Abstract
Genetically encoded tags are of fundamental importance for live cell imaging. We show that small tetracysteine (TetCys) tags can be highly advantageous for the functionality of the host protein compared with large fluorescent protein tags. One to three concatenated small TetCys tags as well as the large green fluorescent protein (GFP) were fused by integrative epitope tagging to the C terminus of β-tubulin (Tub2) in the budding yeast Saccharomyces cerevisiae. The increasing tag size correlated with functional interference to the host protein. Tub2 tagged with either 1×TetCys (10 amino acids [aa]) or 2×TetCys (20 aa) was able to substitute Tub2 in haploid cells. In contrast, C-terminal tagging of Tub2 with 3×TetCys (29 aa) or with GFP (244 aa) resulted in nonviable haploid cells. Cells expressing Tub2-1×TetCys or Tub2-2×TetCys were stained with FlAsH, which selectively binds to the TetCys-tag. The stained cells displayed dynamic FlAsH-labeled microtubules and low cellular background fluorescence. The presented approach to tag open reading frames (ORFs) at their native loci with very small TetCys-tags and the subsequent visualization of the tagged proteins in vivo can be extended in principle to any ORF in S. cerevisiae.
INTRODUCTION
The visualization and quantification of specifically labeled proteins in living cells has greatly facilitated our understanding of cellular networks. The genetic fusion of fluorescent proteins (FPs) to a host protein proved to be a tremendously valuable tool in cell biology. The molecular mass of the currently used FPs, with green fluorescent protein (GFP) being the most prominent exponent, is in the range of 25–30 kDa (Tsien, 1998; Verkhusha and Lukyanov, 2004). Hence, the fused FP is frequently of the same size or even larger than the host protein. The fused FP potentially interferes with the function and/or localization of the host protein because of its large size.
In the budding yeast Saccharomyces cerevisiae such tag-induced problems are encountered with GFP fusions to tubulin. Yeast has four tubulin genes: two α-tubulins (TUB1 and TUB3), one β-tubulin (TUB2), and one γ-tubulin (TUB4). Fusions between GFP and the carboxy terminus of Tub1 or Tub3 can be incorporated into microtubules but fail to complement the tub1 or the tub3 null mutant, respectively (Carminati and Stearns, 1997). TUB2 chromosomally tagged with the sequence coding for GFP results in nonviable haploid cells. Most studies analyzing the yeast microtubule cytoskeleton have been accomplished by fusing GFP to the N terminus of the α-tubulins Tub1 or Tub3 (Carminati and Stearns, 1997; Straight et al., 1997; Maddox et al., 2000). These GFP-fusion proteins were generally coexpressed with the untagged protein. Hence, in this case, the coexpressed untagged α-tubulin possibly conceals subtle but functionally important constraints of the fusion protein. Therefore, smaller fusion tags might be a possibility to quantify and to avoid FP-induced functional constrains to a host protein.
With the biarsenical-tetracysteine system, an ingenious method for in vivo labeling of proteins was recently introduced that promised to overcome the size-related problems of FP-fusions (Griffin et al., 1998). This method is based on the formation of a stable complex between a synthetic fluorescent biarsenical compound, such as the fluorescein derivative fluorescein arsenical helix binder (FlAsH) and a TetCys motif consisting of six amino acids (e.g., Cys-Cys-Pro-Gly-Cys-Cys). This small tag can be fused genetically to a host protein. For in vivo labeling, FlAsH complexed with two 1,2-ethanedithiol (EDT) molecules is administered in the presence of an excess of 1,2-dithiols such as EDT. TetCys-tagged proteins coexpressed from a plasmid have been successfully visualized in living mammalian cells by using fluorescence microscopy (Griffin et al., 1998; Gaietta et al., 2002). In budding yeast, FlAsH-EDT2 has been used to assay the efficiency of targeted gene transfer (Rice et al., 2001a,b).
Here, we establish the biarsenical-tetracysteine system in S. cerevisiae for live cell imaging. Chromosomal genes were tagged by integrative epitope tagging with consecutive Tet-Cys motifs. We analyzed the tag size-related effects on the functionality of the host protein Tub2.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Growth and manipulation of yeast was carried out according to standard procedures (Gietz et al., 1992; Sherman, 2002). Cells were routinely grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or synthetic complete medium (SC) with 2% glucose or 2% galactose or 3% glycerol (Sherman, 2002). The wild-type strains BY4741 and BY4743 were obtained from Euroscarf (Frankfurt, Germany).
Plasmid Construction
The plasmids pU1TetCys, pU2TetCys, pU3TetCys, and pUGFP are modifications of the plasmid pU6H3VSV (De Antoni and Gallwitz, 2000). The latter vector had been created for C-terminal gene tagging by homologous recombination in yeast. We have replaced the sequence for the 6His-3VSV epitope by either the sequence coding for one, two, or three consecutive TetCys motifs (TetCys: Cys-Cys-Pro-Gly-Cys-Cys) or by the GFP cDNA. A short linker sequence was included between the tag and the host protein as well as between the consecutive TetCys motifs.
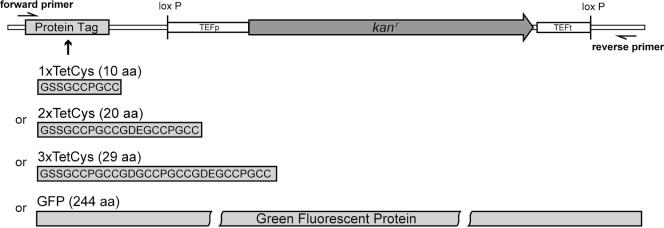
For pU1TetCys, the 6His-3VSV sequence was excised from pU6H3VSV with BsiWI and SalI and replaced by the sequence CGT ACG GGA TCC TCT GGA TGT TGT CCT GGT TGT TGT TAA TGA GTC GAC. For pU2TetCys, the sequence CGT ACG GGA TCC TCT GGA TGT TGT CCT GGT TGT TGT GGT GAC GAA GGA TGC TGC CCA GGT TGT TGT TAA TGA GTC GAC was inserted in the BsiWI and SalI sites. For pU3TetCys, the sequence CGT ACG GGA TCC TCT GGA TGT TGT CCT GGT TGT TGT GGT GAT GGT TGT TGT CCA GGT TGT TGT GGT GAC GAA GGA TGC TGC CCA GGT TGT TGT TAA TGA GTC GAC was inserted in the BsiW1 and SalI sites. For the creation of pUGFP, the coding sequence of GFP was amplified by polymerase chain reaction (PCR) from the plasmid pVT100U-mtGFP (Westermann and Neupert, 2000) and inserted in the BamHI and SalI sites of pU6H3VSV, replacing the 6His-3VSV sequence. For the amplification of the GFP coding sequence, the forward primer CA CGT GGA TCC TCT GGA TGT TGT CCT AGT AAA GGA GAA GAA CTT TTC and the reverse primer GG GTT GTC GAC TCA TTA TTT GTA TAG TTC ATC CAT GCC A were used. For further details of the new tagging vectors, see Figure 1.
Figure 1.
Schematic representation of the new modules in the plasmids pU1TetCys, pU2TetCys, pU3TetCys, and pUGFP. The four plasmids are identical except for the sequences of the epitopes. Therefore, the same set of oligonucleotides (forward primer and reverse primer) can be used to tag an ORF with any of these epitopes.
To produce the PCR product required for epitope tagging, the same set of forward and reverse primers can be used on the four tagging vectors as templates. The forward primer is a chimeric primer of ∼65 nucleotides. Its first ∼44 base pairs are homologous to the 3′ end of the gene of interest, omitting the stop codon, followed by 21 bases (GGATCCTCTGGATGTTGTCCT) complementary to the vector sequence. The reverse primer is ∼65 nucleotides in length. Its first ∼44 nucleotides are derived from a sequence 100- to 200-base pairs downstream of the gene of interest. The remaining 21 nucleotides (ACTATAGGGAGACCGGCAGAT) anneal to the vector.
FlAsH Labeling
Yeast cells expressing recombinant proteins with a TetCys motif as a tag were grown until logarithmic growth phase in SC-medium. One milliliter of the cell culture was harvested by centrifugation and resuspended in ∼20 μl of SC-medium. Of this cell suspension, 1 to 2 μl were used to inoculate 200 μl of SC-medium supplemented with 5 μM EDT (Sigma Chemie, Deisenhofen, Germany) and 2 μM or 4 μM FlAsH-EDT2 (Panvera, Madison, WI) in a single well of a 48-well plate (Nunc, Weisbaden, Germany). Two micromolar FlAsH-EDT2 was sufficient for staining the cells expressing Tub2-TetCys, whereas 4 μM FlAsH-EDT2 seemed to be optimal for cells expressing ATP1-TetCys. Further increases in the dye concentration had no apparent effect on the fluorescence intensity. EDT and FlAsH-EDT2 were added from freshly prepared stock solutions (1 mM EDT in HEPES-buffered saline and 0.2 mM FlAsH-EDT2 in Tris-HCl, pH 7.5, respectively). The amount of cells chosen for inoculation depended on the growth rate of the specific strain. For staining, the cells were incubated in the well plate overnight in a shaking incubator at 30°C in the dark. After overnight incubation, the 200-μl culture was in the logarithmic growth phase. Subsequently, the cells were harvested by centrifugation. To wash out nonspecifically bound FlAsH, the cells were resuspended in 1 ml of SC-medium with 50 μM EDT and incubated on a rotating wheel for ∼10 min in the dark. After a further washing step with SC-medium without EDT, the cells were resuspended in SC-medium. Due to the high affinity of the fluorophore to the TetCys motif, the binding of the label to the tagged protein is stable for several hours.
Growth Rates
Cells were grown in 48-well-plates (Nunc) at 30°C in a rotary shaker. They were inoculated from a preculture into a single well containing 300 μl of SC-medium to a starting OD595 of ∼0.02. Growth was monitored at an OD595 with a microplate spectrophotometer (μQuant; Bio-Tek Instruments, Winooski, VT).
Microscopy
A beam scanning microscope (TCS SP2; Leica Lasertechnik, Heidelberg, Germany) equipped with a 1.2 numerical aperture water immersion lens (Leica 63×, Planapo) was used for image acquisition. The fluorophores were excited at 488 nm and detection was at 500–560 nm. Photomultiplier tubes were used for the fluorescence detection. The imaging was performed at ambient temperature (∼22°C) for technical reasons and the cells were mounted in SC-medium with 1% low melting agarose to inhibit spatial movement of the cells. All confocal images are pseudocolored to obtain the best representation of the intensity distributions.
Miscellaneous
Integrative epitope tagging was performed as described previously (De Antoni and Gallwitz, 2000). Southern blot and tetrad analysis was carried out according to standard procedures (Sambrook and Russel, 2001; Sherman, 2002).
RESULTS
Tub2 Tagged at Its C Terminus with 1×TetCys or 2×TetCys Substitutes Tub2
In the yeast S. cerevisiae, PCR-based strategies to introduce epitope tags to chromosomal loci are well established (Wach et al., 1997; De Antoni and Gallwitz, 2000). We constructed new modules for the C-terminal tagging of chromosomal genes with sequences coding for the FlAsH binding motifs (Figure 1). Owing to the high affinity of FlAsH to the peptide sequence Cys-Cys-Pro-Gly-Cys-Cys, we chose this TetCys motif and multiple forms of it as a tag (Adams et al., 2002). In addition, a module with the coding sequence for GFP as C-terminal tag also was created. The three TetCys modules (1×TetCys, 2×TetCys, and 3×TetCys) and the GFP module were inserted in the tagging vector pU6H3VSV by replacing the sequence coding for the 6×His-3×VSV tag (De Antoni and Gallwitz, 2000). The new vectors differ only in the sequences coding for the epitopes but are otherwise identical. Thus, for epitope tagging, the same set of forward and reverse primers can be used on the four tagging vectors. In addition, the plasmids contain a loxP-kanMX-loxP cassette from the origin vector. This cassette can be used to excise the kanr marker gene from the genome by a transient expression of Cre-recombinase in the cells (De Antoni and Gallwitz, 2000). This feature allows tagging of different genes by using the kanr marker repeatedly.
The mentioned modules were used to tag the TUB2 gene coding for β-tubulin. The sequences of the used forward and reverse primers are given in Table 1. Tub2 is an essential protein and it is the only β-tubulin isoform in S. cerevisiae. At first, we tagged Tub2 successfully in haploid yeast cells with the 1×TetCys motif (10 aa) and the 2×TetCys motif (20 aa) by transforming the cells with the respective PCR product. Despite substantial efforts, we could not create haploid cells expressing Tub2 tagged with either the 3×TetCys-motif (29 aa) or GFP (244 aa), indicating that in the case of Tub2 the size of the C-terminal tag is crucial to maintain functionality.
Table 1.
Oligonucleotides used for construction and verification of tagged strains
| Name | Sequence |
|---|---|
| TUB2-for | 5′-GCTCCACAAAACCAAGATGAACCAATCACTGAGAATTTTGAAGGATCCTCTGGATGTTGTCCT-3′ |
| TUB2-rev | 5′-CCGCTGAAGTGAGGATCAAAAATCAATAGCTCGGAAGGTTAAAGGACTATAGGGAGACCGGCAGAT-3′ |
| TUB2-3′ out | 5′-ATTTGAACAACTTGGTCTCG-3′ |
| ATP1-for | 5′-GGCATCTCTAAAGAGTGCTACTGAATCATTTGTTGCCACTTTTGGATCCTCTGGATGTTGTCCT-3′ |
| ATP1-rev | 5′-GATTTTTCAGGGTTATTGTTGGGCTGCACTTTAAATTTAGTACTATAGGGAGACCGGCAGAT-3′ |
| ATP1-3′ out | 5′-CTGAAGCCGCTCCTCTACAAT-3′ |
| kanB | 5′-CTGCAGCGAGGAGCCGTAAT-3′ |
| pU-R0 | 5′-TATTCTGGGCCTCCATGTC-3′ |
Tub2 C-terminally Tagged with More Than 29 Amino Acids Does Not Functionally Substitute Tub2 But Is Still Incorporated into Microtubules When Coexpressed with Tub2
Next, we decided to create diploid cells heterozygous for the tagged TUB2 gene. In these heterozygous strains, one wild-type TUB2 allele was replaced by TUB2 tagged with either 1×TetCys, 2×TetCys, 3×TetCys, or GFP. All four heterozygous strains were viable. In each case, the proper integration of the modules at the desired locus was verified by PCR on genomic DNA by using the primers TUB2-3′out and kanB (Table 1). The resulting PCR product was sequenced using the primer pU-R0. The number of inserted modules was determined by Southern blotting. In all analyzed strains that were correctly tagged according to PCR analysis, only a single module insertion was detected (Figure 2), indicating that the tagging system works properly.
Figure 2.

Tagging of TUB2 results in unique integration of modules into the genome. For the Southern blot, genomic DNA from diploid cells heterozygous for the tagged TUB2 gene was digested with AsnI and EcoRI, separated on a 1% agarose gel, and probed with a part of the kanMX-cassette. Lane 1, wild type; 2, strain expressing Tub2-1×TetCys; 3, strain expressing Tub2-2×TetCys; 4, strain expressing Tub2-3×TetCys; and 5, strain expressing Tub2-GFP. Expected fragment lengths are 1267 nucleotides (nt), 1297 nt, 1324 nt, and 1969 nt for lanes 2–5, respectively.
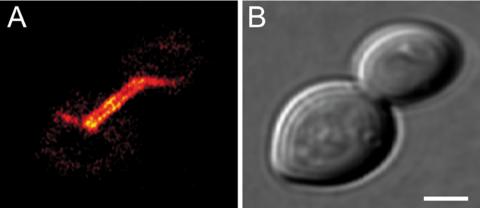
To verify that the tagged Tub2 is incorporated into microtubules, we decided to analyze the Tub2-GFP–expressing strain because GFP is the largest of the tags used. Confocal microscopy reveals that cells coexpressing Tub2 and Tub2-GFP incorporate Tub2-GFP into microtubules (Figure 3). We conclude that Tub2 can tolerate large C-terminal tags without losing its ability to be incorporated into microtubules in vivo. Tetrad dissection of diploid cells heterozygous for the tagged TUB2 resulted in no haploid viable progeny expressing Tub2-3×TetCys or Tub2-GFP (Table 2). This finding further supported our previous finding that already 29 aa, as introduced by the 3×TetCys tag, abolish the full functionality of the host protein Tub2. Hence Tub2 C-terminally tagged with >29 amino acids is incorporated into microtubules but does not functionally substitute the essential protein Tub2. To assess more subtle influences of the smaller 1×TetCy and 2×TetCys-tags, the heterozygous strains were further analyzed for their ability to form viable spores.
Figure 3.
Tub2-GFP is integrated into microtubules when coexpressed with Tub2. Diploid heterozygous cells coexpressing Tub2 and Tub2-GFP were grown to logarithmic growth phase and imaged with a confocal microscope. (A) GFP-fluorescence. Displayed is a single optical plane. (B) Corresponding bright field image. Bar, 2 μm.
Table 2.
Tetrad analysis
| Parental strain | Viable WT progeny [% of dissected WT spores] | Viable mutant progeny [% of dissected mutant spores] |
|---|---|---|
| TUB2/TUB2 | 96 | Not applicable |
| TUB2/TUB2-1×TetCys | 87 | 51 |
| TUB2/TUB2-2×TetCys | 95 | 3 |
| TUB2/TUB2-3×TetCys | 90 | 0 |
| TUB2/TUB2-GFP | 46 | 0 |
WT, wild type.
>30 tetrads from each strain analyzed.
Even Very Small C-terminal Tags Have an Influence on the Functionality of Tub2
The four strains heterozygous for the tagged TUB2 gene, as well as the isogenic diploid wild-type strain, were allowed to sporulate and the haploid progeny was analyzed for viability (Table 2). In the wild-type strain, >95% of all spores were viable and grew out to colonies. After sporulation of the heterozygous diploid cells carrying one TUB2-1×TetCys, TUB2-2×TetCys, or TUB2-3×TetCys allele, ∼90% of the resulting spores expressing native Tub2 grew to colonies (Table 2). Hence, the different TetCys-tags to Tub2 do not seem to strongly interfere with the sporulation process in cells heterozygous for this allele. For cells carrying one TUB2-GFP allele, the fraction of wild-type spores growing out to colonies was reduced to <50%. In cells heterozygous for the tagged TUB2, a large C-terminal GFP-tag, but not the 29 aa 3×TetCys-tag, deteriorates the efficiency of the sporulation process.
Next, we analyzed the colonies growing out of spores expressing a tagged Tub2 protein (Table 2). Fifty-one percent of the spores expressing Tub2-1×TetCys grew out to colonies. Thus, already the 10 aa introduced by the 1×Tet-Cys-tag reduced the capability of mutant spores to form colonies. Fusion of 10 more amino acids to the C terminus by the 2×TetCys-tag (20 aa) further reduced the fraction of surviving mutant spores to ∼3%. None of the spores (>60 each) expressing Tub2-3×TetCys or Tub2-GFP grew out to colonies. Hence, although C-terminal tags of 10 and 20 aa are tolerated for vegetative growth by the cells, these tags reduce the viability of the spores.
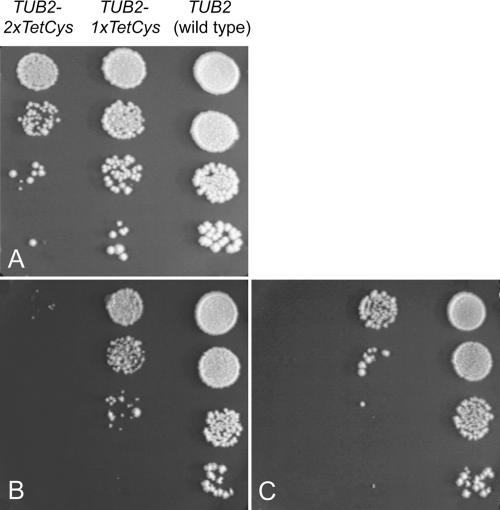
To further investigate the influence of the C-terminal tag size on cell viability, serial dilutions of haploid wild-type or mutant strains expressing either Tub2-1×TetCys or Tub2-2×TetCys were spotted onto plates and incubated at 25, 30, or 37°C (Figure 4). The mutant strain expressing Tub2-1×TetCys displayed a moderate growth defect at 30°C under these conditions, whereas the strain expressing Tub2-2×TetCys had a considerable growth defect. In both strains, the growth defects were increased at reduced as well as at elevated growth temperatures.
Figure 4.
Growth phenotypes. Haploid wild-type and mutant strains expressing Tub2, Tub2-1×TetCys, or Tub2-2×TetCys, respectively, were grown overnight in glucose-containing medium. Then, 10-fold serial dilutions were spotted onto YPD-plates. The plates were incubated for 5 d at different temperatures. (A) 30°C. (B) 25°C. (C) 37°C.
We conclude that already very small C-terminal tags to Tub2 have an immediate effect on the functionality of the protein, the constraints increasing with the length of the C-terminal tag.
In Vivo Labeling with FlAsH-EDT2
Because Tub2-GFP is not fully functional and is not tolerated by haploid cells, we decided to stain cells expressing Tub2-1×TetCys or Tub2-2×TetCys with FlAsH. It has been suggested previously that in budding yeast FlAsH-EDT2 does not have access to the interior of the cell until spheroblasts are formed by removal of the cell wall, thus making this method ineligible for yeast live cell imaging (Griffin et al., 2000). Here, we developed a protocol for labeling proteins in live and intact yeast cells. We concentrated on the labeling of haploid cells expressing Tub2-1×TetCys or Tub2-2×TetCys, because we reasoned that cells expressing only the tagged form of Tub2 would result in higher fluorophore densities after labeling. For efficient labeling, cells were grown overnight in a fluid medium with 2 μM FlAsH-EDT2 and 5 μM EDT. The cell density was chosen so that after overnight incubation, the cell culture was in the logarithmic growth phase. For imaging, the live cells were embedded in SC-medium with 1% low melting agarose. Cells expressing Tub2-1×TetCys (Figure 5, A and E) or Tub2-2×TetCys (Figure 5C) displayed microtubular structures after staining with FlAsH-EDT2. Using this novel staining procedure, the majority of cells displayed labeled microtubules (Figure 5, A and B). We encountered a remarkably low unspecific intracellular background with a favorable signal to noise ratio (Figure 5, C and D). In these cells, every β-tubulin protein had been C-terminally tagged and because a further increase of the FlAsH-EDT2 concentration did not increase the fluorescence intensity, it is likely that the vast majority of the tagged proteins had been labeled with FlAsH. To analyze whether the stained microtubules displayed a dynamic behavior, we used time-lapse confocal microscopy (Figure 5, E and F). The stained microtubules were found to be dynamic, displaying changes in localization as well as microtubule polymerization and depolymerization. However, because the viability of cells expressing Tub2-1×TetCys is slightly reduced (Figure 4), it may still be possible that the dynamical behavior is altered compared with the wild type. Furthermore, the reagents required for labeling might affect the cells. Therefore, we analyzed the influence of FlAsH-EDT2 and EDT on the growth rate of the cells.
Figure 5.
FlAsH-labeled microtubules in live S. cerevisiae are dynamic, and the cells display low cellular background. Shown are haploid yeast cells expressing Tub2-1×TetCys or Tub2-2×TetCys without additional β-tubulin. Yeast cells were incubated overnight with FlAsH-EDT2 and subsequently imaged with a scanning confocal microscope. (A) Fluorescence image of cells expressing Tub2-1×TetCys. The majority of cells have stained microtubules. (B) Corresponding bright field image to A. Bar, 5 μm. (C) Fluorescence image of a budding haploid cell expressing Tub2-2×TetCys after staining with FlAsH-EDT2 overlaid with a bright field image. Bar, 2 μm. (D) Fluorescence intensity profile along the line indicated in C. The arrows indicate the position of the cell wall. (E) Time-lapse series of a cell expressing Tub2-1×TetCys after staining with FlAsH-EDT2. Confocal images were taken at the indicated time points. Displayed are maximum intensity projections of optical sections. The arrow points to a depolymerizing microtubule. (F) Corresponding bright field image to E. Bar, 5 μm.
FlAsH-EDT2 and EDT Have Little Effect on Yeast Growth
We determined the growth rates of wild-type cells and mutant cells expressing Tub2-1×TetCys in growth media supplemented with FlAsH-EDT2 and EDT (Table 3). In synthetic complete medium, wild-type cells had a doubling rate of 97.9 ± 5.3 min, and cells expressing Tub2-1×TetCys a doubling rate of 133.2 ± 5.2 min, confirming the previously observed effect of Tub2-1×TetCys on cell viability (Figure 4). EDT supplemented to the growth medium without FlAsH-EDT2 had no significant influence on the growth rates. In contrast, FlAsH-EDT2 displayed some toxicity on the cells and reduced the growth rates to 114.1 ± 4.7 and 173.7 ± 8.8 min, respectively. EDT has been suggested to be an antidote to FlAsH (Griffin et al., 1998). We did not observe a significant effect of EDT to alleviate the FlAsH-EDT2 induced toxicity, although the statistical spread of the data is large enough to cover a minor antitoxic effect of EDT. We conclude that incubation of S. cerevisiae with FlAsH-EDT2 or FlAsH-EDT2 and EDT has a slight toxic effect on the cells but does not abolish growth.
Table 3.
Doubling times of the wild-type cells and cells expressing Tub2-1×TetCys
| Medium | Wild-Type doubling time [min] | TUB2-1×TetCys doubling time [min] |
|---|---|---|
| SC | 97.9±5.3 | 133.2±5.2 |
| SC + 5 μM EDT | 106.4±8.4 | 134.9±6.2 |
| SC + 2 μM FlAsH-EDT2 | 114.1±4.7 | 173.7±8.8 |
| SC + 2 μM FlAsH-EDT2 + 5 μM EDT | 125.6±4.1 | 160.6±9.3 |
Six independent experiments; errors are SDs.
Concatenated Multiple TetCys-Tags Do Not Increase Fluorescence Intensity
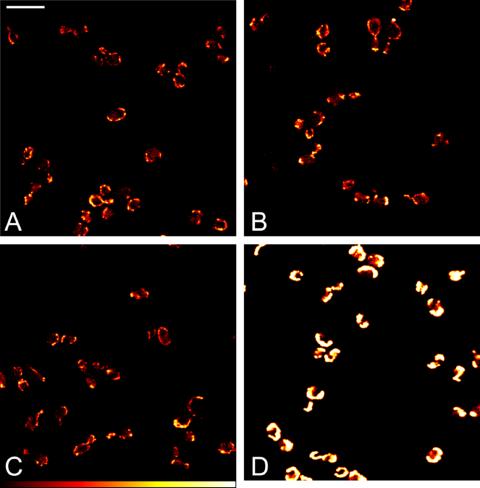
Next, we investigated the influence of multiple TetCys-tags on the fluorescence intensity of FlAsH-labeled cells. For this comparison, we decided to tag ATP1, coding for the α subunit of the mitochondrial F1F0 ATP synthase complex. Atp1 is essential for growth on nonfermentable carbon sources. For integrative epitope tagging, we used the oligonucleotides ATP1-for and ATP1-rev (Table 1). Haploid cells expressing Atp1 tagged with 1×TetCys, 2×TetCys, 3×TetCys, or GFP displayed normal growth on media containing glycerol as the only carbon source. Hence, we assume that the tagged protein is functional, irrespective of the tag. Labeling was performed with 4 μM FlAsH-EDT2. We found this concentration to be optimal for labeling of cells expressing TetCys-tagged Atp1 because a further increase of FlAsH-EDT2 did not enhance fluorescence intensity. Comparison of FlAsH-labeled cells expressing Atp1-1×TetCys, Atp1-2×TetCys, or Atp1-3×TetCys did not reveal noticeable differences in the initial fluorescence intensity of the labeled cells (Figure 6). We note, however, that the fluorescence seemed to be more resistant against photobleaching with increasing numbers of TetCys-tags fused to Atp1, which is likely due to mutual quenching of the neighboring fluorophores (Eggeling et al., 1999; Lakowicz, 1999). Using identical imaging conditions cells expressing Atp1-GFP displayed a stronger fluorescence intensity as well as less pronounced photobleaching than the FlAsH-labeled cells (Figure 6). The higher fluorescence intensity of the GFP-labeled Atp1 is at the expense of the tag size. So, it depends on the specific question, which labeling approach is the most appropriate.
Figure 6.
Comparison of fluorescence intensities. Haploid cells expressing Atp1-1×TetCys (A), Atp1-2×TetCys (B), or Atp1-3×Tet-Cys (C) were grown to the logarithmic growth phase, stained with FlAsH-EDT2, and imaged with a confocal microscope. Haploid cells expressing Atp1-GFP were imaged under identical imaging conditions (D). The same look-up table is used for all images. Shown are single optical sections. Bar, 10 μm.
DISCUSSION
The new tagging modules allow the generation of transcriptional fusions in their normal chromosomal loci under the endogenous promoter. By using integrative epitope tagging in haploid cells, the native ORF is replaced by its TetCys-tagged form. As a consequence, the potential functional artifacts possibly introduced by the tag to the host protein are not masked by additionally expressed native protein.
Heterozygous diploid cells readily incorporated tagged Tub2 into microtubules, irrespective of the size of the C-terminal tag. Similarly, GFP-tagged α-tubulin coexpressed with native α-tubulin has previously been used to study the dynamic behavior of yeast microtubules, giving important insight into microtubule turnover and dynamics (Carminati and Stearns, 1997; Straight et al., 1997; Maddox et al., 2000). However, although the GFP-tagged Tub2 is localized to microtubules when coexpressed with Tub2, we found Tub2-GFP to be functionally restricted. Haploid cells expressing Tub2-GFP from the endogenous promoter were nonviable. This agrees with the finding of Huh et al. (2003) who failed to generate a haploid strain expressing Tub2-GFP. Tub2 tagged with the 3×TetCys motif also did not complement the tub2 null mutant, although the 8 times smaller Tub2-3×TetCys was less disturbing to sporulation than Tub2-GFP in heterozygous cells. Hence, various cellular processes show different reactions, depending on the size of the tag to Tub2. We found that haploid cells expressing Tub2-2×Tet-Cys were viable. Therefore, the 9-aa size difference between the 2×TetCys-tag and the 3×TetCys-tag had a crucial impact on the functionality of the host protein. The C-terminal region of Tub2 seems to be located on the outside of the microtubule (Nogales et al., 1999). This region has been shown to be important for the binding of motor proteins to microtubules (Okada and Hirokawa, 2000). This prominent localization of the C terminus is a reasonable explanation for the observation that small C-terminal tags to Tub2 are better tolerated by the cells than large tags. Still, even the small 1×TetCys-tag apparently interferes with the full functionality of microtubules, because Tub2-1×TetCys-expressing cells have a reduced growth rate, which also might be reflected in the seemingly shorter microtubules in cells expressing Tub2-1×TetCys. It will require further analysis to determine to what extend the dynamical behavior of the microtubules is affected by the 1×TetCys-tag to Tub2.
In general, as in the case of microtubules, a large FP-tag may be especially interfering if the host protein is part of a highly ordered macromolecular complex that does not allow any extra space for a bulky tag. It is reasonable that small tags are also beneficial, for example, for mitochondrial proteins that must be in an unfolded linear conformation to pass through both the outer and inner membrane translocation channels (Truscott et al., 2003). Bulky FPs, which are known to be relatively resistant against unfolding, are likely to cause problems in this process. We assume therefore that a substantial number of proteins could be identified where a small tag would be functionally superior to a large and disturbing FP tag.
With the FlAsH staining procedure presented here, unspecific background fluorescence was almost negligible in haploid cells. This is a surprising finding because high unspecific background has been reported to be the most serious drawback of the method in mammalian cell lines (Griffin et al., 2000; Stroffekova et al., 2001). FlAsH seems to have the highest affinity for the sequence Cys-Cys-Pro-Gly-Cys-Cys (Adams et al., 2002). In the sequenced S. cerevisiae genome only one ORF (YKL131W) can be identified that encodes a protein containing this sequence. However, YKL131W is currently annotated as a hypothetical ORF so the expression level of the corresponding protein is unclear but likely to be low, or the ORF is even not transcribed at all (Dolinski et al., 2004). FlAsH also has been reported to bind, albeit with lower affinities, to other amino acid sequences, namely, to Cys-Cys-Xaa-Xaa-Cys-Cys or Cys-Cys-Xaa-Cys-Cys, where Xaa is a noncysteine amino acid (Adams et al., 2002). The S. cerevisiae genome contains another four ORFs coding for proteins with these motifs. All of them are low expressed proteins. Hence, yeast apparently contains only few endogenous proteins that potentially bind FlAsH. Together with the optimized staining procedure, this is the likely explanation for the low cellular background upon labeling with FlAsH-EDT2.
Albeit its clear advantages, the biarsenical-tetracysteine system has several limitations. Using the tagging modules presented here, it is straightforward to omit the current spacer between the host protein and the 1×TetCys-tag, resulting in the minimal tag size of six aa. Still, even such a minimal tag may interfere with the full functionality of the host protein. Because the addition of EDT and FlAsH-EDT2 reduced the growth rate of the cells by only ∼25%, toxicity of these compounds is not likely a major caveat of the system in S. cerevisiae. The quantum efficiencies and extinction coefficients of FlAsH and GFP are in the same range, whereas FlAsH is more prone to photobleaching (Adams et al., 2002). Therefore, it is not surprising that a direct comparison of cells expressing either Atp1-GFP or Atp1 tagged with 1×TetCys, 2×TetCys, or 3×TetCys resulted, under identical imaging conditions, in a higher fluorescence intensity in case of the GFP-tagged protein. Remarkably, concatenating multiple copies of the TetCys motif did not increase the fluorescence intensity, but the fluorescence seemed to be more resistant against photobleaching with increasing numbers of TetCys-tags. Similar observations have been made previously with multiple GFP-tags to the microtubule binding domain of the microtubule-associated protein E-MAP-115 (Faire et al., 1999). In that study, it was shown that concatenating of GFPs did not result in an increase in fluorescence intensity but lead to a reduction of photobleaching. It is a well know phenomenon that concatenation of fluorophores frequently leads to mutual quenching of their fluorescence emission (Eggeling et al., 1999; Lakowicz, 1999). This may prevent an increase in fluorescence intensity with increasing numbers of attached fluorophores and also might account for the observed photostabilization effect. It can be anticipated that it will be a challenging task to use the biarsenical-tetracysteine system in combination with weakly expressed proteins or for extended time-lapse studies. We expect that other fluorophores with higher quantum efficiencies and less sensitivity to photobleaching would help to overcome this potential restriction of the biarsenical-tetracysteine system.
ReAsH, another TetCys-motif binding fluorophore with a red shifted emission wavelength, has been shown to be useful for correlating live-cell fluorescence microscopy with electron microscopy in mammalian cell lines (Gaietta et al., 2002). Furthermore, it has been recently shown that ReAsH- or FlAsH-mediated chromophore-assisted light inactivation can be used to inactivate selected proteins within a cell (Marek and Davis, 2002; Tour et al., 2003). With the modules for epitope tagging presented here and the described methodology for live cell labeling, it can be anticipated that these concepts can be readily transferred to S. cerevisiae on a genome-wide scale to complement and extend GFP-based approaches.
In summary, we have shown that small TetCys-tags can be highly advantageous for the functionality of the host protein as compared with large fluorescent protein tags. FlAsH-labeled proteins can be used to investigate protein dynamics in time and space. We have substantially enlarged the scope of the biarsenical-tetracysteine system by creating the tools to tag and visualize proteins expressed in S. cerevisiae from their endogenous promoters on a genome-wide scale.
Acknowledgments
We thank Stefan W. Hell for funding this project and fruitful discussions. We thank Anna De Antoni for the donation of the plasmid pU6H3VSV. We thank Jaydev Jethwa for careful reading of the manuscript. We are grateful to Dieter Gallwitz for the use of laboratory infrastructure.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0454. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0454.
Abbreviations used: aa, amino acid; EDT, 1,2-ethanedithiol; FlAsH-EDT2, fluorescein arsenical helix binder, bis-EDT adduct (4′,5′-bis(1,3,2-dithioarsolan-2-yl)fluorescein); FP, fluorescent protein; GFP, green fluorescent protein; ORF, open reading frame; TetCys, tetracysteine.
References
- Adams, S.R., Campbell, R.E., Gross, L.A., Martin, B.R., Walkup, G.K., Yao, Y., Llopis, J., and Tsien, R.Y. (2002). New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124, 6063-6076. [DOI] [PubMed] [Google Scholar]
- Carminati, J.L., and Stearns, T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138, 629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni, A., and Gallwitz, D. (2000). A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246, 179-185. [DOI] [PubMed] [Google Scholar]
- Dolinski, K., et al. (2004). Saccharomyces Genome Database. http://www.yeastgenome.org/.
- Eggeling, C., Widengren, J., Rigler, R., and Seidel, C.A.M. (1999). Photostabilities of fluorescent dyes for single-molecule spectroscopy: mechanisms and experimental methods for estimating photobleaching in aqueous solution. In: Applied Fluorescence in Chemistry, Biology and Medicine, ed. W. Rettig, B. Strehmel, M. Schrader, and H. Seifert, Berlin: Springer, 193-240.
- Faire, K., Waterman-Storer, C.M., Gruber, D., Masson, D., Salmon, E.D., and Bulinski, J.C. (1999). E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J. Cell Sci. 112, 4243-4255. [DOI] [PubMed] [Google Scholar]
- Gaietta, G., Deerinck, T.J., Adams, S.R., Bouwer, J., Tour, O., Laird, D.W., Sosinsky, G.E., Tsien, R.Y., and Ellisman, M.H. (2002). Multicolor and electron microscopic imaging of connexin trafficking. Science 296, 503-507. [DOI] [PubMed] [Google Scholar]
- Gietz, D., Jean, A.S., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, B.A., Adams, S.R., Jones, J., and Tsien, R.Y. (2000). Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 327, 565-578. [DOI] [PubMed] [Google Scholar]
- Griffin, B.A., Adams, S.R., and Tsien, R.Y. (1998). Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269-272. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J.R. (1999). Principles of Fluorescence Spectroscopy, Amsterdam: Kluwer Academic Publishers.
- Maddox, P.S., Bloom, K.S., and Salmon, E.D. (2000). The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2, 36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek, K.W., and Davis, G.W. (2002). Transgenically encoded protein photo-inactivation (FIAsH-FALI): acute inactivation of synaptotagmin I. Neuron 36, 805-813. [DOI] [PubMed] [Google Scholar]
- Nogales, E., Whittaker, M., Milligan, R.A., and Downing, K.H. (1999). High-resolution model of the microtubule. Cell 96, 79-88. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and Hirokawa, N. (2000). Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA 97, 640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, M.C., Bruner, M., Czymmek, K., and Kmiec, E.B. (2001a). In vitro and in vivo nucleotide exchange directed by chimeric RNA/DNA oligonucleotides in Saccharomyces cerevisae. Mol. Microbiol. 40, 857-868. [DOI] [PubMed] [Google Scholar]
- Rice, M.C., Czymmek, K., and Kmiec, E.B. (2001b). The potential of nucleic acid repair in functional genomics. Nat. Biotechnol. 19, 321-326. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D.W. (2001). Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sherman, F. (2002). Getting started with yeast. Methods Enzymol. 350, 3-41. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Marshall, W.F., Sedat, J.W., and Murray, A.W. (1997). Mitosis in living budding yeast - anaphase A but no metaphase plate. Science 277, 574-578. [DOI] [PubMed] [Google Scholar]
- Stroffekova, K., Proenza, C., and Beam, K.G. (2001). The protein-labeling reagent FLASH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Arch. 442, 859-866. [DOI] [PubMed] [Google Scholar]
- Tour, O., Meijer, R.M., Zacharias, D.A., Adams, S.R., and Tsien, R.Y. (2003). Genetically targeted chromophore-assisted light inactivation. Nat. Biotechnol. 21, 1505-1508. [DOI] [PubMed] [Google Scholar]
- Truscott, K.N., Brandner, K., and Pfanner, N. (2003). Mechanisms of protein import into mitochondria. Curr. Biol. 13, R326-R337. [DOI] [PubMed] [Google Scholar]
- Tsien, R.Y. (1998). The green fluorescent protein. Annu. Rev. Biochem. 67, 509-544. [DOI] [PubMed] [Google Scholar]
- Verkhusha, V.V., and Lukyanov, K.A. (2004). The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 22, 289-296. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Albertisegui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous His3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065-1075. [DOI] [PubMed] [Google Scholar]
- Westermann, B., and Neupert, W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427. [DOI] [PubMed] [Google Scholar]