Abstract
Heterotrimeric G proteins have been implicated in the regulation of membrane trafficking, but the mechanisms involved are not well understood. Here, we report that overexpression of the stimulatory G protein subunit (Gαs) promotes ligand-dependent degradation of epidermal growth factor (EGF) receptors and Texas Red EGF, and knock-down of Gαs expression by RNA interference (RNAi) delays receptor degradation. We also show that Gαs and its GTPase activating protein (GAP), RGS-PX1, interact with hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), a critical component of the endosomal sorting machinery. Gαs coimmunoprecipitates with Hrs and binds Hrs in pull-down assays. By immunofluorescence, exogenously expressed Gαs colocalizes with myc-Hrs and GFP-RGS-PX1 on early endosomes, and expression of either Hrs or RGS-PX1 increases the localization of Gαs on endosomes. Furthermore, knock-down of both Hrs and Gαs by double RNAi causes greater inhibition of EGF receptor degradation than knock-down of either protein alone, suggesting that Gαs and Hrs have cooperative effects on regulating EGF receptor degradation. These observations define a novel regulatory role for Gαs in EGF receptor degradation and provide mechanistic insights into the function of Gαs in endocytic sorting.
INTRODUCTION
Heterotrimeric G proteins serve as important molecular switches that relay extracellular signals from G proteincoupled receptors (GPCRs) on the cell membrane to downstream effectors (Gilman, 1987; Neves et al., 2002). Besides their plasma membrane location, heterotrimeric G proteins also are found on membranes of intracellular compartments along both the endocytic and secretory pathways where indirect evidence suggests they play several roles in membrane trafficking (Bomsel and Mostov, 1992; Helms, 1995; Nurnberg and Ahnert-Hilger, 1996; Stow and Heimann, 1998). One of the prototypical heterotrimeric G proteins, Gαs, the stimulatory subunit of heterotrimeric G proteins, has been suggested to regulate endocytic trafficking. Reagents that activate Gαs, e.g., cholera toxin and a peptide mimicking the interacting region of Gαs with the β2-adrenergic receptor, block endosome-endosome and phagosomeendosome fusion in vitro (Colombo et al., 1992, 1994; Beron et al., 1995). Cholera toxin and recombinant Gαs proteins also have been found to promote transcytosis of the polymeric IgA receptor through endosomes in polarized epithelial cells (Bomsel and Mostov, 1993).
Although the molecular basis for the function of Gαs in signal transduction at the plasma membrane has been well characterized, little is known about the mechanisms whereby Gαs influences endocytic trafficking. Our recent discovery of RGS-PX1 has provided a putative link between Gαs and endocytic trafficking (Zheng et al., 2001). RGS-PX1, a member of the regulator of G protein signaling (RGS) protein family (De Vries et al., 2000; Hollinger and Hepler, 2002), functions as a GTPase activating protein (GAP) for Gαs through its conserved RGS domain that interacts specifically with Gαs, but no other Gα protein (Zheng et al., 2001). RGS-PX1 is also known as sorting nexin 13 (SNX13) and serves as an SNX protein, through its phosphatidylinositol-binding phoX (PX) domain. This domain is shared by SNX proteins that are involved in protein sorting in endosomes (Haft et al., 1998; Worby and Dixon, 2002). We showed previously that RGS-PX1 is a functional SNX protein that is localized on endosomes and delays epidermal growth factor (EGF) receptor degradation, probably at the steps of endosome sorting and lysosome targeting (Zheng et al., 2001). The fact that RGS-PX1 can bind Gαs and affect EGF receptor trafficking suggested that Gαs also might be involved in regulating of EGF receptor endocytosis and downregulation.
The EGF receptor represents the classical model system to study mechanisms of ligand-induced receptor endocytosis and down-regulation in mammalian cells (Carpenter, 2000; Katzmann et al., 2002; Sorkin and Von Zastrow, 2002). On ligand binding, EGF receptors are rapidly internalized via clathrin-coated pits and delivered to early endosomes where the majority of the receptors are sorted into the lumenal vesicles of late endosomes or multivesicular bodies (MVBs) and targeted for degradation in lysosomes (Wishart et al., 2001; Katzmann et al., 2002; Sorkin and Von Zastrow, 2002; Stahl and Barbieri, 2002; Raiborg et al., 2003). Hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) has been demonstrated to be a central player in this sorting pathway (Raiborg and Stenmark, 2002; Clague and Urbe, 2003; Raiborg et al., 2003). Hrs is found in specialized clathrin-coated microdomains of early endosomes that are enriched in mono-ubiquitinated receptors targeted for lysosomal degradation (Raiborg et al., 2001b; Sachse et al., 2002). Hrs also recruits the endosomal sorting complexes required for transport (ESCRT) complexes to endosomes through its interaction with Tsg101 in ESCRT complex I and regulates the formation of MVBs (Bache et al., 2003a; Katzmann et al., 2003; Lu et al., 2003). In this study, we have investigated the effects of Gαs on EGF receptor degradation. Our findings establish a novel role for Gαs in regulating endocytic trafficking and sorting.
MATERIALS AND METHODS
Materials
Mammalian expression vector pcDNA3.1 containing Gαs long (L) and short (S) splicing variants were obtained from Guthrie cDNA Resource Center (Sayre, PA). Mammalian expression vector pXER-EGFR encoding the EGF receptor was obtained from Dr. Gordon Gill (University of California, San Diego, CA). pGαs-green fluorescent protein (GFP) construct, expressing a Gαs-GFP fusion protein with GFP inserted between the helical and GTPase domains, was obtained from Dr. Mark Rasenick (University of Illinois, Chicago, IL) (Yu and Rasenick, 2002). The pcDNA3-myc-Hrs construct was obtained from Dr. A Beans (University of Texas Medical School, Houston, TX). The GFP-tagged RGS-PX1 construct containing residues 257–957 of human RGS-PX1 was described previously (Zheng et al., 2001). The FLAG-tagged RGS-PX1 construct was prepared by inserting the cDNA encoding human RGS-PX1 (residues 51–957) into p3XFLAG-CMV-10 (Sigma-Aldrich, St. Louis, MO).
Antibodies
Affinity-purified rabbit IgG against Gαs used for immunoblotting was obtained from Calbiochem (San Diego, CA). Rabbit antibodies against Rab5 were provided by Dr. Angela Wandinger-Ness (University of New Mexico, Albuquerque, NM). Other antibodies were obtained from the following sources: monoclonal antibodies (mAbs) against actin and FLAG (M2) (Sigma-Aldrich), myc (Cell Signaling Technology, Beverly, MA), and GFP (BD Biosciences Clontech, Palo Alto, CA), and polyclonal antibodies against EGF receptor (Santa Cruz Biotechnology, Santa Cruz, CA) and GFP and Hrs (Alexis Biochemicals, San Diego, CA).
Cell Culture and Transfection
Human embryonic kidney (HEK)293T cells (obtained from Dr. Alexandra Newton, University of California, San Diego, CA), and Cos7 cells were maintained in Dulbecco's modified Eagle's high glucose medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (FCS) (Hyclone Laboratories, Logan, UT), penicillin, and streptomycin. HEK293 cells were transfected using calcium phosphate as described previously (Zheng et al., 2000). Cos7 cells were transfected using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions.
Epidermal Growth Factor Receptor (EGFR) Degradation Assays
HEK293 cells in six-well plates were transfected with pXER-EGFR together with pcDNA3.1-Gαs-L and pcDNA3.1-Gαs-S (1:1), or pcDNA3.1 empty vector. Twenty-four hours after transfection, cells were serum starved overnight in DMEM with 0.5% fetal bovine serum (FBS) and then incubated in the presence or absence of 100 nM EGF (Molecular Probes, Eugene, OR) at 37°C. Cells were lysed in Laemmli sample buffer or in 50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton, and Complete protease inhibitor (Roche Diagnostics) followed by immunoblotting with antibodies to EGFR, Gαs, and actin. EGF receptor degradation was quantified by densitometry (three independent experiments) by using Quantity One software (Bio-Rad, Hercules, CA).
Immunofluorescence
Cos7 cells were fixed in 3% paraformaldehyde (PFA) in 100 mM phosphate buffer, pH 7.4, for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 10% fetal calf serum for 30 min, and incubated with primary antibodies for 1 h at 25°C, followed by Alexa Fluor 594-conjugated goat anti-mouse F(ab′)2 and/or Alexa 488 goat anti-rabbit F(ab′)2 (Molecular Probes) for 1 h. Some specimens were permeabilized with 0.05% saponin for 1 min at 4°C before fixation. Specimens were analyzed using a Zeiss Axiophot equipped with a Hamamatsu Orca ER charge-coupled device (CCD) or by deconvolution microscopy by using an Applied Precision (Issaquah, WA) Delta Vision imaging system coupled to an S100 fluorescence microscope (Carl Zeiss, Thornwood, NY). For cross-sectional images of cells, stacks were obtained with 200-nm step width. Deconvolution was done on an SGI workstation (Mountain View, CA) by using Delta Vision reconstitution software, and images were processed as Tiff files by using Photoshop 7.0 (Adobe Systems, San Jose, CA).
Uptake of Texas Red EGF
Cos7 cells were transfected with pCDNA3, Gαs-GFP, or myc-Hrs for 12 h. After serum starvation for 3 h, cells were incubated in DMEM containing 0.4 μg/ml Texas Red EGF (Molecular Probes) in 0.5% FBS for 10 min at 37°C and washed and incubated in DMEM containing 0.5% FBS for up to 1 h at 37°C.
For semiquantitative analysis of bound and internalized Texas Red EGF, all images were captured with the exact same settings. Control cells and cells expressing the GFP constructs were traced using Photoshop. For each cell, the number and intensity of positive pixels (pixels with grayscale values between 75 and 255) was determined using Image J software (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij). Results were calculated as the total number of positive pixels per condition multiplied by the cumulative pixel intensity divided by the number of cells. Ten to 30 cell profiles were measured for each condition, and the results are displayed as the mean of three separate experiments.
Immunogold Labeling
HEK293 cells were fixed in 4% PFA alone or 4% PFA containing 0.2% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, overnight, pelleted in 10% gelatin in phosphate buffer, cryoprotected, and snap frozen in liquid nitrogen. Ultrathin cryosections (70–90 nm) were cut at –100°C on a Leica Ultracut UCT with an EM FCS cryoattachment (Leica, Bannockburn, IL) by using a Diatome diamond knife (Diatome US, Fort Washington, PA), picked up with a 1:1 mixture of 2.3 M sucrose and 2% methyl cellulose (15 cp), and transferred onto Formvarand carbon-coated copper grids. Sections were blocked and incubated with primary antibodies for 2 h at room temperature, followed by gold conjugated goat anti-rabbit IgG and gold conjugated goat anti-mouse IgG (Amersham Biosciences, Piscataway, NJ) for 1 h. Sections were contrasted for 10 min with 2% neutral uranyl acetate and stained for 10 min with 0.2% uranyl acetate in 1.8% methyl cellulose on ice. Grids were viewed and photographed using a Philips CM-10 transmission electron microscope (FEI, Hilsboro, OR) equipped with a 794 Multiscan CCD camera (Gatan, Pleasanton, CA).
Coimmunoprecipitation
HEK293 cells were plated in 60-mm plates and transfected with various constructs. After 48 h, cells were lysed in 1% Triton X-100 in phosphatebuffered saline (PBS) buffer containing protease inhibitors (0.12 mg/ml phenylmethylsulfonyl fluoride, 2 mg/ml leupeptin, and 1 mg/ml aprotinin) at 4°C for 30 min and centrifuged at 15,000 × g for 10 min. Cell lysates were incubated with primary antibodies overnight at 4°C, followed by incubation with protein Aor G-Sepharose (Oncogene, San Diego, CA) for an additional 1 h at 4°C. Beads were washed (three times) with lysis buffer and boiled in Laemmli sample buffer, and bound immune complexes were analyzed by SDS-PAGE and immunoblotting.
For coimmunoprecipitation of Hrs and Gαs from cytosolic and membrane fractions, HEK293 cells were scraped into cold PBS containing protease inhibitors and homogenized by 10 passages through a 28 1/2-gauge needle. Nuclei and unbroken cells were removed by centrifugation, and postnuclear supernatants were centrifuged at 100,000 × g for 1 h at 4°C to prepare cytosolic (supernatant, S100) and membrane (pellet, P100) fractions (Zheng et al., 2000). Membrane pellets were lysed in 1% Triton X-100 in PBS containing protease inhibitors for 1 h, centrifuged (15,000 × g for 10 min), and the membrane lysates and cytosolic fractions were used for immunoprecipitation.
Immunoblotting
Protein samples were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). Membranes were blocked in Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 500 mM NaCl) containing 0.1% Tween 20 and 5% nonfat milk and incubated with primary antibodies for 2 h at room temperature or overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit or antimouse IgG (Bio-Rad) and enhanced chemiluminescence detection (Pierce Chemical, Rockford, IL).
In Vitro Glutathione S-Transferase (GST) Pull-Down Assays
Full-length rat Hrs cDNA and a human RGS-PX1 fragment (PXC) containing the PX domain and the C-terminus (residues 526–957) were amplified by polymerase chain reaction (PCR) and subcloned into pGEX-KG (Amersham Biosciences). GST fusion proteins were expressed in Escherichia coli BL21 and purified on glutathione-Sepharose 4B (Pharmacia, Piscataway, NJ) beads according to the manufacturer's instructions. 35S-labeled, in vitro translation products of Gαs or Hrs were prepared by using the TNT T7 rabbit reticulocyte Quick Coupled Transcription/Translation system (Promega, San Luis Obispo, CA) in the presence of [35S]methionine (1000 Ci/mmol, in vivo cell labeling grade; Amersham Biosciences), pcDNA3.1-Gαs-L and pcDNA3.1Gαs-S (1:1 ratio) or pCDNA3-myc-Hrs. For pull-down assays, GST fusion proteins (∼75 μg) immobilized on beads were incubated with in vitro-translated products in 20 mM Tris-HCl, pH 8.0, 2 mM MgSO4, 6 mM β-mercaptoethanol, 5% glycerol, and 0.01% C12E10, in the presence of protease inhibitors for 2 h at 4°C, and washed four times with the same buffer. GST pull-down assays on brain lysates (5 mg) were performed using a lysis buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl, 300 mM sucrose, 1% Triton X-100, and 0.01% C12E10 as described previously (Zheng et al., 2001). Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-PAGE, and visualized by autoradiography.
RNA Interference
The following small-interfering RNA (siRNA) oligos synthesized by Dharmacon Research (Lafayette, CO) were used for RNAi knock-down of Gαs and Hrs (Bache et al., 2003b): Gαs-sense, 5′-GGC GCA GCG UGA GGC CAA CdTdT; Gαs-antisense, 5-GUU GGC CUC ACG CUG CGC CdTdT; and Hrs-sense, 5′ CGA CAA GAA CCC ACA CGU CdTdT; Hrs-antisense, 5′ GAC GUG UGG GUU CUU GUC GdTdT. All oligos were designed based on human sequences. Scrambled RNA oligos (scramble II duplex; Dharmacon Research) were used as controls. Cos7 cells in six-well plates (30% confluent; 1.5 ml of normal culture medium without antibiotics per well) were transfected with 1 μl of 75 μM siRNA duplex and 8 μl of Oligofectamine reagent (Invitrogen) according to the manufacturer's instructions. The cells were analyzed 72 h after transfection. For double RNAi experiments, the total RNAi oligos were kept the same among different wells by addition of scrambled RNAi oligos.
RESULTS
Gαs Overexpression Promotes Degradation of EGF Receptors and Texas Red EGF
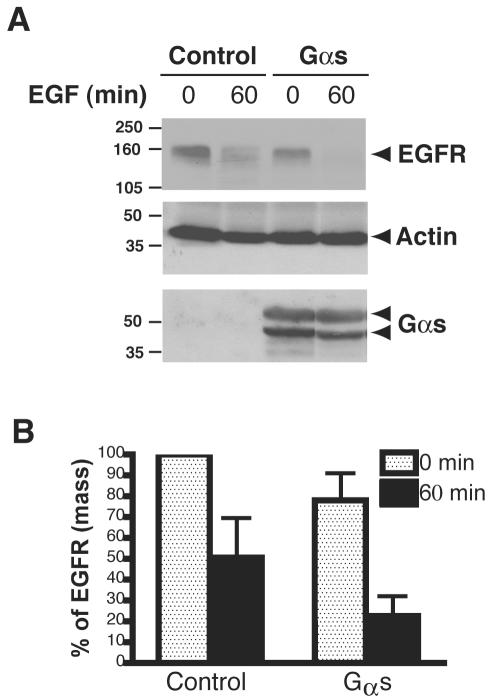
RGS-PX1 acts both as a GAP that regulates the activity of Gαs and as a SNX involved in the down-regulation of the EGF receptor (Zheng et al., 2001). These dual activities suggested that RGS-PX1 could link Gαs to EGF receptor sorting at endosomes and that Gαs also might be involved in EGF receptor down-regulation. To find out whether this is the case, we transiently transfected HEK293 cells with EGF receptor and either Gαs or control vector and determined the kinetics of EGF receptor degradation. As shown in Figure 1, A and B, cells transfected with Gαs contained less EGF receptors (∼20%) at steady state than control cells, suggesting Gαs expression enhances basal turnover of EGFR. Similarly, ligand-induced degradation of the receptor was enhanced in cells transfected with Gαs, because 80% of the receptors were degraded by 60 min after adding EGF (Figure 1, A and B), whereas in cells transfected with control vector, only 50% of the receptors had been degraded.
Figure 1.
Overexpression of Gαs promotes degradation of EGFR in HEK293 cells. (A) HEK293 cells were transfected with pXER-EGFR together with pcDNA3.1-Gαs(Gαs) or control vector (control) for 24 h, serum starved overnight, and then treated with 100 nM EGF for 0 or 60 min, followed by immunoblotting with antibodies against EGF receptor, actin, or Gαs. Gαs is seen as two bands representing the long and short forms of Gαs. Data shown are representative of at least three independent experiments. (B) Quantification of EGF receptor degradation. Results from three independent experiments were analyzed by Quantity One software (Bio-Rad). When cells transfected with control vectors are stimulated with EGF, ∼50% of the receptors seen at 0 min are degraded by 60 min after adding EGF. In cells transfected with Gαs, degradation is enhanced as ∼80% of the receptors are degraded by 60 min. Data presented as percentage of total EGF receptor at 0 min in control cells.
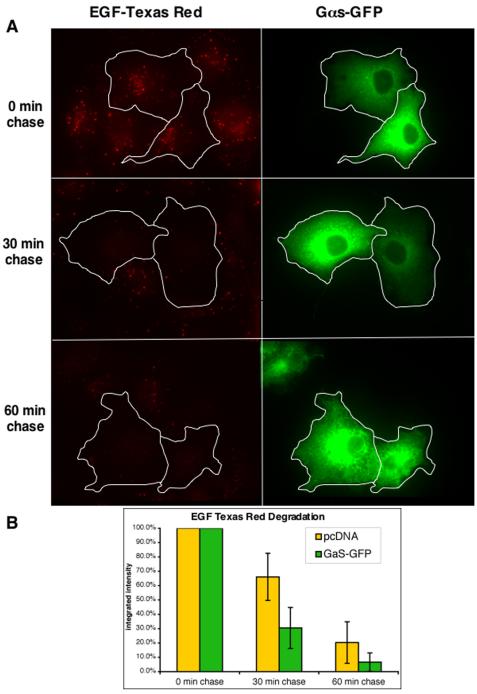
Next, we used immunofluorescence to evaluate the effects of overexpressing Gαs on the uptake and degradation of Texas Red EGF. Cos7 cells transfected with Gαs-GFP (Yu and Rasenick, 2002) or empty vector were incubated with Texas Red EGF for 10 min followed by incubation in the absence of ligand for 30 or 60 min. As shown in Figure 2A, the levels of Texas Red EGF were similar in cells transfected with Gαs-GFP and control vector after 10-min incubation with Texas Red EGF, suggesting that Gαs overexpression does not impair internalization of EGF. However, at 30 and 60 min “chase,” cells expressing Gαs-GFP contained significantly less Texas Red EGF than those transfected with empty vector (Figure 2A) or GFP alone (our unpublished data). Semiquantitative analysis of the amount of Texas Red EGF remaining (Figure 2B) revealed that in cells transfected with control vector, 66% remained at 30 min and 20% at 60 min, whereas only 30 and 7% remained at the same times in cell expressing Gαs-GFP. As a control, we also transfected Hrs into Cos7 cells and found, consistent with previous reports (Raiborg et al., 2001b; Bishop et al., 2002; Urbe et al., 2003), that overexpression of Hrs strongly inhibited Texas Red EGF degradation (our unpublished data). We also examined the effect of Gαs overexpression on the uptake of transferrin-Alexa594 in Cos7 cells and found no difference in transferrin uptake between cells transfected with Gαs-GFP and GFP alone (our unpublished data). Together, these results indicate that overexpression of Gαs promotes specific degradation of EGF receptors and their ligands.
Figure 2.
Overexpression of Gαs-GFP promotes degradation of Texas Red. (A) Cos7 cells transfected with pGαs-GFP or control vector were incubated with Texas Red EGF for 10 min and chased for 30 or 60 min. Cells expressing Gαs-GFP (traced in white) and those expressing control vector showed similar levels of Texas Red EGF at 0-min chase. However, after 30- or 60-min chase there is considerably less Texas Red EGF remaining in cells expressing Gαs-GFP. (B) Semiquantitative representation of the data shown in A. In cells transfected with control vector ∼30% of the Texas Red EGF is degraded at 30 min and 80% by 60 min, whereas in cells expressing Gαs-GFP ∼70% are degraded at 30 min and ∼95% at 60 min. Average integrated intensity of Texas Red EGF pixels per cell were measured as described in Materials and Methods. Data are expressed as the mean ± SE of three experiments.
Depletion of Gαs Delays Degradation of EGF Receptors
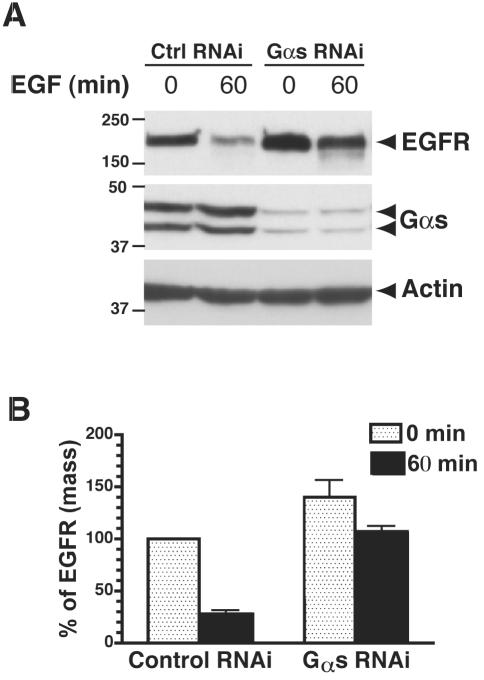
We further evaluated the effects of knocking down endogenous Gαs protein levels in Cos7 cells on ligand-induced degradation of EGF receptors. We found that siRNA oligos designed specifically for Gαs blocked EGF-dependent receptor degradation (Figure 3, A and B). In cells transfected with scrambled siRNA, 70% of the receptors were degraded after 60-min stimulation with EGF, whereas in cells transfected with Gαs siRNA degradation was delayed as only 34% of the EGFR had been degraded (Figure 3, A and B). In addition, cells transfected with Gαs siRNA had higher levels (140%) of EGF receptor at steady state than cells transfected with scrambled siRNA (Figure 3, A and B), suggesting the basal level of EGF receptor turnover is delayed by knockdown of Gαs. Thus, we have shown by three different approaches that Gαs regulates EGF receptor degradation.
Figure 3.
Depletion of Gαs expression by RNAi delays degradation of the EGF receptor. Cos7 cells were transfected with 37.5 nM control or Gαs-specific siRNA oligos by using Oligofectamine. After 3 d, cells were treated with 100 nM EGF for 60 min, lysed, and analyzed by immunoblotting with antibodies against EGF receptor, Gαs, and actin. The level of Gαs is reduced to <5% of control levels in cells transfected with oligos specific for Gαs. In cells transfected with control siRNA, 70% of the EGF receptors are degraded after 60-min stimulation with EGF. In cells transfected with Gαs siRNA, degradation is delayed as only 34% of the receptors are degraded after 60 min. Data presented as percentages of the amount of EGFR at 0 min in each group of cells. Data shown are representative of at least three independent experiments. (B) Quantification of EGF receptor degradation. Results from three independent experiments were analyzed by Quantity One software. Data presented as percent of total EGFR present at 0 min in control cells.
RGS-PX1 Interacts with Hrs In Vivo and In Vitro
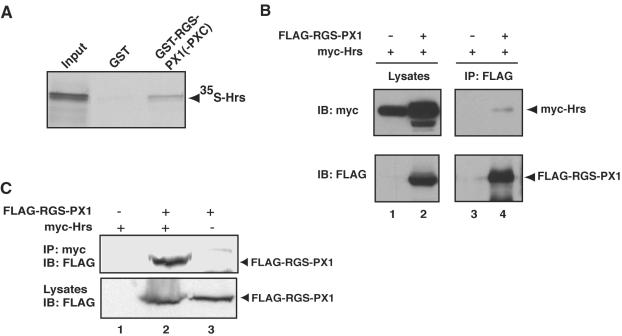
Next, we investigated whether RGS-PX1 or Gαs delays EGF receptor degradation by interacting with components of the endosomal sorting machinery. We reasoned that RGS-PX1 might bind Hrs, an endosomal protein required for efficient degradation of EGF receptors, because Hrs has been shown to interact with SNX1 (Chin et al., 2001), the founding member of the SNX protein family that shares strong sequence homology with the C-terminal PX domain and coiled-coil region of RGS-PX1 (Kurten et al., 1996; Zheng et al., 2001).
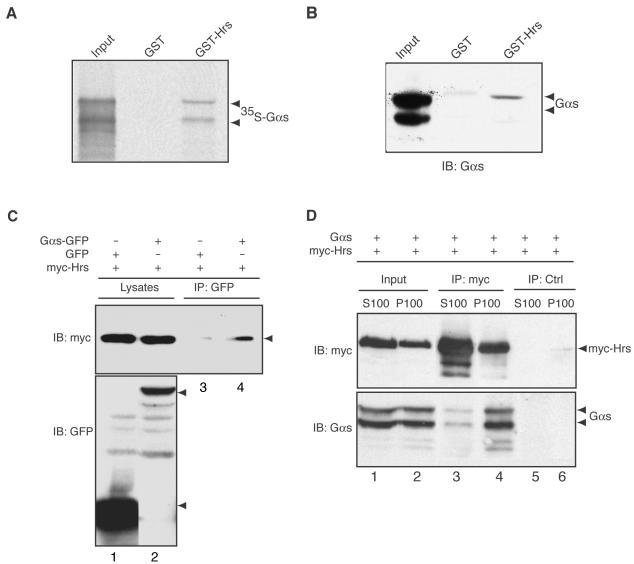
When we incubated 35S-labeled, in vitro-translated Hrs with GST-RGS-PX1(PXC), a GST fusion protein containing the PX domain and C-terminal coiled-coil region of RGS-PX1 that is homologous to SNX1, Hrs bound to GST-RGSPX1(PXC), but not to GST alone (Figure 4A). We further tested whether RGS-PX1 coimmunoprecipitates with Hrs in HEK293 cells transfected with myc-tagged Hrs and FLAGtagged RGS-PX1. We found that when immunoprecipitation was carried out with anti-FLAG IgG, myc-Hrs coprecipitated with FLAG-RGS-PX1 (Figure 4B). Similarly, when antimyc IgG was used, myc-Hrs coprecipitated with FLAG-RGS-PX1 in cells cotransfected with both proteins (Figure 4C). These findings support the conclusion that RGS-PX1 interacts with Hrs both in vitro and in vivo.
Figure 4.
RGS-PX1 interacts with Hrs. (A) In vitro translated, 35S-labeled Hrs binds to GST-RGS-PX1(PXC) but not to GST alone. GST-RGS-PX1(PXC) and GST alone (∼75 μg each) immobilized on glutathione beads were incubated with in vitro-translated Hrs. Bound proteins were separated by SDS-PAGE and detected by autoradiography. Input equals 3% of total in vitro translation product. (B) Myc-Hrs (top, lane 4) coimmunoprecipitates with FLAG-RGS-PX1 in cells transfected with both proteins but not in those transfected with myc-Hrs alone (top, lane 3). HEK293 cells were transfected with FLAG-RGS-PX1 and myc-Hrs or myc-Hrs alone, and immunoprecipitation (IP) was carried out on lysates (lanes 1 and 2) with anti-FLAG mouse IgG, followed by immunoblotting (IB) of immunoprecipitates with anti-myc. (C) FLAG-RGS-PX1 coimmunoprecipitates with myc-Hrs. Immunoprecipitation was carried out with anti-myc on lysates from HEK293 cells transfected with myc-Hrs alone (lane 1), FLAG-RGS-PX1 alone (lane 3), or both FLAG-RGS-PX1 and myc-Hrs (lane 2), followed by immunoblotting with anti-FLAG IgG.
Gαs Interacts with Hrs In Vivo and In Vitro
Given that we have previously shown that RGS-PX1 binds to and serves as a GAP for Gαs (Zheng et al., 2001), we next asked whether Gαs also interacts with Hrs in pull-down and immunoprecipitation assays. We found that 35S-labeled, in vitro-translated Gαs bound to GST-Hrs, but not to GST alone (Figure 5A) and that GST-Hrs, but not GST alone, was able to pull-down endogenous Gαs from brain lysates (Figure 5B). Similarly, when we transfected myc-tagged Hrs together with Gαs-GFP into HEK293 cells and carried out immunoprecipitation with anti-GFP IgG, myc-Hrs coprecipitated with Gαs-GFP (Figure 5C). Because Hrs and Gαs have been found in both membrane and cytosolic fractions, we investigated where they interact. We found that Gαs and Hrs were equally distributed between membrane (P100) and cytosolic fractions (S100) in HEK293 cells expressing myctagged Hrs together with pcDNA3.1-Gαs (Figure 5D, lanes 1 and 2). However, the majority of the Gαs (>95%) coimmunoprecipitated with myc-Hrs from membrane fractions (Figure 5D, lane 4). These results indicate that Gαs interacts with Hrs and that the interaction takes place largely on membranes, presumably on endosomal membranes as both Hrs (Komada et al., 1997; Raiborg et al., 2001a) and RGS-PX1 (Zheng et al., 2001) are localized on early endosomes.
Figure 5.
Interaction between Hrs and Gαs. (A) In vitro-translated, 35S-labeled Gαs binds to GST-Hrs but not to GST alone. GST-Hrs and GST proteins (∼75 μg each) immobilized on glutathione beads were incubated with in vitro-translated, [35S]Gαs as in Figure 4. Input equals 3% of total in vitro translation product. (B) Endogenous Gαs from rat brain lysates binds to GST-Hrs but not to GST. GST-Hrs and GST immobilized on glutathione beads were incubated with rat brain lysates (∼5 mg). Bound proteins were immunoblotted with anti-Gαs IgG. Input equals 3% of total brain lysate. (C) Myc-Hrs coimmunoprecipitates with Gαs-GFP (lane 4). Lysates (lanes 1 and 2) from HEK293 cells transfected with Gαs-GFP or GFP together with myc-Hrs were immunoprecipitated with anti-GFP, followed by immunoblotting with anti-myc and anti-GFP antibodies. (D) Gαs and Hrs are found in approximately equal amounts in both membrane (P100, lane 2) and cytosolic (S100, lane 1) fractions. Gαs coimmunoprecipitates with myc-Hrs predominantly (>95%) from membrane fractions (lane 4, bottom). Very little Gαs is coprecipitated with myc-Hrs from the cytosolic fraction (lane 3, bottom). Cytosolic (S100, lane 1) and membrane (P100, lane 2) fractions prepared from HEK293 cells transfected with Gαs and myc-Hrs were immunoprecipitated with anti-myc (myc, lanes 3 and 4) or control (ctrl, lanes 5 and 6) mouse IgGs, followed by immunoblotting with anti-Gαs and anti-myc antibodies.
Gαs, RGS-PX1, and Hrs Colocalize on Early Endosomes
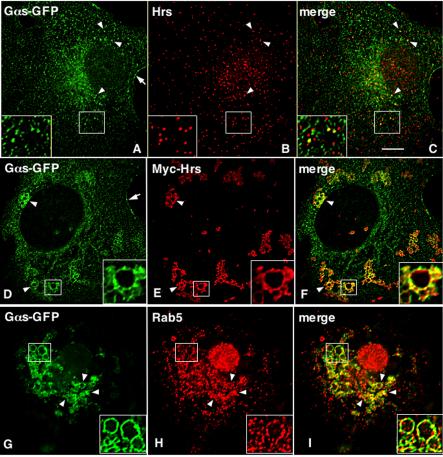
To determine the localization of Gαs and whether it colocalizes with Hrs and RGS-PX1 on endosomes, we carried out indirect immunofluorescence and deconvolution analysis on Cos7 cells expressing Gαs-GFP alone or Gαs together with RGS-PX1 and Hrs. Because roughly 50% of both Gαs and Hrs are found in cytosolic fractions (Figure 5D), we permeabilized the cells before fixation to release cytosolic proteins and facilitate the detection of membrane-associated pools of Gαs and Hrs. In cells transfected with Gαs-GFP alone, GαsGFP showed fine, punctate staining throughout the cytoplasm that partially overlapped with the early endosome markers Hrs (Figure 6, A–C) and Rab5 (our unpublished data).
Figure 6.
Colocalization of Gαs-GFP with myc-Hrs on early endosomes. (A–C) In Cos7 cells transfected with Gαs-GFP alone, Gαs is distributed on the plasma membrane (arrow, A) and on small vesicular structures (arrowheads, A). Hrs is distributed on early endosomes throughout the cell (B). Merged image (yellow) shows occasional overlap in the vesicular distribution of Gαs-GFP and Hrs (arrowheads and inset, C). (D–I) In cells transfected with Myc-Hrs, which promotes formation of large, clustered endosomes Gαs-GFP is distributed on the plasma membrane (arrow, D) and on the enlarged endosomes (arrowheads and inset, D and G). Myc-Hrs (arrowheads and inset, E) and Rab5 (arrowheads and inset, H) are also present on these endosomes. Gαs-GFP colocalizes (yellow) with Myc-Hrs (arrowheads and inset, F) and Rab5 (arrowhead and inset, I). Cos7 cells were transfected with Gαs-GFP alone (A–C) or together with Myc-Hrs (D–I) and permeabilized with saponin before fixation to release the cytosolic proteins and facilitate the detection of membrane-associated pools of Gαs and Hrs. Cells were then fixed with 3% PFA, permeabilized, and double labeled with mouse anti-GFP (A, D, and G), anti-Hrs (B), or anti-myc (E) IgG or rabbit anti-rab5 (H) IgG and analyzed by deconvolution immunofluorescence microscopy. Bar, 2 μm.
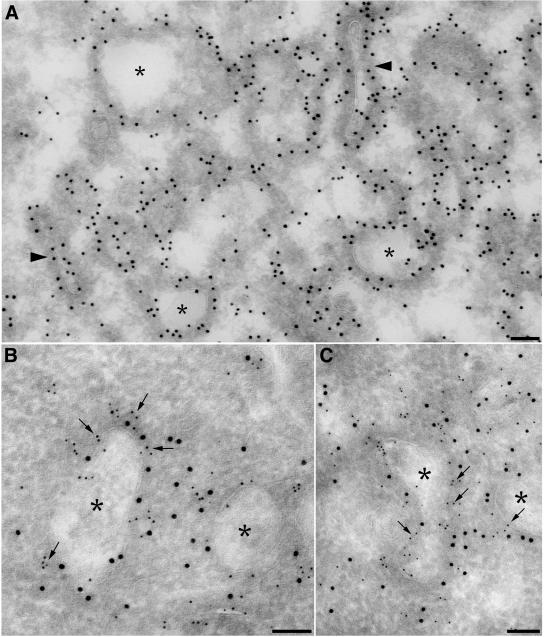
In cells expressing both Gαs-GFP and myc-Hrs, these two proteins strongly colocalized in endosomes (Figure 6, D–F). In agreement with previous reports (Raiborg et al., 2001b; Bishop et al., 2002; Urbe et al., 2003), overexpression of Hrs resulted in enlarged, clustered endosomes. Furthermore, Gαs-GFP and Rab5 colocalized on these enlarged endosomes (Figure 6, G–I). More Gαs colocalized with Hrs on these endosomes (Figure 6, D–F) compared with cells transfected with Gαs-GFP alone (Figure 6, A–C), suggesting expression of Hrs causes more Gαs to translocate to early endosomes. By immunogold labeling, Gαs-GFP and myc-Hrs colocalized in coated microdomains of these enlarged endosomes (Figure 7). Thus, the immunofluorescence results and the coimmunoprecipitation assays together indicate the Gαs binds Hrs on early endosomes.
Figure 7.
Immunogold localization of Hrs and Gαs in HEK293 cells. (A). Gαs-GFP (10-nm gold) is localized to numerous coated tubules (arrowheads) and endosomes (asterisks). (B and C). Myc-Hrs (5-nm gold, arrows) and Gαs-GFP (10-nm gold) colocalize in coated domains of early endosomes (asterisks). HEK293 cells transfected with myc-Hrs and Gαs-GFP were fixed either in 4% PFA (A) or a mixture of 4% PFA and 0.2% glutaraldehyde (B and C) and prepared for ultrathin cryosectioning as described in Materials and Methods. Ultrathin cryosections were labeled with anti-myc mAb (5-nm gold) and polyclonal anti-GFP (10-nm gold). Bar, 100 nm.
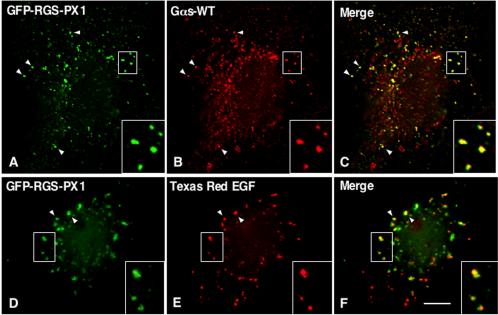
In cells cotransfected with untagged Gαs and GFP-RGSPX1, GFP-RGS-PX1 and Gαs colocalized on endosomes (Figure 8, A–C) that also were labeled with Texas Red EGF after 15 min uptake (Figure 8, D–F). This is consistent with its early endosome localization reported previously. Again, compared with cells expressing Gαs-GFP alone (Figure 6, A–C), more Gαs seemed to be localized on early endosomes (Figure 8, A–C), suggesting expression of RGS-PX1, as well as Hrs, causes more Gαs to translocate to early endosomes.
Figure 8.
Colocalization of Gαs with RGS-PX1 on early endosomes. GFP-RGS-PX1 is found on endosomes (arrowheads and insets, A and D) and partially colocalizes with Gαs-WT (arrowheads and inset, B and C) on endosomes loaded with Texas Red EGF (arrowheads and inset, E and F). Cos7 cells were transfected with GFP-RGS-PX1 and Gαs-WT. In D–F, cells also were incubated with Texas Red EGF for 15 min at 37°C. Cells were permeabilized with saponin, fixed with 3% PFA, double labeled with mouse anti-GFP mAb (A and D), and rabbit anti-Gαs (B) IgG, and analyzed as described in Figure 6. Bar, 2 μm.
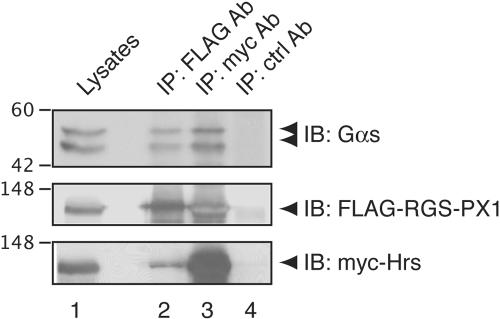
Gαs, RGS-PX1, and Hrs Form a Coprecipitatable Complex
The ability of both RGS-PX1 and Gαs to interact with Hrs and the colocalization of these three proteins on early endoxsomes suggested that they might be present in the same protein complex. To test this possibility, we performed coimmunoprecipitation experiments by using HEK293 cells transfected with myc-Hrs, FLAG-RGS-PX1, and Gαs. When immunoprecipitation was carried out with an anti-myc IgG, both FLAG-RGS-PX1 and Gαs coprecipitated with myc-Hrs (Figure 9, lane 3). Similarly, anti-FLAG IgG was able to bring down both myc-Hrs and Gαs (Figure 9, lane 2). These results suggest that Gαs, RGS-PX1, and Hrs form a coprecipitable protein complex.
Figure 9.
Gαs, FLAG-RGS-PX1, and myc-Hrs form a coprecipitable complex. HEK293 cells were transfected with pcDNA3-Gαs, FLAG-RGS-PX1, and myc-Hrs. Lysates were immunoprecipitated with anti-FLAG (lane 2), anti-myc (lane 3), or control (ctrl) (lane 4) mouse IgGs, followed by immunoblotting with anti-Gαs (top), anti-FLAG (middle), or anti-myc (bottom) IgG. Gαs (top) coprecipitates with both FLAG-RGS-PX1 (lane 2) and myc-Hrs (lane 3).
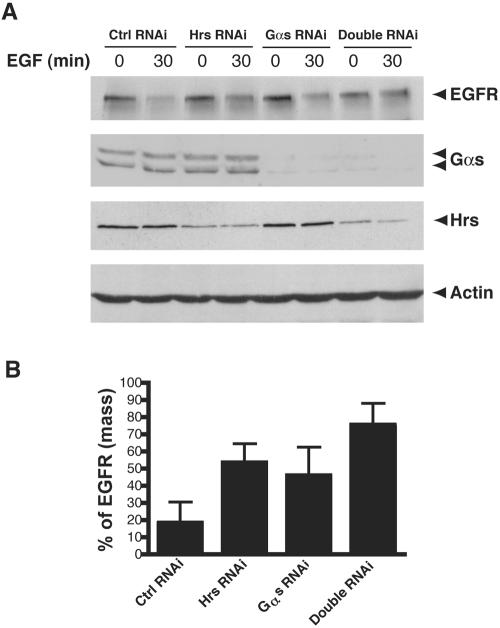
Knockdown of Both Gαs and Hrs Further Delays EGF Receptor Degradation
The interaction between Gαs and Hrs suggests Gαs may function together with Hrs in the endosomal sorting and down-regulation of the EGF receptor. To test this hypothesis, we performed double RNAi experiment to knock-down the expression of both Gαs and Hrs (Figure 10, A and B). In cells transfected with both Gαs and Hrs RNAi oligos, ∼25% of the EGFR was degraded at 30 min after adding EGF, whereas in cells transfected with either Gαs or Hrs RNAi oligos alone, ∼50% of the EGF receptors had been degraded at 30 min. These results together with the interaction between Hrs and Gαs strongly suggest that Gαs cooperates with Hrs in regulating EGF receptor degradation.
Figure 10.
Simultaneous knockdown of both Gαs and Hrs causes a delay in EGF receptor degradation greater than knock-down of either Gαs or Hrs alone. Cos7 cells were transfected with 75 nM, Gαs siRNA alone, Hrs siRNA alone, both Gαs and Hrs siRNA, or with control siRNA oligos by using Oligofectamine. After 3 d, cells were treated with 100 nM EGF for 30 min, lysed, and analyzed by immunoblotting with antibodies against EGF receptor, Gαs, Hrs, and actin. In cells transfected with Hrs or Gαs RNAi alone, 50–55% of the EGF receptors have been degraded after 30 min, whereas in those transfected with both Gαs and Hrs siRNA, degradation is delayed as only 25% of the receptors have been degraded. Data shown are representative of at least three independent experiments. (B) Quantification of EGF receptor degradation. Results from three independent experiments were analyzed by Quantity One software. Data presented as percent of total EGFR at 0 min in each group of cells.
DISCUSSION
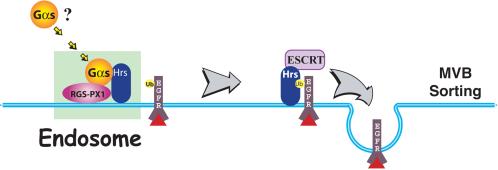
Our work presented here demonstrates a regulatory role for the heterotrimeric Gαs protein in EGF receptor trafficking and down-regulation. We find that expression of Gαs accelerates degradation of both EGF receptors and Texas Red EGF, whereas depletion of Gαs by RNAi delays their degradation. We also show that Gαs forms a complex with RGS-PX1 and Hrs that seems to cause more Gαs to translocate to early endosomes. Based on these findings, we propose the following model (Figure 11) for the function of Gαs on endosomes: 1) in the presence of RGS-PX1, Gαs translocates from the plasma membrane or cytoplasm to early endosomes after EGF binding, where it forms a complex with RGS-PX1 and Hrs; and 2) through interaction with Hrs, Gαs regulates endosomal sorting and hence modulates down-regulation of the EGF receptor.
Figure 11.
Model of the proposed function of Gαs in EGF receptor degradation. After EGF stimulation Gαs is recruited to early endosomes where it forms a complex with RGS-PX1 and Hrs on the endosomal membrane. Together with Hrs, Gαs promotes the sorting of ubiquitinated (Ub) EGFRs into the luminal vesicles of MVBs and hence facilitates their degradation.
Traditionally, heterotrimeric G proteins have been considered to be largely associated with the cell membrane. Our study indicates early endosomes represent a novel intracellular location for Gαs. This is in keeping with previous implications that Gαs plays a role in endosomal functions, such as early endosome fusion, phagosome-endosome fusion, and transcytosis of pIgR (Colombo et al., 1992, 1994; Bomsel and Mostov, 1993; Beron et al., 1995). Consistent with our localization data, more recently it was shown that endogenous Gαs also can be found in rat liver endosomes based on cell fractionation and immunofluorescence studies (Van Dyke, 2004).
How translocation of Gαs to endosomes is triggered is still an open question. There are two possible scenarios. First, activation of a GPCR linked to Gαs could stimulate translocation. It has been reported that activation of β-adrenergic receptors or cholera toxin treatment, Gαs dissociates from the cell membrane into the cytoplasm (Ransnas et al., 1989; Levis and Bourne, 1992; Wedegaertner and Bourne, 1994; Wedegaertner et al., 1996; Yu and Rasenick, 2002). Alternatively, activation of the EGF receptor by EGF could trigger the translocation of Gαs. It has been shown that Gαs is tyrosine phosphorylated by the EGF receptor in vitro and in response to EGF stimulation in vivo (Liebmann et al., 1996; Poppleton et al., 1996). Conceivably, this phosphorylation event might be related to the change in the subcellular localization of Gαs. Furthermore, Gαs has been shown to interact directly with the juxtamembrane region (50 aa) of the EGF receptor in both yeast two-hybrid and coimmunoprecipitation assays; this interaction was suggested to be responsible for the activation of adenylyl cyclase by EGF stimulation in cardiomyocytes (Nair et al., 1990; Sun et al., 1997). Intriguingly, the juxtamembrane region of the EGF receptor contains a dileucine motif that is required for efficient sorting of receptors to lysosomes (Lin et al., 1986; Kil et al., 1999; Bao et al., 2000). The juxtamembrane region also includes a protein kinase C phosphorylation site, and phosphorylation of the EGF receptor by protein kinase C has been shown to switch receptors from the degradation to recycling pathways (Lin et al., 1986; Kil et al., 1999; Bao et al., 2000). It would be interesting to know whether these sorting motifs are involved in the binding of Gαs to the EGF receptor.
In this work, we have used both overexpression and RNAi knock-down approaches to demonstrate the role of Gαs in EGF receptor degradation. Previously, it has been shown that overexpression of Gαi1 inhibited internalization of low-density lipoprotein and transferring, possibly by binding free Gβγ subunits and forming inactive heterotrimers (Lin et al., 1998). Although overexpression of Gαs may cause similar sequestration of Gβγ subunits, our RNAi knock-down results strongly suggest that Gαs plays a direct role in regulating EGF receptor degradation. Whether free Gβγ also is involved in the degradation of EGF receptor directly remains to be investigated.
As a core component of the endosome sorting machinery, Hrs is evolutionarily conserved in eukaryotes. In budding yeast Saccharomyces cerevisiae, the Hrs homolog Vps27 is one of the “class E” vacuolar protein sorting (Vps) proteins required for formation of MVBs, sorting of membrane proteins into MVBs, or budding into MVBs (Vida and Emr, 1995; Katzmann et al., 2003). It is noteworthy that S. cerevisiae does not seem to have a Gαs homolog. The two heterotrimeric G proteins encoded in S. cerevisiae, GPA1 and GPA2, are closer to the Gαi rather than the Gαs subfamily of mammalian G proteins in amino acid sequence. As for RGS-PX1, its putative homolog in S. cerevisiae, Mdm1, contains a PX-associated (PXA) domain of unknown function and a PX domain that binds to phosphatidylinositol-3-phosphate (McConnell and Yaffe, 1992; Yu and Lemmon, 2001). However, Mdm1 does not have a homologous RGS domain, and no functional link between Mdm1 and MVB sorting has been reported to date. The absence of a Gαs homolog in S. cerevisae and the lack of an RGS domain in Mdm1 lead us to propose that Gαs serves as a regulatory module in the endosome sorting machinery in higher organisms, rather than a evolutionarily conserved core component like Hrs.
Depletion of Hrs by RNAi in mammalian cells was shown to decrease the membrane association of the ESCRT complex, reduce the number of MVBs, and disrupt lysosomal targeting of EGF receptors, leading to impaired EGF receptor down-regulation (Bache et al., 2003a,b). We report here that depletion of Gαs by RNAi, similar to Hrs, delays degradation of EGF receptors. Moreover, simultaneous depletion of Gαs and Hrs by double RNAi further inhibited EGF receptor degradation compared with depletion of Gαs or Hrs alone. These results, together with our observation that Gαs interacts with Hrs, suggest that Gαs and Hrs act together to promote ligand-dependent degradation of EGF receptors. We have previously found that overexpression of RGS-PX1 slowed EGF receptor degradation (Zheng et al., 2001), an effect of RGS-PX1 that could be explained by its GAP activity on Gαs. Alternatively, overexpression of RGS-PX1 might have a dominant-negative effect through its interaction with Hrs. Although we have shown that Hrs can form a coimmunoprecipitable complex with RGS-PX1 and Gαs, there is also the possibility that some complexes may contain Hrs and RGS-PX1 only, or Hrs and Gαs only, and that Gαs may promote EGF receptor degradation by competing RGS-PX1 from Hrs.
Hrs has more recently been shown to regulate degradation of other receptors, including the G protein-coupled receptors CXCR4 (Marchese et al., 2003) and DOR (Hislop et al., 2004) and Drosophila Notch and Patched receptors (Jekely and Rorth, 2003), supporting a general role of Hrs in regulating endosomal sorting and degradation of cell surface receptors. It would be of interest to investigate whether the regulatory function of Gαs in sorting EGF receptors can be extended to other receptors, especially those coupled to heterotrimeric G proteins.
Unlike its positive role in endosomal sorting, Hrs has recently been suggested to prevent endosome fusion. Recombinant Hrs proteins were found to inhibit homotypic fusion of early endosomes, probably by binding to SNAP-25, thereby inhibiting the formation of a SNARE protein complex containing syntaxin 13, SNAP-25, and VAMP2 (Sun et al., 2003; Yan et al., 2004). Gαs has similarly been suggested to negatively regulate endosomal fusion based on the observation that activation of Gαs by either cholera toxin or a Gαs stimulatory peptide blocked endosomal fusion in vitro (Colombo et al., 1994).
In summary, our findings support a previously unappreciated role of Gαs in endocytic trafficking and down-regulation of the EGF receptor. Further studies are required to define the precise role of Gαs in endosomal sorting in general, to understand the mechanisms involved in the translocation of Gαs to early endosomes, and to unravel the differences in the regulation of Gαs functions at the plasma membrane and early endosomes.
Acknowledgments
We thank Drs. Gordon Gill, Mark Rasenick, and Angela Wandinger-Ness for kindly providing reagents. This work is supported by National Institutes of Health grants CA-100768 and DK-17780 to M.G.F. B.Z. and C.L. were supported in part by postdoctoral fellowships from the American Heart Association and the Canadian Institute of Health Research, respectively. T.T. and A.B. are members of the Molecular Pathology and Biomedical Sciences Graduate Programs, respectively, University of California, San Diego.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0446. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0446.
Abbreviations used: EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; ESCRT, endosomal sorting complexes required for transport; GAP, GTPase activating protein; GPCR, G protein-coupled receptor; Hrs, hepatocyte growth factor-regulated tyrosine kinase substrate; MVB, multivesicular body; PFA, paraformaldehyde; PX, phoX domain; RGS, regulator of G protein signaling; RNAi, RNA interference; siRNA, small-interfering RNA; SNX, sorting nexin.
References
- Bache, K.G., Brech, A., Mehlum, A., and Stenmark, H. (2003a). Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162, 435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache, K.G., Raiborg, C., Mehlum, A., and Stenmark, H. (2003b). STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513-12521. [DOI] [PubMed] [Google Scholar]
- Bao, J., Alroy, I., Waterman, H., Schejter, E.D., Brodie, C., Gruenberg, J., and Yarden, Y. (2000). Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J. Biol. Chem. 275, 26178-26186. [DOI] [PubMed] [Google Scholar]
- Beron, W., Colombo, M.I., Mayorga, L.S., and Stahl, P.D. (1995). In vitro reconstitution of phagosome-endosome fusion: evidence for regulation by heterotrimeric GTPases. Arch. Biochem. Biophys. 317, 337-342. [DOI] [PubMed] [Google Scholar]
- Bishop, N., Horman, A., and Woodman, P. (2002). Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal proteinubiquitin conjugates. J. Cell Biol. 157, 91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel, M. and Mostov, K.E. (1992). Role of heterotrimeric G proteins in membrane traffic. Mol. Biol. Cell 3, 1317-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel, M. and Mostov, K.E. (1993). Possible role of both the alpha and beta gamma subunits of the heterotrimeric G protein, Gs, in transcytosis of the polymeric immunoglobulin receptor. J. Biol. Chem. 268, 25824-25835. [PubMed] [Google Scholar]
- Carpenter, G. (2000). The EGF receptor: a nexus for trafficking and signaling. Bioessays 22, 697-707. [DOI] [PubMed] [Google Scholar]
- Chin, L.S., Raynor, M.C., Wei, X., Chen, H.Q., and Li, L. (2001). Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 276, 7069-7078. [DOI] [PubMed] [Google Scholar]
- Clague, M.J. and Urbe, S. (2003). Hrs function: viruses provide the clue. Trends Cell Biol. 13, 603-606. [DOI] [PubMed] [Google Scholar]
- Colombo, M.I., Mayorga, L.S., Casey, P.J., and Stahl, P.D. (1992). Evidence of a role for heterotrimeric GTP-binding proteins in endosome fusion. Science 255, 1695-1697. [DOI] [PubMed] [Google Scholar]
- Colombo, M.I., Mayorga, L.S., Nishimoto, I., Ross, E.M., and Stahl, P.D. (1994). Gs regulation of endosome fusion suggests a role for signal transduction pathways in endocytosis. J. Biol. Chem. 269, 14919-14923. [PubMed] [Google Scholar]
- De Vries, L., Zheng, B., Fischer, T., Elenko, E., and Farquhar, M.G. (2000). The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40, 235-271. [DOI] [PubMed] [Google Scholar]
- Gilman, A.G. (1987). G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615-649. [DOI] [PubMed] [Google Scholar]
- Haft, C.R., de la Luz Sierra, M., Barr, V.A., Haft, D.H., and Taylor, S.I. (1998). Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 18, 7278-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J.B. (1995). Role of heterotrimeric GTP binding proteins in vesicular protein transport: indications for both classical and alternative G protein cycles. FEBS Lett. 369, 84-88. [DOI] [PubMed] [Google Scholar]
- Hislop, J.N., Marley, A., and Von Zastrow, M. (2004). role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J. Biol. Chem. 279, 22522-22531. [DOI] [PubMed] [Google Scholar]
- Hollinger, S. and Hepler, J.R. (2002). Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54, 527-559. [DOI] [PubMed] [Google Scholar]
- Jekely, G. and Rorth, P. (2003). Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 4, 1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Stefan, C.J., Babst, M., and Emr, S.D. (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162, 413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil, S.J., Hobert, M., and Carlin, C. (1999). A leucine-based determinant in the epidermal growth factor receptor juxtamembrane domain is required for the efficient transport of ligand-receptor complexes to lysosomes. J. Biol. Chem. 274, 3141-3150. [DOI] [PubMed] [Google Scholar]
- Komada, M., Masaki, R., Yamamoto, A., and Kitamura, N. (1997). Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 272, 20538-20544. [DOI] [PubMed] [Google Scholar]
- Kurten, R.C., Cadena, D.L., and Gill, G.N. (1996). Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science 272, 1008-1010. [DOI] [PubMed] [Google Scholar]
- Levis, M.J. and Bourne, H.R. (1992). Activation of the alpha subunit of Gs in intact cells alters its abundance, rate of degradation, and membrane avidity. J. Cell Biol. 119, 1297-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann, C., Graness, A., Boehmer, A., Kovalenko, M., Adomeit, A., Steinmetzer, T., Nurnberg, B., Wetzker, R., and Boehmer, F.D. (1996). Tyrosine phosphorylation of GSalpha and inhibition of bradykinin-induced activation of the cyclic AMP pathway in A431 cells by epidermal growth factor receptor. J. Biol. Chem. 271, 31098-31105. [DOI] [PubMed] [Google Scholar]
- Lin, C.R., Chen, W.S., Lazar, C.S., Carpenter, C.D., Gill, G.N., Evans, R.M., and Rosenfeld, M.G. (1986). Protein kinase C phosphorylation at Thr 654 of the unoccupied EGF receptor and EGF binding regulate functional receptor loss by independent mechanisms. Cell 44, 839-848. [DOI] [PubMed] [Google Scholar]
- Lin, H.C., Duncan, J.A., Kozasa, T., and Gilman, A.G. (1998). Sequestration of the G protein beta gamma subunit complex inhibits receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA 95, 5057-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Hope, L.W., Brasch, M., Reinhard, C., and Cohen, S.N. (2003). TSG101 interaction with HRS mediates endosomal trafficking and receptor downregulation. Proc. Natl. Acad. Sci. USA 100, 7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese, A., Raiborg, C., Santini, F., Keen, J.H., Stenmark, H., and Benovic, J.L. (2003). The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell 5, 709-722. [DOI] [PubMed] [Google Scholar]
- McConnell, S.J., and Yaffe, M.P. (1992). Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J. Cell Biol. 118, 385-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, B.G., Parikh, B., Milligan, G., and Patel, T.B. (1990). Gs alpha mediates epidermal growth factor-elicited stimulation of rat cardiac adenylate cyclase. J. Biol. Chem. 265, 21317-21322. [PubMed] [Google Scholar]
- Neves, S.R., Ram, P.T., and Iyengar, R. (2002). G protein pathways. Science 296, 1636-1639. [DOI] [PubMed] [Google Scholar]
- Nurnberg, B., and Ahnert-Hilger, G. (1996). Potential roles of heterotrimeric G proteins of the endomembrane system. FEBS Lett. 389, 61-65. [DOI] [PubMed] [Google Scholar]
- Poppleton, H., Sun, H., Fulgham, D., Bertics, P., and Patel, T.B. (1996). Activation of Gsalpha by the epidermal growth factor receptor involves phosphorylation. J. Biol. Chem. 271, 6947-6951. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K.G., Mehlum, A., Stang, E., and Stenmark, H. (2001a). Hrs recruits clathrin to early endosomes. EMBO J. 20, 5008-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Bremnes, B., Mehlum, A., Gillooly, D.J., D'Arrigo, A., Stang, E., and Stenmark, H. (2001b). FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114, 2255-2263. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Rusten, T.E., and Stenmark, H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446-455. [DOI] [PubMed] [Google Scholar]
- Raiborg, C. and Stenmark, H. (2002). Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell Struct. Funct. 27, 403-408. [DOI] [PubMed] [Google Scholar]
- Ransnas, L.A., Svoboda, P., Jasper, J.R., and Insel, P.A. (1989). Stimulation of beta-adrenergic receptors of S49 lymphoma cells redistributes the alpha subunit of the stimulatory G protein between cytosol and membranes. Proc. Natl. Acad. Sci. USA 86, 7900-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse, M., Urbe, S., Oorschot, V., Strous, G.J., and Klumperman, J. (2002). Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol. Biol. Cell 13, 1313-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, A. and Von Zastrow, M. (2002). Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell. Biol. 3, 600-614. [DOI] [PubMed] [Google Scholar]
- Stahl, P.D. and Barbieri, M.A. (2002). Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sci STKE 2002, PE32. [DOI] [PubMed] [Google Scholar]
- Stow, J.L. and Heimann, K. (1998). Vesicle budding on Golgi membranes: regulation by G proteins and myosin motors. Biochim. Biophys. Acta 1404, 161-171. [DOI] [PubMed] [Google Scholar]
- Sun, H., Chen, Z., Poppleton, H., Scholich, K., Mullenix, J., Weipz, G.J., Fulgham, D.L., Bertics, P.J., and Patel, T.B. (1997). The juxtamembrane, cytosolic region of the epidermal growth factor receptor is involved in association with alpha-subunit of Gs. J. Biol. Chem. 272, 5413-5420. [PubMed] [Google Scholar]
- Sun, W., Yan, Q., Vida, T.A., and Bean, A.J. (2003). Hrs regulates early endosome fusion by inhibiting formation of an endosomal SNARE complex. J. Cell Biol. 162, 125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbe, S., Sachse, M., Row, P.E., Preisinger, C., Barr, F.A., Strous, G., Klumperman, J., and Clague, M.J. (2003). The UIM domain of Hrs couples receptor sorting to vesicle formation. J. Cell Sci. 116, 4169-4179. [DOI] [PubMed] [Google Scholar]
- Van Dyke, R.W. (2004). Heterotrimeric G protein subunits are located on rat liver endosomes. BMC Physiol. 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T.A. and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner, P.B. and Bourne, H.R. (1994). Activation and depalmitoylation of Gs alpha. Cell 77, 1063-1070. [DOI] [PubMed] [Google Scholar]
- Wedegaertner, P.B., Bourne, H.R., and von Zastrow, M. (1996). Activation-induced subcellular redistribution of Gs alpha. Mol. Biol. Cell 7, 1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, M.J., Taylor, G.S., and Dixon, J.E. (2001). Phoxy lipids. revealing PX domains as phosphoinositide binding modules. Cell 105, 817-820. [DOI] [PubMed] [Google Scholar]
- Worby, C.A. and Dixon, J.E. (2002). Sorting out the cellular functions of sorting nexins. Nat. Rev. Mol. Cell. Biol. 3, 919-931. [DOI] [PubMed] [Google Scholar]
- Yan, Q., Sun, W., McNew, J.A., Vida, T.A., and Bean, A.J. (2004). Ca2+ and NSF differentially regulate disassembly of SNARE complexes on early endosomes. J. Biol. Chem. [DOI] [PubMed]
- Yu, J.W. and Lemmon, M.A. (2001). All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179-44184. [DOI] [PubMed] [Google Scholar]
- Yu, J.Z. and Rasenick, M.M. (2002). Real-time visualization of a fluorescent G(alpha)(s): dissociation of the activated G protein from plasma membrane. Mol. Pharmacol. 61, 352-359. [DOI] [PubMed] [Google Scholar]
- Zheng, B., Chen, D., and Farquhar, M.G. (2000). MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc. Natl. Acad. Sci. USA 97, 3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B., Ma, Y.C., Ostrom, R.S., Lavoie, C., Gill, G.N., Insel, P.A., Huang, X.Y., and Farquhar, M.G. (2001). RGS-PX1, a GAP for GalphaS and sorting nexin in vesicular trafficking. Science 294, 1939-1942. [DOI] [PubMed] [Google Scholar]