motif. Disruption of cell-cell adhesion releases ARVCF from the plasma membrane and an increased fraction of the protein localizes to the nucleus. Nuclear localization of ARVCF also requires the PDZ-binding motif and can be mediated by the PDZ domains of ZO-2. Thus, the interaction of ARVCF with distinct PDZ-domain proteins determines its subcellular localization. Interactions with ZO-1 and ZO-2, in particular, may mediate recruitment of ARVCF to the plasma membrane and the nucleus, respectively, possibly in response to cell-cell adhesion cues.
INTRODUCTION
Tight junctions (TJs) and adherens junctions (AJs) are composed of distinct families of transmembrane adhesion proteins and associated cytosolic proteins that link the transmembrane proteins to the actin cytoskeleton. The possibility to assemble different combinations of transmembrane and cytosolic adhesion plaque proteins allows for the formation of different adhesion complexes, each presumably bringing particular structural and functional properties to sites of cell-cell contact (Borrmann et al., 2000). At TJs, the zonula occludens (ZO) proteins ZO-1, ZO-2, and ZO-3 bind to claudins, junction adhesion molecule, and occludin via their PDZ domains (claudins and junction adhesion molecule) or the guanylate kinase homology domain (occludin) (Stevenson and Keon, 1998; Cereijido et al., 2000; Gonzalez-Mariscal et al., 2000). ZO-1 can further heterodimerize with ZO-2 or ZO-3 through PDZ domain interactions (Itoh et al., 1997; Ando-Akatsuka et al., 1999; Wittchen et al., 1999). ZO proteins bind actin filaments (Fanning et al., 1998; Wittchen et al., 1999; Fanning et al., 2002) and several signaling molecules, suggesting that they serve as platforms for the recruitment or assembly of signaling complexes in response to cell-cell adhesion (Zahraoui et al., 2000; Angst et al., 2001).
In AJs, classical cadherins assemble different combinations of α-, β-, γ-catenin (plakoglobin), p120ctn, δ-catenin, and Armadillo-repeat gene deleted in Velo-cardio-facial syndrome (ARVCF) (Marrs and Nelson, 1996; Borrmann et al., 2000; Angst et al., 2001; Nagafuchi, 2001). β- and γ-catenin bind directly and competitively to the membrane distal region of the cytosolic domain of cadherins (Mathur et al., 1994; Staddon et al., 1995). α-catenin associates with β- or γ-catenin to form either α-/γ- or α-/β-catenin complexes (Borrmann et al., 2000). α-catenin binds F-actin directly (Rimm et al., 1995) or through the interaction with the F-actin binding proteins α-actinin and vinculin (Nieset et al., 1997; Weiss et al., 1998).
ARVCF and two other members of the p120ctn family (Peifer et al., 1994), p120ctn itself (Reynolds et al., 1992; Staddon et al., 1995) and δ-catenin/NPRAP (Paffenholz and Franke, 1997; Zhou et al., 1997), associate with the membrane proximal region of the cytosolic tail of classical cadherins (Ozawa and Kemler, 1998; Yap et al., 1998; Lu et al., 1999; Kaufmann et al., 2000). ARVCF can associate with E-, M-, and possibly N-cadherin (Kaufmann et al., 2000; Waibler et al., 2001). Although widely expressed (Sirotkin et al., 1997), ARVCF is generally present in very low amounts compared with p120ctn (Mariner et al., 2000), suggesting that high levels of expression are either not required for its function or that they are temporally or spatially restricted. δ-catenin, for example, is brain specific, expressed highest during brain development and only present in low amounts in adult neuronal tissues (Paffenholz and Franke, 1997; Zhou et al., 1997).
ARVCF and p120ctn bind to cadherins via their armadillo-repeat domains in a mutually exclusive manner (Mariner et al., 2000), presumably competing for the same binding site (Kaufmann et al., 2000). However, due to the low abundance of ARVCF compared with p120ctn in most cells and tissues (Mariner et al., 2000), ARVCF is unlikely to function solely as a competitive binding protein, but may rather confer distinct properties to AJs. Consistent with the notion that the two members of the p120ctn family mediate different functions, ARVCF lacks the cellular branching activity induced by p120ctn when overexpressed in fibroblasts (Mariner et al., 2000). Xenopus laevis ARVCF, however, mediates branching via the RhoA and Rac pathways (Fang et al., 2004). Nuclear localization of ARVCF has been reported previously (Mariner et al., 2000), the functional relevance of which is unclear.
In polarized epithelial cells, ZO proteins are generally restricted to TJs and absent from AJs. However, in nonepithelial cells that lack TJs, ZO-1 predominantly localizes to AJ, where it binds to α-catenin (Itoh et al., 1991, 1993, 1997). In cardiomyocytes, ZO-1 is found in intercalated discs (Barker et al., 2002), regions of intimate contact between adjacent myocytes that also contain N-cadherin and ARVCF (Itoh et al., 1991; Toyofuku et al., 1998; Kaufmann et al., 2000). Even in epithelial cells, ZO proteins colocalize and associate with AJ markers under certain circumstances. For example, in the initial stages during the establishment of apico-basal polarity in Madin-Darby canine kidney (MDCK) cells, ZO-1 and E-cadherin colocalize at sites where adjacent cells establish contact with each other (Yonemura et al., 1995; Rajasekaran et al., 1996; Adams et al., 1998; Ando-Akatsuka et al., 1999; Vasioukhin et al., 2000). As polarization proceeds, these contact zones mature into TJ by the gradual recruitment of occludin and the segregation of E-cadherin from ZO-1 to form AJs (Ando-Akatsuka et al., 1999).
Here, we characterize novel interactions of ARVCF with ZO-1 and ZO-2. ARVCF and ZO-1 form a complex with E-cadherin and the three proteins colocalize at sites of initial cell-cell adhesion and, in polarized cells, to a region adjacent to the apical adhesion complex. Membrane recruitment as well as nuclear localization of ARVCF is regulated by cell-cell adhesion and the interaction with PDZ-domain proteins. ZO-1, in particular, may regulate plasma membrane localization of ARVCF, whereas ZO-2 may play a role in localizing ARVCF to the nucleus.
MATERIALS AND METHODS
Antibodies
The following commercial primary antibodies were used: rabbit polyclonal anti-ZO-1 and ZO-2 (Zymed Laboratories, South San Francisco, CA), rat monoclonal anti-hemagglutinin (HA) (Roche Diagnostics, Indianapolis, IN), mouse monoclonal anti-E-cadherin (BD Biosciences Transduction Laboratories, Lexington, KY), goat polyclonal anti-ARVCF (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-Xpress (Invitrogen, Carlsbad, CA), and mouse anti-glutathione S-transferase (GST) (Santa Cruz Biotechnology). Horseradish peroxidase (HRP)-labeled secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA), Pierce Chemical (Rockford, IL), or Bio-Rad (Hercules, CA), and fluorescently labeled (Alexa 488 and 594) secondary antibodies were from Molecular Probes (Eugene, OR).
Yeast Two-Hybrid Screen
The screening of a pretransformed mouse 17-d embryo cDNA library by using the N-terminal three PDZ domains of human ZO-1 (amino acids 1-507) and bait vectors carrying the three PDZ domains of canine ZO-2 (amino acids 1-591) and ZO-3 (amino acids 1-467) have been described previously (Kausalya et al., 2001).
Plasmid Constructs
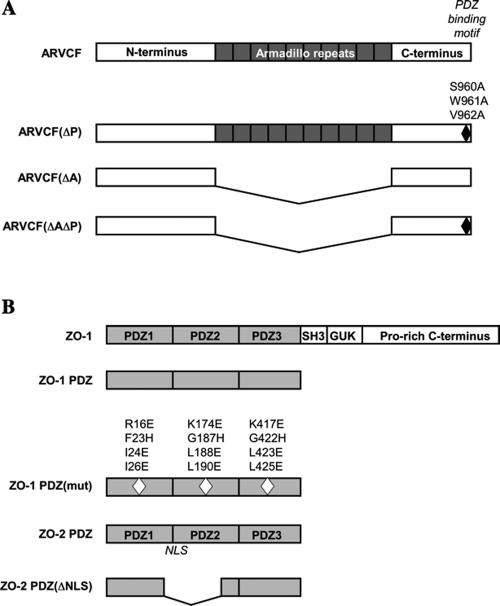
The full-length human ARVCF cDNA was obtained by reverse transcription-polymerase chain reaction of testis cDNA library (BD Biosciences Clontech, Palo Alto, CA) by using suitable primers covering the 5′ and 3′ coding region of the cDNA. The primers were designed to introduce a 5′ EcoR I restriction site followed by an N-terminal HA-epitope tag and a 3′ XhoI restriction site. To generate a mutant ARVCF with an inactivated PDZ-binding motif [ARVCF(ΔP)], the codons for the C-terminal serine-tryptophane-valine (SWV) were changed to code for alanines. Mutants lacking the armadillo repeats (amino acids 336-782) were generated in the background of the wild-type [ARVCF(ΔA)] or PDZ-binding motif mutant [ARVCF(ΔAΔP)] constructs. The ARVCF constructs were subcloned into the pCDNA3 vector for expression in mammalian cells and for in vitro translation. Myc-tagged ZO-1 and ZO-2 constructs carrying only the three PDZ domains (ZO-1 PDZ, ZO-2 PDZ) have been described previously (Reichert et al., 2000b; Jaramillo et al., 2004). To inactivate the binding capacity of the PDZ domains in the context of the ZO-1 PDZ construct [ZO-1 PDZ(mut)], suitable overlapping primer pairs were used to generate substitutions in amino acids predicted to contact C-terminal interacting peptides (Doyle et al., 1996) in PDZ1 (R16E, F23H, I24E, and I26E), PDZ2 (K174E, G187H, L188E, and L190E) and PDZ3 (K417E, G422H, L423E, and L425E). Plasmids for Xpress-tagged ZO-2 PDZ domains (ZO-2 PDZ) or a ZO-2 PDZ mutant with an inactivated nuclear localization signal [ZO-2 PDZ(ΔNLS)] were obtained from Dr. Gonzalez-Mariscal (Jaramillo et al., 2004). To generate GST-ZO-1 PDZ fusion protein, a cDNA encoding the N-terminal three PDZ domains of human ZO-1 (amino acids 1-507) was cloned into pGEX4T to obtain a C-terminal in frame fusion to the GST cDNA. An expression vector encoding the cytosolic tail of E-cadherin fused to GST was as described previously (Müller et al., 1999). The sequences of the primers used are available upon request, a schematic representation of the proteins expressed by the different cDNA constructs is provided in Figure 1.
Figure 1.
Schematic diagrams of ARVCF, ZO-1, and ZO-2. (A) Domain structure of ARVCF. The amino acid substitutions that abolish binding to PDZ-domains [ARVCF(ΔP); black diamond] and the deletion of the armadillo domains (ARVCF(ΔA) are indicated. (B) Domain organization of ZO-1. Truncated ZO-1 and ZO-2 proteins carrying only the three PDZ domains (ZO-1 PDZ, ZO-2 PDZ) are depicted and amino acid substitutions in the PDZ domains of ZO-1 that abolish binding to C-terminal peptide [ZO-1 PDZ(mut), white diamonds] and the deletion of the nuclear localization signal in ZO-2 PDZ (ZO-2 (ΔNLS) are shown. Diagrams are not to scale.
Cell Culture and Transfection of MDCK Cells
MDCK strain II cells were cultured as described previously (Honing and Hunziker, 1995; Reichert et al., 2000a; Mauro et al., 2001). Wild-type and mutant human ARVCF cDNAs in the pcDNA3 expression vector were transfected into MDCK cells by using LipofectAMINE (Invitrogen). MDCK cell lines stably expressing ARVCF were selected in G418 and expressing clones identified by immunofluorescence or Western blot analysis. MDCK cells were grown on permeable Transwell polycarbonate filter units (Costar, Cambridge, MA) to obtain polarized cell monolayers (Reichert et al., 2000a). MCF7 cells were transiently transfected with different cDNA constructs using LipofectAMINE (Invitrogen).
GST Pull-Down
GST, GST-ZO-1 PDZ, GST-ARVCF, or GST-E-cadherin tail fusion proteins were produced, purified, and bound to glutathione Sepharose-4B (Amersham Biosciences, Piscataway, NJ) following standard protocols. Bound proteins were quantified by SDS-PAGE by using known amounts of BSA as standards. Beads carrying 2-10 μg of GST or GST fusion proteins were incubated with 5-10 μl of in vitro translated (Quick Coupled T7 TNT; Promega, Madison, WI) wild-type, or mutant proteins in binding buffer (25 mM Tris, pH 7.5, 50 mM NaCl, 0.1% Tween 20 or Triton X-100, 20 mM MgCl2, and 1 mM dithiothreitol [DTT] for GST-ZO-1-PDZ fusions or 20 mM HEPES, pH 7.9, 60 mM NaCl, 6 mM MgCl2, 8.2% glycerine, 0.1 mM EDTA, and 0.5% CHAPS for GST-E-cadherin tail fusion) for 2 h at 4°C. The beads were washed with washing buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20 or Triton X-100, 20 mM MgCl2, and 1 mM DTT for GST-ZO-1-PDZ fusions or 20 mM Tris-HCl, pH 8.8, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40 for GST-E-cadherin tail fusions), resuspended in SDS sample buffer, and bound proteins analyzed by SDS-PAGE, autoradiography, and densitometry.
Coimmunoprecipitations
Control MDCK cells or transfected cells expressing either wild-type or mutant ARVCF were washed with cold phosphate-buffered saline (PBS) and lysed on ice in lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% deoxycholate, and supplemented with a protease inhibitor cocktail). The postnuclear supernatant was precleared and immunoprecipitated either with anti-ZO-1 (Zymed Laboratories), anti-HA (Roche Diagnostics), or anti-E-cadherin (BD Biosciences Transduction Laboratories) antibodies and protein G-Sepharose (Pharmacia, Uppsala, Sweden). Immunoprecipitates were washed with lysis buffer, fractionated by SDS-PAGE (10% acrylamide), blotted onto polyvinylidene difluoride (PVDF) membranes, and probed with antibodies to HA, ARVCF (Santa Cruz Biotechnology), ZO-1 (BD Biosciences Transduction Laboratories and Zymed Laboratories) followed by suitable HRP-labeled secondary antibodies (Jackson ImmunoResearch Laboratories and Pierce Chemical) and chemiluminescence (Super Signal West Pico; Pierce Chemical). Autoradiographs were analyzed by densitometry. Alternatively, different wild-type or mutant ARVCF and ZO-1 forms were cotranslated in vitro as detailed above. After immunprecipitation of either ARVCF or ZO-1, the proteins were fractionated by SDS-PAGE, the gels were dried, and radio-actively labeled proteins were visualized by autoradiography.
Blot Overlay
In vitro-translated ARVCF or ARVCF(ΔP) (20-50 μl of TNT reaction) was fractionated by SDS-PAGE and transferred to PVDF membranes. Blots were incubated with a purified GST-ZO-1 PDZ domain fusion protein (75 nM), followed by a mouse anti-GST antibody and HRP-labeled goat anti-mouse antibodies, all in PBS with 0.2% Tween 20.
Immunofluorescence Labeling
MDCK cells grown on glass coverslips or 0.4-μm Transwell filters (Costar) were fixed with either cold methanol (2.5 min; –20°C) or paraformaldehyde (PFA) (3.7%; 30 min), and, in the case of PFA fixation, permeabilized with Triton X-100 (0.2% in PBS). After blocking in 10% goat serum (Invitrogen), cells were incubated with rat anti-HA, rabbit anti-ZO-1 or ZO-2, or monoclonal mouse anti-E-cadherin or anti-Xpress (Invitrogen) antibodies (5 μg/ml) followed by suitable, fluorescently labeled secondary antibodies (1:1000; Molecular Probes). Images were acquired using a confocal laser scanning microscope (Bio-Rad) and IMARIS software (Bitplane, Zurich, Switzerland).
Calcium-Switch and Detection of Cell-Cell Contact Protocols
Disassembly and resealing of TJs and AJs was essentially carried out as described previously (Lawrence et al., 2002). Briefly, filter grown MDCK cells were incubated for 45 min at 37°C in 2 mM EDTA in PBS to chelate calcium. Cells were then washed and either fixed or incubated for 2 h at 37°C in complete media before fixation. To analyze cell-cell adhesion in MCF7 cells, a day after transient transfection the cells were plated on coverslips at an appropriate density and fixed after contact formation was observed.
Cytochalasin D treatment
Cells were incubated for 1 h at 37°C with 20 μg/ml cytochalasin D (Sigma-Aldrich, St. Louis, MO) in culture media and then washed with cold PBS and processed for immunoprecipitation or immunofluorescence labeling as described above. Labeling with rhodamine-phalloidin was used to confirm disruption of F-actin by the drug.
RESULTS
ARVCF Interacts with ZO-1 and ZO-2 but Not with ZO-3
Proteins that interact with the PDZ domains of ZO-1 were identified in a yeast two-hybrid screen of a mouse 17-d embryo library by using the three PDZ domains of human ZO-1 as a bait (Kausalya et al., 2001). Sequencing of positive clones identified cDNAs encoding the C-terminal region of ARVCF. A cDNA clone encoding amino acids 670-893 of mouse ARVCF was selected for further characterization of the interaction with the PDZ-domain of different ZO proteins by using the two-hybrid assay (Table 1). In addition to the PDZ domains of ZO-1, the ARVCF construct interacted with the PDZ domains of ZO-2 but not with those of ZO-3. The interaction of the C terminus of Connexin-45 with ZO-1 and ZO-3 (Kausalya et al., 2001), and p53 with T-Ag served as positive controls, whereas lamin and the empty library vector were used as negative controls, confirming the specificity of the observed associations.
Table 1.
Interaction of the PDZ domains of ZO-1 and ZO-2 with a construct encoding the C-terminal region of ARVCF (amino acids 670-893) or a mutant thereof (ARVCFΔP) in which the SWV amino acids encoding a putative PDZ-binding motif were changed to alanines
| ARVCF | ARVCF(ΔP) | Cx-45 | Library vector | Laminin | T-Ag | |
|---|---|---|---|---|---|---|
| ZO-1 PDZ | +++ | - | +++ | - | - | nd |
| ZO-2 PDZ | +++ | - | - | - | - | nd |
| ZO-3 PDZ | - | - | +++ | - | - | nd |
| Bait vector | - | - | - | - | - | - |
| p53 | - | - | - | - | - | +++ |
Interactions were determined by monitoring growth of cotransformed yeast on selective media and β-galactosidase activity. Similar results were obtained under low and high stringency conditions. Empty bait (pGBKT7) and library (pACT2) plasmids and a lamin construct served as negative controls. T-antigen (T-Ag) and p53, two known interaction partners, were used as a positive control. Growth on dropout media and β-galactosidase activities for yeast coexpressing T-Ag and p53 (positive control) were similar for clones coexpressing the C-terminus of ARVCF and the PDZ-domains of ZO-1 and ZO-2. Since the ZO-3 bait can interact with the PDZ-motif in Cx-45, the lack of an interaction with ARVCF was not due to the experimental set up. +++, interaction; -, no interaction; nd, not determined.
PDZ domains generally interact with short C-terminal amino acid sequences and the C-terminal SWV in ARVCF conforms to a class I PDZ-binding motif (Sheng and Sala, 2001). Mutation of the S, W, and V amino acids to alanine (A) abolished the interaction with the PDZ domains of ZO-1 and ZO-2 (Table 1), showing that the SWV sequence is a functional PDZ-domain binding motif that mediates the observed interactions.
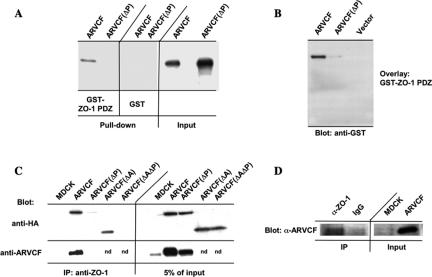
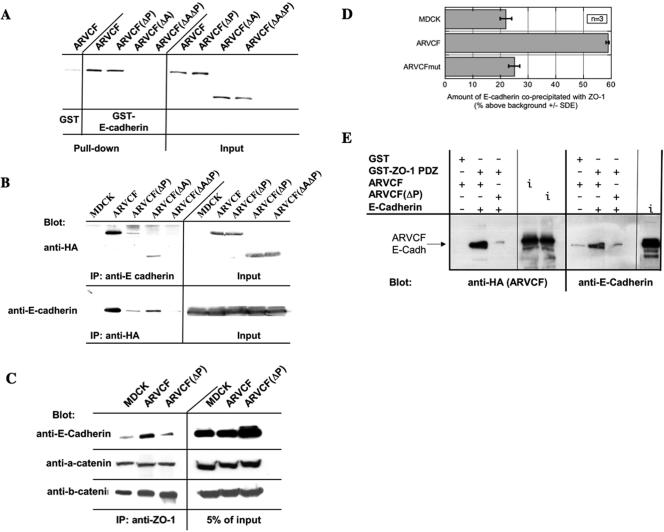
Because ARVCF was initially identified using the PDZ domains of ZO-1 as bait, subsequent experiments focused on the characterization of the binding between ARVCF and ZO-1. A GST fusion protein carrying the three PDZ domains of ZO-1 was analyzed for its ability to bind full-length in vitro-translated ARVCF or a mutant with a destroyed PDZ-binding motif [ARVCF(ΔP)] (Figure 1A). Confirming the yeast two-hybrid results, the GST-ZO-1 PDZ bound to ARVCF but not ARVCF(ΔP) (Figure 2A). To further establish that the association was direct, in vitro-translated ARVCF and ARVCF(ΔP) were fractionated by SDS-PAGE, transferred to PVDF membranes, and the blot overlaid with GST-ZO-1 PDZ. As shown in Figure 2B, GST-ZO-1 PDZ bound to ARVCF but not ARVCF(ΔP), consistent with a direct interaction.
Figure 2.
Binding of ARVCF and ZO-1. (A) In vitro-translated ARVCF binds to a GST fusion protein containing the PDZ domains of ZO-1. GST or GST-ZO-1 PDZ fusion proteins were coupled to glutathione beads and incubated with in vitro-translated, radioactively labeled ARVCF or ARVCF(ΔP). Protein bound to the beads (pull-down) was analyzed by SDS-PAGE and autoradiography. An aliquot (5%) of the in vitro translated material was directly analyzed to confirm that similar amounts of the in vitro-translated proteins were added to the binding reaction. (B) ZO-1 PDZ domains bind directly to ARVCF in blot overlays. In vitro-translated ARVCF or ARVCF(ΔP) were fractionated by SDS-PAGE, transferred to PVDF membranes, and incubated with ZO-1 PDZ domains fused to GST. Bound GST-ZO-1 PDZ was detected with a labeled anti-GST antibody. (C) ARVCF coprecipitates with ZO-1 from transfected MDCK cells. Control cells (MDCK) or cells expressing ARVCF, ARVCF(ΔP), ARVCF(ΔA), or ARVCF(ΔAΔP) were lysed and equal amounts of total protein used to immunoprecipitate ZO-1. Precipitates were blotted to detect ARVCF (anti-HA and anti-ARVCF) that was bound to ZO-1. An aliquot of the cell lysate (5%) was directly blotted to determine the amount of wild-type and mutant ARVCF present in the lysates. Untransfected cells served as a control for the specificity of the precipitation; nd, not determined. (D) Endogenous ARVCF and ZO-1 coprecipitate from MDCK cells. Endogenous ZO-1 was immunoprecipitated from lysates of MDCK cells and precipitates were analyzed by Western blot by using an anti-ARVCF antibody to detect associated ARVCF. Beads coated with an irrelevant antibody (IgG) failed to precipitate ARVCF. An aliquot of the MDCK cell lysate (5%), or for comparision cells transfected with ARVCF, was directly blotted. Approximately 20× the amount of protein used in C was used for the coprecipitation of endogenous proteins.
These results thus establish a direct association between the PDZ-domains of ZO-1 and a C-terminal PDZ-binding motif in ARVCF.
ARVCF and ZO-1 Interact In Vivo and Partially Colocalize in Polarized MDCK Cells
To determine whether full-length ZO-1 and ARVCF interact in vivo, epithelial MDCK cells, which endogenously express ZO-1 and low levels of ARVCF (Figure 2C), were stably transfected with cDNAs coding for wild-type or mutant human ARVCF constructs carrying an N-terminal HA-tag.
To analyze whether ZO-1 and ARVCF associate in vivo, endogenous ZO-1 was immunoprecipitated and precipitates blotted with antibodies to HA or, as a control, ARVCF. ARVCF but not ARVCF(ΔP) coprecipitated with ZO-1 (Figure 2C). Similarly, an ARVCF mutant lacking the armadillo repeats [ARVCF(ΔA); Figure 1A] coprecipitated with ZO-1, but the association was lost if the PDZ-binding motif was mutated [ARVCF(ΔAΔP); Figure 1A]. Furthermore, endogenous ARVCF coprecipitated with ZO-1 from MDCK cells (Figure 2D), which express low levels of ARVCF (Figure 2C; Kaufmann et al., 2000), validating the coprecipitation experiments from cells overexpressing the tagged protein.
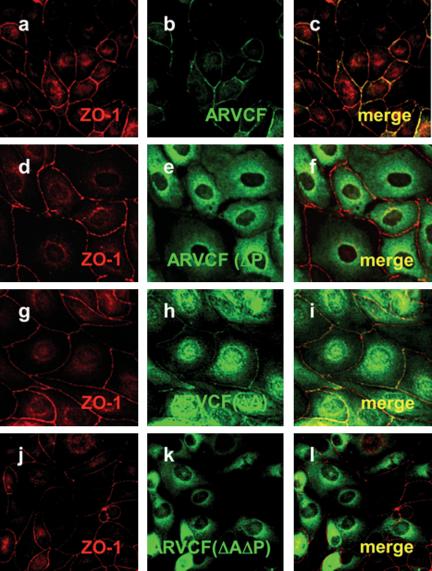
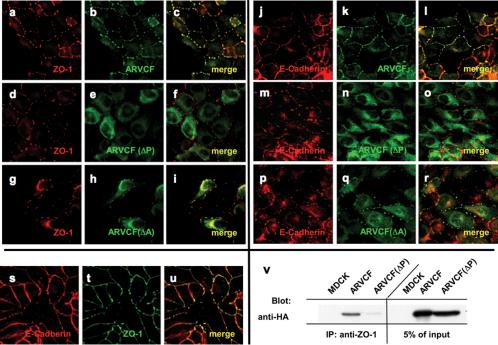
To determine whether ZO-1 and ARVCF colocalize, MDCK cells expressing wild-type or mutant ARVCF were stained with antibodies to ZO-1 and HA and analyzed by confocal immunofluorescence microscopy (Figure 3). In cells grown on coverslips, ARVCF was present at the plasma membrane at sites of cell-cell contact, where it extensively colocalized with ZO-1 (a-c). In contrast, ARVCF(ΔP) was cytoplasmic and absent from the plasma membrane and the nucleus (d-f). Compared with wild-type ARVCF, the amount of ARVCF(ΔA) at the plasma membrane was reduced but still detectable, with an increased fraction found in the cytoplasm and nucleus (g and h). The fraction of ARVCF(ΔA) present on the plasma membrane extensively colocalized with ZO-1. Similar to ARVCF(ΔP), ARVCF(ΔAΔP) was absent from the plasma membrane and excluded from the nucleus (j-l).
Figure 3.
Colocalization of ARVCF and ZO-1 in transfected MDCK cells. MDCK cells transfected with wild-type ARVCF (a-c) or mutant ARVCF(ΔP) (d-f), ARVCF(ΔA) (g-i), or ARVCF(ΔAΔP) (j-l) were grown on glass coverslips, fixed, permeabilized, and stained for ZO-1 (red, a, d, g, and j) and ARVCF (green, b, e, h, and k). Confocal images of the ZO-1 and ARVCF labeling were merged to show regions of colocalization (yellow, c, f, i, and l).
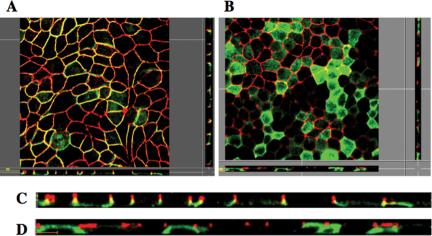
The colocalization of ARVCF and ZO-1 was confirmed in polarized MDCK cell monolayers grown on permeable polycarbonate filters (Figure 4). Cells were stained for ZO-1 and ARVCF, and images of horizontal sections at the height of the TJ (Figure 4, A and B) or vertical sections along the apico-basal axis, c and d) of the cell monolayer were acquired. Although ARVCF (green, A and C) predominantly localized to the lateral plasma membrane, ARVCF(ΔP) was absent from the plasma membrane (B and C), consistent with the results for cells grown on coverslips (Figure 3). Although ARVCF was found along the length of the lateral membrane, a significant degree of colocalization with ZO-1 (red) was observed only in the apical region of the lateral membrane (yellow). In contrast, ARVCF(ΔP) did not colocalize with ZO-1. Interestingly, in contrast to the characteristic and well localized punctuate staining for ZO-1 in control cells (our unpublished data) or cells expressing ARVCF(ΔP) (D), the ZO-1 staining extended further down into the lateral membrane in cells expressing ARVCF (C). Expression levels of endogenous ARVCF in MDCK cells were too low to be detected by immunostaining (Mariner et al., 1999).
Figure 4.
ARVCF partially colocalizes with ZO-1 to a discrete region between the lateral plasma membrane and TJ of polarized MDCK cells. MDCK cells transfected with ARVCF (a and c) or ARVCF(ΔP) (b and d) were grown on polycarbonate filters to obtain polarized cell monolayers. Cells were fixed, permeabilized and stained for ARVCF (green) and ZO-1 (red). Confocal images corresponding to horizontal sections at the height of TJ (a and b) or to vertical sections along the apico-basal axis of the monolayer (c and d) were acquired and merged. Yellow denotes regions where ARVCF and ZO-1 colocalize. Bar, 10 μm.
In conclusion, ARVCF and ZO-1 associate and partially colocalize in vivo, with both the interaction and colocalization being dependent on the presence of an intact PDZ-binding motif in ARVCF.
Colocalization and Association of ARVCF with ZO-1 Does Not Require an Intact Actin Cytoskeleton
Because ZO-1 and ARVCF are found in adhesion complexes that link to F-actin, we analyzed whether compromising the integrity of the actin cytoskeleton by using cytochalasin D altered the colocalization and association of the two proteins. Cytochalasin D severely disrupted the normal organization of the actin cytoskeleton in MDCK cells (our unpublished data) and the contiguous localization of ZO-1 and ARVCF to areas of cell-cell contact (Figure 2) was broken up into strings of patches were ZO-1 and ARVCF extensively colocalized (Figure 5, a-c). The cytoplasmic localization of ARVCF(ΔP) was not altered by the drug (d-f). ARVCF(ΔA) showed a punctuate staining pattern similar to ARVCF, but the fraction present in the cytosol was increased in drug-treated cells (g-i).
Figure 5.
Colocalization and coprecipitation of ARVCF and ZO-1 or E-cadherin in transfected MDCK cells treated with cytochalasin D. (a-u) Colocalization of ZO-1, E-cadherin, and ARVCF. MDCK cells transfected with wild-type ARVCF (a-c and j-l) or mutant ARVCF(ΔP) (d-f and m-o) or ARVCF(ΔA) (g-i or p-r) were fixed, permeabilized, and stained for ZO-1 (red, a, d, and g), E-cadherin (red, j, m, and p), and wild-type or mutant ARVCF (green, b, e, h, k, n, and q). Confocal images of the ZO-1 or E-cadherin and ARVCF labeling were merged to show regions of colocalization (yellow, c, f, i, l, o, and r). Colocalization of E-cadherin (red, s) and ZO-1 (green, t) in cytochalasin-treated cells is shown in yellow (u). v, ARVCF coprecipitates with ZO-1 from cytochalasin D-treated MDCK cells. Control cells (MDCK) or cells expressing ARVCF or ARVCF(ΔP) were lysed, and equal amounts of total protein were used to immunoprecipitate ZO-1. Precipitates were blotted to detect ARVCF that was bound to ZO-1. An aliquot of the cell lysate (5%) was directly blotted to determine the amount of wild-type and mutant ARVCF present in the lysates. Untransfected cells served as a control for the specificity of the precipitation.
As expected from their known association, also ARVCF and E-cadherin extensively colocalized in cytochalasin D-treated cells (j-l). Whereas ARCVF(ΔP) did not colocalize with E-cadherin (m-o), a detectable fraction of ARVCF(ΔA) was present in E-cadherin-containing patches (p-r). As expected from the colocalization of ARVCF with ZO-1 as well as with E-cadherin, also ZO-1 and E-cadherin colocalized in cytochalasin D-treated cells (s-u). Furthermore, ZO-1 and ARVCF but not ARVCF(ΔP) coprecipitated from cytochalasin D-treated cells (v).
Thus, the colocalization and interaction of ZO-1 with ARVCF does require an intact actin cytoskeleton.
ARVCF Can Associate with E-Cadherin via Its Armadillo Domains or through Binding to PDZ-Domain Proteins
Intriguingly, the PDZ binding motif seemed to be critical for plasma membrane localization of ARVCF in MDCK (Figures 2, 3, 4), 293T (our unpublished data) and MCF7 (see below) cells, whereas deletion of the armadillo repeats, which have been shown to mediate the interaction between ARVCF and M-cadherin (Kaufmann et al., 2000), only partially affected plasma membrane localization (Figures 4 and 5). We therefore analyzed the contributions of the armadillo repeats in binding of ARVCF to E-cadherin, both in vitro and in vivo.
An immobilized GST-fusion protein carrying the cytosolic domain of E-cadherin bound in vitro-translated ARVCF (Figure 6A), confirming the direct interaction of ARVCF with cadherins (Kaufmann et al., 2000; Mariner et al., 2000). Deletion of the armadillo domains in ARVCF abolished the interaction, similar to the results obtained for M-cadherin (Kaufmann et al., 2000). The interaction of ARVCF(ΔP) with the GST-E-cadherin fusion protein was comparable with that of wild-type ARVCF, indicating that the PDZ-binding motif is dispensable for E-cadherin binding in vitro.
Figure 6.
Binding of ARVCF and E-cadherin. (A) Binding of in vitro-translated wild-type and mutant ARVCF to a GST fusion protein containing the cytosolic domain of E-cadherin. GST or GST-E-cadherin tail fusion proteins were coupled to glutathione beads and incubated with in vitro translated, radioactively labeled, ARVCF, ARVCF(ΔP), ARVCF(ΔA), or ARVCF(ΔAΔP). Protein bound to the beads (pull-down) was analyzed by SDS-PAGE and autoradiography. An aliquot (5%) of the in vitro-translated material was directly analyzed to confirm that similar amounts of the in vitro translated proteins were added to the binding reaction. (B) Coprecipitation of wild-type or mutant ARVCF with E-cadherin. Control cells (MDCK) or cells expressing ARVCF, ARVCF(ΔP), ARVCF(ΔA), or ARVCF(ΔAΔP) were lysed, and equal amounts of total protein were used to immunoprecipitate E-cadherin. Precipitates were blotted to detect ARVCF (anti-HA) that was bound to E-cadherin. Alternatively, wild-type or mutant ARVCF was precipitated with the anti-HA antibody, and precipitates were blotted to detect bound E-cadherin. An aliquot of the cell lysate (5%) was directly blotted to determine the total amount of ARVCF or E-cadherin, respectively, present in the lysates. Untransfected cells served as a control for the specificity of the precipitations. (C) Coprecipitation of ZO-1 and E-cadherin. Control cells (MDCK) or cells expressing ARVCF or ARVCF(ΔP) were lysed, and equal amounts of total protein were used to immunoprecipitate ZO-1. Precipitates were then analyzed by Western blot to detect E-cadherin that was bound to ZO-1. An aliquot of the cell lysate (5%) was directly blotted to determine the amount of E-cadherin present in the lysate. (D) Quantification. The relative amounts of E-cadherin that coprecipitated with ZO-1 from control cells or those expressing wild-type or mutant ARVCF was quantified. (E) In vitro association of ZO-PDZ, ARVCF, and E-cadherin. Immoblized GST or GST-ZO-1 PDZ domain fusion proteins were incubated with in vitro translated ARVCF or ARVCF(ΔP) and E-cadherin. Proteins bound to the beads were then identified by Western blot. An aliquot of the in vitro translation reaction was directly blotted (i; input) to monitor the amount of ARVCF, ARVCF(ΔP), and E-cadherin added to the reaction.
In vivo, however, the association of ARVCF(ΔP) with endogenous E-cadherin was severely impaired compared with wild-type ARVCF (Figure 6B). ARVCF(ΔA), on the other hand, retained E-cadherin binding activity, which was abolished only if the PDZ-binding motif also was mutated. The ability of the different ARVCF constructs to coprecipitate with E-cadherin in vivo therefore correlated with the extent of their colocalization with E-cadherin (Figure 4) and their presence at the plasma membrane (Figures 3, 4, 5). Thus, in vivo, the armadillo domains as well as the C-terminal PDZ-binding motif contribute to the plasma membrane localization of ARVCF and its association with E-cadherin.
Given the interaction of ARVCF with cadherins and ZO-1, we next analyzed whether E-cadherin coprecipitated with ZO-1 and whether the association was affected by overexpressing ARVCF. E-cadherin was detected in ZO-1 precipitates from control cells or cells expressing ARVCF or ARVCF(ΔP) (Figure 6C). Interestingly, however, significantly more E-cadherin coprecipitated with ZO-1 from cells expressing ARVCF compared with either control cells or cells expressing ARVCF(ΔP) (Figure 6D), suggesting that ARVCF can link ZO-1 to E-cadherin. Basal levels of E-cadherin present in the ZO-1 precipitates from control cells or cells expressing ARVCF(ΔP) likely reflect the known association of ZO-1 with E-cadherin via α-catenin (Itoh et al., 1993, 1997; Rajasekaran et al., 1996; Ando-Akatsuka et al., 1999).
To confirm complex formation between E-cadherin, ARVCF, and ZO-1 in vitro, ARVCF and the cytosolic domain of E-cadherin were translated in vitro and incubated with immobilized GST-ZO-1 PDZ. ARVCF and the E-cadherin tail bound to the ZO-1 PDZ domains (Figure 6E), and binding of either protein required an intact PDZ-binding motif in ARVCF. This experiment confirms the ability of ARVCF to link E-cadherin to ZO-1 and shows that a complex containing the three proteins can assemble in vitro.
In conclusion, ARVCF can link ZO-1 to E-cadherin, and in vivo, the armadillo domains of ARVCF as well as its interaction with PDZ domain proteins contribute to the association of ARVCF with E-cadherin.
Plasma Membrane and Nuclear Localization of ARVCF Require the Interaction with PDZ-Domain Proteins and Are Regulated by Cell-Cell Adhesion
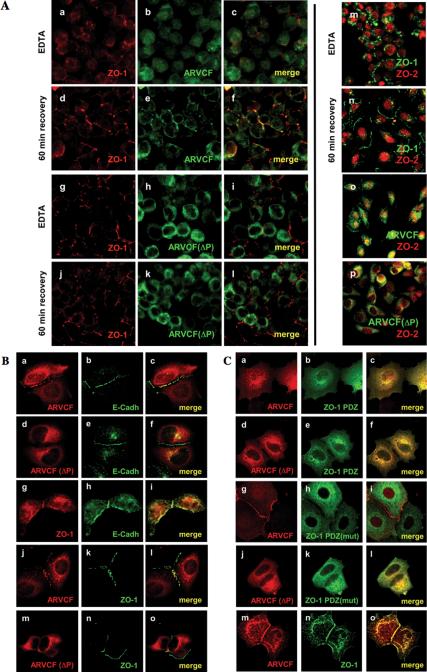
Variable degrees of nuclear localization have been reported for ARVCF in different cell types (Kaufmann et al., 2000; Mariner et al., 2000; Waibler et al., 2001). To determine whether nuclear localization of ARVCF was regulated by cell-cell adhesion, a “calcium-switch” protocol was used to reversibly disassemble and reseal AJs and TJs (Lawrence et al., 2002). Briefly, filter-grown MDCK cells expressing ARVCF were treated with EDTA to break down cell-cell adhesion and the subcellular distribution of ARVCF and ZO proteins was analyzed, either immediately after the EDTA treatment or after recovery to allow the cells to reestablish cell-cell contact.
Calcium depletion resulted in the loss of ARVCF and ZO-1 from the plasma membrane (Figure 7A, a-c). Although ARVCF was observed in both the cytoplasm and the nucleus, ARVCF(ΔP) was excluded from the nucleus (g-i). ZO-1 remained in the cytosol showing a punctuate staining similar to that previously reported under these conditions (Ivanov et al., 2004). After a 60-min recovery, ARVCF and ZO-1 were recruited to sites of initial cell-cell contact, where they extensively colocalized (d-f). The amount of ARVCF present in the nucleus was notably reduced after recovery. Although ZO-1 relocalized to sites of cell-cell adhesion in cells expressing ARVCF(ΔP), the latter remained cytosolic (j-i).
Figure 7.
Plasma membrane recruitment and nuclear localization of ARVCF require the PDZ-binding motif and are regulated by cell-cell adhesion. (A) Calcium switch experiments. MDCK cells grown on filters and expressing either ARVCF (a-f) or ARVCF(ΔP) (g-l) were incubated with EDTA to deplete calcium and disrupt cell-cell adhesion. The distribution of ARVCF (green) and ZO-1 (red) was analyzed after EDTA-treatment or after the incubation for 60 min in calcium-containing media to allow reestablishment of cell-cell adhesion, by staining with suitable antibodies and visualization by confocal microscopy. In panels where staining for ARVCF and ZO-1 was merged, yellow indicates colocalization of the two proteins. In m-p, ZO-2 (red) was colocalized with ZO-1 (green, m and n), or ARVCF or ARVCF(ΔP) (green, o and p, respectively), either after calcium depletion or a 60-min recovery. (B) Recruitment of ARVCF to sites of cell-cell contact in MCF7 cells. Sparsely plated MCF7 cells either transfected with cDNAs for ARVCF (a-c and j-l), ARVCF(ΔP) (d-f and m-o), or not transfected (g-i) were processed for immunofluorescence staining, and the localization of ARVCF, ARVCF(ΔP), and endogenous E-cadherin and ZO-1 visualized by confocal microscopy. Colocalization of the respective proteins is yellow in the merged images. (C) Effect of ZO-1 mutants on ARVCF localization in MCF7 cells. Sparse MCF7 cells coexpressing ARVCF or ARVCF(ΔP) (a-c, g-i and d-f, j-l, respectively) and either ZO-1 PDZ domains (a-f), a ZO-1 PDZ carrying inactivated PDZ-domains [PDZ(mut); g-l] or a full-length ZO-1 construct were analyzed by immunofluorescence staining and confocal microscopy. ZO-1 constructs carried an N-terminal myc tag. Colocalization of the respective proteins is yellow in the merged images.
ZO-1 released from the plasma membrane was excluded from the nucleus after calcium depletion, whereas ZO-2 efficiently accumulated in the nucleus (m). Redistribution of ZO-2 from the nucleus back to sites of cell-cell contact lagged behind that of ZO-1 or ARVCF (n-p) and required a longer recovery (our unpublished data; Islas et al., 2002).
Thus, disruption of cell-cell adhesion results in the release of ARVCF from the plasma membrane and increased appearance in the nucleus. Furthermore, nuclear localization of ARVCF apparently requires the interaction with a PDZ-domain protein.
ARVCF, ZO-1, and E-Cadherin Are Recruited to Sites of Initial Cell-Cell Contact
Given the presence of ARVCF at sites of cell-cell contact after recovery from calcium depletion, we further characterized the recruitment of ARVCF, ZO-1, and E-cadherin to sites of initial cell-cell adhesion in sparse cultures of transiently transfected MCF7 cells.
As shown in Figure 7B and similar to MDCK cells, ARVCF and E-cadherin (a-c), ZO-1 and E-cadherin (g and h), as well as ARVCF and ZO-1 extensively colocalized to sites of cell-cell contact. In cells expressing ARVCF(ΔP), which remained cytoplasmic, E-cadherin (d-f) and ZO-1 (m-o) were still recruited to contact sites.
Because plasma membrane localization of ARVCF required an intact PDZ-binding motif and thus the ability to interact with ZO-1 (or other PDZ-domain proteins), we analyzed whether overexpressing ZO-1 mutants affects the recruitment of ARVCF to regions of cell-cell adhesion. Overexpressing an N-terminal portion of ZO-1 encoding the three PDZ domains (Figure 1B, ZO-1 PDZ) interfered with the recruitment of ARVCF to sites of cell-cell contact (Figure 7C, a-c). As observed in MDCKI cells (Reichert et al., 2000b), the ZO1 PDZ domains remained mostly cytoplasmic and presumably prevented recruitment of ARVCF to the plasma membrane by competing for its interaction with wild-type ZO-1 and/or other PDZ-domain proteins. In contrast, overexpression of a ZO-1 PDZ construct in which the ability of the PDZ domains to interact with ligand had been obliterated [Figure 1B, ZO-1 PDZ(mut)], did not interfere with plasma membrane recruitment of ARVCF (g-i). As expected, neither ZO-1 PDZ nor ZO-1 PDZ(mut) altered the cytoplasmic distribution of ARVCF(ΔP) (d-f and j-l) nor was membrane localization of ARVCF affected by overexpressing wild-type ZO-1 (m-o). Coimmunoprecipitation experiments confirmed that in vitro-translated [35S]methionine-labeled ARVCF and ZO-1 PDZ domains associated with each other and that the binding was abolished by mutation of either the PDZ-binding motif in ARVCF (ARVCF(ΔP) or the PDZ domains in ZO-1 PDZ [ZO-1 PDZ(mut)] (Figure 8, v).
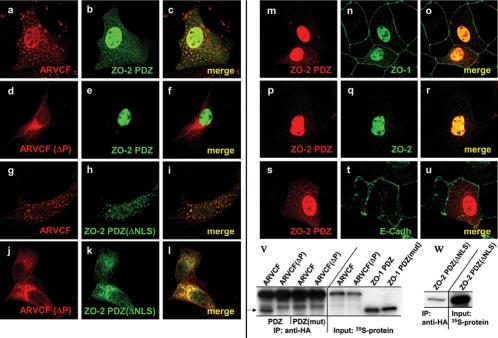
Figure 8.
Role of ZO-2 in nuclear localization of ARVCF. MDCK cells expressing ARVCF (a-c and g-i) or ARVCF(ΔP) (d-f and j-l) were transfected with cDNAs encoding the ZO-2 PDZ domains (a-f) or ZO-2 PDZ domains with an inactivated NLS [ZO-2 PDZ(ΔNLS), g-l] and analyzed by immunofluorescence staining and confocal microscopy. Colocalization of the respective proteins is yellow in the merged images. In m-u, the distribution of endogenous ZO-1, ZO-2, and E-cadherin was analyzed in MDCK cells transfected with a plasmid encoding the ZO-2 PDZ domains. ZO-2 constructs carried an N-terminal Xpress tag. Colocalization of the respective proteins is yellow in the merged images. v, mutation of the PDZ domains in ZO-1 PDZ abolishes the interaction with ARVCF. ARVCF or ARVCF(ΔP) and ZO-1 PDZ or ZO-1 PDZ(mut) were in vitro cotranslated in the presence of [35S]methionine. ARVCF or ARVCF(ΔP) was then immunoprecipitated, the precipitates fractionated by SDS-PAGE, and the dried gels used for autoradiography. w, deletion of the NLS in ZO-2 PDZ(ΔNLS) does not affect the interaction with ARVCF. ARVCF and ZO-2 PDZ(ΔNLS) were in vitro cotranslated in the presence of [35S]methionine, ARVCF was then immunoprecipitated, the precipitates fractionated by SDS-PAGE, and the dried gels used for autoradiography. An aliquot of the in vitro translation reaction was directly analyzed to monitor the amount of protein added to the reaction. The arrow shows the location of ZO-1 PDZ (v).
Thus, E-cadherin, ZO-1, and ARVCF are recruited to sites of initial cell-cell contact. Binding of the ZO-1 PDZ domains per se does not facilitate membrane recruitment of ARVCF, indicating a requirement for the intact ZO-1 and possibly its association with membrane proteins and/or the cytoskeleton for this process.
The PDZ Domains of ZO-2 Mediate Efficient Nuclear Localization of ARVCF
Given the interaction between the ZO-2 PDZ-domains and ARVCF (Table 1) and the nuclear localization of the two proteins after disruption of cell-cell adhesion (Figure 7A), we explored the possible role for ZO-2 in nuclear localization of ARVCF. ZO-2 has several nuclear localization signals (NLSs) within its N-terminal part (Jaramillo et al., 2004), and we therefore analyzed the subcellular distribution of ARVCF in MDCK cells cotransfected with cDNAs for ARVCF and ZO-2 PDZ (Figure 1B) or a mutant with impaired nuclear localization capacity [ZO-2 PDZ(ΔNLS); Figure 1B].
Overexpression of ZO-2 PDZ displaced ARVCF from the plasma membrane into punctate cytoplasmic structures and strongly enhanced nuclear accumulation of ARVCF (Figure 8, a-c). ARVCF and ZO-2 PDZ colocalized not only in the nucleus but also in the cytoplasmic structures. The ZO-2 PDZ induced displacement of ARVCF from the plasma membrane as well as the colocalization support the interaction between the two proteins. ARVCF(ΔP) was excluded from the nucleus (d and e), showing that nuclear localization of ARVCF required its ability to interact with ZO-2 PDZ.
ZO-2 PDZ(ΔNLS) was still able to relocalize ARVCF from the plasma membrane to the cytoplasmic structures (g-l), indicating that deletion of the NLS did not affect the interaction with ARVCF (see below). ARVCF, however, was effectively excluded from the nucleus in cells expressing ZO-2 PDZ(ΔNLS), directly implicating ZO-2 PDZ in nuclear translocation and/or retention of ARVCF.
Interestingly, the ZO-2 PDZ domains also partially relocalized endogenous ZO-1 from sites of cell-cell contact to the nucleus (m-o), likely reflecting association between ZO-1 and ZO-2 through PDZ domain interactions (Wittchen et al., 1999). Consistent with previous observations (Jaramillo etal., 2004), endogenous ZO-2 accumulated in the nucleus of sparse cell cultures (p-r). The plasma membrane localization of E-cadherin, in contrast, was not altered by the expression of ZO-2 PDZ (s-u). As expected from the functional effect of ZO-2 PDZ(ΔNLS) on ARVCF localization and its codistribution with ARVCF, coimmunoprecipitation experiments using vitro-translated [35S]methionine-labeled ARVCF and ZO-2 PDZ(ΔNLS) confirmed the interaction between the two proteins (Figure 8, w).
In conclusion, the ZO-2 PDZ domains mediate nuclear accumulation of ARVCF in a process that requires both a PDZ-based interaction and the NLS in ZO-2, consistent with a role for ZO-2 in nuclear translocation and/or retention of ARVCF.
DISCUSSION
We identified ARVCF as a binding partner of ZO-1 and ZO-2 and characterized the role of PDZ-domain proteins in plasma membrane and nuclear localization of ARVCF. Based on several assays, including yeast two-hybrid, pull-down, and ligand overlay, ARVCF was shown to bind directly to ZO-1 through the interaction of a C-terminal type 1 PDZ-binding motif (SWV) in ARVCF and PDZ domains in ZO-1. ARVCF also interacts with PDZ domains of ZO-2, but not ZO-3, confirming the notion that different ZO proteins recruit distinct partners to TJs.
Coprecipitation and colocalization experiments showed that ARVCF and ZO-1 also interact in vivo. Interestingly, the two proteins only colocalized in the apical most region of the lateral plasma membrane of fully polarized MDCK cells, thus defining two distinct pools of plasma membrane-associated ARVCF, one in the region of the apical junctional complex, the second along the length of the lateral domain. Because inactivation of the PDZ-binding motif led to the loss of the bulk of ARVCF from the plasma membrane, PDZ-domain proteins other than ZO-1 such as Erbin (Laura et al., 2002) are likely to be involved in localizing ARVCF to the lateral plasma membrane. Cell type-specific expression of different PDZ-domain proteins promoting the interaction of ARVCF with cadherins or other membrane-associated proteins may thus explain why colocalization of ARVCF and cadherins is often dependent on the cellular context (Waibler et al., 2001). Interestingly, overexpression of ARVCF, but not ARVCF(ΔP), resulted in a more lateral distribution of ZO-1, consistent with the two proteins interacting. Because endogenous ARVCF levels are too low to be detected in MDCK cells (see also Mariner et al., 1999), it is not clear whether the endogenous protein preferentially localizes to the more apical or basal region, or whether it is distributed along the length of the lateral plasma membrane. In any case, because the bulk of the overexpressed ARVCF localized to the plasma membrane, the mechanism(s) that recruit ARVCF were not saturated in the transfected MDCK cells.
ARVCF can directly interact through its armadillo repeats with cadherins (Kaufmann et al., 2000; Waibler et al., 2001), and it has been proposed that membrane localization of ARVCF is mediated through this association. It was therefore surprising that inactivation of the PDZ-binding motif resulted in the loss of ARVCF from the plasma membrane, in particular because cadherins do not contain PDZ domains. We confirmed that in vitro, the armadillo repeats are indeed required for binding ARVCF to the cytosolic tail of E-cadherin. Under these conditions, inactivation of the PDZ-binding motif does not affect the association of the two proteins. In vivo, however, ARVCF lacking the armadillo domains was still present at the plasma membrane, albeit at reduced levels. A fraction of the mutant lacking the armadillo repeats also coprecipitated with E-cadherin and only the inactivation of the PDZ-binding motif in ARVCF(ΔA) led to the complete loss of membrane localization and E-cadherin coprecipitation. This suggests that ARVCF can bind to E-cadherin via different mechanisms (Figure 9A). On one hand, ARVCF may associate with cadherins via the armadillo repeats and with ZO-1 via the PDZ-binding motif, thereby linking ZO-1 to E-cadherin. Indeed, a complex between ZO-1 PDZ domains, E-cadherin and ARVCF could form in vitro and overexpression of ARVCF increased the amount of ZO-1 that coprecipitated with E-cadherin. Such a complex also may explain the redistribution of ZO-1 from TJs to a more lateral location. Alternatively, based on the known association of ZO-1 with the cadherin complex via α-catenin (Itoh et al., 1993, 1997; Rajasekaran et al., 1996; Ando-Akatsuka et al., 1999), ZO-1 could link ARVCF indirectly to E-cadherin. Such an interaction could explain the displacement of ARVCF from the plasma membrane after overexpression of the ZO-1 (or ZO-2) PDZ domains. In addition, it also is possible that a fraction of ARVCF is recruited to tight junctions by directly binding to ZO-1 or ZO-2.
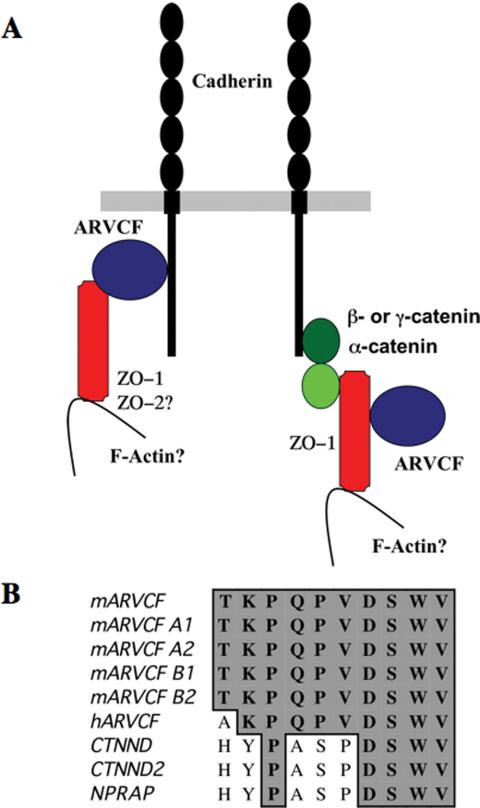
Figure 9.
Putative E-cadherin/ARVCF/ZO-1 associations and C-terminal PDZ binding motifs in members of the p120ctn protein family. (A) Schematic model of possible E-cadherin complexes. Cadherins may associate directly with the armadillo repeats of ARVCF, which in turn may bind to the ZO-1 PDZ domains via its PDZ-binding motif. Alternatively, cadherins may bind ARVCF indirectly via their known interaction with β- or γ-catenin, α-catenin, and ZO-1. ZO-2 also may associate with ARVCF. (B) Alignment of the C terminus of different members of the p120ctn protein family. The C-terminal 10 amino acids of ARVCF splice variants and the different members of the p120ctn family are aligned. Identical amino acids are boxed and shaded. With the exception of p120ctn itself, the C-terminal PDZ-binding SWV motif identified in ARVCF is conserved, indicating that other members of the p120ctn family also may bind to ZO-1, ZO-2, and possibly other PDZ-domain proteins.
Plasma membrane localization of ARVCF not only required a functional PDZ-binding motif but also cell-cell adhesion. Dissociation of cell-cell adhesion or sparse culture conditions led to the release of ARVCF from the plasma membrane into the cytoplasm and the nucleus. Although a presence of ARVCF in the nucleus has been reported (Mariner et al., 2000; Waibler et al., 2001), a cell-cell adhesion-dependent shuttling between plasma membrane and nuclear localizations has not been documented. Even without affecting cell-cell adhesion, nuclear localization of ARVCF(ΔA) was increased, correlating with the less efficient membrane localization of ARVCF after deletion of the armadillo repeats. Under any condition, mutation of the PDZ-binding motif resulted in the effective exclusion of ARVCF from the nucleus, showing that nuclear translocation and/or retention requires a functional PDZ-binding motif and hence the interaction with a PDZ-domain protein.
ZO-1 remained cytoplasmic in sparse cell cultures or after disruption of cell-cell adhesion, suggesting that it does not participate in nuclear localization of ARVCF. ZO-2, in contrast, relocated to the nucleus with ARVCF, and, given the interaction between the ZO-2 PDZ domains and ARVCF, raised the possibility that ZO-2 may play a role in nuclear localization of ARVCF. Such a role for ZO-2 is indeed supported by the ability of the ZO-2 PDZ domain to efficiently relocate ARVCF from the plasma membrane to the nucleus in a process that required the ability of the two proteins to interact and the presence of a functional NLS in the ZO-2 PDZ domains. Thus, ZO-2 could be involved in nuclear translocation and/or retention of ARVCF and play a role in regulating postulated functions of ARVCF in gene expression. Interestingly, in addition to the nuclear relocalization, the PDZ domains of ZO-2 induced cytosolic patches that contained ARVCF. Although the nature of these patches remains to be elucidated, they did not contain E-cadherin nor did they colocalize with a lysosomal marker (our unpublished data).
Little is known about the role of ARVCF in cell-cell adhesion. Members of the p120ctn family bind laterally to cadherins and have been postulated to regulate, positively or negatively, the lateral clustering of adhesion complexes (Ozawa and Kemler, 1998) and thus adhesive strength (Ozawa and Kemler, 1998; Yap et al., 1998). More recently, several p120ctn family members, including ARVCF, have been implicated in cadherin turnover (Davis et al., 2003). Despite a common role in cadherin turnover, ARVCF lacks the cellular branching activity associated with the expression of p120ctn in fibroblasts (Reynolds et al., 1996; Mariner et al., 2000), or the induction of a fibroblastoid phenotype observed for δ-catenin in MDCK cells (Lu et al., 1999). Consistent with these studies, we did not observe morphological changes by overexpressing wild-type or mutant ARVCF in several cell lines, including 3T3 adipocytes, where Xenopus laevis ARVCF has been reported to induce branching (Fang et al., 2004). This apparently contradictory result may reflect the fact that Xenopus ARVCF is structurally more closely related to mammalian p120ctn than to mammalian ARVCF. Although E-cadherin, ZO-1, and ARVCF may play a role in initial stages of cell-cell adhesion, assembly of junctional components was not visibly altered by the overexpression of wild-type or mutant ARVCF. Also other functional parameters, such as transepithelial resistance, cyst formation in Matrigel, and wound healing were not significantly altered, possibly reflecting functional redundancy between ARVCF and the more abundant p120ctn (Davis et al., 2003; Fang et al., 2004). Because ZO-1 links to the actin cytoskeleton (Fanning et al., 1998, 2002; Wittchen et al., 1999), one function of the interaction of ARVCF with ZO-1 may be to anchor cadherins, possibly in the vicinity of the apical adhesion complex, to F-actin.
In conclusion, although the precise cellular functions of ARVCF remain to be elucidated, its recruitment to either the plasma membrane or the nucleus, and hence postulated functions in adhesion and transcription, respectively, seem to be tightly regulated by interactions with PDZ-domain proteins. The C-terminal PDZ-binding motif in ARVCF is conserved among the different alternative splice variants and, with the exception of p120ctnitself, is also found in the other p120ctn family members (Figure 9B), suggesting that they may also bind ZO-1, ZO-2, or other PDZ-domain proteins. Whether these interactions regulate the subcellular localization of other members of the p120ctn family remains to be determined.
Acknowledgments
We thank Pascal Beguin for critically reading the manuscript, Sachdev Sidhu and Anna Starzinski-Powitz for helpful discussions, and Werner Franke and Lorenza Gonzalez-Mariscal for providing antibodies to ARVCF and the ZO-2 PDZ constructs, respectively. This work was supported by the Agency for Science, Technology and Research, Singapore.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-04-0350. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0350.
References
- Adams, C.L., Chen, Y.T., Smith, S.J., and Nelson, W.J. (1998). Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando-Akatsuka, Y., Yonemura, S., Itoh, M., Furuse, M., and Tsukita, S. (1999). Differential behavior of E-cadherin and occludin in their colocalization with ZO-1 during the establishment of epithelial cell polarity. J. Cell. Physiol. 179, 115-125. [DOI] [PubMed] [Google Scholar]
- Angst, B.D., Marcozzi, C., and Magee, A.I. (2001). The cadherin superfamily: diversity in form and function. J. Cell Sci. 114, 629-641. [DOI] [PubMed] [Google Scholar]
- Barker, R.J., Price, R.L., and Gourdie, R.G. (2002). Increased association of ZO-1 with connexin43 during remodeling of cardiac gap junctions. Circ. Res. 90, 317-324. [DOI] [PubMed] [Google Scholar]
- Borrmann, C.M., Mertens, C., Schmidt, A., Langbein, L., Kuhn, C., and Franke, W.W. (2000). Molecular diversity of plaques of epithelial-adhering junctions. Ann. N.Y. Acad. Sci. 915, 144-150. [DOI] [PubMed] [Google Scholar]
- Cereijido, M., Shoshani, L., and Contreras, R.G. (2000). Molecular physiology and pathophysiology of tight junctions. I. Biogenesis of tight junctions and epithelial polarity. Am. J Physiol. 279, G477-G482. [DOI] [PubMed] [Google Scholar]
- Davis, M.A., Ireton, R.C., and Reynolds, A.B. (2003). A core function for p-120-catenin in cadherin turnover. J. Cell Biol. 163, 525-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D.A., Lee, A., Lewis, J., Kim, E., Sheng, M., and MacKinnon, R. (1996). Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Fang, X., Ji, H., Kim, S.W., Park, J.I., Vaught, T.G., Anastasiadis, P.Z., Ciesiolka, M., and McCrea, P.D. (2004). Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. J. Cell Biol. 165, 87-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, A.S., Jameson, B.J., Jesaitis, L.A., and Anderson, J.M. (1998). The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273, 29745-29753. [DOI] [PubMed] [Google Scholar]
- Fanning, A.S., Ma, T.Y., and Anderson, J.M. (2002). Isolation and functional characterization of the actin binding region in the tight junction protein ZO-1. FASEB J. 16, 1835-1837. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal, L., Betanzos, A., and Avila-Flores, A. (2000). MAGUK proteins: structure and role in the tight junction. Semin. Cell Dev. Biol. 11, 315-324. [DOI] [PubMed] [Google Scholar]
- Honing, S., and Hunziker, W. (1995). Cytoplasmic determinants involved in direct lysosomal sorting, endocytosis, and basolateral targeting of rat lgp120 (lamp-I) in MDCK cells. J. Cell Biol. 128, 321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas, S., Vega, J., Ponce, L., and Gonzalez-Mariscal, L. (2002). Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp. Cell Res. 274, 138-148. [DOI] [PubMed] [Google Scholar]
- Itoh, M., Nagafuchi, A., Moroi, S., and Tsukita, S. (1997). Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J. Cell Biol. 138, 181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Nagafuchi, A., Yonemura, S., Kitani-Yasuda, T., and Tsukita, S. (1993). The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J. Cell Biol. 121, 491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Yonemura, S., Nagafuchi, A., and Tsukita, S. (1991). A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell-cell adhesion sites. J. Cell Biol. 115, 1449-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, A.I., Nusrat, A., and Parkos, C.A. (2004). Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol. Biol. Cell 15, 176-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo, B.E., Ponce, A., Moreno, J., Betanzos, A., Huerta, M., Lopez-Bayghen, E., and Gonzalez-Mariscal, L. (2004). Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp. Cell Res. 297, 247-258. [DOI] [PubMed] [Google Scholar]
- Kaufmann, U., Zuppinger, C., Waibler, Z., Rudiger, M., Urbich, C., Martin, B., Jockusch, B.M., Eppenberger, H., and Starzinski-Powitz, A. (2000). The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J. Cell Sci. 113, 4121-4135. [DOI] [PubMed] [Google Scholar]
- Kausalya, P.J., Reichert, M., and Hunziker, W. (2001). Connexin45 directly binds to ZO-1 and localizes to the tight junction region in epithelial MDCK cells. FEBS Lett. 505, 92-96. [DOI] [PubMed] [Google Scholar]
- Laura, R.P., Witt, A.S., Held, H.A., Gerstner, R., Deshayes, K., Koehler, M.F., Kosik, K.S., Sidhu, S.S., and Lasky, L.A. (2002). The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277, 12906-12914. [DOI] [PubMed] [Google Scholar]
- Lawrence, D.W., Comerford, K.M., and Colgan, S.P. (2002). Role of VASP in reestablishment of epithelial tight junction assembly after Ca2+ switch. Am. J. Physiol. 282, C1235-C1245. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Paredes, M., Medina, M., Zhou, J., Cavallo, R., Peifer, M., Orecchio, L., and Kosik, K.S. (1999). delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144, 519-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner, D.J., Sirotkin, H., Daniel, J.M., Lindman, B.R., Mernaugh, R.L., Patten, A.K., Thoreson, M.A., and Reynolds, A.B. (1999). Production and characterization of monoclonal antibodies to ARVCF. Hybridoma 18, 343-349. [DOI] [PubMed] [Google Scholar]
- Mariner, D.J., Wang, J., and Reynolds, A.B. (2000). ARVCF localizes to the nucleus and adherens junction and is mutually exclusive with p120(ctn) in E-cadherin complexes. J. Cell Sci. 113, 1481-1490. [DOI] [PubMed] [Google Scholar]
- Marrs, J.A., and Nelson, W.J. (1996). Cadherin cell adhesion molecules in differentiation and embryogenesis. Int. Rev. Cytol. 165, 159-205. [DOI] [PubMed] [Google Scholar]
- Mathur, M., Goodwin, L., and Cowin, P. (1994). Interactions of the cytoplasmic domain of the desmosomal cadherin Dsg1 with plakoglobin. J. Biol. Chem. 269, 14075-14080. [PubMed] [Google Scholar]
- Mauro, L., Bartucci, M., Morelli, C., Ando, S., and Surmacz, E. (2001). IGF-I receptor-induced cell-cell adhesion of MCF-7 breast cancer cells requires the expression of junction protein ZO-1. J. Biol. Chem. 276, 39892-39897. [DOI] [PubMed] [Google Scholar]
- Müller, T., Choidas, A., Reichmann, E., and Ullrich, A. (1999). Phosphorylation and free pool of beta-catenin are regulated by tyrosine kinases and tyrosine phosphatases during epithelial cell migration. J. Biol. Chem. 274, 10173-10183. [DOI] [PubMed] [Google Scholar]
- Nagafuchi, A. (2001). Molecular architecture of adherens junctions. Curr. Opin. Cell Biol. 13, 600-603. [DOI] [PubMed] [Google Scholar]
- Nieset, J.E., Redfield, A.R., Jin, F., Knudsen, K.A., Johnson, K.R., and Wheelock, M.J. (1997). Characterization of the interactions of alpha-catenin with alpha-actinin and beta-catenin/plakoglobin. J. Cell Sci. 110, 1013-1022. [DOI] [PubMed] [Google Scholar]
- Ozawa, M., and Kemler, R. (1998). The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J. Cell Biol. 142, 1605-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz, R., and Franke, W.W. (1997). Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation 61, 293-304. [DOI] [PubMed] [Google Scholar]
- Peifer, M., Berg, S., and Reynolds, A.B. (1994). A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76, 789-791. [DOI] [PubMed] [Google Scholar]
- Rajasekaran, A.K., Hojo, M., Huima, T., and Rodriguez-Boulan, E. (1996). Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J. Cell Biol. 132, 451-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert, M., Muller, T., and Hunziker, W. (2000a). The PDZ domains of zonula occludens-1 induce an epithelial to mesenchymal transition of Madin-Darby canine kidney I cells - Evidence for a role of beta-catenin/Tcf/Lef signaling. J. Biol. Chem. 275, 9492-9500. [DOI] [PubMed] [Google Scholar]
- Reichert, M., Muller, T., and Hunziker, W. (2000b). The PDZ domains of zonula occludens-1 induce an epithelial to mesenchymal transition of Madin-Darby canine kidney I cells. Evidence for a role of beta-catenin/Tcf/Lef signaling. J. Biol. Chem. 275, 9492-9500. [DOI] [PubMed] [Google Scholar]
- Reynolds, A.B., Daniel, J.M., Mo, Y.Y., Wu, J., and Zhang, Z. (1996). The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp. Cell Res. 225, 328-337. [DOI] [PubMed] [Google Scholar]
- Reynolds, A.B., Herbert, L., Cleveland, J.L., Berg, S.T., and Gaut, J.R. (1992). p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene 7, 2439-2445. [PubMed] [Google Scholar]
- Rimm, D.L., Koslov, E.R., Kebriaei, P., Cianci, C.D., and Morrow, J.S. (1995). Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA 92, 8813-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, M., and Sala, C. (2001). PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24, 1-29. [DOI] [PubMed] [Google Scholar]
- Sirotkin, H., et al. (1997). Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics 41, 75-83. [DOI] [PubMed] [Google Scholar]
- Staddon, J.M., Smales, C., Schulze, C., Esch, F.S., and Rubin, L.L. (1995). p120, a p120-related protein (p100), and the cadherin/catenin complex. J. Cell Biol. 130, 369-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, B.R., and Keon, B.H. (1998). The tight junction: morphology to molecules. Annu. Rev. Cell Dev. Biol. 14, 89-109. [DOI] [PubMed] [Google Scholar]
- Toyofuku, T., Yabuki, M., Otsu, K., Kuzuya, T., Hori, M., and Tada, M. (1998). Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J. Biol. Chem. 273, 12725-12731. [DOI] [PubMed] [Google Scholar]
- Vasioukhin, V., Bauer, C., Yin, M., and Fuchs, E. (2000). Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100, 209-219. [DOI] [PubMed] [Google Scholar]
- Waibler, Z., Schafer, A., and Starzinski-Powitz, A. (2001). mARVCF cellular localisation and binding to cadherins is influenced by the cellular context but not by alternative splicing. J. Cell Sci. 114, 3873-3884. [DOI] [PubMed] [Google Scholar]
- Weiss, E.E., Kroemker, M., Rudiger, A.H., Jockusch, B.M., and Rudiger, M. (1998). Vinculin is part of the cadherin-catenin junctional complex: complex formation between alpha-catenin and vinculin. J. Cell Biol. 141, 755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittchen, E.S., Haskins, J., and Stevenson, B.R. (1999). Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. J. Biol. Chem. 274, 35179-35185. [DOI] [PubMed] [Google Scholar]
- Yap, A.S., Niessen, C.M., and Gumbiner, B.M. (1998). The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 141, 779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura, S., Itoh, M., Nagafuchi, A., and Tsukita, S. (1995). Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J. Cell Sci. 108, 127-142. [DOI] [PubMed] [Google Scholar]
- Zahraoui, A., Louvard, D., and Galli, T. (2000). Tight junction, a platform for trafficking and signaling protein complexes. J. Cell Biol. 151, F31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Liyanage, U., Medina, M., Ho, C., Simmons, A.D., Lovett, M., and Kosik, K.S. (1997). Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport 8, 2085-2090. [DOI] [PubMed] [Google Scholar]