Abstract
Polymorphisms have been identified in the Xq28 locus as risk loci for rheumatoid arthritis (RA). Here, we investigated the association between three polymorphisms in the Xq28 region containing TMEM187 and IRAK1 (rs13397, rs1059703, and rs1059702) in two unstudied populations: Tunisian and French. The rs13397 G and rs1059703 T major alleles were significantly increased in RA patients (n = 408) compared with age-matched controls (n = 471) in both Tunisian and French women. These results were confirmed by a meta-analysis replication study including two independent Greek and Korean cohorts. The rs1059702 C major allele was significantly associated with RA, only with French women. In the French population, the GTC haplotype displayed a protective effect against RA, while the ATC, GCC, and GTT haplotypes conferred significant risk for RA. No association for these haplotypes was found in the Tunisian population. Our results replicated for the first time the association of the three Xq28 polymorphisms with RA risk in Tunisian and French populations and suggested that RA susceptibility is associated with TMEM187-IRAK1 polymorphisms in women. Our data further support the involvement of X chromosome in RA susceptibility and evidence ethnicities differences that might be explained by differences in the frequencies of SE HLA-DRB1 alleles between both populations.
1. Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease. It is characterized by chronic destructive inflammation in synovial joints. The prevalence of RA is about 1% in the adult European population and is three times more common in women than in men [1–4]. The aetiology of this complex disease is still poorly understood but is considered as a result of interaction between susceptibility genes and environmental factors [5]. The genetic contribution to RA has been estimated to be about 50–60% [6], with the HLA (Human leukocyte antigen) classes II molecules remaining the most powerful known genetic factor [7]. There is extensive evidence for the association between certain HLA-DRB1 alleles that contain a conserved sequence of five amino acids (Q/RK/RRAA) in the third hypervariable region of the DRβ1 chain, the so-called shared epitope (SE), and RA susceptibility and severity [8–10].
Genome-wide association studies (GWAS) and high-density array Immunochip studies for single nucleotide polymorphisms (SNPs) genotyping of populations have identified over 101 RA risk loci involved among individuals of European and Asian ancestry [11–15]. Among these, IRAK1 (interleukin 1 receptor associated kinase) was the first X chromosome locus reported as associated with RA susceptibility and is thus of importance given the female predominance of the disease. IRAK1 is a serine-threonine protein kinase and an essential component of the toll/interleukin 1 receptor (TIR) signaling pathway involved in the pathogen-mediated inflammation [16]. Interestingly, the IRAK1 gene is located on the Xq28 region that harbours several SNPs that have also been associated with susceptibility to autoimmune diseases. A recent case-control study investigated numerous SNPs located on the Xq28 locus and identified rs1059703 and rs1059702, encoding for pSer532Leu and pPhe196Ser, as two IRAK1 SNPs most significantly associated with RA susceptibility in Korean families [17]. These authors also showed that the major haplotype (rs1059702 T and rs1059703 C) was associated with increased IRAK1 activity. Of note, upstream IRAK1 and within the Xq28 risk locus, Eyre et al. reported the SNP rs13397 associated with RA risk among individuals of northern European ancestry [13]. This polymorphism is located within the TMEM187 gene, which encodes a transmembrane protein of unknown function.
The relationship between these three polymorphisms on the Xq28 region and RA risk in different population remains unaddressed. Interestingly, a clear separation of different ethnic and regional populations was evidenced when considering northern and southern Europe groups separately in genetic studies, suggesting the existence of European population genetic substructures [18]. In addition, there are also clear differences between northern and southern Europe for the main genetic risk factor for RA, HLA-DRB1∗04:04, and DRB1∗04:01 genotypes being the most frequent in northern populations, while HLA-DRB1∗10:01 and DRB1∗04:05 alleles are more frequent in southern Europe, as well as in Maghreb [19, 20]. Therefore, the present study aimed at investigating the association between critical polymorphisms in Xq28, from rs13397 (A/G) through rs1059703 (C/T) and rs1059702 (C/T) in the TMEM187 and IRAK1 locus, respectively, with risk for RA in two not previously studied populations displaying different genetic background: Tunisian and French. We also examined genetic differences for the HLA-DRB1 alleles in both RA populations. A meta-analysis including two other independent cohorts was then performed to test the overall effect of these TMEM187-IRAK1 polymorphisms on RA.
2. Patients and Methods
2.1. Study Subjects
For RA patients and healthy controls, informed consents were provided in accordance with national procedures. French studies were approved by local human ethical committees: sample collection and analysis (DC-2008-327) were approved by the “Cellule Bioéthique, Direction Générale pour la Recherche et l'Innovation, Ministère de l'Enseignement Supérieur et de la Recherche” (Ministry Bioethics Unit) and by Comité de Protection des Personnes Sud Méditerrannée IV (ID RCB 2008-A01087-48). Blood samples were collected from a total of 879 unrelated female participants: 408 patients with RA and 471 controls from Tunisia and France. Detailed size of Tunisian and French cohort is shown in Table 1. RA patients fulfilled the 2010 ACR/EULAR classification criteria for RA [21]. The clinical examination consisted of questionnaire and a physical examination: age, geographical origin, family history, body mass index, smoking habits, the presence of hypertension, diabetes, levels of physical activity, and alcohol consumption. The 471 controls matched with age were recruited from Voluntary Bone Marrow Donor (VBMD). Less than 1% of healthy VBMD women come from Maghreb. All patients were positive for anti-citrullinated peptide antibodies (ACPA) as determined by commercially available ELISA kits (Orgentec®, Hamburg, Germany) and Euroimmun® for Tunisian cases and CCP2 enzyme-linked immuno sorbent assay (ELISA) (Immunoscan RA, Euro-Diagnostica, Arnhem, Netherlands) for French cases.
Table 1.
Characteristics of female samples from Tunisian and French cohort.
| Characteristic | Tunisian population | French population | ||
|---|---|---|---|---|
| Controls | RA patients | Controls | RA patients | |
| Size of the cohort | 131 | 119 | 340 | 289 |
| Age (mean ± SD years) |
56.65 ± 17.37 | 52.16 ± 13.61 | 46.24 ± 8.92 | 63.15 ± 20.71 |
| Disease duration (years) | NA | 15.3 ± 5.6 | NA | 14.8 ± 6.9 |
| Positive ACPA, n (%) | NA | 119 (100) | NA | 289 (100) |
| Positive RF, n (%) | NA | 98 (74.7) | NA | 200 (69.20) |
| C-reactive protein (mg/L) | NA | 12.2 ± 22.7 | NA | 13.6 ± 21.8 |
| DAS28 | NA | 6.3 ± 1.2 | NA | 5.2 ± 1.4 |
| Smoking status% | 66 | 55 | 76 | 47 |
2.2. Genotyping
Selection for Xq28 polymorphisms from previously reported SNPs was conducted using the following criteria: (a) the minor allele frequency (MAF) was >0.05 in the Caucasian population, according to the international HapMap project databank (http://www.hapmap.org) and to the NCBI SNP database (https://www.ncbi.nlm.nih.gov), (b) it was a tagging SNP, and (c) it has been reported to be associated with RA or other autoimmune diseases in different populations or other types of studies as GWAS. Genomic DNA was extracted from total blood leukocytes using standard methods including proteinase K digestion, followed by phenol–chloroform extraction and ethanol precipitation. Finally, all subjects were genotyped for three SNPs (rs1059702 and rs1059703 and rs13397) using direct PCR sequencing with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and an Applied Biosystems (ABI) 3130xL genetic analyzer (Applied Bio Systems, Foster City, CA, USA). PCR reactions were performed in a Bio-Rad thermal cycler using Taq Polymerase (Prim 5 by Fisher Scientific) with initial denaturing conditions at 96°C for 5 min, followed by 30 cycles of 96°C for 30 s, 64°C for 30 s, abd 72°C for 45 s and a final extension of 72°C for 10 min.
2.3. HLA-DRB1 Genotyping
Genomic DNA from whole blood was extracted as described in the subsection “Genotyping.” HLA-DRB1 typing was carried out according to the manufacturer's specification for LAB type SSO HD (One Lambda Inc., USA) and the retrieved output was analyzed by HLA Fusion v 1.2.1 software (One lambda Inc., USA) for allele information. The following alleles or group of alleles were genotyped: HLA-DRB1∗01, HLA-DRB1∗01:03, HLA-DRB1∗03, HLA-DRB1∗04:01, HLA-DRB1∗04:02, HLA-DRB1∗04:03, HLA-DRB1∗04:04, HLA-DRB1∗04:05, HLA-DRB1∗04:06, HLA-DRB1∗04:07, HLA-DRB1∗04:08, HLA-DRB1∗04:11, HLA-DRB1∗07, HLA-DRB1∗08, HLA-DRB1∗09, HLA-DRB1∗1001, HLA-DRB1∗11, HLA-DRB1∗12, HLA-DRB1∗13, HLA-DRB1∗14, HLA-DRB1∗15, and HLA-DRB1∗16 (Table 2).
Table 2.
Shared epitope frequency and HLA-DRB1 alleles frequency in Tunisian and French RA.
| Alleles HLA-DRB1 | Tunisian RA Allele number (N = 264) |
Tunisian allele frequency (%) | French RA Allele number (N = 346) |
French allele frequency (%) | OR [95% CI] | p value | pvalue∗ |
|---|---|---|---|---|---|---|---|
| #DRB1∗01:XX | 16 | 6.06 | 59 | 22.35 | 0.31 [0–0.51] | 1.6E−05 | 35.2E−05 |
| DRB1∗01:03 | 0 | 0.00 | 1 | 0.38 | 0 [0–4.55] | 0.32 | NS |
| DRB1∗03:XX | 42 | 15.91 | 15 | 5.68 | 4.17 [2.48–+∞] | 1E−06 | 22E−06 |
| #DRB1∗04:01 | 11 | 4.17 | 48 | 18.18 | 0.26 [0–0.47] | 1.5E−05 | 33E−05 |
| DRB1∗04:02 | 1 | 0.38 | 1 | 0.38 | 1.31 [0.20–+∞] | 0.39 | NS |
| DRB1∗04:03 | 3 | 1.14 | 4 | 1.52 | 0.98 [0–3.23] | 0.51 | NS |
| #DRB1∗04:04 | 8 | 3.03 | 30 | 11.36 | 0.32 [0–0.64] | 0.001 | 0.0022 |
| #DRB1∗04:05 | 19 | 7.20 | 12 | 4.55 | 2.15 [1.16–+∞] | 0.01 | NS |
| DRB1∗04:06 | 6 | 2.27 | 0 | 0.00 | +∞ [2.49–+∞] | 0.002 | 0.044 |
| DRB1∗04:07 | 0 | 0.00 | 1 | 0.38 | 0 [0–4.56] | 0.32 | NS |
| #DRB1∗04:08 | 3 | 1.14 | 9 | 3.41 | 0.43 [0–1.27] | 0.11 | NS |
| DRB1∗04:11 | 1 | 0.38 | 0 | 0.00 | +∞ [0.37–+∞] | 0.18 | NS |
| DRB1∗07:XX | 41 | 15.53 | 46 | 17.42 | 1.19 [0.81–+∞] | 0.21 | NS |
| DRB1∗08:XX | 3 | 1.14 | 1 | 0.38 | 3.96 [0.68–+∞] | 0.11 | NS |
| DRB1∗09:XX | 1 | 0.38 | 2 | 0.76 | 0.65 [0–3.98] | 0.41 | NS |
| #DRB1∗10:01 | 33 | 12.50 | 13 | 4.92 | 3.65 [2.09–+∞] | 2.3E−05 | 50.6E−05 |
| DRB1∗11:XX | 19 | 7.20 | 25 | 9.47 | 0.99 [0–1.66] | 0.5 | NS |
| DRB1∗12:XX | 0 | 0.00 | 5 | 1.89 | 0 [0–0.83] | 0.03 | NS |
| DRB1∗13:XX | 40 | 15.15 | 24 | 9.09 | 2.39 [1.52–+∞] | 0.0005 | 0.011 |
| DRB1∗14:XX | 4 | 1.52 | 4 | 1.52 | 1.31 [0.43–+∞] | 0.33 | NS |
| DRB1∗15:XX | 12 | 4.55 | 33 | 12.50 | 0.45 [0–0.80] | 0.009 | NS |
| DRB1∗16:XX | 1 | 0.38 | 13 | 4.92 | 0.09 [0–0.49] | 0.002 | 0.044 |
#Shared epitope, XX means all known alleles, DRB1∗14: XX apart from DRB1∗14:02
∗ p value corrected with Bonferroni correction.
NS: not significant.
2.4. Meta-Analysis Study
A literature search using PubMed, Science Direct, and Web of Science databases was conducted to identify publications that examine the association of TMEM187 rs13397, IRAK1 rs1059703, IRAK1 rs1059702, and IRAK1 rs3027898 polymorphisms with RA. Our research strategy was based on a combination of the following keywords: variation, variant, polymorphism, SNP, Rheumatoid arthritis, RA, TMEM187, IRAK1, rs13397, rs1059703, rs1059702, and rs3027898. We also reviewed the references mentioned in the identified articles for any additional relevant studies. Association studies included in our meta-analyses had to meet the following criteria: (1) evaluation of association between TMEM187 rs13397, IRAK1 rs1059703, IRAK1 rs1059702, or IRAK1 rs3027898 polymorphisms and the susceptibility to RA, (2) use of case-control design, and (3) inclusion of available genotype frequencies or sufficient information for calculation. The following information was extracted from the selected studies: first author, years of publication, characteristics of cases and controls (mean age, distribution of gender, and ethnicity), number of cases and controls, and genotype/allele frequency information. The strength of the association between the three polymorphisms and RA was measured by pooled OR with 95% CI. The Z test was used to determine the significance (p < 0.05) of the pooled OR. The heterogeneity between studies was checked by Q test and I2 statistics. We used a fixed effect model when p value of the Q test was less than 0.10 or I2 was equal to or less than 50%; otherwise, random effect model was used. Finally this meta-analysis was performed using Review Manager 5.3 software (Cochrane Collaboration, London, UK).
2.5. Statistical Methods
In this study, the Bayesian statistical method was used to evaluate the confidence intervals (called in the Bayesian framework credibility intervals) and to compute the p values associated with our tests, as in [20]. Indeed, the Bayesian statistical methods are more appropriate in case of small samples and the “approximate value” of the variance of the logarithm of the Odds Ratio. In the Bayesian framework, the unknown parameter is considered as the realization of a random variable, which forces us to choose an a priori probability distribution for that random variable. We chose an “a priori” distribution for a pair of frequencies and decided that the two random variables are independent, each with the probability density equal to 1 on the interval (0.1). It is fair to say that our prior is uninformative: it is the uniform distribution on the interval (0.1). The “a posteriori” distribution of each parameter is easily shown from the Bayes formula to be a Beta distribution. More precisely, if while observing n realizations of 1 s and 0 s, we get n 1 s and n′ 0 s, then the a posteriori distribution of the proportion is the Beta (n + 1, n′ + 1) probability distribution. Since the two samples come from completely unrelated sources, it is fair to assume that the two proportions are conditionally independent, given the data. Consequently a posteriori distribution of the pair of parameters (corresponding to the proportions of the given allele among the patients and among the control individuals) is the product of two Beta distributions whose density is explicitly known. Our method involves simulating one million realizations of that pair of random variables, from which we deduce one million realizations of the Odds Ratio. The p value of the test is then obtained by looking at which proportion of those values is smaller (resp., larger) than 1; this gives us t. Finally we want to discuss the issue of multiple testing. We have used our Bayesian test method 6 times (Table 5) where the probability of an error among six successive tests is not controlled by the probability of an error in each single test. The p values were corrected for multiple comparisons using the Bonferroni correction in Table 2. PLINK 1.7 version software was used for analysis of association between haplotypes and disease, and Haploview 4.2 software was used for LD analysis. All haplotypes with frequencies less than 5% were ignored in analysis. Distributions of data were tested with the Shapiro-Wilk test. Chi2 or Fisher test was used for categorical data; Yates correction was performed. For numerical data, comparisons of medians were performed using Mann–Whitney or Student's t-tests. For correlation study, statistical analyses were performed at the conventional two-tailed α level of 0.05 using R software version 3.0.2.10. Odds ratios (OR) with 95% confidence interval (CI) were also calculated. A p value less than 0.05 was considered statistically significant.
Table 5.
TMEM187-IRAK1 genotypes for samples of RA and healthy controls in Tunisian and French female populations.
| Gene/SNPs | Population | Genotype | Controls (%) |
RA cases (%) |
OR (95% CI) | p values |
|---|---|---|---|---|---|---|
| TMEM 187 rs13397 | Tunisian | GG | 92 (70) | 66 (53) | 0.48 (0.12–0.73) | 0.0025 |
| AG | 23 (18) | 46 (41) | ||||
| AA | 16 (12) | 7 (6) | ||||
| French | GG | 220 (65) | 131 (45) | 0.45 (0.21–0.59) | 1.0e − 6 | |
| AG | 97 (28) | 137 (47) | ||||
| AA | 23 (7) | 21 (8) | ||||
|
| ||||||
| IRAK 1 rs1059703 | Tunisian | CC | 15 (11) | 9 (7) | 0.46 (0.13–0.71) | 0.0017 |
| CT | 27 (21) | 51 (43) | ||||
| TT | 89 (68) | 59 (50) | ||||
| French | CC | 27 (8) | 34 (12) | 0.36 (0.15–0.47) | 1.0e − 6 | |
| CT | 85 (25) | 133 (46) | ||||
| TT | 228 (67) | 122 (42) | ||||
|
| ||||||
| IRAK 1 rs1059702 | Tunisian | CC | 81 (62) | 62 (52) | 0.67 (0.18–1.02) | 0.0608 |
| CT | 34 (26) | 43 (36) | ||||
| TT | 16 (12) | 14 (12) | ||||
| French | CC | 237 (70) | 157 (54) | 0.52 (0.24–0.68) | 3.7e − 5 | |
| CT | 83 (24) | 100 (35) | ||||
| TT | 20 (6) | 32 (11) | ||||
∗ p values compared genotypes according to the dominant/recessive model.
3. Results
3.1. Demographics and Phenotypes of the Study Subjects
Characteristics of the 408 ACPA (anti-citrullinated protein antibody) positive RA and 471 controls from Tunisia and South Eastern France (Marseille and Montpellier) are shown in Table 1. All samples were females. The average age was 52 years for the Tunisian RA and 63 years for French RA women. Age was not significantly different between patients and control females. There were more presently smokers in controls than in patients group.
3.2. Frequency of HLA-DRB1 Alleles and SE
Allelic frequencies of the HLA-DRB1 alleles in Tunisian and French RA patients are presented in Table 2. Due to the size of cohorts, we applied Bayesian statistical method to define susceptibility or protective genotypes and calculate accurate confidence intervals for the associated Odds Ratios. HLA-DRB1∗01, HLA-DRB1∗03, HLA-DRB1∗04:01, HLA-DRB1∗04:04, HLA-DRB1∗04:05, HLA-DRB1∗04:06, HLA-DRB1∗10, HLA-DRB1∗13, HLA-DRB1∗15, and HLA-DRB1∗16 showed statistical differences between the French and the Tunisian RA women. No statistical differences were found for HLA-DRB1∗01:03, HLA-DRB1∗04:02, HLA-DRB1∗04:03, HLA-DRB1∗04:07, HLA-DRB1∗04:08, HLA-DRB1∗04:11, HLA-DRB1∗07, HLA-DRB1∗08, HLA-DRB1∗09, HLA-DRB1∗11, HLA-DRB1∗12, and HLA-DRB1∗14 between both populations. The RA associated HLA-DRB1 alleles that were significantly more present in the Tunisian samples compared to those in the French samples were, by order of Odds Ratios, HLA-DRB1∗04:05 (OR = 2.15), HLA-DRB1∗13 (OR = 2.39), HLA-DRB1∗10 (OR = 3.65), HLA-DRB1∗03 (OR = 4.17), and HLA-DRB1∗04:06 (OR = +∞). Similarly, RA associated HLA-DRB1 alleles that were significantly more observed in the French than in the Tunisian samples were, by order of Odds Ratios, HLA-DRB1∗16 (OR = 0.09), HLA-DRB1∗04:01 (OR = 0.26), HLA-DRB1∗01 (OR = 0.31), HLA-DRB1∗04:04 (OR = 0.32), and HLA-DRB1∗15 (OR = 0.45). Three HLA-DRB1 alleles (DRB1∗04:05, DRB1∗12:XX, and DRB1∗15:XX) did not show significant differences between Tunisian and French RA women after the Bonferroni correction.
Two (HLA-DRB1∗04:05 and HLA-DRB1∗10) out of seven shared SE alleles were overrepresented in the Tunisian RA population, and three SE alleles (HLA-DRB1∗01, HLA-DRB1∗04:01, and HLA-DRB1∗04:04) were overrepresented in the French population. SE single dose showed similar frequency in both Tunisian and French RA women (50% and 57.80%, resp.), while French RA population displayed higher SE double dose than Tunisian RA population (20.34% and 9.09%, resp.) (Table 3).
Table 3.
Shared epitope single dose/double dose frequency in Tunisian and French RA women.
| Shared epitope (SE) | Tunisian RA N = 132 |
Tunisian SE frequency (%) |
French RA N = 173 |
French SE frequency (%) |
|---|---|---|---|---|
| SE single dose | 66 | 50.00 | 100 | 57.80 |
| SE double dose | 12 | 9.09 | 35 | 20.34 |
|
| ||||
| Total | 78 | 59.09 | 135 | 78.48 |
3.3. Association between Three TMEM187-IRAK1 Polymorphisms and RA Susceptibility in Women Samples from Tunisian and French Populations
Genotype distributions of all three investigated SNPs were in Hardy-Weinberg equilibrium in both patient and control groups (χ2 < 3.84). Among the genotype frequencies of the three SNPs analyzed using the dominant modelling, the rs13397 and rs1059703 demonstrated a significant association with RA susceptibility in both populations. The rs13397 G allele is present in 75% of RA Tunisian patients and in 69% RA French (Table 4). The frequency of the rs13397 G major allele was significantly lower in RA patients than in controls in both Tunisian (OR 0.48 [95% CI 0.12–0.73], p = 0.0025) and French (OR = 0.45 [95% CI 0.21–0.59], p < 0.001) women populations. The rs1059703 T major allele was significantly decreased in patients compared with controls in both Tunisian (OR = 0.46 [95% CI 0.13–0.71], p < 0.001) and French (OR = 0.36 [95% CI 0.15–0.47], p < 0.001) women populations. The SNP rs1059702 C showed a significant association with RA in French patients as compared with control subjects (OR 1.97 [95% CI 1.22–11.52], p < 0.001), but none in the Tunisian population. Of note, no correlation was found between polymorphisms and the SE (data not shown).
Table 4.
TMEM187-IRAK1 allelic frequencies in samples of RA and healthy controls from Tunisian and French female populations.
| Gene/SNPs | Populations | Alleles | Controls (%) | RA cases (%) |
|---|---|---|---|---|
| TMEM 187 rs13397 | Tunisian | G | 207 (79) | 178 (75) |
| A | 55 (21) | 60 (25) | ||
| French | G | 537 (79) | 399 (69) | |
| A | 143 (21) | 179 (31) | ||
|
| ||||
| IRAK 1 rs1059703 | Tunisian | C | 57 (22) | 69 (29) |
| T | 205 (78) | 169 (71) | ||
| French | C | 139 (20) | 201 (35) | |
| T | 541 (80) | 377 (65) | ||
|
| ||||
| IRAK 1 rs1059702 | Tunisian | C | 196 (75) | 167 (70) |
| T | 66 (25) | 71 (30) | ||
| French | C | 557 (82) | 414 (72) | |
| T | 123 (18) | 164 (28) | ||
3.4. Linkage Disequilibrium of TMEM187-IRAK1 Locus with RA in the Samples of the Tunisian and French Populations
To determine the extent of linkage disequilibrium (LD) among the three polymorphisms enclosed in the TMEM187-IRAK1 locus in both Tunisian and French populations, standardized LD coefficient r2 was calculated for all pairs of the three polymorphisms. Our study showed that the IRAK1 SNPs rs1059703 and rs1059702 did not belong to the same linkage disequilibrium locus, both in Tunisian and French populations (Figure 1). We also showed that rs13397 and rs1059703 polymorphisms were in moderate linkage disequilibrium (r2 = 0.5) in the Tunisian population (Figure 1(a)), but not in the French population (Figure 1(b), r2 = 0.11).
Figure 1.
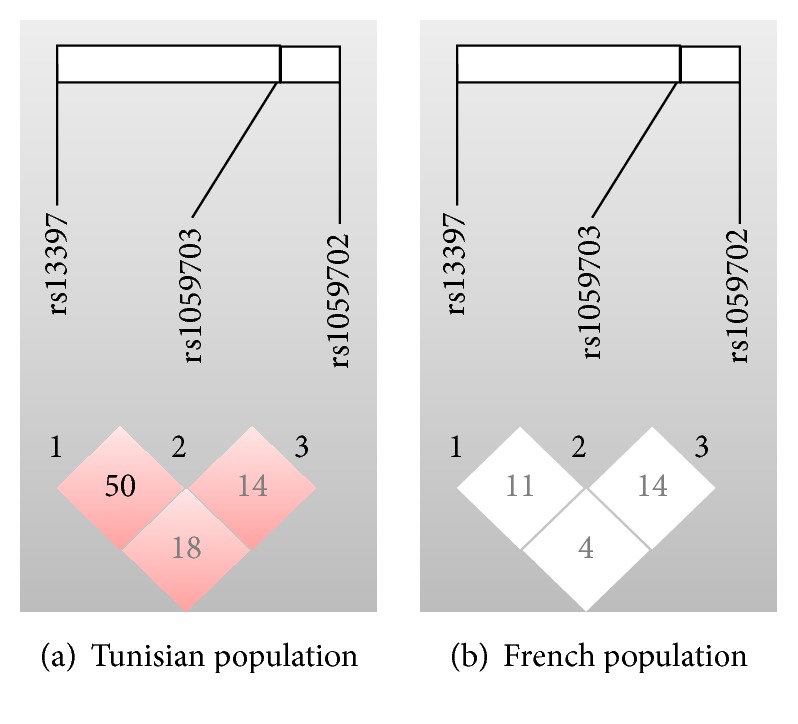
Linkage disequilibrium (LD) between TMEM187-IRAK1 SNPs. Correlations (r2 values) among all 3 single nucleotide polymorphisms (SNPs) located in the TMEM187-IRAK1 locus are indicated in the diamonds for the Tunisian (a) and French (b) populations. Haploview 4.2 software was used for LD analyses.
3.5. Haplotype Analysis
To examine the combined effect of three variants within the TMEM187-IRAK1 locus, the haplotype frequencies defined by the three SNPs rs13397, rs1059703, and rs1059702 were analyzed. Among the potential haplotypes, four common haplotypes were identified in our populations with frequencies greater than 5% in both cases and controls (Table 6).
Table 6.
Association of SNP haplotypes in the TMEM187-IRAK1 region with RA susceptibility.
| Chromosome | Populations | Haplotypes | Frequencies | OR (95%CI) | p values | Effects | |
|---|---|---|---|---|---|---|---|
| Controls | Cases | ||||||
| X | Tunisian | [ATC] | 0.15 | 0.15 | 1.00 (0.63–1.59) | 0.995 | NA |
| French | 0.09 | 0.19 | 3.97 (2.57–6.13) | <0.001 | Risk | ||
| Tunisian | [GCC] | 0.14 | 0.17 | 1.21 (0.79–1.89) | 0.379 | NA | |
| French | 0.06 | 0.15 | 2.39 (1.52–3.77) | <0.001 | Risk | ||
| Tunisian | [GTT] | 0.15 | 0.18 | 1.09 (0.70–1.71) | 0.692 | NA | |
| French | 0.04 | 0.10 | 2.05 (1.16–3.62) | 0.009 | Risk | ||
| Tunisian | [GTC] |
0.57 | 0.50 | 0.77 (0.57–1.04) | 0.083 | NA | |
| French | 0.68 | 0.39 | 0.23 (0.16–0.33) | <0.001 | Protector | ||
Order of SNPs: [rs13397, rs1059703, rs1059702]
NA: no association.
Results of the analysis suggested that the GTC haplotype, which carries the rs13397 G variant, displayed a protective effect against RA in our sample of the French population (OR = 0.23 (95% CI) [0.16–0.33], p < 0.001) while the ATC, GCC, and GTT haplotypes conferred significant risk for RA in the same population. French women with the ATC, GCC, or GTT haplotypes had 3.97-, 2.39-, and 2.05-fold higher risk of having RA compared to controls. In contrast, association between all 4 detected haplotypes and disease was not significant in the sample of the Tunisian women. Overall, these data further support the involvement of the Xq28 locus in RA susceptibility in the French population and underscore ethnical differences.
3.6. Meta-Analysis Study
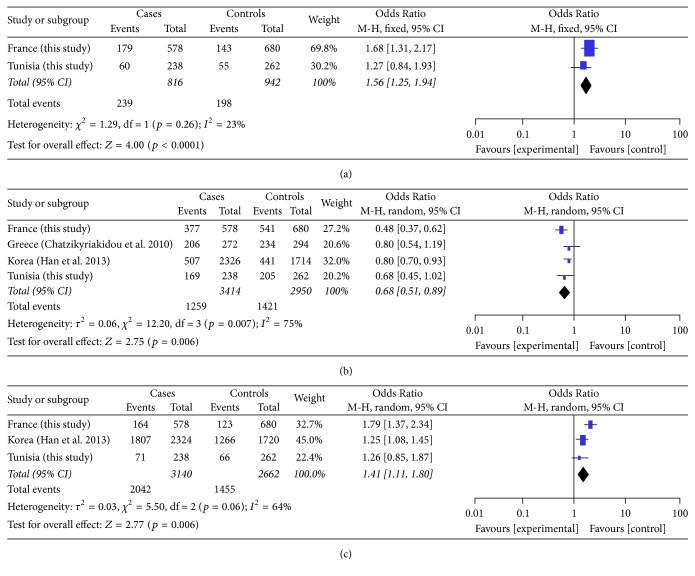
A meta-analysis was performed and included datasets from 2076 RA cases and 2112 healthy controls. To carry out the meta-analysis, we combined our findings with three independent studies that fit the inclusion criteria and included a total of 1668 RA patients and 1641 ethnically matched healthy controls from Greece and Korea [17, 22, 23]. The samples size of these studies varied from 119 to 1318 cases and from 131 to 1016 controls. The characteristics of all studies were summarized in Table 7. Meta-analysis results showed a significant association between TMEM187 rs13397 (OR = 1.56, 95% CI = 1.25–1.94, and p value < 0.0001) and IRAK1 rs1059702 (OR = 1.41, 95% CI = 1.11–1.80, and p value = 0.006) polymorphisms and susceptibility to RA (Figures 2(a) and 2(c)). The IRAK1 rs1059703 polymorphism showed a significant protective effect in RA (OR = 0.68, 95% CI = 0.51–0.89, and p value = 0.006) (Figure 2(b)). The IRAK1 rs3027898 polymorphism that was available in the three independent studies from Korea, China, and Greece showed no significant association with RA risk (OR = 0.96, 95% CI = 0.67–1.39, and p value = 0.83).
Table 7.
Characteristics of all studies included in meta-analysis within the TMEM187-IRAK1 locus.
| Study | Population | Group | Subjects | Women's sex% | Average age in years | Genotype |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TMEM 187 | IRAK 1 | IRAK 1 | IRAK 1 | ||||||||||||||||||||||
| rs13397 | rs1059703 | rs1059702 | rs3027898 | ||||||||||||||||||||||
| GG | AG | AA | G | A | CC | CT | TT | C | T | CC | CT | TT | C | T | AA | AC | CC | A | C | ||||||
| This study | Tunisia | Case | 119 | 100% | 52.16 ± 13.61 | 66 | 46 | 7 | 178 | 60 | 9 | 51 | 59 | 69 | 169 | 62 | 43 | 14 | 167 | 71 | — | — | — | — | — |
| Control | 131 | 100% | 56.65 ± 17.37 | 92 | 23 | 16 | 207 | 55 | 15 | 27 | 89 | 57 | 205 | 81 | 34 | 16 | 196 | 66 | — | — | — | — | — | ||
| This study | French | Case | 289 | 100% | 63.15 ± 20.71 | 131 | 137 | 21 | 399 | 179 | 34 | 133 | 122 | 201 | 377 | 157 | 100 | 32 | 414 | 164 | — | — | — | — | — |
| Control | 340 | 100% | 46.24 ± 8.92 | 220 | 97 | 23 | 537 | 143 | 27 | 85 | 228 | 139 | 541 | 237 | 83 | 20 | 557 | 123 | — | — | — | — | — | ||
| Chatzikyriakidou et al. (2010) | Greece | Case | 136 | 80% | 60.8 ± 12.7 | — | — | — | — | — | 7 | 52 | 77 | 66 | 206 | — | — | — | — | — | 71 | 45 | 20 | 187 | 85 |
| Control | 147 | 78% | 56.23 ± 5.13 | — | — | — | — | — | 7 | 46 | 94 | 60 | 234 | — | — | — | — | — | 91 | 47 | 9 | 229 | 65 | ||
| Han et al. (2013) | Korea | Case | 1,318 | 100% | 51.8 ± 12.2 | — | — | — | — | — | 715 | 389 | 59 | 1819 | 507 | 62 | 393 | 707 | 517 | 1807 | 719 | 383 | 56 | 1821 | 495 |
| Control | 1,016 | 100% | 36.7 ± 12.5 | — | — | — | — | — | 468 | 337 | 52 | 1273 | 441 | 59 | 336 | 465 | 454 | 1266 | 478 | 321 | 50 | 1277 | 421 | ||
| Zhang et al. (2013) | China | Case | 214 | 71% | 54.63 (±15.77) | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 28 | 42 | 141 | 98 | 324 |
| Control | 478 | 71% | 54.84 (±10.45) | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 337 | 103 | 35 | 173 | 777 | ||
Figure 2.
Forest plot of the association between TMEM187 rs13397 (a), IRAK1 rs1059703 (b), and IRAK1 rs1059702 (c) polymorphisms and RA. Meta-analyses were performed for the rs13397 (a), rs1059703 (b), and rs1059702 (c) polymorphisms, respectively, using Review Manager 5.3 software.
4. Discussion
In the present study, we aimed at investigating the association between three critical polymorphisms in the TMEM187-IRAK1 locus and risk for ACPA positive RA female patients in two not previously studied populations: Tunisian and French. We found that the rs13397 A and rs1059703 C risk alleles were significantly associated with RA susceptibility for both populations. The rs1059702 T allele was only found associated with risk for RA in the French sample. Four haplotypes were detected in both populations. In the French population, the major GTC haplotype displayed a protective effect against RA, whereas the GCC, GTT, and ATC haplotypes conferred significant increased risk for RA. No association for these haplotypes was however found in the Tunisian population. Finally, a strong genetic association of RA susceptibility exists for the rs13397/rs1059703 block only in women Tunisian subjects.
Among the many loci that have been identified as associated with RA risk using either candidate gene association studies or GWAS, IRAK1 is the only gene located on the X chromosome. The IRAK1 gene encodes the interleukin 1 receptor-associated kinase 1, a serine/threonine kinase that mediates TLRs and IL-1 receptor signaling. Upon receptor stimulation, IRAK1 is recruited, phosphorylated, and responsible for TLR- and IL1-induced activation of the NF-κB signaling cascade, thus playing a critical role in the early inflammatory response [24]. The IRAK1 gene is located within the Xq28 locus that contains several polymorphisms associated with increased susceptibility to autoimmunity, including systemic lupus erythematous [25], systemic sclerosis [26, 27], and RA [13, 17, 22, 23]. Altogether, these studies demonstrated that the Xq28 locus is a shared locus for several complex diseases, thereby highlighting its role in inflammatory disease susceptibility. Recently, two studies devoted attention to the TMEM187-IRAK1 locus and identified two SNPs in a Korean population (rs1059702 and rs1059703) [17] and one SNP (rs13397) in a large cohort of northern European ancestry [13], respectively, associated with increased disease susceptibility. In an attempt to investigate new, not previously studied populations and to replicate the above studies, we examined the distribution of these three SNPs and found statistical differences between RA patients and controls for both Tunisian and French populations. Although risk alleles for both rs1059702 and rs1059703 were different, they were also associated with risk for RA in both Tunisian and French populations. Indeed, the rs1059702 T and rs1059703 C polymorphisms were in strong association with RA in Korean women while we found the rs1059702 C and rs1059703 T alleles in our cohorts. In agreement with the study of Eyre et al. [13], we found the minor rs13397 A allele to be the risk allele in both the French and Tunisian cohorts. Earlier work performed on a small Greek cohort found no association between the rs1059703 and RA [22]. These differences might have several explanations, including the sample size of cohorts used, the Bayesian statistical method being better suited than the SPSS statistical package to decide about susceptibility in small samples, the classification criteria used to select RA patients (ACR 1987 versus 2010 ACR/EULAR that discriminate or not between ACPA positive RA patients), and different ethnic and regional populations.
Concerning the haplotype and LD studies, we are the first to study the combination of LD and haplotypes of these three SNPs with RA. We found that, with the exception of rs13397 and rs1059703 polymorphisms, which were in moderate linkage disequilibrium in the Tunisian population (r2 = 0.5), other polymorphisms were not in linkage disequilibrium in both population. This finding seems to be in contradiction with the Korean study that previously reported a linkage disequilibrium block on Xq28 enclosing both rs1059702 and rs1059703 [17]. Discrepancies between Korean, Tunisian, and French populations might be due to differences in genetic background. Interestingly, a comparative study reported that none of the susceptibility loci identified by GWAS in Caucasian patients with RA contributed significantly to disease in Koreans [28].
We found differences between French and Tunisian populations for association of SNP haplotypes in the TMEM187-IRAK1 locus with RA susceptibility. The CTG haplotype displayed a protective effect against RA, while the ATC, GCC, and GTT haplotypes conferred significant susceptibility to RA, in French women. All four haplotypes showed no association in the Tunisian women sample. In both Tunisian and French cohorts, we found the GTC common haplotype in 57 and 68% of the control populations, respectively. This GTC haplotype carries the major alleles for the three SNPs rs13397 G, rs1059703 T, and rs1059702 C.
Although alleles and haplotypes identified among Tunisian and French subjects display differences with previous studies performed with Korean and northern European cohorts, our present work confirms significant association of the three SNPs with RA susceptibility. The minor discrepancy found for the rs1059702 in the Tunisian RA women population might be due to the size of the cohort. Indeed, pooled results for the three different ethnical groups including Tunisian confirm a significant association between rs1059702 and risk for RA.
Our meta-analysis including a larger case-control data set of two independent studies from Korea and Greece in addition to our data sets confirmed that TMEM187 rs13397, IRAK1 rs1059702, and IRAK1 rs1059703 are associated with RA. Unfortunately, data sets on the TMEM187 rs13397 from populations of European ancestry [13] were not available for inclusion in our meta-analysis. The IRAK1 rs3027898 that was not investigated in our study but available in the three independent studies from Korea, China, and Greece showed no significant association in the meta-analysis (data not shown). To the best of our knowledge, this is the first meta-analysis study on the association between RA and these four polymorphisms in TMEM187-IRAK1.
Many studies have documented that one of the key genetic elements closely linked to RA susceptibility is the HLA complex locus located in the chromosome 6 [15]. Our present study evaluated the frequency of HLA-DRB1 alleles in Tunisian and French RA women, all ACPA positive, and confirmed that both populations belonging to different ethnical groups do not share the same HLA susceptibility alleles. As expected for the French population, we find the usual association between RA and HLA-DRB1∗01, HLA-DRB1∗04:01, and HLA-DRB1∗04:04 [20]. We show that two SE alleles, HLA-DRB1∗04:05 and HLA-DRB1∗10, are overrepresented in the RA Tunisian population. This is in agreement with a previous report showing their association with RA susceptibility in Tunisia [19]. The HLA-DRB1∗04 allele has also been reported in RA populations from Morocco [29], Saudi Arabia [30], and Turkey [31]. The HLA-DRB1∗10 allele has been reported to be associated with RA in Saudi Arabia [30], Italy [32], Spain [33], Turkey [34], and Asian Indian [35] populations.
A meta-analysis performed in Latin America RA population (Colombia, Peru, Brazil, Argentina, Mexico, and Chili) showed that HLA-DRB1∗04:04 allele is associated with disease risk [36]. Moreover, this whole genome scan study also shows that the second most consistently linked region to RA is the Xq28 locus. The absence of the HLA-DRB1∗01:03, HLA-DRB1∗04:07, and HLA-DRB1∗12 alleles in our RA Tunisian population sample is in accordance with the protective effect suggested by previous study [19]. Considering the observed differences between ethnical groups for HLA-DRB1, our findings suggest that the association of the TMEM187-IRAK1 locus with RA risk may play a role independently of the major genetic risk locus for RA.
5. Conclusion
In conclusion, the present study illustrates the involvement of the Xq28 locus in RA susceptibility in both Tunisian and French populations with ethnical differences. Finally, the meta-analysis combining our findings with two independent studies from Greece and Korea further supports the fact that the three polymorphisms are important in the susceptibility for RA. Although a growing number of studies suggest the contribution of the X chromosome to RA, as to many other autoimmune diseases, future works shall be performed to compare female and male RA populations to identify the exact contribution of this locus, with a focus on TMEM187-IRAK1 polymorphisms. Furthermore, functional studies on the impact of these polymorphisms on both gene expressions in RA populations might help defining new therapeutic perspectives in RA.
Acknowledgments
This work is supported by the Inserm, University of Montpellier, the EU funding IMI- BTCure (no. 115142) and ERASMUS-Mundus (no. 2888). The authors thank Elodie Sanchez and Rosana Ferreira-Lopez for technical and clinical support. They are grateful to Nikoleta Vrcelj for English editing.
Abbreviations
- ACPA:
Anti-citrullinated protein antibody
- ACR:
American College of Rheumatology
- AR:
Androgen receptor
- CCP2:
Cyclic-citrullinated peptide
- CD40L:
Cluster of differentiation 40 ligand
- CI:
Confidence interval
- ELISA:
Enzyme-linked Immunosorbent assay
- EULAR:
European League against Rheumatism
- FOXP3:
Forkhead box P3
- GWAS:
Genome-wide association studies
- HLA-DRB1:
Human leukocyte antigen-DRB1
- IL2RG:
Interleukin-2 receptor gamma chain
- IRAK1:
Interleukin 1 receptor associated kinase 1
- LD:
Linkage disequilibrium
- MAF:
Minor allele frequency
- NF-kB:
Nuclear factor-kappa B
- OR:
Odds Ratio
- PCR:
Polymerase chain reaction
- RA:
Rheumatoid arthritis
- SNP:
Single nucleotide polymorphism
- SE:
Shared epitope
- TLR:
Toll-like receptors
- TMEM187:
Transmembrane protein 187
- VBMD:
Voluntary bone marrow donor.
Competing Interests
The authors declaring that they have no competing interests are Olfa Khalifa, Nathalie Balandraud, Nathalie Lambert, Isabelle Auger, Jean Roudier, Audrey Sénéchal, David Geneviève, Christophe Picard, Gérard Lefranc, Hicham Charoute, Isabelle Touitou, Bakridine M'Madi Mrenda, Cécilia Benedito, Etienne Pardoux, Anne-Laure Gagez, Yves-Marie Pers, Christian Jorgensen, Touhami Mahjoub, and Florence Apparailly.
Authors' Contributions
Olfa Khalifa carried out the molecular genetic studies, recruited the Tunisian RA patients and healthy donors, performed the sequence alignment, analyzed data, wrote the manuscript and performed the meta-analysis study. Nathalie Balandraud, Nathalie Lambert, Jean Roudier, Isabelle Auger recruited the French RA patients and healthy donors, collected materials, analyzed data, and participate in writing the manuscript. Audrey Sénéchal performed the sequence alignment and participated in writing the manuscript. David Geneviève, Gérard Lefranc, Isabelle Touitou, Yves-Marie Pers, Christian Jorgensen overviewed the project and participated in writing the manuscript. Christophe Picard provided materials of healthy donors, participated in writing the manuscript and performed the HLA-DRB1 typing. Bakridine M'Madi Mrenda, Etienne Pardoux, Cécilia Benedito, Anne-Laure Gagez developed mathematic tools, performed statistical analyses, and participated in writing the manuscript. Touhami Mahjoub recruited the Tunisian healthy donors, overviewed the project and participated in writing the manuscript. Florence Apparailly conceived of the study, overviewed the design and coordination of the project, and participated in writing the manuscript. All authors read and approved the final manuscript.
References
- 1.Linos A., Worthington J. W., O'fallon M., Kurland L. T. The epidemiology of rheumatoid arthritis in Rochester, Minnesota: a study of incidence, prevalence, and mortality. American Journal of Epidemiology. 1980;111(1):87–98. doi: 10.1093/oxfordjournals.aje.a112878. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen P. K., Amos C. I., Lee A. T., et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nature Genetics. 2009;41(7):820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Vollenhoven R. F. Sex differences in rheumatoid arthritis: more than meets the eye... BMC Medicine. 2009;7, article 12 doi: 10.1186/1741-7015-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona L., Cross M., Williams B., Lassere M., March L. Rheumatoid arthritis. Best Practice and Research: Clinical Rheumatology. 2010;24(6):733–745. doi: 10.1016/j.berh.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Dieudé P. Rheumatic diseases: environment and genetics. Joint Bone Spine. 2009;76(6):602–607. doi: 10.1016/j.jbspin.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Viatte S., Plant D., Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nature Reviews Rheumatology. 2013;9(3):141–153. doi: 10.1038/nrrheum.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perricone C., Ceccarelli F., Valesini G. An overview on the genetic of rheumatoid arthritis: a never-ending story. Autoimmunity Reviews. 2011;10(10):599–608. doi: 10.1016/j.autrev.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 8.Scally S. W., Petersen J., Law S. C., et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. Journal of Experimental Medicine. 2013;210(12):2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregersen P. K., Silver J., Winchester R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis and Rheumatism. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 10.Lundström E., Källberg H., Alfredsson L., Klareskog L., Padyukov L. Gene-environment interaction between the DRB1 shared epitope and smoking in the risk of anti-citrullinated protein antibody-positive rheumatoid arthritis: all alleles are important. Arthritis and Rheumatism. 2009;60(6):1597–1603. doi: 10.1002/art.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl E. A., Raychaudhuri S., Remmers E. F., et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature Genetics. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padyukov L., Seielstad M., Ong R. T. H., et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Annals of the Rheumatic Diseases. 2011;70(2):259–265. doi: 10.1136/ard.2009.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyre S., Bowes J., Diogo D., et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nature Genetics. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K., Bang S.-Y., Lee H.-S., et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Annals of the Rheumatic Diseases. 2015;74(3, article no. e13) doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada Y., Terao C., Ikari K., et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nature Genetics. 2012;44(5):511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 16.Martin M. U., Wesche H. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochimica et Biophysica Acta—Molecular Cell Research. 2002;1592(3):265–280. doi: 10.1016/s0167-4889(02)00320-8. [DOI] [PubMed] [Google Scholar]
- 17.Han T.-U., Cho S.-K., Kim T., Joo Y. B., Bae S.-C., Kang C. Association of an activity-enhancing variant of IRAK1 and an MECP2-IRAK1 haplotype with increased susceptibility to rheumatoid arthritis. Arthritis and Rheumatism. 2013;65(3):590–598. doi: 10.1002/art.37804. [DOI] [PubMed] [Google Scholar]
- 18.Tian C., Kosoy R., Nassir R., et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Molecular Medicine. 2009;15(11-12):371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamad M. B., Mahfoudh N., Marzouk S., et al. Association study of human leukocyte antigen-DRB1 alleles with rheumatoid arthritis in south Tunisian patients. Clinical Rheumatology. 2012;31(6):937–942. doi: 10.1007/s10067-012-1954-z. [DOI] [PubMed] [Google Scholar]
- 20.Balandraud N., Picard C., Reviron D., et al. HLA-DRB1 genotypes and the risk of developing anti citrullinated protein antibody (ACPA) positive rheumatoid arthritis. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0064108.e64108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aletaha D., Neogi T., Silman A. J., et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and Rheumatism. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 22.Chatzikyriakidou A., Voulgari P. V., Georgiou I., Drosos A. A. A polymorphism in the 3'-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine. 2010;77(5):411–413. doi: 10.1016/j.jbspin.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Pu J., Wang X., et al. IRAK1 rs3027898 C/A polymorphism is associated with risk of rheumatoid arthritis. Rheumatology International. 2013;33(2):369–375. doi: 10.1007/s00296-012-2379-3. [DOI] [PubMed] [Google Scholar]
- 24.Sun X.-F., Zhang H. NFKB and NFKBI polymorphisms in relation to susceptibility of tumour and other diseases. Histology and Histopathology. 2007;22(12):1387–1398. doi: 10.14670/HH-22.1387. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman K. M., Zhao J., Kelly J. A., et al. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancestral groups. Annals of the Rheumatic Diseases. 2013;72(3):437–444. doi: 10.1136/annrheumdis-2012-201851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David Carmona F., Carmen Cénit M., Diaz-Gallo L.-M., et al. New insight on the Xq28 association with systemic sclerosis. Annals of the Rheumatic Diseases. 2013;72(12):2032–2038. doi: 10.1136/annrheumdis-2012-202742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dieudé P., Bouaziz M., Guedj M., et al. Evidence of the contribution of the X chromosome to systemic sclerosis susceptibility: association with the functional IRAK1 196Phe/532Ser haplotype. Arthritis and Rheumatism. 2011;63(12):3979–3987. doi: 10.1002/art.30640. [DOI] [PubMed] [Google Scholar]
- 28.Lee H.-S., Korman B. D., Le J. M., et al. Genetic risk factors for rheumatoid arthritis differ in caucasian and Korean populations. Arthritis and Rheumatism. 2009;60(2):364–371. doi: 10.1002/art.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atouf O., Benbouazza K., Brick C., et al. HLA polymorphism and early rheumatoid arthritis in the Moroccan population. Joint Bone Spine. 2008;75(5):554–558. doi: 10.1016/j.jbspin.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Al-Swailem R., Al-Rayes H., Sobki S., Arfin M., Tariq M. HLA-DRB1 association in Saudi rheumatoid arthritis patients. Rheumatology International. 2006;26(11):1019–1024. doi: 10.1007/s00296-006-0119-2. [DOI] [PubMed] [Google Scholar]
- 31.Uçar F., Karkucak M., Alemdaroğlu E., et al. HLA-DRB1 allele distribution and its relation to rheumatoid arthritis in eastern Black Sea Turkish population. Rheumatology International. 2012;32(4):1003–1007. doi: 10.1007/s00296-010-1792-8. [DOI] [PubMed] [Google Scholar]
- 32.Bongi S. M., Porfirio B., Rombolà G., Palasciano A., Beneforti E., Bianucci G. Shared-epitope HLA-DRB1 alleles and sex ratio in Italian patients with rheumatoid arthritis. Joint Bone Spine. 2004;71(1):24–28. doi: 10.1016/S1297-319X(03)00099-X. [DOI] [PubMed] [Google Scholar]
- 33.Yelamos J., Garcia-Lozano J. R., Moreno I., et al. Association of HLA-DR4-Dw15 (DRB1∗0405) and DR10 with rheumatoid arthritis in a Spanish population. Arthritis and Rheumatism. 1993;36(6):811–814. doi: 10.1002/art.1780360611. [DOI] [PubMed] [Google Scholar]
- 34.Kinikli G., Ateş A., Turgay M., Akay G., Kinikli S., Tokgöz G. HLA-DRB1 genes and disease severity in rheumatoid arthritis in Turkey. Scandinavian Journal of Rheumatology. 2003;32(5):277–280. doi: 10.1080/03009740310003901. [DOI] [PubMed] [Google Scholar]
- 35.Taneja V., Giphart M. J., Verduijn W., Naipal A., Malaviya A. N., Mehra N. K. Polymorphism of HLA-DRB, -DQA1, and -DQB1 in rheumatoid arthritis in Asian Indians: association with DRB1∗0405 and DRB1∗1001. Human Immunology. 1996;46(1):35–41. doi: 10.1016/0198-8859(95)00165-4. [DOI] [PubMed] [Google Scholar]
- 36.Delgado-Vega A. M., Anaya J.-M. Meta-analysis of HLA-DRB1 polymorphism in Latin American patients with rheumatoid arthritis. Autoimmunity Reviews. 2007;6(6):402–408. doi: 10.1016/j.autrev.2006.11.004. [DOI] [PubMed] [Google Scholar]