Abstract
The ESCRT-I, -II, and -III protein complexes function to create multivesicular bodies (MVBs) for sorting of proteins destined for the lysosome or vacuole. Prior studies with Saccharomyces cerevisiae have shown that the ESCRT-III protein Snf7p interacts with the MVB pathway protein Bro1p as well as its homolog Rim20p. Rim20p has no role in MVB formation, but functions in the Rim101p pH-response pathway; Rim20p interacts with transcription factor Rim101p and is required for the activation of Rim101p by C-terminal proteolytic cleavage. We report here that ESCRT-III proteins Snf7p and Vps20p as well as all ESCRT-I and -II proteins are required for Rim101p proteolytic activation in S. cerevisiae. Mutational analysis indicates that the Rim20p N-terminal region interacts with Snf7p, and an insertion in the Rim20p “Bro1 domain” abolishes this interaction, as determined with two-hybrid assays. Disruption of the MVB pathway through mutations affecting non-ESCRT proteins does not impair Rim101p processing. The relationship between the MVB pathway and Rim101p pathway is conserved in Candida albicans, because mutations in four ESCRT subunit genes abolish alkaline pH-induced filamentation, a phenotype previously seen for rim101 and rim20 mutants. The defect is suppressed by expression of C-terminally truncated Rim101-405p, as expected for mutations that block Rim101p proteolytic activation. These results indicate that the ESCRT complexes govern a specific signal transduction pathway and suggest that the MVB pathway may provide a signal that regulates pH-responsive transcription.
INTRODUCTION
The endosomal trafficking pathway plays a pivotal role in the interaction of cells with their environment (Conner and Schmid, 2003). It provides a means to take up extracellular fluid and particles, permitting acquisition of nutrients and entry of pathogens. It is crucial for down-regulation of surface receptors by conveying them to the lysosome or vacuole for degradation (Katzmann et al., 2002; Raiborg et al., 2003). Defects in endosomal trafficking thus have profound effects on signal transduction and cell growth control.
Many components of the endosomal trafficking machinery are well conserved among eukaryotes, including the subunits of three multiprotein complexes called ESCRT-I, -II, and -III (Lemmon and Traub, 2000; Katzmann et al., 2002; Raiborg et al., 2003). The ESCRT subunits are among a large group of gene products called “class E” vacuolar protein sorting (VPS) gene products in the yeast Saccharomyces cerevisiae (Bryant and Stevens, 1998; Katzmann et al., 2002). Mutations in these genes cause accumulation of an aberrant endosome called a class E compartment. Class E VPS gene products direct formation of and trafficking through multivesicular bodies (MVBs). MVBs form through invagination of the limiting endosomal membrane, permitting lumenal sequestration of membrane proteins that may be degraded upon fusion of the endosome with the lysosome or vacuole. Much of the same MVB formation machinery is also used in lysosome/vacuole biogenesis for trafficking of biosynthetic cargoes from the Golgi body (Bryant and Stevens, 1998; Katzmann et al., 2002). This machinery is also exploited by HIV and other viruses to produce capsids through membrane budding, a reaction that is topologically equivalent to endosomal membrane invagination (Raiborg et al., 2003). Therefore, the MVB formation machinery is used for diverse purposes.
Several recent observations indicate that the yeast MVB formation machinery may be used in sensing or responding to the pH of the surrounding medium. First, the MVB trafficking protein Bro1p (Nikko et al., 2003; Odorizzi et al., 2003) is a homolog of the pH-response regulators PalA (Penalva and Arst, 2002) of Aspergillus nidulans and Rim20p (Davis et al., 2000; Xu and Mitchell, 2001) of the yeasts S. cerevisiae and Candida albicans. Rim20p acts in the Rim101p pathway, which is described below. Second, several protein interaction studies have indicated that Rim20p interacts with ESCRT-III subunit Snf7p/Vps32p and with Vps4p, an AAA-ATPase required for ESCRT-III dissociation and recycling (Ito et al., 2001; Vincent et al., 2003; Bowers et al., 2004). The interaction with Snf7p homologues is a conserved property of Bro1p family members, as revealed by studies of A. nidulans PalA and human AIP1/Alix (Strack et al., 2003; Vincent et al., 2003; von Schwedler et al., 2003; Peck et al., 2004). Third, Snf7p/Vps32p also interacts with Rim13p, another component of the Rim101p pathway (Ito et al., 2001). Fourth, large-scale functional profiling studies indicate that many MVB-defective mutants, like Rim101p pathway-defective mutants, grow poorly at alkaline pH (Giaever et al., 2002). Several MVB-defective mutants are also sensitive to LiCl and NaCl, as are Rim101p pathway-defective mutants (Lamb et al., 2001; Giaever et al., 2002; Bowers et al., 2004). Finally, the MVB pathway gene VPS28 is required for expression of the alkaline pH-induced XPR2 gene of the yeast Yarrowia lipolytica (Gonzalez-Lopez et al., 2002). These observations suggest that the Rim101p pH-response pathway and the MVB formation machinery may have a close functional relationship.
Yeast Rim101p is a zinc finger protein that functions as a transcriptional repressor (Lamb and Mitchell, 2003). Rim101p is translated as a functionally inactive protein that requires proteolytic removal of the C-terminal region to yield the active Rim101p repressor (Li and Mitchell, 1997; Lamb and Mitchell, 2003). A number of gene products are required for proteolytic activation of Rim101p, including a cysteine protease, Rim13p/Cpl1p, a C-terminal region binding protein, Rim20p, putative membrane proteins, Rim9p and Rim21p, and a soluble protein, Rim8p (Penalva and Arst, 2002; Davis, 2003). Proteolytic activation of Rim101p is more efficient during growth in alkaline medium than in acidic medium (Li and Mitchell, 1997), and it is required for expression of several alkaline pH-induced genes (Lamb et al., 2001; Serrano et al., 2002). The mechanism of Rim101p cleavage is not known in detail, but we have proposed that Rim20p and Snf7p form a scaffold that brings protease Rim13p and substrate Rim101p in close proximity (Xu and Mitchell, 2001).
In this study, we set out to determine the functional relationship of Rim20p and Snf7p. Our findings indicate that there is a close relationship between many ESCRT subunits and Rim101p proteolytic activation and suggest that the conserved Bro1 domain of Rim20p and Bro1p family members is the region of interaction with Snf7p. Our genetic analysis of C. albicans MVB pathway mutants argues that the relationship between ESCRT components and Rim101p cleavage is conserved in this fungal pathogen. The interaction between Snf7p and Rim20p, along with the properties of MVB pathway mutants that do not affect the ESCRT complex directly, suggest that the regulation of Rim101p processing by ESCRT subunits is functionally distinct from their role in plasma membrane protein turnover.
MATERIALS AND METHODS
Strains and Media
S. cerevisiae strains were derived from the SK-1 background (Kane and Roth, 1974) except for the two-hybrid strains Y187 and Y190 (Durfee et al., 1993). The epitope-tagged RIM101-HA2 allele has been described (Li and Mitchell, 1997). The rim101Δ::His3MX6, rim8Δ::His3MX6, rim9Δ::His3MX6, rim13Δ::His3MX6, rim20Δ::His3MX6, rim21Δ::His3MX6, snf7Δ::His3MX6, and bro1Δ::His3MX6 mutations remove each entire open reading frame (ORF) and were introduced by PCR product-directed gene disruption, using plasmid pFA6a-His3MX6 (Longtine et al., 1998) as the His3MX6 template. The vps27Δ::kanMX4, vps28Δ::kanMX4, vps37Δ::kanMX4, vps23Δ::kanMX4, vps36Δ::kanMX4, vps25Δ::kanMX4, snf8Δ::kanMX4, snf7Δ::kanMX4, vps20Δ::kanMX4, vps24Δ::kanMX4, vps2Δ::kanMX4, vps4Δ::kanMX4, bro1Δ::kanMX4, vps60Δ::kanMX4, and doa4Δ::kanMX4 mutations remove each entire ORF and were introduced by PCR product-directed gene disruption, using genomic DNA from respective yeast deletion clones from Invitrogen (Carlsbad, CA) as the template. PCR with outside primers confirmed genotypes; primer sequences are available by request. S. cerevisiae strain construction involved standard methods including mating, meiotic crosses, and transformations (Kaiser et al., 1994).
For experiments in Figures 1B and 3, all strains are derived from WXY169 (MATα RIM101-HA2 ura3 leu2 trp1 his3 lys2 gal80::LEU2 rme1Δ5::LEU2 ho::LYS2). For experiments in Figure 2, all strains are derived from WXY22 (MATα RIM101-HA2 rim20Δ::URA3 ura3 leu2 trp1 his3 lys2 gal80::LEU2 rme1Δ5::LEU2 ho::LYS2). For growth assays in Tables 1 and 2, all strains are derived from WXY263 (MATa rim20Δ::HIS3MX6 ura3 leu2 trp1 his3 lys2 gal80::LEU2 ho::LYS2).
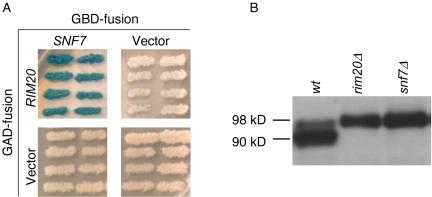
Figure 1.
(A) Two-hybrid assays of Snf7p-Rim20p interaction. Strain Y190, carrying pWX287 (GBD-Snf7p fusion) or pAS2-1 (GBDvector), was mated with strain Y187, carrying pWX36 (GAD-Rim20p fusion) or pACT2 (GAD-vector), two-hybrid reporter GAL1-lacZ expression was visualized on X-gal plates. (B) Processing of epitope-tagged Rim101-HA2p in wild-type and mutant strains. Extracts from yeast strains grown in YPD medium were analyzed on an anti-HA Western blot to visualize Rim101-HA2p. The masses of Rim101-HA2p forms, indicated along the right margin, were estimated by comparison with size standards. The strains included WXY169 (RIM101-HA2), WXY219 (RIM101-HA2 rim20Δ), and WXY306 (RIM101-HA2 snf7Δ).
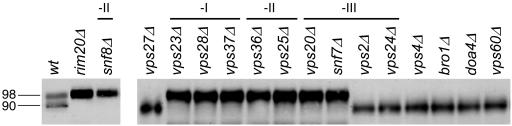
Figure 3.
Processing of epitope-tagged Rim101-HA2p in wild-type and mutant strains. Extracts from yeast strains grown in YPD medium were analyzed on an anti-HA Western blot to visualize Rim101-HA2p. The strains included the wild-type WXY169 (lane wt), and mutants derived from WXY169 by deletion of the entire ORF of each respective gene as labeled. ESCRT-I, -II, and -III complex subunits are designated -I, -II, and -III, respectively.
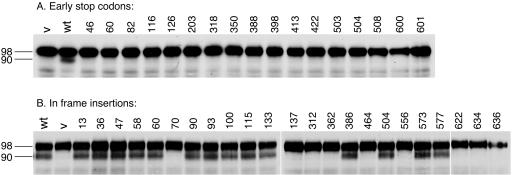
Figure 2.
Processing of epitope-tagged Rim101-HA2p in strain WXY219 (RIM101-HA2 rim20Δ) carrying a complementing wild-type RIM20 plasmid pWX272 (lane wt), pRS314 (vector control, lane v), or various rim20 insertion mutant plasmids. The RIM20 insertion mutations include those causing premature nonsense mutations (A; mutants described in Table 1) or 5 codon in-frame insertions (B; mutants described in Table 2).
Table 1.
RIM20 structure-function study: nonsense insertion mutants
| Expressiona
|
Function (complementation)b
|
Two-hybrid interactionc
|
|||||
|---|---|---|---|---|---|---|---|
| Insertion sited | Rim20-V5p | GAD-Rim20p | Rim101p processing | Growth on 0.4M NaCl | Growth on 20 mM LiCl | Rim101p | Snf7p |
| 46 | - | + | - | - | - | - | - |
| 60 | - | + | - | - | - | - | - |
| 82 | - | + | - | - | - | - | - |
| 116 | - | weak | - | - | - | - | - |
| 126 | - | + | - | - | - | - | - |
| 203 | - | weak | - | - | - | - | - |
| 318 | - | + | - | - | - | - | + |
| 350 | - | + | - | - | - | - | + |
| 388 | - | + | - | - | - | - | + |
| 398 | - | + | - | - | - | - | - |
| 413 | - | weak | - | - | - | - | + |
| 422 | - | + | - | - | - | - | + |
| 503 | - | weak | - | - | - | - | + |
| 504 | - | weak | - | - | - | - | + |
| 508 | - | + | - | - | - | - | + |
| 600 | - | + | - | - | - | - | + |
| 601 | - | + | - | - | - | - | + |
| WT | + | + | + | + | + | + | + |
| Vector | - | - | - | - | - | - | - |
Expression from two sets of RIM20 constructs was confirmed by anti-V5 (for Rim20-V5p) or anti-HA (for GAD-Rim20p) Western blots. +, wild-type level expression; -, no detectable expression; weak, weaker than the wild-type level expression
The functional assays tested whether plasmids carrying various rim20 mutants (pWX272-ixxx) can complement the rim20 deletion in the genome. +, wild-type level of functionality (comparable to pWX272, the wild-type control in row WT); -, no complementation (comparable to the vector control, in row Vector)
The two-hybrid assays were carried out as describe in Materials and Methods. +, wild-type level of interaction; -, no interaction; weak, weaker than wild type, but significant above background levels of interaction. All negative controls (interaction with an empty vector) were done in parallel, and none turned blue in 48 h (unpublished data)
The residue number given in the table indicates the position of the first altered amino acid (or stop codon) caused by the insertion
Table 2.
RIM20 structure-function study: in-frame insertion mutants
| Expressiona
|
Function (complementation)b
|
Two-hybrid interactionc
|
|||||
|---|---|---|---|---|---|---|---|
| Insertion sited | Rim20-V5p | GAD-Rim20p | Rim101p processing | Growth on 0.4 M NaCl | Growth on 20 mM LiCl | Rim101p | Snf7p |
| 13 | + | + | + | + | + | + | + |
| 36 | + | + | + | + | + | + | + |
| 47 | + | + | + | + | + | + | + |
| 58 | + | + | + | + | + | + | + |
| 60 | + | + | + | + | + | + | + |
| 70 | + | + | - | - | - | - | - |
| 90 | + | + | + | + | + | + | + |
| 93 | + | + | + | + | + | + | + |
| 100 | + | + | + | + | + | + | + |
| 115 | + | + | + | + | + | + | + |
| 133 | + | + | + | + | + | + | + |
| 137 | + | + | - | - | - | + | + |
| 312 | + | + | - | - | - | weak | + |
| 362 | + | + | - | - | - | - | + |
| 386 | + | + | + | + | + | + | + |
| 464 | + | + | - | - | - | - | + |
| 504 | + | + | + | + | + | + | + |
| 556 | + | + | - | - | - | + | + |
| 573 | + | + | + | + | + | + | + |
| 577 | + | + | + | + | + | + | + |
| 622 | weak | weak | - | - | - | - | weak |
| 634 | - | weak | - | - | - | - | + |
| 636 | weak | weak | - | - | - | - | weak |
| WT | + | + | + | + | + | + | + |
| Vector | - | - | - | - | - | - | - |
Expression from two sets of RIM20 constructs was confirmed by anti-V5 (for Rim20-V5p) or anti-HA (for GAD-Rim20p) Western blots. +, wild-type level expression; -, no detectable expression; weak, weaker than the wild-type level expression
The functional assays tested whether plasmids carrying various rim20 mutants (pWX272-ixxx) can complement the rim20 deletion in the genome. +, wild-type level of functionality (comparable to pWX272, the wild-type control in row WT); -, no complementation (comparable to the vector control, in row Vector)
The two-hybrid assays were carried out as describe in Materials and Methods. +, wild-type level of interaction; -, no interaction; weak, weaker than wild type, but significant above background levels of interaction. All negative controls (interaction with an empty vector) were done in parallel, and none turned blue in 48 h (unpublished data)
The residue number given in the table indicates the position of the first altered amino acid (or stop codon) caused by the insertion
All C. albicans strains were derived from strain BWP17 (genotype: ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) through standard transformation methods (Wilson et al., 1999). To create C. albicans insertion mutants, we cloned PCR products corresponding to each gene in vector pGEM-T-Easy (Promega, Madison, WI) and subjected them to in vitro transposon mutagenesis with Tn7-UAU1 donor plasmid pAED98 (Davis et al., 2002). Insertions near the middle of each ORF were identified by restriction analysis. These insertions were used to create heterozygous C. albicans mutants through Arg+ selection, which were then subjected to Arg+ Ura+ selection to isolate mitotic recombinants (Enloe et al., 2000). Genotypes were verified by PCR as described (Davis et al., 2002). For each mutation, three independent homozygotes (each from an independent transformant) were analyzed. Suppression studies were conducted with plasmids pDDB61 (RIM101) and pDDB71 (RIM101-405) as described (Davis et al., 2000). The specific genes cloned for insertional disruption included BRO1 (ORF19.1670), VPS28 (ORF19.212), VPS24 (ORF19.2031), VPS23 (ORF19.2343), VPS36 (ORF19.8414), VPS2 (ORF19.945), and SNF7 (ORF19.6040). Because the BRO1 ORF is large, we cloned only the 5′ 1398 base pairs of the coding region for insertional mutagenesis and used an insertion in the middle of this segment to create mutant strains.
Media composition followed standard recipes (Kaiser et al., 1994; Davis et al., 2002).
Plasmid Construction
Construction of pWX272 (pRS314-RIM20-V5) was previously described (Xu and Mitchell, 2001). The pWX272 plasmid was subject to transposon insertion mutagenesis using New England Biolabs GPS-LS system (Beverly, MA), following the manufacturer's instructions. The resulting plasmids were digested with the restriction enzyme PmeI to remove the transposon and then religated to generate plasmids with a 15-base pair insertion within the RIM20 coding region. These plasmids were sequenced through the entire RIM20 coding region to determine the insertion sites and to confirm that there were no secondary mutations. The plasmids were then named pWX272-ixxx, where the “xxx” indicates the position of the first altered amino acid (or stop codon) caused by the insertion. A total of 40 different 15-base pair insertion mutants of RIM20 were generated and listed in Tables 1 and 2.
The construction of pWX14 (GBD-Rim101p fusion) and pWX36 (GAD-Rim20p fusion) was previously described (Xu and Mitchell, 2001). To generate fusions of Gal4p DNA-binding domain (GBD) to Snf7p, an NcoI site was engineered in forward primers (pSNF7-F: 5′-atggccatggaggccatgtggtcatcactttttgg-3′), and a BamHI site in reverse primers (pSNF7-R: 5′-tatggatcctcaaagccccatttctgcttg-3′). The “ATG” within the NcoI site is in-frame with the coding sequences. Genomic DNA from S. cerevisiae strain AMP108 (genotype: MATα ura3 leu2 trp1 lys2 ho::LYS2) was used as a PCR template. PCR products were first cloned into pGEM-t-Easy vectors (Promega). Inserts released by NcoI-BamHI double digestions were moved into NcoI-BamHI-digested vector pAS1-CYH2. The nucleotide sequence of each entire ORF was verified by sequencing.
To generate fusions of Gal4p activation domain (GAD) to the collection of mutant Rim20p derivatives, an NcoI site was engineered in forward primers (pRIM20-F3: 5′-gctctagatgccatggacatgagtgaactgcttgc-3′), and a BamHI site in reverse primers (pRIM20-R: 5′-tgcaggatccaagcttatcattggtcttcaaatgtc-3′). The “ATG” within the NcoI site is in-frame with the coding sequences. Plasmid DNA of pWX272-ixxx was used as PCR template. PCR products were first cloned into pGEM-t-Easy vectors (Promega). Inserts released by NcoI-BamHI double digestions were moved into NcoI-BamHI-digested pWX36, replacing the wild-type allele. The nucleotide sequence of each entire ORF was verified by sequencing. This collection of plasmids was named in the same manner as their respective PCR templates; pWX36-ixxx, where the “xxx” indicates the first altered amino acid (or stop codon) caused by the insertion (i.e., same as their respective PCR templates).
Growth Assays
Wild-type strain KB395 (MATa ura3 leu2 trp1 his3 lys2 gal80::LEU2 ho::LYS2) and otherwise isogenic strains WXY263 (rim20Δ::His3MX6) transformed with various plasmids (pWX272, pWX272-TI-01-41, pRS314) were grown in liquid SC-Trp media to approximately the same cell density. Each culture was diluted 1:5, 1:25, 1:125, 1:625 with H2O. Three microliters of each of the dilutions was spotted onto an unsupplemented YPD plate, or YPD plates supplemented with 0.4 M NaCl, or 20 mM LiCl. The plates were incubated at 30°C for 1-4 d before the growth was scored.
Two-hybrid Interaction Assays
Strain Y190, carrying various GBD fusion plasmids, was mated with strain Y187, carrying various GAD fusion plasmids, and diploids were selected on SC-Trp-Leu plates. The diploids were transferred to SC-Trp-Leu plates containing 100 mg/l X-gal, and let grow for 48 h at 30°C to visualize the expression from a GAL1-lacZ reporter. For quantitative assays, 24-h cultures in SC-Trp-Leu were diluted 1/25 into fresh SC-Trp-Leu medium and harvested after three doublings. β-galactosidase assays were conducted on permeabilized cells as previously described (Xu and Mitchell, 2001).
Western Blot Assays
Cells from 10-ml midlog phase culture in YPD were pelleted, resuspended in 100 μl 3× Laemmli buffer, and boiled for 5 min. After centrifugation, 5-20 μl of the supernatants was fractionated on a 9% SDS-polyacrylamide gel and transferred to nitrocellulose. For HA epitope detection, the filter was probed with anti-HA-peroxidase mAb (3F10, Roche, Diagnostics, Indianapolis, IN; 10-4 dilution in TBST), and was developed with ECL detection reagents (Amersham, Piscataway. NJ). For V5 epitope detection, the filter was probed with anti-V5-HRP antibody (Invitrogen, 1:5000 dilution in TBST) and was developed with ECL detection reagents (Amersham).
RESULTS
Requirement for Snf7p in Rim101p Processing
Interaction between Snf7p and Rim20p has been detected through two-hybrid analysis with nutritional gene reporters (Ito et al., 2001; Bowers et al., 2004). We confirmed this interaction using a lacZ reporter system to test interaction in cells carrying GAD-RIM20 and GBD-SNF7 test plasmids. Cells carrying both plasmids expressed the lacZ reporter at high levels; cells carrying plasmids lacking either insert did not (Figure 1A). These results confirm that Snf7p and Rim20p are capable of interaction.
Rim20p is required for proteolytic activation of the transcription factor Rim101p, directing removal of a C-terminal ∼8-kDa Rim101p segment. Thus, epitope-tagged Rim101-HAp is detectable in wild-type cells as both 98-kDa (unprocessed) and 90-kDa (processed) forms, but in rim20 mutant cells as only a 98-kDa form (Xu and Mitchell, 2001; Figure 1B, lanes 1 and 2). In a snf7 mutant, only a 98-kDa form of Rim101-HAp was detectable (Figure 1B, lane 3). We conclude that Snf7p is required for Rim101p processing.
Structural Analysis of Rim20p
Rim20p interacts with both Snf7p and Rim101p, which have in common a highly acidic C-terminal region. This region of Rim101p is sufficient to interact with Rim20p (Xu and Mitchell, 2001). To determine whether Snf7p and Rim101p interact with the same region of Rim20p, we used two-hybrid analysis to identify the region of Rim20p that interacts with Snf7p. The subject of our analysis was a panel of 15-nucleotide RIM20 insertion alleles, some yielding C-terminally truncated products (nonsense insertions; Table 1) and others yielding products with insertions of 5 amino acids (in-frame insertions; Table 2).
We first used Western analysis to determine the expression levels of each product in two genetic contexts. First, each allele was generated in a RIM20-V5 low copy-number plasmid, in which a V5 epitope is specified at the 3′ end of the RIM20 ORF. Expression of RIM20-V5 is directed by the RIM20 promoter region. The nonsense insertion products were not detectable (Table 1), as expected. Most of the in-frame insertion products were detectable at levels comparable to the control wild-type Rim20-V5p (Table 2). The exceptions were the three most C-terminal in-frame insertion products (following codons 622, 634, and 636), which accumulated at substantially lower levels than wild-type Rim20-V5p. We found previously that a site-directed mutation in this region (codons 623-624) also impaired Rim20-V5p accumulation (Xu and Mitchell, 2001). Second, each allele was transferred to a high copy-number GAD-RIM20 plasmid, in which an HA epitope is specified at the 5′ end of the RIM20 segment. Expression of GAD-RIM20 is directed at high levels by a modified ADH1 promoter. All of the nonsense insertion products were detectable, though five truncations displayed reduced accumulation (Table 1). Accumulation of the in-frame insertion GAD-RIM20 products was similar to that of the corresponding RIM20-V5 products (Table 2). These results help to guide the interpretation of assays of mutant product interaction and function below.
The GAD-RIM20 insertions were used to map the Snf7p-interaction region in two-hybrid assays. Ten of 11 nonsense insertions that produced at least the 318 N-terminal residues of Rim20p were capable of Snf7p interaction (Table 1). Nonsense insertions producing 203 or fewer N-terminal residues of Rim20p failed to interact with Snf7p (Table 1). Among in-frame insertions, we found one allele that disrupted Rim20p-Snf7p interaction without affecting GAD-Rim20p accumulation (Table 2). This insertion (designated RIM20-i70) lies between codons 70 and 71, affecting a region called the Bro1 domain (Odorizzi et al., 2003) that is well conserved among Rim20p and Bro1p homologues. These results indicate that the N-terminal 318 residues of Rim20p are sufficient for interaction with Snf7p and that the region near residue 70 is required for interaction with Snf7p.
Similar assays were used to map the Rim101p-interacting region of Rim20p. The nonsense insertion mutants showed no detectable interaction with Rim101p (Table 1). Four in-frame insertion mutants that yielded stable products were impaired in interaction with Rim101p as well (Table 2). One was the RIM20-i70 insertion that also caused a Snf7p interaction defect. A second insertion, RIM20-i312, caused weak though detectable Rim101p interaction. Two additional insertions, RIM20-i362 and RIM20-i464, yielded no detectable Rim101p interaction. We have shown previously that the Rim20p C-terminal segment, comprising residues 353-661, is sufficient for interaction with Rim101p in two-hybrid assays (Xu and Mitchell, 2001). The properties of RIM20-i362 and RIM20-i464 as well as the nonsense insertion mutants are consistent with the idea that this Rim20p C-terminal segment has a major role in Rim20p-Rim101p interaction.
Functional Analysis of Rim20p Mutant Derivatives
To assess the functional significance of Rim20p interactions, we examined Rim101-HAp processing in strains expressing each Rim20p insertion mutant derivative. These mutants were expressed from low copy-number RIM20-V5 plasmids in strains with a chromosomal rim20Δ allele. The nonsense insertion mutants were all defective in Rim101-HAp processing (Table 1 and Figure 2A). This result is consistent with the fact that these mutants were all defective in interaction with Rim101p. Among in-frame insertion mutants, nine alleles caused Rim101p processing defects. Three defective alleles, RIM20-i622, -i634, and -i636, caused reduced Rim20p accumulation. Three defective alleles, RIM20-i312, -i362, and -i464, were associated with reduced Rim20p-Rim101p interaction. One defective allele, RIM20-i70, was associated with reduced Rim20p-Rim101p and Rim20p-Snf7p interaction. Finally, the two defective alleles RIM20-i137 and -i556 were not measurably impaired in protein-protein interactions or Rim20p accumulation. As an independent assay for Rim20p function, we assayed the ability of each mutant RIM20 allele to promote growth in the presence of 0.4 M NaCl or 20 mM LiCl. Growth on these media requires expression of the efflux pump gene ENA1 (Garciadeblas et al., 1993), which in turn depends on Rim101p activity (Lamb et al., 2001; Lamb and Mitchell, 2003). We found that defects in Rim101p processing correlated with poor growth on NaCl and LiCl media for all RIM20 mutants. Therefore, all Rim20p insertions that impair Rim20p-Rim101p interaction as well as the one mutation that impairs both Rim20p-Rim101p and Rim20p-Snf7p interaction lead to defects in Rim101p processing and in Rim101p functional activity. These results support the model that interaction between Rim101p and Rim20p is required for Rim101p processing (Xu and Mitchell, 2001). They are consistent with the model that Rim20p-Snf7p interaction is also required for Rim101p processing, but the dual Rim20-i70p interaction defects do not permit a stronger conclusion.
Relationship of Multivesicular Body Formation and Rim101p Processing
Snf7p is a component of the ESCRT-III complex, which associates transiently with vesicles to promote invagination of the limiting membrane (Babst et al. 2002a). ESCRT-III complex formation and vesicle association depends on the ESCRT-I and -II protein complexes (Babst et al. 2002b; Katzmann et al., 2002). To determine whether all ESCRT components are required for Rim101p processing, we examined mutants in each ESCRT component for presence of the processed form of epitope-tagged Rim101-HAp (Figure 3). Rim101p processing was dependent on ESCRT-III subunits Snf7p and Vps20p, but independent of Vps2p and Vps24p. Rim101p processing was also dependent on ESCRT-II subunits Snf8p(Vps22p), Vps25p, and Vps36p, and on ESCRT-I subunits Vps23p, Vps28p, and Vps37p. Therefore, the majority of ESCRT subunits are required for Rim101p processing.
One explanation for ESCRT-dependence of Rim101p processing is that ESCRT function may be required for stable accumulation of Snf7p. However, examination of V5 epitope-tagged Snf7p levels by Western analysis showed that most ESCRT-defective mutations had no effect on Snf7p accumulation (unpublished data). The one exception was that three independently constructed vps28Δ mutants all showed reduced Snf7p levels. Expression of GAL1-SNF7 from a multicopy vector did not restore Rim101p processing in one vps28Δ mutant tested (unpublished data). These results indicate that ESCRT deficiencies do not block Rim101p processing simply as a result of reduced Snf7p levels.
Several additional class E VPS gene products are required along with ESCRT subunits for MVB function. We found that Rim101p processing was independent of all such gene products tested (Figure 3), including Vps27p, Vps4p, Doa4p, Vps60p, and (as previously reported; Xu and Mitchell, 2001) Bro1p. The dependence of Rim101p processing on class E VPS gene products thus distinguishes the majority of ESCRT subunits from non-ESCRT proteins.
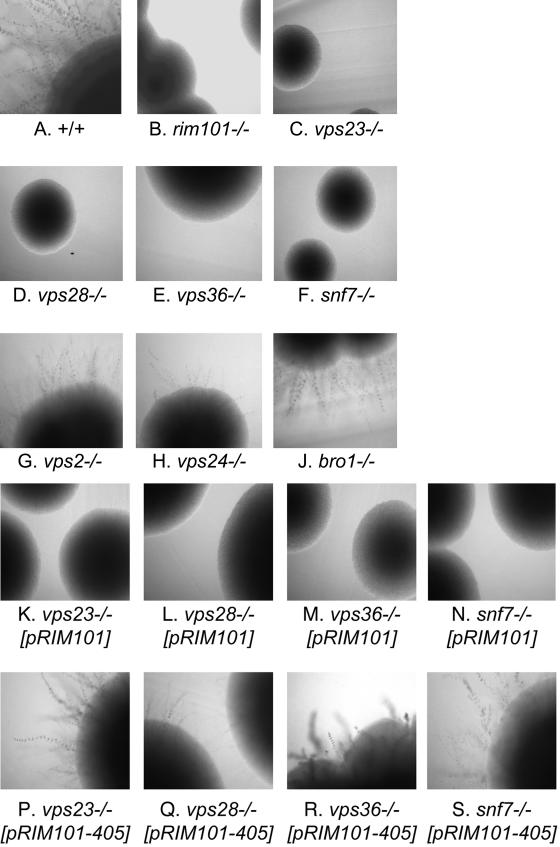
The relationship between the MVB pathway and Rim101p processing may be evolutionarily conserved. To gain insight into this possibility, we turned to the fungal pathogen C. albicans. Prior studies have shown that C. albicans has a Rim101p-processing pathway (Porta et al., 1999; Ramon et al., 1999; Davis et al., 2000; Davis, 2003; Li et al., 2004). Mutants defective in Rim101p processing or activity are defective in alkaline pH-induced hyphal formation, which may be assayed through examination of colony perimeters for hyphae on pH 8 plates, as illustrated by wild-type and rim101/rim101 strains (Figure 4, A and B). We used this assay to test C. albicans mutants homozygous for insertions in several class E VPS genes. As expected from the S. cerevisiae studies, we observed that C. albicans Vps23p, Vps28p, Vps36p, and Snf7p are required for alkaline pH-induced filamentation, and that Vps2p, Vps24p, and Bro1p are not (Figure 4, C-J). Many C. albicans mutations are known to alter filamentation responses (Liu, 2001), including mutations affecting vesicle trafficking (Bruckmann et al., 2000; Palmer et al., 2003). We used the genetic criterion of suppression to determine whether the class E mutants described here are defective in filamentation because of a Rim101p-processing defect. We showed previously that alkaline pH-induced filamentation is restored to Rim101p-processing defective mutants by introduction of a RIM101405 allele, which specifies a C-terminally truncated product, but not by introduction of a wild-type RIM101 allele (Davis et al., 2000). We observed that alkaline pH-induced filamentation was restored to mutants defective in Vps23p, Vps28p, Vps36p, and Snf7p by RIM101405, but not by RIM101 (Figure 4, K-S). These results support the idea that the relationship between the MVB pathway and Rim101p processing is conserved in both S. cerevisiae and C. albicans.
Figure 4.
Filamentation of C. albicans wild-type and mutant strains. Colonies grown for 3 d at 37°C on M199 pH 8 plates were photographed. Similar results were obtained with three independently isolated insertion mutants for each gene. Hisreference strain DAY286 (A) was compared with Hismutant strains carrying insertions in RIM101 (B) and other genes as indicated (C-J). His+ derivatives of each mutant were created with integrated HIS1:RIM101 plasmids (K-N) or integrated HIS1:RIM101-405 plasmids (P-S). The RIM101-405 allele expresses a truncated Rim101p derivative that permits alkaline pH-induced filamentation in the absence of upstream processing pathway components (Davis et al., 2000).
DISCUSSION
Function of MVB Pathway Components in Rim101p Processing
The MVB pathway is known to interface with signal transduction pathways through receptor sequestration and proteolytic turnover; mutations that block receptor sorting into MVBs prolong signaling (Bilodeau et al., 2002; Katzmann et al., 2002). This paradigm might be extended to the Rim101p pathway with the proposal that a plasma membrane recep tor functions as a negative regulator of the Rim101p pathway and that the MVB pathway promotes its turnover by conveying it to the vacuole. In this model, Snf7p has an indirect role in Rim101p processing.
This model is consistent with the finding that most ESCRTdefective mutants are defective in Rim101p processing, because these mutants would be defective in turnover of the hypothetical receptor. However, our observations with other MVB pathway mutants limit the plausibility of the model. For example, Vps27p is not required for Rim101p processing. Vps27p binds to peptidyl-ubiquitin, which targets membrane proteins for the MVB pathway, and to endosomal membrane phosphatidylinositol 3-phosphate (Bilodeau et al., 2002; Shih et al., 2002; Katzmann et al., 2003). Thus, it serves as an anchor to recruit ESCRT-I to the endosomal membrane and initiate MVB formation (Katzmann et al., 2003). Defects in Vps27p cause accumulation of both surface proteins and biosynthetic proteins from the Golgi in a class E compartment (Piper et al., 1995; Gong and Chang, 2001). A hypothetical receptor that inhibited Rim101p processing would have to reach the vacuole for degradation independently of Vps27p function, unlike all other known MVB cargo proteins. In addition, the deubiquitinating enzyme Doa4p (Amerik et al., 2000) is required for rapid degradation of membrane proteins Pma1-10p (Gong and Chang, 2001), Ste3p (Chen and Davis, 2002), and Ste6p (Losko et al., 2001); the hypothetical receptor would have to be degraded through an independent mechanism. Finally, Vps2p, Vps24p, and Vps4p are not required for Rim101p processing. Vps4p is required for release of ESCRT-III from the vesicle surface and for MVB formation; Vps2p and Vps24p recruit Vps4p to the endosomal surface (Katzmann et al., 2002), probably in conjunction with Vps20p (Yeo et al., 2003). Vps4p is required for degradation of membrane protein Ste6p (Kranz et al., 2001); Vps24p is required for degradation of membrane protein Smf1p (Paidhungat and Garrett, 1998). The ability of the hypothetical receptor to reach the vacuole for degradation independently of Vps27p, Doa4p, Vps2p, Vps24p, and Vps4p would be unprecedented, particularly if its degradation depends on the ESCRT complexes.
A second model is that Rim20p must interact with Snf7p to direct Rim101p cleavage. In this model, Snf7p has a direct role in Rim101p processing that is not exerted through its role in endocytic vesicle metabolism. We are not aware of precedents for this model among MVB pathway proteins, but it accounts for our observations and is consistent with many known properties of MVB pathway proteins.
Snf7p and Vps20p, the two ESCRT-III subunits that are required for Rim101p processing, form a subcomplex of ESCRT-III (Babst et al. 2002a). This subcomplex can associate with class E compartments independently of the other ESCRT-III subunits, Vps2p and Vps24p. The finding that Snf7p and Vps20p are required for Rim101p processing, whereas Vps2p and Vps24p are not, is consistent with the characterization of ESCRT-III subcomplexes in MVB formation.
Our findings are consistent with current proposals for the relationship of ESCRT-I and -II to the Snf7p-Vps20p subcomplex. One proposal is that ESCRT-I and -II act in succession to promote formation of the Snf7p-Vps20p subcomplex and its transient association with the endosomal surface (Babst et al. 2002b; Katzmann et al., 2002). This idea is based on the isolation of separable ESCRT complexes, the finding that mutations in ESCRT-II subunits disrupt colocalization of Snf7p and Vps20p, and the finding that mutations in ESCRT-I subunits prevent association of ESCRT-II with the endosomal surface. According to this view, all ESCRT-I and -II subunit mutations should prevent Rim101p processing by preventing Snf7p-Vps20p subcomplex assembly. A second proposal is that ESCRT-I and -II subunits interact with Snf7p and Vps20p to form a “core ESCRT complex” (Bowers et al., 2004). This idea is based on the detection of multiple inter-ESCRT two-hybrid interactions and shared ion sensitivities of mutants defective in ESCRT-I, -II, Snf7p, or Vps20p. According to this view, a single multi-protein core ESCRT complex may interact with Rim20p to promote Rim101p processing.
It is possible that association of Snf7p-Vps20p with the endosomal membrane is required for Rim101p processing. In support of the idea, we note that vps2Δ and vps4Δ mutations result in endosomal association of almost all of the Snf7p pool as well as the bulk of the Vps20p pool (Babst et al. 2002a). The fact that these mutations do not impair Rim101p processing suggests that endosome-associated Snf7p-Vps20p is competent to direct processing. This view fits with the general idea that Snf7p-Vps20p serves as a platform for recruitment of multiple proteins (Babst et al. 2002a; Bowers et al., 2004), including Vps2p-Vps24p, Rim20p, Bro1p, and Doa4p. On the other hand, Vps27p binds to ESCRT-I subunit Vps23p (Bilodeau et al., 2003; Bowers et al., 2004) and is required to recruit ESCRT-I to the endosome membrane (Katzmann et al., 2003). Thus a vps27Δ mutation should abolish association of Snf7p-Vps20p with the endosomal membrane, yet this mutation does not impair Rim101p processing. This observation may indicate that nonendosomal Snf7p-Vps20p is competent to direct Rim101p processing. A second possibility comes from the suggestion of Katzmann et al. (2003) that HIV Gag may mimic Vps27p to recruit ESCRT-I to a novel membrane site; perhaps a Rim101p pathway-specific protein substitutes for Vps27p for specialized ESCRT-I recruitment.
Relationship between Rim20p and Bro1p
Rim20p and Bro1p are homologues, but the functional significance of that homology has been elusive. Bro1p functions in the MVB pathway and not in the Rim101p pathway; Rim20p functions in the Rim101p pathway and not in the MVB pathway (Xu and Mitchell, 2001; Nikko et al., 2003; Odorizzi et al., 2003; Bowers et al., 2004). No synthetic rim20 bro1 double mutant phenotype has been detected (Xu and Mitchell, 2001; Odorizzi et al., 2003; Bowers et al., 2004). We found that the N-terminal region of Rim20p is sufficient to interact with Snf7p, and the RIM20-i70 mutation in this region disrupts Rim20p-Snf7p interaction. The RIM20-i70 mutation inserts residues in the middle of the conserved Bro1 domain (Odorizzi et al., 2003), lying between the conserved sequences KFP and EFTW. Thus we suggest that the Bro1 domain is the region of interaction between Rim20p-Bro1p family members and Snf7p family members. A recent report shows that the N-terminal half of human AIP1, which includes the Bro1 domain, is sufficient for interaction with human Snf7p (Peck et al., 2004), in keeping with this model.
The RIM20-i70 mutation also blocks Rim20p-Rim101p interaction. This defect is unexpected because the C-terminal region of Rim20p—far from the Bro1 domain—is sufficient for efficient interaction with Rim101p (Xu and Mitchell, 2001). The defect does not appear to be a trivial consequence of Rim20-i70p instability based on Western analysis. One simple explanation is that full-length Rim20p cannot interact with Rim101p unless the Bro1 domain is first bound to Snf7p, so that a Snf7p binding defect would cause a Rim101p binding defect. This model predicts that Rim20p-Rim101p interaction will be impaired in a snf7 mutant background. However, we have observed that Rim20p-Rim101p interaction is equivalent in SNF7 and snf7 hosts (unpublished results). A second explanation is that the Bro1 domain of Rim20p binds independently to Snf7p and Rim101p. In keeping with this model, we note that Bro1p-Rim101p twohybrid interaction is detectable, though weak (Xu and Mitchell, unpublished results), as expected if a region conserved in Bro1p and Rim20p is capable of Rim101p binding.
The interaction between Rim20p and Vps4p reported by Bowers et al. (2004) and Ito et al. (2001) remains enigmatic. We found here that Vps4p is not required for Rim101p processing, although it is possible that Rim20p-Vps4p interaction influences the rate of Rim101p processing. Rim20p-Vps4p interaction may also provide a means for Rim20p to influence Vps4p activity. Whether either of these speculative possibilities is true remains to be determined.
The MVB Pathway, Rim101p Function, and pH Sensing
We have shown that ESCRT subunits are required for Rim101p processing in S. cerevisiae, and we have provided genetic support for this conclusion in C. albicans. These observations suggest that there may be a regulatory relationship between the MVB pathway and the Rim101p pathway (Figure 5). One may imagine a scenario in which increases in ESCRT activity favor both flux through the MVB pathway and the rate of Rim101p processing. A second scenario is that increased flux through the MVB pathway sequesters the ESCRT machinery and blocks Rim101p processing. We do not have direct evidence in favor of either idea, but we note that Rim101p is a transcriptional repressor of PRB1 (Lamb and Mitchell, 2003), which specifies a major vacuolar endopeptidase (Moehle et al., 1987). We expect Prb1p to be required for degradation of MVB pathway cargo, and thus that flux through the MVB pathway might lift repression of PRB1 by blocking Rim101p processing.
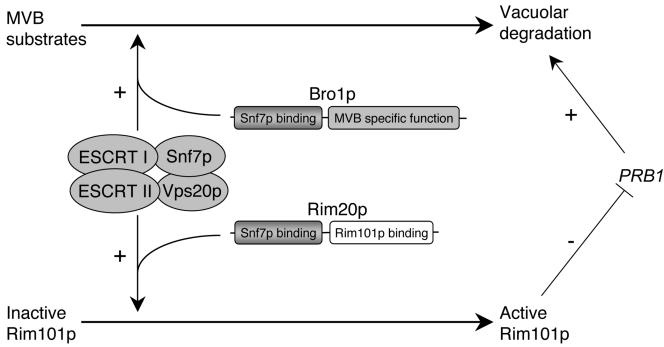
Figure 5.
Relationship between the Rim101p and MVB pathways. The ESCRT-I, ESCRT-II, and Snf7p-Vps20p complexes are required for both MVB formation and Rim101p activation. Homologous proteins Bro1p and Rim20p interact with Snf7p and function in the MVB and Rim101p pathways, respectively. The MVB pathway delivers cargo proteins to the vacuole that include membrane proteins destined for degradation and proteases. Rim101p represses diverse genes including PRB1, which specifies a major vacuolar endopeptidase. Changes in overall ESCRT activity may modulate Rim101p and MVB pathway activity in parallel. In addition, the two pathways may compete for limiting ESCRT activity.
The Rim101p pathway is required for alkaline pH responses in S. cerevisiae, C. albicans, and Y. lipolytica (Lambert et al., 1997; Porta et al., 1999; Ramon et al., 1999; Davis et al., 2000; Lamb et al., 2001; Gonzalez-Lopez et al., 2002; Serrano et al., 2002; Davis, 2003), as expected from the pioneering work on the homologous PacC pathway of A. nidulans by Penalva, Arst, and coworkers (Penalva and Arst, 2002). The fact that there is a connection between the Rim101p and MVB pathways raises the interesting possibility that external pH, a signal that regulates Rim101p processing in vivo (Li and Mitchell, 1997; Li et al., 2004), influences MVB pathway activity as well. Formation of MVB-like liposomes in vitro is dependent on a pH gradient across the liposome membrane (Matsuo et al., 2004), so it seems possible that MVB formation in vivo is a pH-sensitive process. A speculative possibility is that MVB pathway activity may function in place of a specific surface receptor as a regulator of Rim101p processing.
Acknowledgments
We thank Vincent Bruno and Clarissa Nobile for their help with C. albicans genetics and Jacob Boysen, Karen Barwell, Vincent Bruno, and Clarissa Nobile for many helpful discussions and comments on the manuscript. This work was supported by RO1 grant GM39531 from the National Institutes of Health.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-08-0666. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0666.
References
- Amerik, A.Y., Nowak, J., Swaminathan, S., and Hochstrasser, M. (2000). The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar proteinsorting and endocytic pathways. Mol. Biol. Cell 11, 3365-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., Estepa-Sabal, E.J., Meerloo, T., and Emr, S.D. (2002a). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271-282. [DOI] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., Snyder, W.B., Wendland, B., and Emr, S.D. (2002b). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283-289. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., Urbanowski, J.L., Winistorfer, S.C., and Piper, R.C. (2002). The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534-539. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., Winistorfer, S.C., Kearney, W.R., Robertson, A.D., and Piper, R.C. (2003). Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163, 237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. et al. (2004). Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5, 194-210. [DOI] [PubMed] [Google Scholar]
- Bruckmann, A., Kunkel, W., Hartl, A., Wetzker, R., and Eck, R. (2000). A phosphatidylinositol 3-kinase of Candida albicans influences adhesion, filamentous growth and virulence. Microbiology 146(Pt 11), 2755-2764. [DOI] [PubMed] [Google Scholar]
- Bryant, N.J., and Stevens, T.H. (1998). Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62, 230-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., and Davis, N.G. (2002). Ubiquitin-independent entry into the yeast recycling pathway. Traffic 3, 110-123. [DOI] [PubMed] [Google Scholar]
- Conner, S.D., and Schmid, S.L. (2003). Regulated portals of entry into the cell. Nature 422, 3744. [DOI] [PubMed] [Google Scholar]
- Davis, D. (2003). Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44, 1-7. [DOI] [PubMed] [Google Scholar]
- Davis, D., Wilson, R.B., and Mitchell, A.P. (2000). RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20, 971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D.A., Bruno, V.M., Loza, L., Filler, S.G., and Mitchell, A.P. (2002). Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162, 1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T. et al. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555-569. [DOI] [PubMed] [Google Scholar]
- Enloe, B., Diamond, A., and Mitchell, A.P. (2000). A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182, 5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblas, B. et al. (1993). Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 236, 363-368. [DOI] [PubMed] [Google Scholar]
- Giaever, G. et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- Gong, X., and Chang, A. (2001). A mutant plasma membrane ATPase, Pma110, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA 98, 9104-9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lopez, C.I., Szabo, R., Blanchin-Roland, S., and Gaillardin, C. (2002). Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 160, 417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. et al. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Kane, S.M., and Roth, R. (1974). Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118, 8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Stefan, C.J., Babst, M., and Emr, S.D. (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162, 413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, A., Kinner, A., and Kolling, R. (2001). A family of small coiled-coilforming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12, 711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, T.M., and Mitchell, A.P. (2003). The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, T.M., Xu, W., Diamond, A., and Mitchell, A.P. (2001). Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276, 1850-1856. [DOI] [PubMed] [Google Scholar]
- Lambert, M., Blanchin-Roland, S., Le Louedec, F., Lepingle, A., and Gaillardin, C. (1997). Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17, 3966-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, S.K., and Traub, L.M. (2000). Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12, 457-466. [DOI] [PubMed] [Google Scholar]
- Li, M., Martin, S.J., Bruno, V.M., Mitchell, A.P., and Davis, D.A. (2004). Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3, 741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., and Mitchell, A.P. (1997). Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145, 63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. (2001). Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4, 728-735. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S. et al. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Losko, S., Kopp, F., Kranz, A., and Kolling, R. (2001). Uptake of the ATPbinding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12, 1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, H. et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531-534. [DOI] [PubMed] [Google Scholar]
- Moehle, C.M., Tizard, R., Lemmon, S.K., Smart, J., and Jones, E.W. (1987). Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol. Cell. Biol. 7, 4390-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko, E., Marini, A.M., and Andre, B. (2003). Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J. Biol. Chem. 278, 50732-50743. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Katzmann, D.J., Babst, M., Audhya, A., and Emr, S.D. (2003). Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116, 1893-1903. [DOI] [PubMed] [Google Scholar]
- Paidhungat, M., and Garrett, S. (1998). Cdc1 is required for growth and Mn2+ regulation in Saccharomyces cerevisiae. Genetics 148, 1777-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, G.E., Cashmore, A., and Sturtevant, J. (2003). Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot. Cell 2, 411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, J.W., Bowden, E.T., and Burbelo, P.D. (2004). Structure and function of human Vps20 and Snf7 proteins. Biochem. J. 377, 693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva, M.A., and Arst, H.N., Jr. (2002). Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66, 426-446, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131, 603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta, A., Ramon, A.M., and Fonzi, W.A. (1999). PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181, 7516-7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Rusten, T.E., and Stenmark, H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446-455. [DOI] [PubMed] [Google Scholar]
- Ramon, A.M., Porta, A., and Fonzi, W.A. (1999). Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181, 7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, R., Ruiz, A., Bernal, D., Chambers, J.R., and Arino, J. (2002). The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol 46, 1319-1333. [DOI] [PubMed] [Google Scholar]
- Shih, S.C. et al. (2002). Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389-393. [DOI] [PubMed] [Google Scholar]
- Strack, B., Calistri, A., Craig, S., Popova, E., and Gottlinger, H.G. (2003). AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689-699. [DOI] [PubMed] [Google Scholar]
- Vincent, O., Rainbow, L., Tilburn, J., Arst, H.N., Jr., and Penalva, M.A. (2003). YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23, 1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler, U.K. et al. (2003). The protein network of HIV budding. Cell 114, 701-713. [DOI] [PubMed] [Google Scholar]
- Wilson, R.B., Davis, D., and Mitchell, A.P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., and Mitchell, A.P. (2001). Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183, 6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, S.C. et al. (2003). Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 116, 3957-3970. [DOI] [PubMed] [Google Scholar]