Abstract
Septins are GTP binding proteins important for cytokinesis in many eukaryotes. The Schizosaccaromyces pombe genome sequence predicts orthologues of four of five Saccharomyces cerevisiae septins involved in cytokinesis and these are named Spns1-4p. That spns1-4 are not essential genes permitted the application of a combined genetic and proteomics approach to determine their functional relationships. Our findings indicate that Spns1-4p are present throughout interphase as a diffusely localized ∼8.5S complex containing two copies of each septin linked together as a chain in the order Spn3p-Spn4p-Spn1p-Spn2p. Septin recruitment to the medial region of the cell is genetically separable from ring formation, and whereas it is normally restricted to mitosis, it can be promoted without activation of the mitotic cell cycle machinery. Coalescence into ring structures requires Spn1p and Spn4p associate with at least one other septin subunit and the expression of Mid2p that is normally restricted to mitosis. This study establishes the functional requirements for septin complex organization in vivo.
INTRODUCTION
Septins are a family of guanine nucleotide binding proteins. They were first identified in Saccharomyces cerevisiae for their role in cytokinesis (Hartwell, 1971). In the past 10 years, it has become evident that septins are highly conserved cytoskeletal elements found in most eukaryotes (Faty et al., 2002; Macara et al., 2002). Although many septins have been implicated in the process of cell division because of their localization to the cell division site and/or because the disruption of their function leads to cytokinesis defects, septins are likely involved in additional biological processes because they are present in nondividing cells such as neurons and in complexes with components of the secretory apparatus (reviewed in Field and Kellogg, 1999; Kartmann and Roth, 2001; Faty et al., 2002).
Septins typically exist in heterooligomeric complexes (Field et al., 1996; Frazier et al., 1998; Hsu et al., 1998; Sheffield et al., 2003). In Drosophila, three septins form a stable complex that likely consists of a linear array of each septin homodimer (Field et al., 1996). When expressed in Escherichia coli, three mammalian septins can form a complex of a size consistent with a hexamer, i.e., two of each subunit (Sheffield et al., 2003). In S. cerevisiae, four septins form a stable complex that has been predicted to consist of two molecules each of Cdc3p, Cdc10p, and Cdc12p and 1 molecule of Cdc11p (Frazier et al., 1998). Although clearly existing in complexes, the organization of septins within these complexes has not been investigated previously.
In S. cerevisiae, Cdc3p, Cdc10p, Cdc11p, and Cdc12p together with another septin, Shs1/Sep7, (Mino et al., 1998) assemble into a patch at the incipient bud site during G1. As the cell cycle progresses, septins reorganize at the mother-bud neck into an hourglass-shaped collar of cortical filaments (Frazier et al., 1998). Recombinant septins have the capacity to form filaments in vitro in a GTP-dependent manner (Mendoza et al., 2002; Versele and Thorner, 2004). However, although GTP-binding is important for in vitro filament assembly and ring formation in vivo (Versele and Thorner, 2004), nucleotide turnover on septins in vivo is so slow that GTP hydrolysis is unlikely to play a role in septin dynamics during the cell cycle (Vrabioiu et al., 2004). Rather, other events regulated by the GIN4 family of kinases (Longtine et al., 1998; Mortensen et al., 2002; Dobbelaere et al., 2003) and/or Cla4p (Dobbelaere et al., 2003; Schmidt et al., 2003; Versele and Thorner, 2004) in S. cerevisiae have been shown to be important for assembly of functional septin rings in vivo. However, exactly how septin complexes are assembled into higher order structures such as rings remains unknown.
Once organized into a ring structure in S. cerevisiae, septins are thought to perform multiple functions at the mother-daughter bud neck. These include providing a boundary that restricts certain determinants to particular cortical domains (reviewed by Faty et al., 2002) and acting as a bud neck scaffold necessary for the localization of many factors involved in polarity and cell division (reviewed by Gladfelter et al., 2001). Whether septins perform all of these roles in other organisms is yet to be established.
Septins have a conserved structure. They contain a central GTP-binding domain flanked by a basic region at the amino terminus, and most septins contain a coiled-coil domain at the carboxy terminus. With the completion of the Schizosaccharomyces pombe genome sequence (Wood et al., 2002), homologues of the S. cerevisiae septins Cdc3p, Cdc10p, Cdc11p, and Cdc12p were identified and named Spn1p, Spn2p, Spn3p, and Spn4p, respectively. spn1-4 are not essential, but the absence of any or all of them (Berlin et al., 2003; Tasto et al., 2003; our unpublished data) causes a delay in the completion of cell division, resulting in a chained cell phenotype. Spn1p and Spn3p are known to organize at the site of cell division late in mitosis where they form a ring that splits as the septum forms, but this ring does not constrict with actomyosin ring invagination; rather, the ring dissipates after cell cleavage (Berlin et al., 2003; Tasto et al., 2003).
Mid2p is a S. pombe protein related at its C-terminus to S. cerevisiae Bud4p, Candida albicans Int1p, and anillins present in multicellular eukaryotes (Field and Alberts, 1995; Sanders and Herskowitz, 1996; Oegema et al., 2000; Gale et al., 2001). Anillin is able to direct septin filament assembly along actin bundles in an in vitro assay containing only purified components (Kinoshita et al., 2002). Mid2p lacks an obvious F-actin binding domain as is present in anillin, but Mid2p is similarly required for the proper organization of septin rings at the site of cell division (Berlin et al., 2003; Tasto et al., 2003). Like septins, mid2+ is not an essential gene, but its loss causes a chained cell phenotype (Berlin et al., 2003; Tasto et al., 2003). Fluorescence recovery after photobleaching (FRAP) analysis of Spn1p-GFP has shown that septin rings are quite dynamic in mid2Δ cells, whereas in wild-type cells they are very stable (Berlin et al., 2003). Mid2p production is normally restricted to anaphase and overproduction of a stabilized truncation mutant of Mid2p leads to persistence of septin rings through multiple cell divisions (Tasto et al., 2003). These observations suggest that Mid2p interacts either directly or indirectly with septins to promote their organization into stable ring structures in late mitosis.
To begin to address how Mid2p and its relatives affect septin ring organization, we have investigated how Spns 1-4p are organized in S. pombe cells using a combination of proteomic and genetic approaches. Our data suggest that the building blocks of septin rings are ∼8.5S complexes containing two of each septin subunit linked together in an almost linear array of specific order. These units are assembled into a functional ring structure in at least two steps. Recruitment to the medial region can occur if septation is forced during interphase but coalescence into a ring structure requires Mid2p that is normally produced only in mitosis. Our data also indicate that each septin subunit contributes uniquely to the formation of septin structures.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
S. pombe strains used in this study are listed in Supplementary Table 1 and were grown in YE or minimal medium with the appropriate supplements as described (Moreno et al., 1991). DNA transformations were done by electroporation (Prentice, 1992) or lithium acetate transformation (Keeney and Boeke, 1994). To arrest cells in S phase, cultures were grown in 12 mM hydroxyurea at 25°C for 4 h.
The spn1, spn2, spn3, and spn4 open reading frames (ORFs) were each tagged at their 3′ ends with the HA3-KanR, myc13-KanR, 2X CTAP-KanR, CFP-KanR, YFP-KanR, and EGFP-KanR cassette as previously described (Bahler et al., 1998). KanR transformants were screened by whole cell PCR and then by immunoblotting to confirm the accurate integration and expression of the fusion protein.
The spn1 ORF was replaced with ura4+ by homologous recombination in a diploid strain (Bahler et al., 1998). Ura+ transformants were screened for the proper gene disruptant by whole-cell PCR. A heterozygous diploid strain was then sporulated at 25°C followed by tetrad dissection to isolate the haploid spn1:: ura4 strain.
Molecular Biology Techniques
PCR amplifications were carried out with TaqPlus Precision polymerase (Stratagene, La Jolla, CA) according to manufacturer's instructions. Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Purification and Analyses of Septin TAP Complexes
Eight-liter cultures of strains producing TAP-tagged proteins were grown to log phase and the tagged proteins were isolated as described (Tasto et al., 2001). For silver staining, one quarter of the eluate from the purification was precipitated with TCA, resuspended in LDS-PAGE buffer and resolved on a 4–12% NuPAGE using MOPS buffer (Novex, Encinitas, CA). Silver staining was carried out using the Plus One kit as recommended by the manufacturer (Amersham Pharmacia Biotech, Piscataway, NJ). The TAP complexes were analyzed by DALPC-tandem mass spectrometry as described previously (Ohi et al., 2002; Sanders et al., 2002).
Immunoprecipitations, Sucrose Gradients, and Immunoblots
Whole cell lysates were prepared in NP-40 buffer followed by anti-HA or anti-Myc immunoprecipitations as previously described (McDonald et al., 1999). Denatured lysates were prepared as described (Burns et al., 2002). For sucrose gradient analysis, a 200-μl volume of protein lysate was layered onto a 5-ml 10–30% sucrose gradient prepared in NP-40 buffer. Gradients were ultracentrifuged at 38,000 rpm for 18 h in a Beckman SW50.1 rotor (Berkeley, CA). Sedimentation markers were fractionated on gradients prepared and spun in parallel. Fractions were collected from the bottom of the gradient and resolved on 4–12% NuPAGE gels in MOPS buffer and subsequently transferred by electroblotting to PVDF membrane (Immobilon P; Millipore, Bedford, MA). Immunoblotting was done with anti-HA (12CA5; 2 μg/ml), anti-Myc (9E10; 2 μg/ml), or anti-GFP (1:1000 dilution of serum) antibodies. Primary antibodies were detected with horseradish peroxidase–conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (0.4 mg/ml; Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:50,000 followed by ECL visualization using SuperSignal (Pierce, Rockford, IL).
Yeast Two-hybrid Assays
The yeast two-hybrid system used in this study was described previously (James et al., 1996). spn3 and spn1 cDNAs were cloned into the bait plasmid pGBT9 (Clontech, Palo Alto, CA), spn1+, spn2+, spn3+, and spn4+ cDNAs were cloned into the prey plasmid pGAD424 (Clontech) and sequenced to ensure the absence of PCR-induced mutations and that the correct reading frame had been retained.
To test for protein interactions, both bait and prey plasmids were contransformed into S. cerevisiae strain PJ69-4A. β-galactosidase reporter enzyme activity in the two-hybrid strains was measured using the Galacto-StarTM chemiluminescent reporter assay system according to the manufacturer's instructions (Tropix, Bedford, MA) with the exception that cells were lysed by glass bead disruption. Each sample was measured in triplicate. Reporter assays were recorded either on the BMG luminometer (Bartlett-Williams Scientific, Chapel Hill, NC) or the Mediator Phl luminometer (Aureon Biosystems, Vienna, Austria).
Microscopy
Strains producing GFP-, YFP-, or CFP-tagged proteins were grown in YE medium and visualized in live cells or in cells fixed with ethanol. Microscopy was performed at room temperature using a spinning disk confocal microscope (Ultraview LCI; PerkinElmer, Norwalk, CT) and Ultraview LCI software (v5.2; PerkinElmer) for image acquisition. Images were processed using Velocity software (v1.4.2; Improvision, Lexington, MA). Z-series optical sections were taken at 0.5-μm spacing for 3D reconstructions.
RESULTS
The Four S. pombe Septins Form a Linear Complex
To define the building blocks of S. pombe septin rings, we analyzed how S. pombe Spn1p, Spn2p, Spn3p, and Spn4p interacted. For these experiments, spn1-4 were tagged at their endogenous loci with sequences encoding a variety of epitopes (TAP, HA, myc, GFP, CFP, and/or YFP) to create C-terminal–tagged variants under control of their own promoters. Unless otherwise noted, only strains that exhibited wild-type phenotypes were used for these studies. spn1-4 were also deleted from the genome either alone or in combination. Although none of these S. pombe proteins is essential, the absence of any or all of them causes a delay in the completion of cell division resulting in a chained cell phenotype, although the severity of this defect varies (Berlin et al., 2003; Tasto et al., 2003; see below and our unpublished observations).
First, we examined the composition of Spn3p-TAP, Spn4p-TAP, and Spn2p-TAP complexes purified from asynchronous populations of cells. The spn1-TAP strain was not used because it displayed a loss of function phenotype (unpublished data). Spns 1-4p were detected by silver staining and mass spectrometry in all three purifications (Figure 1, A–C and Table 1). No additional proteins were identified by mass spectrometry as specific components of these septin complexes. All other identified proteins were detected in mock TAP eluates from strains lacking TAP-tagged proteins and/or from unrelated TAP complexes (unpublished data).
Figure 1.
Composition of S. pombe septin complexes. (A–H) Silver-stained gels of a portion of Spn-TAP complexes from the indicated strains. Spn proteins are indicated with arrows. Note that the calmodulin-binding portion of the TAP tag remains after the purification and changes the mass of the proteins. Other bands are nonspecific contaminants. (I) Protein lysates prepared in denaturing conditions from the indicated strains were resolved by SDS-PAGE and blotted with anti-GFP serum. (J) anti-myc (top panel) and anti-HA (bottom panel) immunoprecipitates from the indicated strains were blotted with anti-myc antibodies.
Table 1.
TAP/DALPC results
| Strains
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Protein | ||||||||
| Spn1p | 14 | 15 | 17 | 12 | 0 | 0 | 16 | 15 |
| Spn2p | 14 | 12 | 12 | 0 | 0 | 0 | 13 | 11 |
| Spn3p | 14 | 13 | 15 | 11 | 17 | 13 | 0 | 0 |
| Spn4p | 10 | 10 | 13 | 9 | 12 | 0 | 10 | 0 |
Strains: 1) spn3-TAP, 2) spn4-TAP, 3) spn2-TAP, 4) spn3-TAP spn2Δ, 5) spn3-TAP spn1Δ, 6) spn3-TAP spn4Δ, 7) spn4-TAP spn3Δ, and 8) spn2-TAP spn4Δ. Numbers in columns below the line are unique tryptic peptides identified from each protein by mass spectrometry.
Next, the composition of septin TAP complexes purified from strains lacking one of the septin proteins was determined by silver staining and mass spectrometry. In Spn3-TAP complexes purified from spn2Δ cells, Spn3p, Spn4p, and Spn1p were identified (Figure 1D and Table 1). In Spn3-TAP complexes purified from spn1Δ cells, only Spn3p and Spn4p were present (Figure 1E and Table 1). Although Spn2p did not copurify with Spn3p from spn1Δ cells, Spn2p-GFP was readily detected in an spn1Δ strain suggesting that it is not degraded in the absence of Spn1p (Figure 1I). Spn3p-TAP did not copurify with any other septin from an spn4Δ strain (Figure 1F and Table 1). These data suggested that septins interacted in a linear order of Spn3p-Spn4p-Spn1p-Spn2p (see Figure 2A). To confirm this, we examined the composition of the Spn4p-TAP complexes from spn3Δ cells and Spn2p-TAP complexes from spn4Δ cells. As predicted, Spn4p, Spn1p, and Spn2p were detected in Spn4p-TAP complexes from spn3Δ cells (Figure 1G and Table 1) and in the Spn2p-TAP complex isolated from spn4Δ cells, Spn1p and Spn2p but not Spn3p were detected (Figure 1H and Table 1). Spn3p was present, however, in cells lacking Spn4p, so degradation did not explain the lack its association with other septins (Figure 1I). Finally, we examined whether Spn4p and Spn1p remained associated in the absence of both Spn2p and Spn3p. Consistent with the proposed Spn3p-Spn4p-Spn1p-Spn2p arrangement, Spn1p-myc and Spn4p-HA coimmunoprecipitated from lysates prepared from a spn2Δ spn3Δ strain (Figure 1J). Based on the lower recovery of coimmunoprecipitated proteins, this Spn1p-Spn4p complex appears to be less stable than in the presence of Spn2p or Spn3p. Consistent with the coimmunoprecipitation results, Spn4p-TAP complexes purified from spn2Δspn3Δ cells contained Spn1p as determined by mass spectrometry as well as significantly more heat shock proteins than the TAP complexes described above (unpublished data). We interpret this as an indicator of complex instability and/or aggregation.
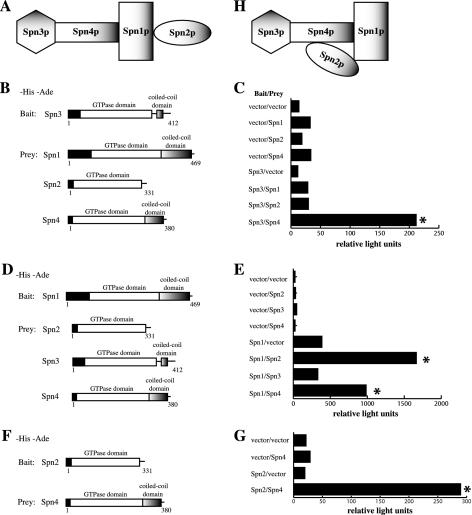
Figure 2.
Protein-protein interactions mapped between septin complex components. (A and H) Models of S. pombe septin interactions. (B) The PJ69-4A strain was transformed with pGBT9 or pGBT9spn3, and pGAD424 or pGAD424 carrying spn1, spn2, or spn4, which are shown schematically. (C) β-galactosidase activity (represented in relative light units) of the stains described in B. (D) The PJ69-4A stain was transformed with pGBT9 or pGBT9spn1 and pGAD424 or pGAD424 carrying spn2, spn3, or spn4, which are shown schematically. (E) β-galactosidase activity (represented in relative light units) of the stains described in D. (F) The PJ69-4A stain was transformed with pGBT9 or pGBT9spn2 and pGAD424 or pGAD424 carrying spn4, which are shown schematically. (G) β-galactosidase activity (represented in relative light units) of the stains described in D. Asterisk indicates positive interactions.
To examine the ability of septins to interact with each other in a different manner, directed two-hybrid analysis was performed. As would be predicted by a linear model of interaction derived from the TAP experiments (Figure 2A), Spn3p interacted robustly with Spn4p, but not with Spn1p or Spn2p (Figure 2, B and C). Also, Spn1p interacted with both Spn2p and Spn4p, but not Spn3p (Figure 2, D and E). We next addressed the final possibility of whether Spn2p could interact with Spn4p. Inconsistent with the linear model of assembly derived from the TAP experiments (Figure 2A), significant interaction was observed between Spn2p and Spn4p (Figure 2, F and G). Although we considered the possibility that this interaction might be bridged by a S. cerevisiae septin, this seems unlikely because we did not detect any other bridging interactions in our analysis (for example, spn2 with spn3, or spn3 with spn1). Therefore, we conclude that most likely 1) Spn2p can interact directly with Spn4p and 2) Spn1p is required to stabilize this association during a biochemical purification of the complex. Thus, our model was refined by these experiments to encompass the likelihood of a Spn2p-Spn4p interaction stabilized by Spn2p-Spn1p and Spn4p-Spn1p associations (Figure 2H).
S. pombe Septin Complexes Contain at Least Two Copies of Each Subunit
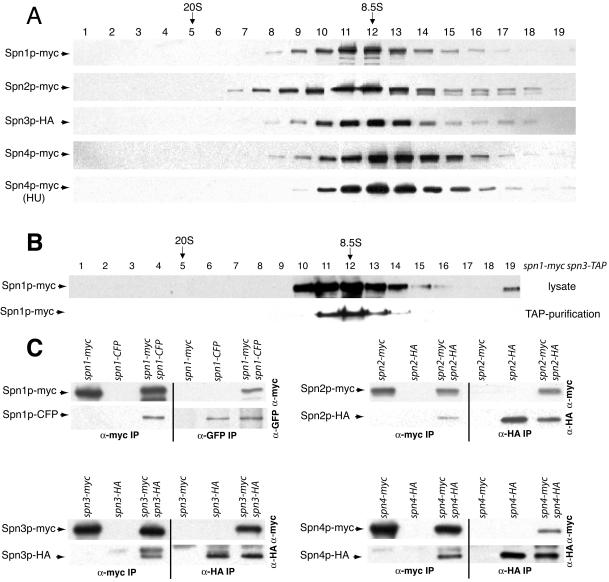
We next asked whether each septin protein existed primarily in a complex with other septins by sucrose gradient analysis. Each myc- or HA-epitope–tagged septin present in lysates from asynchronously growing cells migrated with a single peak of 8.5S (Figure 3A), indicating that the majority of each septin exists primarily as a component of a larger septin complex. Because septin rings would be expected to sediment much faster than 8.5S, we reasoned that the 8.5S complex corresponds to the septin ring organizational unit present throughout the remainder of the cell cycle and does not correspond to higher order ring structures that must disassemble under our lysis conditions. Indeed, Spn4p-myc was found primarily in the 8.5S fraction in hydroxyurea-treated cells (Figure 3A) that are arrested in S-phase (unpublished data). We have so far been unable to purify a larger complex containing septins (unpublished data). We also confirmed that the 8.5S complex is what is purified by tandem affinity purification because Spn1p-myc purified by tandem affinity purification from a spn1-myc spn3-TAP strain cosedimented with unpurified Spn1p-myc from a protein lysate (Figure 3B).
Figure 3.
Characterization of S. pombe septin complexes. (A) Lysates prepared in NP-40 buffer from each myc-epitope– or HA-epitope–tagged septin protein from asynchronously growing cells or a hydroxyurea (HU)-arrested culture were resolved on sucrose gradients. Fractions were collected from the bottom of the gradient (1) and immunoblotted with 9E10 or 12CA5 to detect myc or HA-tagged proteins. The peaks of thyroglobulin (20S) and aldolase (8.5S) collected from gradients prepared and run in parallel are indicated. (B) A TAP and a lysate prepared in NP-40 buffer from spn1-myc spn3-TAP cells were resolved on sucrose gradients. Fractions were collected from the bottom of the gradient and immunoblotted with 9E10 to detect Spn1p-myc. The peaks of thyroglobulin (20S) and aldolase (8.5S) collected from gradients prepared and run in parallel are indicated. (C) anti-myc (left side of panels) and anti-GFP or anti-HA (right side of panels) immunoprecipitates from the indicated strains were blotted with anti-myc (top panels) and anti-GFP or anti-HA (bottom panels) antibodies.
The sedimentation value of the S. pombe septin complex at 8.5S is similar to that reported for a S. cerevisiae septin complex (9S), which is predicted to contain two each of three subunits and one of a fourth (Frazier et al., 1998). To determine if S. pombe septins are organized similarly, diploid strains containing two different epitope-tagged alleles of each septin were constructed. These diploid strains were phenotypically wild-type (unpublished data) but the two epitope tags could introduce some instability to the complex. In the cases of spn2-HA/spn2-myc, spn3-HA/spn3-myc, and spn4-HA/spn4-myc strains, anti-HA immunoprecipitates contained the myc-tagged septins in addition to the HA-tagged septins (Figure 3C). Coprecipitating proteins were not observed in the single HA-tagged strains (Figure 3C). Conversely, anti-myc immunoprecipitates from the double-tagged strains but not the single myc–tagged strains contained HA-tagged proteins in addition to myc-tagged proteins. In lysates prepared from spn1-CFP/spn1-myc diploid cells, antibodies to GFP precipitated both Spn1p-CFP and Spn1p-myc and conversely anti-myc immunoprecipitations contained both Spn1p-myc and Spn1p-CFP. Coimmunoprecipitation was not observed in strains producing single epitope–tagged proteins (Figure 3C). From these data, we conclude that S. pombe septin complexes contain at least two copies of each subunit. Considering the sedimentation value of 8.5S, the proteomics data, and the similarity with the S. cerevisiae septin complex (Frazier et al., 1998), it is probable that each S. pombe septin complex contains just two copies of each subunit, although this remains to be proven rigorously.
S. pombe Septins Associate in a Cooperative Manner
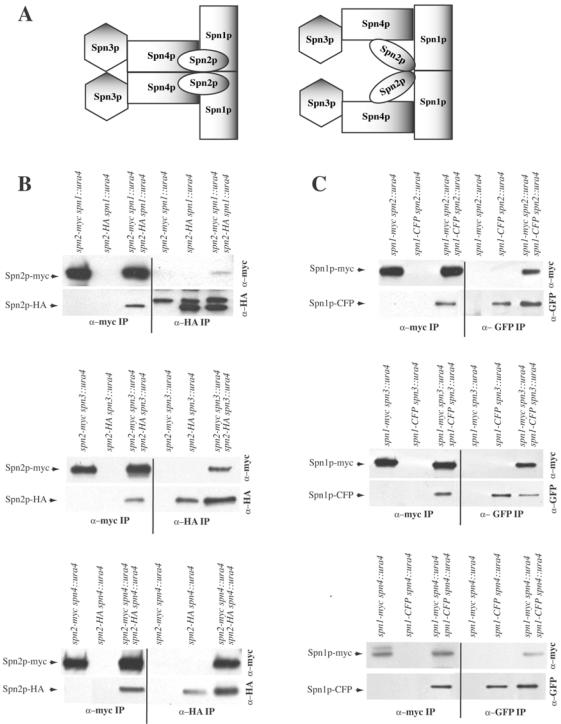
Thus far, our data supported a model in which two copies of Spn3p-Spn4p-Spn1p-Spn2p are arrayed in an almost linear order in which each S. pombe septin directly contacts its identical partner as well as one or two nonidentical subunits (Figure 4A, left panel). To determine whether there were cooperative interactions between septins to facilitate homotypic interactions, we examined which septins maintained homotypic interactions in the absence of other components. Again, diploid strains producing two different epitope-tagged alleles of each septin in all combinations of deletion strains were used in immunoprecipitation experiments.
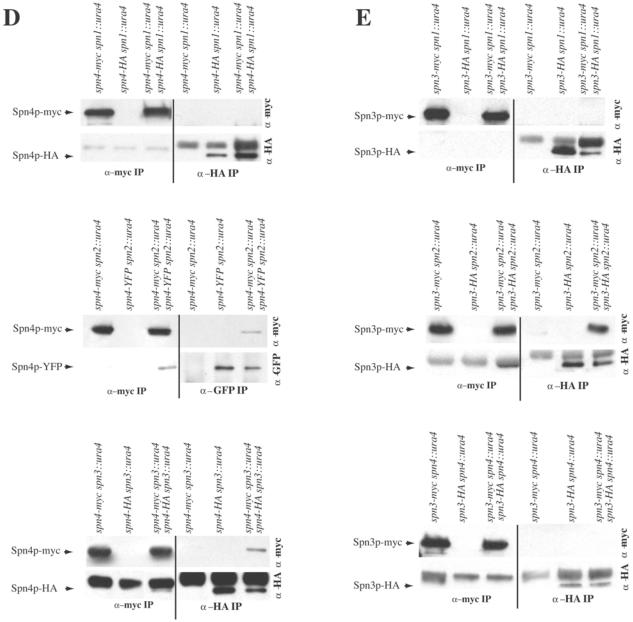
Figure 4.
S. pombe septins interact in cooperative manner. (A) Possible models of S. pombe septin complex organization. (B–E) anti-myc (left side of panels) and anti-GFP or anti-HA (right side of panels) immunoprecipitates from the indicated strains were blotted with anti-myc (top panels) and anti-GFP or anti-HA (bottom panels) antibodies.
First, the role of Spn1p, Spn3p, and Spn4p in the Spn2p-Spn2p interaction was examined. In the absence of spn1, spn3, or spn4, anti-HA immunoprecipitates from the double-tagged strains but not the single-tagged strains contained Spn2p-myc in addition to Spn2p-HA (Figure 4B). Conversely, anti-myc immunoprecipitates from the double-tagged strains but not the single-tagged strains contained Spn2p-HA in addition to Spn2p-myc (Figure 4B). Therefore, we conclude that the Spn2p-Spn2p interaction is independent of the other three septins.
Next, we examined the Spn1p-Spn1p interaction in the absence of spn2, spn3, or spn4. In lysates prepared from spn1-myc spn2::ura4/spn1-CFP spn2::ura4, spn1-myc spn3::ura4/spn1-CFP spn3::ura4, or spn1-myc spn4::ura4/spn1-CFP spn4::ura4 diploid cells, antibodies to GFP precipitated both Spn1p-CFP and Spn1p-myc, and conversely anti-myc immunoprecipitations contained both Spn1p-myc and Spn1p-CFP. Coprecipitating proteins that interacted specifically with the antibodies were not observed in strains producing single epitope–tagged proteins (Figure 4C). Therefore, the Spn1p-Spn1p interaction, like the Spn2p-Spn2p interaction, is independent of other septins.
In contrast, we found that the ability of Spn3p and Spn4p to self-associate depended on certain of the other septins. In the absence of spn1, anti-HA immunoprecipitates from the spn4-myc spn1::ura4/spn4-HA spn1::ura4 double-tagged strain did not contain Spn4p-myc; the only protein detected in either the single- or double-tagged strains was Spn4p-HA (Figure 4D). Reciprocally, anti-myc immunoprecipitates from the spn4-myc spn1::ura4/spn4-HA spn1::ura4 double-tagged strain did not contain Spn4-HA either; the only protein detected in either the single- or double-tagged strains was Spn4p-myc (Figure 4D). However, in lysates prepared from spn4-mycspn2::ura4/spn4-YFPspn2::ura4 or spn4-myc spn3::ura4/spn4-HA spn3::ura4 diploid cells, antibodies to GFP or HA did precipitate Spn4p-myc in addition to either Spn4p-YFP or Spn4p-HA, and conversely anti-myc immunoprecipitations contained both Spn4p-YFP or Spn4p-HA in addition to Spn4p-myc. Therefore, the Spn4p-Spn4p interaction did require Spn1p but did not require Spn2p or Spn3p.
Lastly, the Spn3p-Spn3p interaction was found to be independent of spn2 but dependent on spn1 and spn4 (Figure 4E). Anti-HA immunoprecipitates from the spn3-myc spn1::ura4/spn3-HA spn1::ura4 or spn3-myc spn4::ura4/spn3-HA spn4::ura double-tagged strain did not pull down Spn3p-myc; the only protein detected in either the single- or double-tagged strains was Spn3p-HA (Figure 4E). Reciprocally, anti-myc immunoprecipitates from the spn3-myc spn1::ura4/spn3-HA spn1::ura4 or spn3-myc spn4::ura4/spn3-HA spn4::ura double-tagged strain did not pull down Spn3-HA; the only protein detected in either the single- or double-tagged strains was Spn3p-myc (Figure 4E). In contrast, anti-HA immunoprecipitates from the spn3-myc spn2::ura4/spn3-HA spn2::ura4 double-tagged strain but not the single-tagged strain contained Spn3p-myc in addition to Spn3p-HA (Figure 4E). Conversely, anti-myc immunoprecipitates from the spn3-myc spn2::ura4/spn3-HA spn2::ura4 double-tagged strain but not the single-tagged strain contained Spn3p-HA in addition to Spn3p-myc (Figure 4E).
In summary, the model consistent with all of our biochemical data is illustrated in Figure 4A, right panel, which indicates a branch in the complex because we have gathered no evidence that Spn3p and Spn4p are able to directly associate with themselves. However, the reduced recoveries of coprecipitating proteins in some cases (Figure 4B, top panel, and D, middle panel) and the absence of coprecipitating proteins in others indicate that most heterotypic septin interactions within the complex have either stabilizing effects or are absolutely required for homotypic septin interactions. Considering this and the straight edges of septin filaments observed by electron microscopy (Field et al., 1996; Frazier et al., 1998; Kinoshita et al., 2002; Mendoza et al., 2002; Versele and Thorner, 2004), we must also consider the model illustrated in Figure 4A, left panel, for septin organization and ultrastructural analysis will be required to distinguish between the two.
Interdependence of Septins for Ring Formation
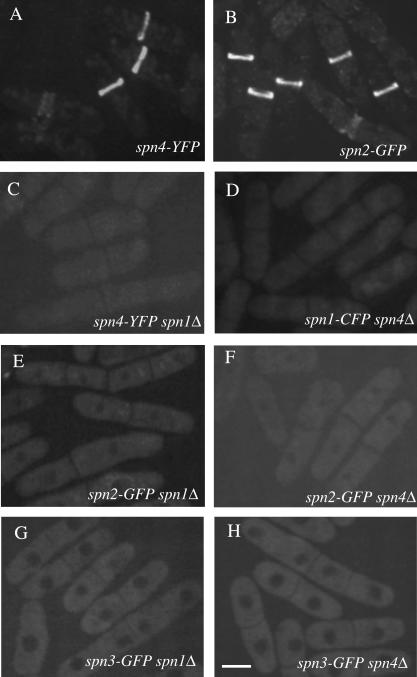
Spn3p-GFP and Spn1p-GFP/CFP were shown previously to be localized diffusely during interphase and then organized into a ring structure at the end of anaphase (Berlin et al., 2003; Tasto et al., 2003). This ring splits as the septum forms but does not constrict with actomyosin ring invagination; rather, the ring dissipates after cell cleavage (Berlin et al., 2003; Tasto et al., 2003). Spn4p-YFP and Spn2p-GFP localized identically to Spn1p and Spn3p (Figure 5, A and B). To determine which septin molecules or assemblies of molecules were critical for this localization pattern, we used GFP/CFP/YFP-tagged strains in combination with deletion alleles.
Figure 5.
Septin rings are absent in spn1Δ and spn4Δ cells. The (A) spn4-YFP (KGY496), (B) spn2-GFP (KGY4230), (C) spn4-YFP spn1Δ (KGY842), (D) spn1-CFP spn4Δ (KGY4417), (E) spn2-GFP spn1Δ (KGY4258), (F) spn2-GFP spn4Δ (KGY4259), (G) spn3-GFP spn1Δ (KGY4260), and (H) spn3-GFP spn4Δ (KGY2281) strains were grown in YE medium at 25°C, and GFP-tagged proteins were visualized in live cells. Scale bar, 5 μm.
In the absence of Spn1p, the remaining septins showed no specific localization (Figure 5, C, E, and G). Likewise, Spn1p, Spn2p, and Spn3p failed to concentrate at the medial cortex in spn4Δ cells (Figure 5, D, F, and H). When these observations are considered in light of the model arising from our biochemical data (Figure 4A), we conclude that neither Spn3p or Spn2p alone, nor the subcomplexes of Spn3p-Spn4p or Spn2p-Spn1p can be recruited to the medial region of cells or form ring structures. It is not surprising then that the cell separation defect of spn1Δ and spn4Δ cells is as severe as the loss of all spns (unpublished data).
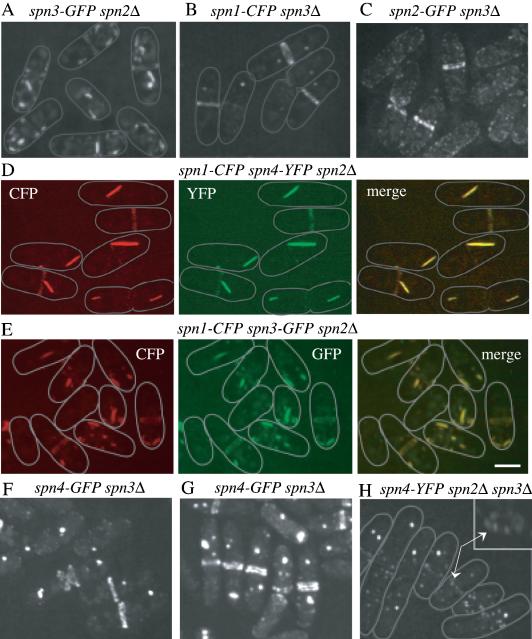
In spn2Δ cells, we found that Spn1p, Spn4p, and Spn3p localized to ring structures (Figure 6, A, D, and E). We infer that these rings are functional because most spn2Δ cells undergo normal cell separation (Figure 6, A, D, and E) in contrast to spn1Δ or spn4Δ cells (see Figure 5, C–H). Interestingly, Spn1p, Spn4p, and Spn3p were also detected in ectopic structures in the cytoplasm of spn2Δ cells (Figure 6, A, D, and E). To determine whether these structures contained more than one septin, we constructed spn2Δ strains expressing Spn1p-CFP and Spn4p-YFP or, Spn1p-CFP and Spn3p-GFP. In both strains, the septins colocalized in rings and ectopic structures (Figure 6, D and E), results consistent with the biochemical analysis of septin complexes in spn2Δ cells presented above that indicated Spn1p, Spn3p, and Spn4p remained together in the absence of Spn2p. Similarly, Spn1p-CFP, Spn2p-GFP, and Spn4p-GFP localized to rings and ectopic structures in spn3Δ cells and spn3Δ cells also displayed a milder cell separation phenotype (Figure 6, B, C, F, and G) than spn1Δ or spn4Δ cells (see Figure 5, C–H). Because we used mild fixation for one of our colocalization experiments (Figure 6D), we compared live and fixed cells directly (Figure 6, F and G, and unpublished data) and found no significant difference in septin localization patterns. Septin localization patterns appear to resist ethanol fixation very well. Lastly, we examined the localizations of Spn1p-CFP or Spn4p-YFP in cells lacking both Spn2p and Spn3p. In these strains, Spn1p-CFP and Spn4p-YFP did not form rings (Figure 6F and unpublished data). Consistent with this, the phenotype of spn2Δ spn3Δ cells is as severe as spn1Δ or spn4Δ cells (Figure 6F and unpublished data). Most cells contained ectopic septin structures and in some cells, Spn1p-CFP and Spn4p-YFP formed a ring of medially placed “dots” (Figure 6F and unpublished data). Combining this observation with our biochemical data, we conclude that a subcomplex of Spn1p-Spn4p is sufficient for formation of ectopic structures and localizing to the medial cortex, but at least one other septin is required for assembly of a ring structure.
Figure 6.
Interdependence of septins for ring formation. (A) spn3-GFP spn2Δ (KGY3211), (B) spn1-CFP spn3Δ (KGY730), or (C) spn2-GFP spn3Δ (KGY3169) cells were grown at 25°C, and GFP-tagged proteins were visualized in live cells. (D) spn1-CFP spn4-YFP spn2Δ (KGY761) cells were fixed in ethanol and both Spn1p-CFP and Spn4p-YFP were photographed separately, and then the images merged. (E) spn1-CFP spn3-GFP spn2Δ (KGY887) cells were grown at 25°C, and Spn1p-CFP and Spn3p-GFP were photographed separately from live cells, and the images were merged. (F and G) spn4-GFP spn3Δ cells (KGY5000) were grown at 25°C, and either visualized live in (F) or fixed in ethanol before visualization (G). (H) Spn4p-YFP was visualized in live spn4-YFP spn2Δ spn3Δ cells (KGY1178). The inset shows the middle cell rotated on its Z-axis. Arrows point to the arrangement of Spn4p-YFP “dots” in the medial region of the cell. Gray lines represent the outlines of individual cells. Scale bar, 5 μm.
S. pombe Septin Ring Formation Requires Two Steps
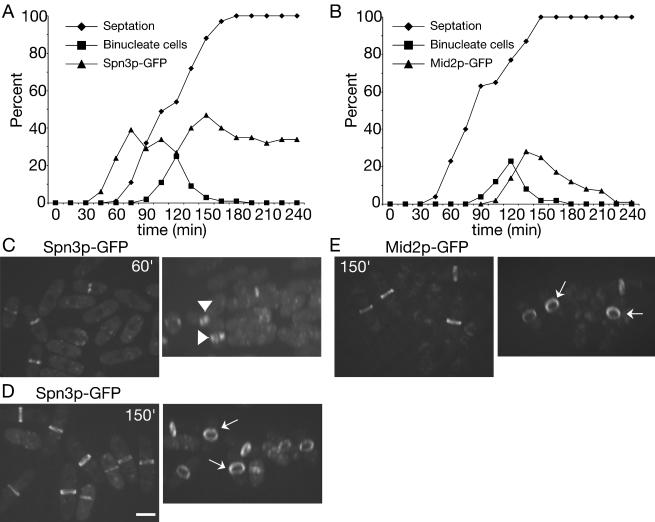
The results presented above suggest that S. pombe septins 1-4 are present in 8.5S complexes during all stages of the cell cycle except late mitosis when they are recruited to the medial cortex and organize into higher order oligomers that are visualized as rings. To determine whether septin organization into higher order structures requires entry into mitosis, we examined the ability of synchronized cultures of cdc16-116 cells to form septin rings when shifted to their nonpermissive temperature. Cdc16p is a component of the bipartite GAP for the small GTPase, Spg1p, whose activation triggers the septation initiation network (SIN; Schmidt et al., 1997; Furge et al., 1998). The SIN induces actin ring constriction and septum formation normally as cells exit mitosis (McCollum and Gould, 2001). When Cdc16p function is abrogated during interphase, septation can occur before mitosis (Minet et al., 1979), and this allowed us to ask whether septin rings could form during interphase. cdc16-116 spn3-GFP cells synchronized in early G2 phase were obtained by centrifugal elutriation and shifted to 36°C. Samples were collected periodically to examine nuclear division, septum formation, and formation of Spn3p-GFP rings (Figure 7A). At 60 min after shift to 36°C, Spn3p-GFP had accumulated in the medial region of cells and septa began to form in a population of interphase cells. However, close examination of these cells revealed that Spn3p-GFP staining was more diffuse than in wild-type cells and extended across the entire septum in a disk rather than being restricted to the cortex (Figure 7C). It was never visualized as a split ring in these cells (Figure 7C). In contrast, Spn3p-GFP rings were tight, focused, and cortically restricted in cells that had passed into and through mitosis before septum formation; in these cells split rings were easily detected (Figure 7D). We also examined the recruitment of Mid2p to the medial cortex in cdc16-116 cells at restrictive temperature. Consistent with the cell cycle regulation of Mid2p abundance and localization observed previously (Tasto et al., 2003), Mid2p rings were detected only in cells that had passed through mitosis before septum formation (Figure 7, B and E). Further, the appearance of Mid2p at the cortex coincided with the assembly of organized septin rings (compare 150 min time points; Figure 7, D and E). These findings indicate that ring formation and septation can drive recruitment of septin complexes to the medial region of the cell but another factor(s) available only in mitosis, such as Mid2p, alone or in combination with other factors is required for proper septin ring organization.
Figure 7.
Septin recruitment to the cortex, but not stable ring formation, can be induced during interphase. (A) spn3-GFP cdc16-116 (KGY738) and (B) mid2-GFP cdc16-116 (KGY741) cells were grown to midlog phase at 25°C, synchronized in early G2 phase by centrifugal elutriation, and then shifted to 36°C. Samples of cells were collected every 15 min. One portion of each sample was imaged immediately to determine the percentage of septated cells and the percentage of cells with Spn3p-GFP and Mid2p-GFP medial localization, respectively. Another portion was fixed with ethanol immediately and then stained with DAPI to determine the percentage of binucleate cells. (C and D) Spn3p-GFP localization in live cells from (C) 60 min or (D) 150 min time points (left panels of each). Images in the left panels were also rotated in the Z-axis to better reveal septin ring organization (right panels). Short arrows point to disorganized septin rings at 60 min. Longer arrows point to organized rings at 150 min. (E) Mid2p-GFP localization in live cells from 150 min time point. Cells are also rotated in the Z-axis to better show Mid2p ring organization. Arrows point to organized Mid2p rings. Scale bar, 5 μm.
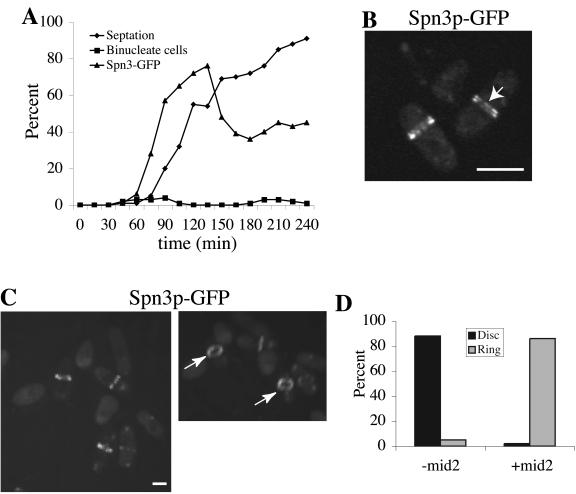
To determine whether Mid2p is the sole additional factor required for septin ring organization in interphase cdc16-116 cells, we mildly overproduced HA-Mid2p from the nmt41 promoter before selecting G2 phase cells by centrifugal elutriation. In this case, Spn3p-GFP was recruited to the medial region before mitosis as before (Figure 8A) and it was now assembled into highly organized ring structures that were easily visualized as split rings once septa had formed (Figure 8, B and C). Virtually no diffuse disk structures were observed (Figure 8, B–D). We conclude from this experiment that Mid2p is solely responsible directly or indirectly for regulating septin ring coalesence in S. pombe. Another note-worthy consequence of mild Mid2p overexpression was the relative persistence of septin rings and the inhibition of mitotic progression, as determined by monitoring the formation of binucleates (Figure 8A). This result is consistent with our previous results, indicating that prolonged expression of Mid2p stabilizes septin ring structures and influences cell cycle progression (Tasto et al., 2003).
Figure 8.
Coalescence of septin rings can be induced during interphase by mid2+ expression. (A) spn3-GFP cdc16-116 (KGY738) cells were transformed with pREP41-HA-mid2+. Transformants obtained in the presence of thiamine in the medium were then grown to midlog phase in the absence of thiamine at 25°C for 15 h. They were then synchronized in early G2 phase by centrifugal elutriation and immediately shifted to 36°C. Samples of cells were collected every 15 min. One portion of each sample was imaged immediately to determine the percentage of septated cells and the percentage of cells with Spn3p-GFP medial localization. Another portion was fixed with ethanol and stained with DAPI to determine the percentage of binucleate cells. (B and C) Spn3p-GFP localization in live cells at the 75-min time point. In B, cells imaged with a single Z-series stack (0.49 μm) are shown to provide the best visualization of split septin rings as indicated by the arrow. In C, the image in the left panel represents a 3D reconstruction of Z-series stacks. This image was rotated in the Z-axis (right panel) to illustrate the cortical restriction of the septin rings. The arrows point to organized septin rings. (C) Quantitation of disorganized septin rings (disk) and organized septin rings (ring) in interphase cdc16-116 cells in the presence or absence of Mid2p. Percentages were determined by visual inspection of Z-series stacks of live cells that contained medially placed septin structures from the experiment described in Figure 7A, 75-min time point, 93 cells examined (-mid2) and from the experiment described in part A of this figure, 75-min time point, 88 cells examined (+mid2). Scale bar, 5 μm.
DISCUSSION
In this study, we have determined the basic organization of S. pombe septin complexes that are used to build septin rings during cytokinesis and defined two discrete steps in septin ring assembly. Because the four septins that participate in S. pombe cell division are not essential for viability, we were able to assess the interdependence and function of each septin in the absence of any one or combination of the others. The model of septin complex organization arising from these studies indicates that Spn1-4p form an almost linear array of a specific order that is held together by a series of interdependent associations (Figure 4A). Further, different subunits within this complex are not equivalent in their functions. Our results have implications for understanding the nature of septin complexes and their properties in other eukaryotes.
First, our data argue that each septin does not possess the intrinsic ability to localize to its site of action or to form ring structures, a possibility raised by the finding that an individual septin subunit (Xenopus laevis Sept2) can polymerize into filaments in vitro (Mendoza et al., 2002). Not only do our findings indicate that individual septin subunits do not normally exist on their own in yeast (in S. pombe, the minimal unit comprises 8 polypeptides), our data also indicate that ring formation in S. pombe requires a minimum of three complexed septins, either Spn3p-Spn4p-Spn1p or Spn4p-Spn1p-Spn2p. That the complexes of three septins are sufficient to form ring structures explains why the absence of spn2+ or spn3+ generates a mild cell separation defect relative to the deletion of spn4+ or spn1+ or both spn2+ and spn3+.
Interestingly, our biochemical and cell biological analyses of various septin deletion strains indicate that the Spn4p-Spn1p subcomplex but not the Spn3p-Spn4p or Spn1p-Spn2p subcomplexes is able to accumulate as dot structures in the medial region of cells. Although a conserved polybasic region within septins binds phosphatidyl inositol biphosphate in vitro and has been implicated in septin localization (Zhang et al., 1999; Casamayor and Snyder, 2003), our results suggest that the Spn1p-Spn4p subcomplex contains the targeting information for the complex and it will be interesting in the future to illuminate its nature. We also observed that subcomplexes of two (Spn4p-Spn1p) or three (Spn3p-Spn4p-Spn1p or Spn4p-Spn1p-Spn2p) septins formed ectopic structures that were not observed in wild-type cells expressing the full complement of the four cytokinetic septins. This raises the possibility that subunits on the end of the array (Spn2p or Spn3p) might normally act in interphase to prevent spurious assembly into aggregates or higher order structures. Purified septins have been observed to polymerize into filaments in vitro (Field et al., 1996; Frazier et al., 1998; Kinoshita et al., 2002; Mendoza et al., 2002). If the ectopic structures observed in vivo are due to septin filament formation rather than aggregation, our observations may be relevant to the findings that individual septins and complexes of septins display differential abilities to polymerize in vitro (reviewed in Mitchison and Field, 2002). Because ectopic structures were only observed with the particular subcomplexes described above, the outcome of in vitro studies might depend on which septin or septin subcomplex is chosen for examination. On that note, our model of septin organization and function suggests that the integrity of the complex as a whole should be examined as part of any structure/function analysis performed on individual subunits. For example, any mutation of either Spn2p (Cdc10p) or Spn3p (Cdc11p) that disturbs its interaction with Spn1p (Cdc3p) or Spn4p (Cdc12p), respectively, would be expected to negatively influence its ability to localize appropriately but not necessarily because an intrinsic targeting domain was disturbed.
Several observations suggest that the organization of septins we have found in S. pombe will be relevant to other septin complexes, at least to those found in S. cerevisiae. First, S. cerevisiae cells lacking Cdc10p or Cdc11p (Spn2p and Spn3p homologues, respectively) contain complexes of the other three septins (Frazier et al., 1998). Second, such complexes retained some function because the cells divided albeit with reduced efficiency (Frazier et al., 1998), a situation similar to what is observed in spn2Δ and spn3Δ cells. Third, subsets of S. cerevisiae proteins produced in E. coli comparable to those we have found associate with one another in S. pombe were able to complex with one another and form filamentous structures (Versele et al., 2004; Versele and Thorner, 2004). Lastly, two-hybrid results testing S. cerevisiae septin interactions support our working models (Figure 4A), although they should be interpreted with caution because of the strong possibility of bridging interactions. These analyses suggest that Cdc11p (Spn3p homolog) interacts with Cdc12p (Spn4p homolog; Lee et al., 2002; Casamayor and Snyder, 2003), Cdc12p interacts strongly with Cdc3p (Spn1p homolog) (Lee et al., 2002), and Cdc3p interacts with Cdc10p (Spn2p homolog; Lee et al., 2002), and there is an absence of strong interaction between Cdc3p and Cdc11p (Lee et al., 2002; Casamayor and Snyder, 2003).
Precocious activation of the septation initiation network (SIN) induced septins to concentrate at the position of the actomyosin ring in interphase cells. However these septin structures were not organized properly. Indeed, they were very reminiscent of septin structures formed in cells lacking Mid2p (Berlin et al., 2003; Tasto et al., 2003) and in fact Mid2p-GFP was not visualized at rings induced by activation of the SIN during interphase. Thus, we conclude that septin accumulation at the medial region of the cell and coalescence into a ring structure are genetically separable events. The first requires an event(s) that can be initiated by actomyosin ring formation and SIN activity but is independent of mitotic entry. By supplying Mid2p artificially to interphase cdc16-116 cells, coalescence into ring structures was induced. This result indicates that Mid2p, directly or indirectly, is solely responsible for septin ring coalescence. The two-step assembly of S. pombe septin rings described here may be analogous to the situation in S. cerevisiae in which septin rings in unbudded cells are relatively unstable and become stabilized as cells proceed to bud (Caviston et al., 2003; Dobbelaere et al., 2003). Mid2p has a homolog in S. cerevisiae, Bud4p (Sanders and Herskowitz, 1996), which is similarly cell cycle regulated, but it is not known whether Bud4p participates in septin ring organization.
An established regulatory step for S. cerevisiae septin ring formation is septin phosphorylation, and several protein kinases have been implicated in septin ring dynamics including the GIN4 family, Cdk1p, and Cla4p (Cvrckova et al., 1995; Longtine et al., 1998; Weiss et al., 2000; Mortensen et al., 2002; Tang and Reed, 2002; Dobbelaere et al., 2003; Schmidt et al., 2003; Versele and Thorner, 2004). Although the S. pombe homologues of the GIN4 kinases, Cdr1p and Cdr2p, do not influence septin ring organization (Morrell et al., 2004), it will be interesting to determine whether S. pombe septin ring assembly is regulated by phosphorylation and whether Mid2p acts in concert with protein kinases to control this process. Another factor implicated in the organization of S. cerevisiae septin rings is a fifth septin, Shs1p/Sep7 (Mortensen et al., 2002; Dobbelaere et al., 2003). S. pombe does not have an ortholog of Shs1p/Sep7 and we did not detect a fifth septin copurifying with Spn1-4p-TAP complexes. The S. pombe genome does encode an additional three septins but their expression is restricted to meiosis (Mata et al., 2002; Rustici et al., 2004). Thus, it is unclear whether the role of Shs1p/Sep7 in S. cerevisiae septin ring organization will extend to other organisms.
As both recombinant and purified septins can polymerize into filaments in vitro (Field et al., 1996; Frazier et al., 1998; Kinoshita et al., 2002; Mendoza et al., 2002; Versele and Thorner, 2004), it is interesting to consider how the complex described here is assembled into such higher order oligomers. It will be informative in future studies to link the ability of particular septin complexes to form filamentous structures in vitro with the in vivo requirements of ring formation we have established here.
Supplementary Material
Acknowledgments
We thank Joseph Tasto for numerous strain constructions, John Pringle for spn2 and spn3 deletion strains and for sharing unpublished information, and Tony Hunter, Dan McCollum, and John Pringle for stimulating discussions. J.L.J. was supported by National Institutes of Health (NIH) grants GM64779 and HL68744. A.J.L. is supported by NIH grants GM64779, HL68744, NS43952, ES11993, and CA098131. Work in the Gould lab was supported by the Howard Hughes Medical Institute, of which K.L.G. is an Investigator.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–07–0640. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–07–0640.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bahler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., 3rd, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Berlin, A., Paoletti, A., and Chang, F. (2003). Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 160, 1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C.G., Ohi, R., Mehta, S., O'Toole, E.T., Winey, M., Clark, T.A., Sugnet, C.W., Ares, M., Jr., and Gould, K.L. (2002). Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor, A. and Snyder, M. (2003). Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol. Cell. Biol. 23, 2762-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J.P., Longtine, M., Pringle, J.R., and Bi, E. (2003). The Role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., De Virgilio, C., Manser, E., Pringle, J.R., and Nasmyth, K. (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9, 1817-1830. [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M.S., Hallberg, R.L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345-357. [DOI] [PubMed] [Google Scholar]
- Faty, M., Fink, M., and Barral, Y. (2002). Septins: a ring to part mother and daughter. Curr. Genet. 41, 123-131. [DOI] [PubMed] [Google Scholar]
- Field, C.M., al-Awar, O., Rosenblatt, J., Wong, M.L., Alberts, B., and Mitchison, T.J. (1996). A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C.M. and Alberts, B.M. (1995). Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 131, 165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C.M. and Kellogg, D. (1999). Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 9, 387-394. [DOI] [PubMed] [Google Scholar]
- Frazier, J.A., Wong, M.L., Longtine, M.S., Pringle, J.R., Mann, M., Mitchison, T.J., and Field, C. (1998). Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge, K.A., Wong, K., Armstrong, J., Balasubramanian, M., and Albright, C.F. (1998). Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8, 947-954. [DOI] [PubMed] [Google Scholar]
- Gale, C., Gerami-Nejad, M., McClellan, M., Vandoninck, S., Longtine, M.S., and Berman, J. (2001). Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12, 3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A.S., Pringle, J.R., and Lew, D.J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681-689. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H. (1971). Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69, 265-276. [DOI] [PubMed] [Google Scholar]
- Hsu, S.C., Hazuka, C.D., Roth, R., Foletti, D.L., Heuser, J., and Scheller, R.H. (1998). Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron 20, 1111-1122. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartmann, B., and Roth, D. (2001). Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J. Cell Sci. 114, 839-844. [DOI] [PubMed] [Google Scholar]
- Keeney, J.B. and Boeke, J.D. (1994). Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics 136, 849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M., Field, C.M., Coughlin, M.L., Straight, A.F., and Mitchison, T.J. (2002). Self- and actin-templated assembly of Mammalian septins. Dev. Cell 3, 791-802. [DOI] [PubMed] [Google Scholar]
- Lee, P.R. et al. (2002). Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6906-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., Fares, H., and Pringle, J.R. (1998). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. et al. (2002). Mammalian septins nomenclature. Mol. Biol. Cell 13, 4111-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., Lyne, R., Burns, G., and Bahler, J. (2002). The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143-147. [DOI] [PubMed] [Google Scholar]
- McCollum, D. and Gould, K.L. (2001). Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11, 89-95. [DOI] [PubMed] [Google Scholar]
- McDonald, W.H., Ohi, R., Smelkova, N., Frendewey, D., and Gould, K.L. (1999). Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19, 5352-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, M., Hyman, A.A., and Glotzer, M. (2002). GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 12, 1858-1863. [DOI] [PubMed] [Google Scholar]
- Minet, M., Nurse, P., Thuriaux, P., and Mitchison, J.M. (1979). Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 137, 440-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino, A., Tanaka, K., Kamei, T., Umikawa, M., Fujiwara, T., and Takai, Y. (1998). Shs1p: a novel member of septin that interacts with spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 251, 732-736. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J. and Field, C.M. (2002). Cytoskeleton: what does GTP do for septins? Curr. Biol. 12, R788-R790. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Morrell, J.L., Nichols, C.B., and Gould, K.L. (2004). The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J. Cell Sci. 117, 5293-5302. [DOI] [PubMed] [Google Scholar]
- Mortensen, E.M., McDonald, H., Yates, J., 3rd, and Kellogg, D.R. (2002). Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13, 2091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Savoian, M.S., Mitchison, T.J., and Field, C.M. (2000). Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 150, 539-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, M.D., Link, A.J., Ren, L., Jennings, J.L., McDonald, W.H., and Gould, K.L. (2002). Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22, 2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice, H.L. (1992). High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 20, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici, G. et al. (2004). Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 36, 809-817. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., and Herskowitz, I. (1996). The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J. Cell Biol. 134, 413-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S.L., Jennings, J., Canutescu, A., Link, A.J., and Weil, P.A. (2002). Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22, 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M., Varma, A., Drgon, T., Bowers, B., and Cabib, E. (2003). Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell 14, 2128-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S., Sohrmann, M., Hofmann, K., Woollard, A., and Simanis, V. (1997). The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11, 1519-1534. [DOI] [PubMed] [Google Scholar]
- Sheffield, P.J., Oliver, C.J., Kremer, B.E., Sheng, S., Shao, Z., and Macara, I.G. (2003). Borg/Septin interactions and the assembly of Mammalian septin heterodimers, trimers, and filaments. J. Biol. Chem. 278, 3483-3488. [DOI] [PubMed] [Google Scholar]
- Tang, C.S. and Reed, S.I. (2002). Phosphorylation of the septin cdc3 in g1 by the cdc28 kinase is essential for efficient septin ring disassembly. Cell Cycle 1, 42-49. [PubMed] [Google Scholar]
- Tasto, J.J., Carnahan, R.H., Hayes McDonald, W., and Gould, K.L. (2001). Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18, 657-662. [DOI] [PubMed] [Google Scholar]
- Tasto, J.J., Morrell, J.L., and Gould, K.L. (2003). An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., Gullbrand, B., Shulewitz, M.J., Cid, V.J., Bahmanyar, S., Chen, R.E., Barth, P., Alber, T., and Thorner, J. (2004). Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 4568-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M. and Thorner, J. (2004). Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164, 701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu, A.M., Gerber, S.A., Gygi, S.P., Field, C.M., and Mitchison, T.J. (2004). The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J. Biol. Chem. 279, 3111-3118. [DOI] [PubMed] [Google Scholar]
- Weiss, E.L., Bishop, A.C., Shokat, K.M., and Drubin, D.G. (2000). Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2, 677-685. [DOI] [PubMed] [Google Scholar]
- Wood, V. et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871-880. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Kong, C., Xie, H., McPherson, P.S., Grinstein, S., and Trimble, W.S. (1999). Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 9, 1458-1467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.