Abstract
Background: Malnutrition affects body growth, size, and composition of children. Yet, few functional biomarkers are known to be associated with childhood morphology.
Objective: This cross-sectional study examined associations of anthropometric indicators of height, musculature, and fat mass with plasma proteins by using proteomics in a population cohort of school-aged Nepalese children.
Methods: Height, weight, midupper arm circumference (MUAC), triceps and subscapular skinfolds, upper arm muscle area (AMA), and arm fat area (AFA) were assessed in 500 children 6–8 y of age. Height-for-age z scores (HAZs), weight-for-age z scores (WAZs), and body mass index–for-age z scores (BAZs) were derived from the WHO growth reference. Relative protein abundance was quantified by using tandem mass spectrometry. Protein-anthropometry associations were evaluated by linear mixed-effects models and identified as having a false discovery rate (q) <5%.
Results: Among 982 proteins, 1, 10, 14, and 17 proteins were associated with BAZ, HAZ, MUAC, and AMA, respectively (q < 0.05). Insulin-like growth factor (IGF)-I, 2 IGF-binding proteins, and carnosinase-1 were associated with both HAZ and AMA. Proteins involved in nutrient transport, activation of innate immunity, and bone mineralization were associated with HAZ. Several extracellular matrix proteins were positively associated with AMA alone. The proteomes of MUAC and AMA substantially overlapped, whereas no proteins were associated with AFA or triceps and subscapular skinfolds. Myosin light-chain kinase, possibly reflecting leakage from muscle, was inversely associated with BAZ. The proteome of WAZ was the largest (n = 33) and most comprehensive, including proteins involved in neural development and oxidative stress response, among others.
Conclusions: Plasma proteomics confirmed known biomarkers of childhood growth and revealed novel proteins associated with lean mass in chronically undernourished children. Identified proteins may serve as candidates for assessing growth and nutritional status of children in similar undernourished settings. The antenatal micronutrient supplementation trial yielding the study cohort of children was registered at clinicaltrials.gov as NCT00115271.
Keywords: plasma proteome, anthropometry, mass spectrometry, children, Nepal, insulin-like growth factor I, insulin-like growth factor–binding proteins
Introduction
Child growth restriction is the result of complex metabolic adjustments to acute or prolonged malnutrition (1). However, our understanding of the biological pathways underlying decelerated linear growth and reduced lean and fat mass remains inadequate. Although anthropometry is the conventional method for assessing growth because of its simplicity, quantitative nature, interpretability, widespread use, and low cost, it offers little biological insight. It may not be sensitive to changes in many aspects of nutritional status and fails to discern complex causes of growth deficit (2). We suggest that novel, valid, and reliable panels of biomarkers can be discovered by exploiting omics capabilities. These molecules may reflect nutritionally regulated biological pathways affecting growth, body composition, function, and development that may be suitable for public health application.
Plasma proteomics is the study of detecting, quantifying, and analyzing proteins present in the plasma (3). It may offer the potential to reveal biological pathways and discover novel protein markers of childhood growth and body composition. The unbiased (i.e., hypothesis-generating) approach of plasma proteomics can allow comprehensive and simultaneous analysis of many circulating and some tissue-leaked proteins reflecting anabolic or catabolic tissue metabolism (3). For example, the growth hormone–insulin-like growth factor (IGF)10-I axis has long been considered the main regulatory system of childhood growth (4). It has endocrine, autocrine, and paracrine growth-promoting effects in most tissues in the body and shows high sensitivity and specificity to indicators of nutritional status (5). However, many other growth factors and intracellular and extracellular matrix proteins are also critical to regulate cellular activity, providing structural and biochemical support for local tissue growth (6, 7). In addition to hormonal and local control, growth and body composition of growing children living in impoverished areas are likely to be affected by a complex interplay between nutrient metabolism and the immune and neurologic systems (8). Thus, the global discovery mode of plasma proteomics may help to advance our understanding of dynamic structural or regulatory mechanisms of tissue growth associated with nutritional or other environmental exposures.
We have assessed nutritional, health, and cognitive status of a population-based birth cohort of school-aged children in the rural plains of Nepal (9–12). In this region, growth restriction starts in fetal life (13) and persists through the preschool- and school-aged years, manifested by low z score distributions in height-for-age (HAZ), BMI-for-age (BAZ), and weight-for-age (WAZ) (10). As a substudy, we have applied quantitative proteomics to reveal plausible biological associations between circulating plasma protein abundance and the nutritional and health status of children. We have demonstrated predictive biomarkers of current micronutrient status and inflammation (14–17) and have identified proteins prospectively associated with cognitive function (18). The strength, stability and broad nature of associations suggest that the plasma proteome may have the potential to identify functional biomarkers of depressed childhood linear and ponderal growth as well as alterations in body composition. Therefore, the proteome may provide unique opportunities to reveal underlying biological mechanisms of malnutrition and poor growth.
In this study, we hypothesized that proteins associated with attained skeletal bone length, musculature, and subcutaneous fat deposition assessed by anthropometric measures and that indicators exist and can be detected and quantified in plasma via an untargeted proteomics approach. Our findings may offer novel insights into pathways and networks that affect growth and body composition and reveal biological links to risk factors and health consequences of child undernutrition in a rural South Asian setting.
Methods
Study population and design.
In 1999–2001, a population-based, randomized controlled trial of antenatal micronutrient supplementation was carried out to improve birth outcomes and infant survival in the Sarlahi District, a rural southeastern region of Nepal (NCT00115271) (13, 19). In 2006–2008, the surviving children born during the trial were followed up to examine the effects of the maternal intervention on growth, body composition, and metabolic health. Details of the child follow-up study and results have been previously reported (10, 11). Children in the present study comprised a stratified, random subset of the trial cohort (Supplemental Figure 1) (14). Among 4130 live-born infants during the maternal micronutrient supplementation trial, 3524 children were followed up at 6–8 y of age. Of 3305 children with available blood samples, 2130 (64%) met imposed inclusion requirements for this analysis of having adequate plasma sample volume and complete epidemiologic data from both the original trial and follow-up study to permit full exploration of admitted child records. We stratified data from eligible children by the original 5 maternal supplement allocation groups and ordered them by calendar date of blood draw in the field during the follow-up study. Two hundred children per supplementation group were systematically selected for micronutrient assessment (n = 1000). Their comparability to the larger group of children with respect to distributions of nutritional and health variables has been described (9). Of these, 50% (n = 500 samples), balanced across maternal intervention groups (n = 100 each) were randomly selected for proteomics analysis. All research activities were restricted to children whose parents provided informed consent. Institutional Review Boards at the Institute of Medicine of Tribhuvan University, Kathmandu, Nepal, and at Johns Hopkins University, Baltimore, Maryland, reviewed and approved the research protocols of both the original trial and follow-up study.
Field data collection and anthropometric measurements.
Standardized, home-based anthropometric measurements obtained on children have been described elsewhere (10). Briefly, standing height (in centimeters) was measured by using Harpenden portable stadiometers (Harpenden), weight (in kilograms) was measured with children lightly clothed by using an electronic scale (Model 881; Seca), and midupper arm circumference (MUAC) (in centimeters) was measured at the midpoint between the acromion and olecranon processes of the left upper arm with an insertion tape (20). Triceps skin fold (in millimeters) was measured at the midpoint of a back upper arm, and subscapular skin fold (in millimeters) was measured ∼2 inches (5.08 cm) below the lateral angle of the shoulder blade with Holtain precision calipers (Holtain Ltd). All measurements were repeated 3 times, and median values were used for analysis. HAZs, BAZs, and WAZs were calculated based on the WHO growth reference for children and young adults 5–19 y of age (21). Arm muscle area (AMA; in square centimeters) and arm fat area (AFA; in square centimeters) were calculated by using equations described by Stewart et al. (10). During another home visit near the time of anthropometric assessment, children were asked to fast overnight after which trained phlebotomists drew early-morning blood samples (11). At a field laboratory center, the samples were centrifuged, and plasma was extracted, aliquoted, and stored at −20°C in freezers. The frozen samples were shipped in liquid-nitrogen dry-vapor shippers to the Center for Human Nutrition at the Johns Hopkins Bloomberg School of Public Health and stored at −80°C.
Proteomics analysis.
Details of quantitative proteomics processes have been reported elsewhere (14). In brief, 6 high-abundance proteins (albumin, IgG, IgA, transferrin, haptoglobin, and antitrypsin) were immune depleted from the plasma samples of children (40 μL) by using a Human-6 Multiple Affinity Removal System LC column to enhance detection of low-abundance proteins. At the MS and Proteomics Core in the Johns Hopkins School of Medicine samples were digested with trypsin overnight. Seven samples and 1 pooled sample were labeled with isobaric Tags for Relative and Absolute Quantitation (iTRAQ) 8-plex reagents. The pool of all 8 samples was fractionated into 24 fractions by strong cation exchange chromatography and loaded to a reverse-phase nanobore column. Eluted peptides were sprayed into an LTQ orbitrap Velos mass spectrometer interfaced with a NanoAcquity ultra-HPLC. Precursor and the fragment ions were analyzed, and MS/MS spectra were extracted and searched against the RefSeq 40-protein database by using Mascot through Proteome Discoverer Software. Peptides with ≥95% confidence were filtered for peptide identification. A total of 72 iTRAQ experiments were carried out for 500 samples in this study.
Statistical analysis.
Distributions of anthropometric measurements and z score indexes were checked for normality, and extreme values (<1%) were excluded from the analysis. Procedures for estimating relative abundance of proteins from reporter ion intensities across all MS spectra have been published elsewhere (22). Linear mixed-effects (LME) models were used to take into account the variability in proteomics data from multiple iTRAQ experiments. Random-intercept models were fitted with each anthropometric measurement or index as a dependent variable, proteins as fixed variables, and each iTRAQ experiment as a random effect. We did not adjust for maternal micronutrient supplementation because it did not significantly affect either plasma protein or anthropometric profiles in this small subset of children (SE Lee, unpublished results, 2015). Parameters were estimated by using restricted maximum likelihood estimation (23). P values were calculated by using a 2-sided test of a null hypothesis that there is no association between individual proteins and an anthropometric outcome. Multiple-hypothesis testing was corrected by controlling the false discovery rate (24). Proteins passing a false discovery rate threshold of 5% (q < 0.05) were considered significant. R2 was estimated by squaring a correlation coefficient (r) between an outcome variable and its respective best linear unbiased prediction from the LME models (25). In addition to proteins linearly associated with each anthropometric outcome, we identified proteins differentially abundant by dichotomized undernutrition status by fitting LME models with relative abundance of proteins as dependent variables, each undernutrition indicator as a fixed variable, and each iTRAQ experiment as a random variable.
Because data for proteins related to height-for-age were most complete, we generated a heatmap to illustrate each child’s LME model-predicted relative abundance for multiple proteins associated with the outcome. Because there were missing values for proteins of interest, we imputed values using multiple imputation 10 times (26) using predicted values from regression models between proteins and the outcome variable. We computed a row dendrogram from hierarchical clustering using a Pearson correlation method to calculate pairwise distances between proteins.
Because coregulated proteins in shared biological pathways can be expected to be correlated with each other, we have constructed a correlation matrix of all identified proteins in this study. We separately present a correlation matrix of proteins associated with AMA to illustrate potential biological clusters specific to arm musculature. We calculated Pearson correlation coefficients of pairwise protein-protein values in each iTRAQ experiment, and averaged correlation coefficients across all iTRAQ experiments were used. The order of proteins in the correlation matrix was determined by optimal leaf ordering that reorganizes proteins that are more correlated to be adjacent. The datasets of anthropometric measurements and indexes and relative abundance of plasma proteins used for analyses are available in Supplemental Table 1. All statistical analyses were performed by using the R Environment for Statistical Computing (version 3.1.2; R Development Core Team).
Protein annotation.
Human Genome Organization gene symbols of proteins were used in tables and text as abbreviations of protein names (27). Functional annotation clustering of proteins associated with anthropometric measurements was carried out to list common biological process terms through the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) by using Biological Process ontology of the Gene Ontology database (28–30). Additional information about molecular and biological functions of proteins was derived from the National Center for Biotechnology Information Protein Database and in-depth literature review (31).
Results
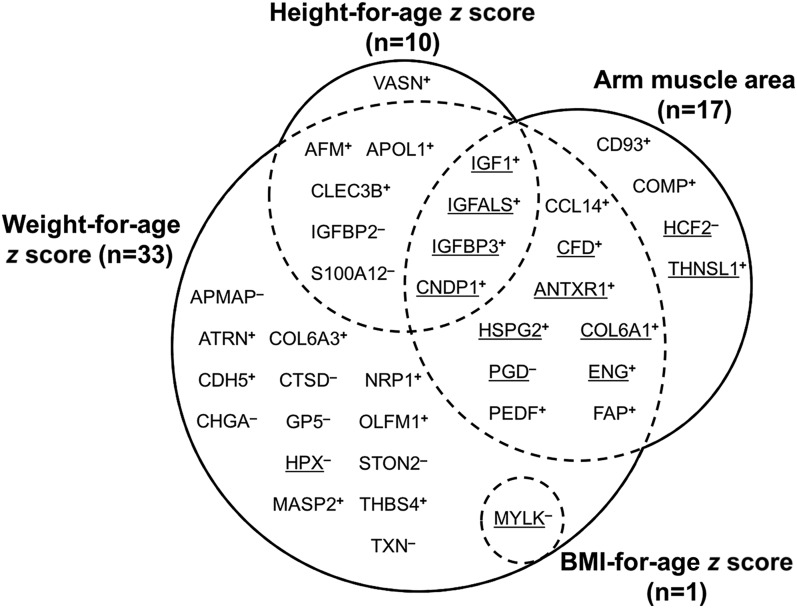
Demographic characteristics and anthropometric measurements of the children in this study are presented in Supplemental Table 2. Children were undernourished as indicated by mean HAZs, BAZs, and WAZs of −1.79, −1.18, and −1.98, respectively, and mean MUAC, triceps skin fold, subscapular skin fold, AMA, and AFA values of 15.4 cm, 5.8 mm, 4.8 mm, 14.8 cm2, and 4.3 cm2, respectively. These are equivalent to mean values of <10th percentile for MUAC, triceps skin fold, and AFA, and <25th percentile for AMA and subscapular skin fold of distributions among well-nourished reference child populations in the United States (32–34). Among 982 proteins that were detected and quantified by MS in >10% of the all plasma samples (n > 50), 10, 14, 17, 1, and 33 plasma proteins were associated with HAZ, MUAC, AMA, BAZ, and WAZ, respectively (q < 0.05) (Figure 1). Four proteins were jointly associated with HAZ and AMA whereas most proteins associated with HAZ, BMZ, and AMA were also associated with WAZ. Except for 2, all proteins associated with MUAC were also associated with AMA. No proteins associated with indicators of fatness (AFA, triceps skin fold, and subscapular skin fold) passed the false discovery rate threshold of 5% (all q ≥ 0.05). Common Gene Ontology terms (categorized as Biological Process) for all identified proteins primarily included regulation of cell proliferation and growth, cell movement, skeletal system and muscle organ development, and inflammatory, defense, and wound healing responses (Supplemental Table 3).
FIGURE 1.
Venn diagram of the number of plasma proteins associated with height-for-age, BMI-for-age, and weight-for-age z scores and arm muscle area in 6- to 8-y-old Nepalese children (q < 0.05). Note: Proteins associated with midupper arm circumference (n = 14), comprising a subset of those associated with arm muscle area, are underlined rather than shown separately. Plasma proteins are indicated as their Human Genome Organization gene symbols. Superscript plus (+) and minus (−) signs indicate positive and negative associations, respectively. AFM, afamin; ANTXR1, anthrax toxin receptor 1; APMAP, adipocyte plasma membrane–associated protein; APOL1, apolipoprotein L1; ATRN, attractin; CCL14, chemokine (C-C motif) ligand 14; CDH5, cadherin 5; CD93, CD93 antigen; CFD, complement factor D; CHGA, chromogranin A; CLEC3B, tetranectin; CNDP1, carnosinase 1; COL6A1, collagen VI, α1; COL6A3, collagen VI, α3; COMP, cartilage oligomeric matrix protein; CTSD, cathepsin D; ENG, endoglin; FAP, fibroblast activation protein; GP5, platelet glycoprotein V; HCF2, heparin cofactor II; HPX, hemopexin; HSPG2, perlecan; IGFALS, insulin-like growth factor, acid labile subunit; IGFBP, insulin like-growth factor–binding protein; IGF1, insulin-like growth factor I; MASP2, mannan-binding lectin serine protease 2; MYLK, myosin light-chain kinase; NRP1, neuropilin 1; OLFM, noelin; PEDF, pigment epithelium-derived factor; PGD, phosphogluconate dehydrogenase; STON2, stonin-2; S100A12, protein S100-A12; THBS4, thrombospondin 4; THNSL1, threonine synthase–like 1; TXN, thioredoxin; VASN, vasorin.
Plasma proteins associated with HAZ or stunting status.
Height-for-age was 0.17–0.80 of a z score higher per 50% increase in relative abundance of positively correlated proteins (Table 1). IGF-I, a growth-promoting factor (36), and 2 IGF-binding proteins [IGF-binding protein, acid labile subunit (IGFALS) and IGF-binding protein 3 (IGFBP3)] were positively associated with height-for-age, although the association was stronger with the binding proteins than IGF-I. Other positive correlates with HAZ included afamin and apo L1, nutrient transport proteins for vitamin E and lipids, respectively (37, 38); carnosinase 1, a carnosine degrading enzyme (39); and tetranectin and vasorin, involved in skeletal system and vascular development, respectively (40, 41).
TABLE 1.
Plasma proteins positively and negatively associated with HAZ in 6- to 8-y-old Nepalese children (q < 0.05)1
| Protein (gene symbol) | n2 | Change in HAZ3 | R2 4 | q5 | Accession6 | Molecular function or biological process7 |
| Positive associations | ||||||
| IGF-binding protein, acid labile subunit (IGFALS) | 498 | 0.80 (0.65, 0.95) | 0.30 | 1.91 × 10−23 | 4826772 | IGF binding |
| IGF-binding protein 3 (IGFBP3) | 498 | 0.59 (0.45, 0.74) | 0.23 | 4.13 × 10−13 | 62243068 | IGF binding |
| Afamin (AFM) | 498 | 0.42 (0.22, 0.62) | 0.15 | 0.0082 | 4501987 | Vitamin E transport |
| IGF I (IGF-I) | 172 | 0.31 (0.15, 0.46) | 0.09 | 0.0128 | 163659901 | Growth factor |
| Tetranectin (CLEC3B) | 498 | 0.55 (0.26, 0.83) | 0.14 | 0.0218 | 156627579 | Bone matrix |
| Apolipoprotein L1 (APOL1) | 498 | 0.32 (0.15, 0.5) | 0.14 | 0.0282 | 211938442 | Lipid transport |
| Carnosinase 1 (CNDP1) | 498 | 0.17 (0.08, 0.27) | 0.14 | 0.0401 | 21071039 | Carnosine hydrolase |
| Vasorin (VASN) | 498 | 0.39 (0.17, 0.62) | 0.14 | 0.0495 | 88702793 | Vasculogenesis |
| Negative associations | ||||||
| Protein S100-A12 (S100A12) | 375 | −0.23 (−0.34, −0.13) | 0.10 | 0.0045 | 5032059 | Immune response |
| IGF-binding protein 2 (IGFBP2) | 498 | −0.28 (−0.44, −0.12) | 0.14 | 0.0495 | 55925576 | IGF binding |
Ten proteins quantified by MS and estimated by linear mixed-effects modeling in >10% of the samples that were positively and negatively associated with HAZ (q < 0.05), listed by the direction and strength of association (in increasing order of q). HAZ, height-for-age z score; IGF, insulin-like growth factor.
Number of child plasma samples in which each protein was detected and quantified by MS. HAZ outliers (n = 2) were excluded; thus, the maximum number was 498.
Estimated change in HAZ (95% CI) of children per 50% (1.5 times) increase in the relative abundance of a protein.
Proportion of variability in HAZ explained by protein.
Multiple hypothesis testing was corrected by using the false discovery rate.
GenInfo sequence number as assigned to all protein sequences by the National Center for Biotechnology Information at the National Library of Medicine, NIH (35).
Represented or known molecular function or biological process of protein.
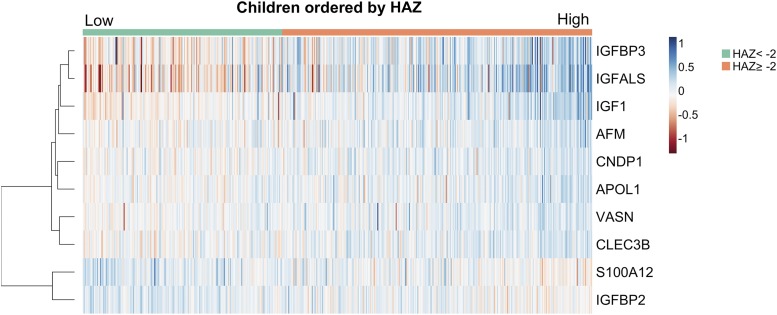
Height-for-age was lower by 0.23–0.28 of a z score per 50% increase in relative abundance of protein S100-A12 (S100A12), a protein involved in the innate immune response (42), and IGF-binding protein 2 (Table 1). In Figure 2, a heatmap displays increases or decreases in modeled relative abundance estimates for the 10 proteins positively and negatively associated with HAZ in increasing order (from the left to right side) among all analyzed children. This illustrates attained height-dependent variation in the plasma protein abundances. In a separate analysis, we also identified plasma proteins differentially abundant by stunting status. Five proteins including IGFALS, IGFBP3, carnosinase 1, and tetranectin, were 4–13% less abundant in the plasma of stunted children than nonstunted children (Supplemental Table 4).
FIGURE 2.
Heatmap of plasma proteins associated with HAZ in 6- to 8-y-old Nepalese children (q < 0.05). Each row and column denotes protein and individual child, respectively. Children (n = 498) were ordered by their HAZ in increasing order from the left to right side. The color of each cell indicates linear mixed-effects model–estimated relative abundance of protein. Blue and red indicate high and low abundance relative to the mean value of each protein, respectively. Row dendrogram (on the left side of heatmap) shows a hierarchical clustering of proteins based on their correlations. The shorter horizontal subbranches represent the higher correlations. AFM, afamin; APOL1, apolipoprotein L1; CLEC3B, tetranectin; CNDP1, carnosinase 1; HAZ, height-for-age z score; IGFALS, insulin-like growth factor, acid labile subunit; IGFBP, insulin-like growth factor–binding protein; IGF1, insulin-like growth factor I; S100A12, protein S100-A12; VASN, vasorin.
Plasma proteins associated with AMA.
Fifteen and 2 proteins, respectively, were positively and negatively associated with AMA (Table 2). AMA was higher by 0.4–1.7 cm2 and lower by ∼1.0 cm2 per 50% increase in relative abundance of positively and negatively correlated proteins, respectively. Although relatively large in number (n = 17), most proteins explained only small percentages of its variation (R2 = 2–6%). We present a correlation matrix to reveal functional clusters of proteins that covary and likely interact within the plasma proteome of AMA in Figure 3. In this matrix we found high positive correlations between the IGF-I, IGFALS, and IGFBP3 proteins reflecting IGF-I ternary complex (r = 0.66–0.81) (43) and moderately high positive correlations (mostly r = 0.24–0.51) among another group of proteins. These include structural components of extracellular matrix, including collagen VI α1, cartilage oligomeric matrix protein, and perlecan (44), and receptors and molecules that interact to regulate extracellular matrix homeostasis, including CD93 molecule, fibroblast activation protein, anthrax toxin receptor 1, and endoglin (45–48).
TABLE 2.
Plasma proteins associated with AMA in 6- to 8-y-old Nepalese children (q < 0.05)1
| Protein (gene symbol) | n2 | Change in AMA3 | R2 4 | q5 | Accession6 | Molecular function or biological process7 |
| Positive associations | ||||||
| IGF-binding protein, acid labile subunit (IGFALS) | 499 | 1.03 (0.64, 1.42) | 0.05 | 1.00 × 10−4 | 4826772 | IGF binding |
| IGF-binding protein 3 (IGFBP3) | 499 | 0.82 (0.45, 1.18) | 0.04 | 0.0043 | 62243068 | IGF binding |
| CD93 antigen (CD93) | 415 | 0.86 (0.42, 1.3) | 0.03 | 0.0238 | 88758613 | Cell adhesion |
| Collagen VI, α1 (COL6A1) | 471 | 0.89 (0.43, 1.34) | 0.03 | 0.0238 | 87196339 | ECM of skeletal muscle |
| Endoglin (ENG) | 430 | 0.89 (0.42, 1.36) | 0.03 | 0.0238 | 4557555 | Regulation of ECM synthesis |
| Pigment epithelium-derived factor (PEDF) | 499 | 1.23 (0.57, 1.88) | 0.03 | 0.0238 | 39725934 | Anti-angiogenesis |
| Anthrax toxin receptor 1 (ANTXR1) | 338 | 0.80 (0.37, 1.23) | 0.04 | 0.0238 | 14149904 | ECM homeostasis |
| Fibroblast activation protein (FAP) | 422 | 0.77 (0.35, 1.19) | 0.03 | 0.0239 | 16933540 | ECM remodeling |
| Chemokine (C-C motif) ligand 14 (CCL14) | 77 | 1.66 (0.7, 2.61) | 0.17 | 0.0369 | 14589961 | Intracellular Ca2+ regulation |
| Complement factor D (CFD) | 499 | 1.24 (0.52, 1.96) | 0.02 | 0.0404 | 42544239 | Complement activation or insulin regulation |
| IGF I (IGF-I) | 173 | 0.66 (0.27, 1.06) | 0.06 | 0.046 | 163659901 | Growth factor |
| Threonine synthase–like 1 (THNSL1) | 166 | 0.96 (0.38, 1.54) | 0.06 | 0.046 | 153792148 | Amino acid biosynthesis |
| Cartilage oligomeric matrix protein (COMP) | 499 | 0.65 (0.26, 1.04) | 0.02 | 0.046 | 40217843 | ECM (noncollagenous glycoprotein) |
| Perlecan (HSPG2) | 492 | 1.26 (0.5, 2.02) | 0.02 | 0.046 | 126012571 | ECM (proteoglycan) |
| Carnosinase 1 (CNDP1) | 499 | 0.39 (0.15, 0.62) | 0.02 | 0.046 | 21071039 | Carnosine hydrolase |
| Negative associations | ||||||
| Heparin cofactor II (HCF2) | 499 | −0.99 (−1.53, −0.46) | 0.03 | 0.0238 | 73858566 | Thrombin inhibitor |
| Phosphogluconate dehydrogenase (PGD) | 97 | −1.05 (−1.66, −0.44) | 0.28 | 0.0369 | 40068518 | Enzyme in the pentose phosphate pathway |
Seventeen proteins quantified by MS and estimated by linear mixed-effects modeling in >10% of the samples that were positively and negatively associated with AMA (q < 0.05), listed by the direction and strength of association (in increasing order of q). AMA, arm muscle area; ECM, extracellular matrix; IGF, insulin-like growth factor.
Number of child plasma samples in which each protein was detected and quantified by MS. One outlier of AMA was excluded; thus, the maximum number of children included in the analysis was 499.
Estimated change in AMA (in square centimeters) (95% CI) of children per 50% (1.5 times) increase in the relative abundance of protein.
Proportion of variability in AMA explained by protein.
Multiple hypothesis testing was corrected by using the false discovery rate.
GenInfo sequence number as assigned to all protein sequences by the National Center for Biotechnology Information at the National Library of Medicine, NIH (35).
Represented or known molecular function or biological process of protein.
FIGURE 3.
Matrix of correlation coefficients among plasma proteins associated with arm muscle area in 6- to 8-y-old Nepalese children (q < 0.05). Blue and red indicate positive and negative correlations, respectively, and darker colors represent a stronger association. All correlation coefficients (r) are presented as r × 102 to improve visualization. Proteins with sample size <100 (n = 2) were not included. ANTXR1, anthrax toxin receptor 1; CD93, CD93 antigen; CFD, complement factor D; CNDP1, carnosinase 1; COL6A1, collagen VI, α1; COMP, cartilage oligomeric matrix protein; ENG, endoglin; FAP, fibroblast activation protein; HCF2, heparin cofactor II; HSPG2, perlecan; IGFALS, insulin-like growth factor, acid labile subunit; IGFBP3, insulin like-growth factor–binding protein 3; IGF1, insulin-like growth factor I; PEDF, pigment epithelium-derived factor; THNSL1, threonine synthase–like 1.
Plasma proteins associated with BMI-for-age.
BMI, reflecting both lean and fat mass, was lower by −1.14 (95% CI: −1.63, −0.66) z scores per 50% increase in relative abundance of myosin light-chain kinase (q = 0.0021). This protein was detected in only 74 of 500 children, which explained 38% of the variability in BAZ. It is an enzyme localized in smooth muscle cells and involved in muscle contraction (49). No other proteins were associated with BAZ or differentially abundant between children with low BMI (BAZ <−2) and normal BMI (BAZ ≥−2).
Plasma proteins associated with WAZ and underweight status.
Weight-for-age was higher by 0.21–0.74 of a z score and lower by 0.21–0.96 of a z score per 50% increase in relative abundance of positively (n = 22) and negatively (n = 11) correlated proteins, respectively (Table 3). IGF-I, IGFALS, and IGFBP3 showed the strongest positive associations (all q < 0.0001), followed by carnosinase 1. In addition to proteins commonly associated with HAZ, BAZ, and AMA, neuropilin and noelin, involved in angiogenesis (50) and neural development (51), respectively, were positively associated with WAZ. Proteins with a variety of functions, including thioredoxin, platelet glycoprotein V, adipocyte plasma membrane-associated protein, chromogranin A, and cathepsin D, were inversely associated with WAZ. In a separate analysis, IGF-I, IGFALS, IGFBP3, carnosinase 1, noelin, and 4 other proteins were 3–18% less abundant and adipocyte plasma membrane-associated protein was 7% more abundant in the plasma of underweight children (WAZ <−2) than children with WAZ ≥−2 (Supplemental Table 5).
TABLE 3.
Plasma proteins positively and negatively associated with WAZ in 6- to 8-y-old Nepalese children (q < 0.05)1
| Protein (gene symbol) | n2 | Change in WAZ3 | R2 4 | q5 | Accession6 | Molecular function or biological process7 |
| Positive associations | ||||||
| IGF-binding protein, acid labile subunit (IGFALS) | 499 | 0.74 (0.6, 0.89) | 0.26 | 6.59 × 10−21 | 4826772 | IGF binding |
| IGF-binding protein 3 (IGFBP3) | 499 | 0.57 (0.43, 0.71) | 0.20 | 3.80 × 10−13 | 62243068 | IGF binding |
| IGF I (IGF-I) | 173 | 0.37 (0.23, 0.51) | 0.17 | 5.13 × 10−05 | 163659901 | Growth factor |
| Carnosinase 1 (CNDP1) | 499 | 0.21 (0.12, 0.31) | 0.11 | 0.0011 | 21071039 | Carnosine hydrolase |
| Pigment epithelium-derived factor (PEDF) | 499 | 0.58 (0.32, 0.84) | 0.11 | 0.0021 | 39725934 | Anti-angiogenesis |
| Perlecan (HSPG2) | 492 | 0.58 (0.28, 0.89) | 0.10 | 0.0133 | 126012571 | ECM (proteoglycan) |
| Noelin (OLFM1) | 464 | 0.44 (0.21, 0.67) | 0.12 | 0.0142 | 17136143 | Neural development |
| Afamin (AFM) | 499 | 0.36 (0.16, 0.56) | 0.10 | 0.0166 | 4501987 | Vitamin E transport |
| Attractin (ATRN) | 499 | 0.51 (0.23, 0.79) | 0.10 | 0.0166 | 21450863 | Inflammatory response |
| Anthrax toxin receptor 1 (ANTXR1) | 338 | 0.31 (0.14, 0.47) | 0.08 | 0.0166 | 14149904 | ECM homeostasis |
| Collagen VI, α1 (COL6A1) | 471 | 0.32 (0.14, 0.51) | 0.11 | 0.0259 | 87196339 | ECM of skeletal muscle |
| Neuropilin 1 (NRP1) | 457 | 0.31 (0.13, 0.49) | 0.13 | 0.0265 | 66864913 | Angiogenesis |
| Complement factor D (CFD) | 499 | 0.5 (0.21, 0.79) | 0.10 | 0.0265 | 42544239 | Complement activation or adipokine |
| Endoglin (ENG) | 430 | 0.32 (0.13, 0.51) | 0.04 | 0.0290 | 4557555 | Regulation of ECM synthesis |
| Apolipoprotein L1 (APOL1) | 499 | 0.28 (0.12, 0.45) | 0.10 | 0.0290 | 211938442 | Lipid transport |
| Thrombospondin 4 (THBS4) | 451 | 0.24 (0.1, 0.37) | 0.08 | 0.0290 | 31543806 | ECM (noncollagenous glycoprotein) |
| Chemokine (C-C motif) ligand 14 (CCL14) | 77 | 0.65 (0.26, 1.05) | 0.13 | 0.0349 | 14589961 | Intracellular Ca2+ regulation |
| Fibroblast activation protein, α subunit (FAP) | 422 | 0.28 (0.11, 0.45) | 0.06 | 0.0354 | 16933540 | ECM remodeling |
| Cadherin 5 (CDH5) | 499 | 0.34 (0.13, 0.55) | 0.10 | 0.0424 | 166362713 | Endothelial cell-cell adhesion |
| Collagen VI, α3 (COL6A3) | 471 | 0.35 (0.13, 0.56) | 0.12 | 0.0424 | 55743106 | ECM of skeletal muscle |
| Tetranectin (CLEC3B) | 499 | 0.43 (0.16, 0.71) | 0.10 | 0.0465 | 156627579 | Bone matrix |
| Mannan-binding lectin serine protease 2 (MASP2) | 112 | 0.67 (0.23, 1.1) | 0.24 | 0.0465 | 21264363 | Complement activation |
| Negative associations | ||||||
| Protein S100-A12 (S100A12) | 376 | −0.21 (−0.31, −0.1) | 0.06 | 0.0077 | 5032059 | Immune response |
| Myosin light-chain kinase, smooth muscle (MLCK) | 74 | −0.96 (−1.44, −0.48) | 0.18 | 0.0077 | 116008190 | Muscle contraction |
| Adipocyte plasma membrane–associated protein (APMAP) | 485 | −0.28 (−0.43, −0.14) | 0.06 | 0.0133 | 24308201 | Adipocyte differentiation |
| IGF-binding protein 2 (IGFBP2) | 499 | −0.28 (−0.44, −0.13) | 0.10 | 0.0166 | 55925576 | IGF binding |
| Chromogranin A (CHGA)20 | 339 | −0.27 (−0.43, −0.11) | 0.08 | 0.0354 | 4502805 | Neuroendocrine secretory protein |
| Thioredoxin (TXN) | 311 | −0.25 (−0.41, −0.1) | 0.07 | 0.0405 | 50592994 | Redox homeostasis |
| Platelet glycoprotein V (GP5) | 445 | −0.24 (−0.39, −0.09) | 0.07 | 0.0424 | 4758460 | Platelet aggregation |
| Stonin-2 (STON2) | 125 | −0.34 (−0.55, −0.12) | 0.09 | 0.0445 | 21361863 | Endocytosis |
| Phosphogluconate dehydrogenase (PGD) | 97 | −0.44 (−0.73, −0.16) | 0.17 | 0.0465 | 40068518 | Enzyme in the pentose phosphate pathway |
| Cathepsin D (CTSD) | 307 | −0.23 (−0.37, −0.08) | 0.13 | 0.0465 | 4503143 | Lysosomal protease |
| Hemopexin (HPX) | 499 | −0.48 (−0.79, −0.17) | 0.10 | 0.0491 | 11321561 | Heme transport |
Thirty-three proteins quantified by MS and estimated by linear mixed-effects modeling in >10% of the samples that were positively and negatively associated with WAZ (q < 0.05), listed by the direction and strength of association (in increasing order of q). ECM, extracellular matrix; IGF, insulin-like growth factor; WAZ, weight-for-age z score.
Number of child plasma samples in which each protein was detected and quantified by MS. One outlier of WAZ was excluded; thus, the maximum number of children included in the analysis was 499.
Estimated change in WAZ (95% CI) of children per 50% (1.5 times) increase in the relative abundance of protein.
Proportion of variability in WAZ explained by protein.
Multiple hypothesis testing was corrected by using false discovery rate.
GenInfo sequence number as assigned to all protein sequences by the National Center for Biotechnology Information at the National Library of Medicine, NIH (35).
Represented or known molecular function or biological process of protein.
A total of 38 plasma proteins were positively or negatively associated with ≥1 evaluated anthropometric measurement or index. A full correlation matrix of all proteins is provided in Supplemental Figure 2.
Discussion
In this typical, rural-plains region of Nepal, chronic undernutrition manifests as short stature and thinness in the early school-aged years. We explored the plasma proteome of children in this area to discover proteins associated with anthropometric measures of skeletal bone and muscular growth and subcutaneous fatness. Our results revealed that certain plasma proteins were associated with HAZ, WAZ, BAZ, and upper AMA, although no associations were observed between plasma protein abundance and indicators of subcutaneous fat (i.e., AFA and triceps and subscapular skinfolds) below a false discovery threshold of 5%. These results collectively suggest that the plasma proteome may reflect underlying biological processes of regulating lean tissue growth in this population of undernourished children.
Strong positive associations between IGF-I and its 2 binding proteins (IGFALS and IGFBP3) and height-for-age, weight-for-age, and AMA support the known anabolic effects of IGF-I axis on skeletal muscle and bone tissue growth (52, 53). IGF-I is known to be responsive to both acute and chronic nutritional status (54). This is consistent with our observation that IGF-I and the 2 binding proteins were associated with skeletal muscle mass and the long-term process of linear bone growth (5). Interestingly, the 2 IGF-I–binding proteins showed stronger associations than IGF-I. Unbound IGF-I can only briefly remain in the bloodstream before forming a ternary complex with its circulating binding proteins to extend its half-life (43). Thus, we postulate that abundance of the 2 IGF-I–binding proteins may be more sensitive to long-term nutritional status than plasma abundance of IGF-I in this chronically undernourished child population.
To a lesser extent than the IGF-I ternary complex, carnosinase 1 was also positively associated with attained height, weight, and musculature. It is an enzyme that hydrolyzes the dipeptide carnosine into its constituent amino acids, β-alanine and histidine (39). Carnosine is an antioxidant that is abundantly found in skeletal muscle and rapidly hydrolyzed in the plasma. Although little is known about the role of carnosinase 1 in bone growth, studies have noted its reduced activity or a low concentration in alcoholic patients with metabolic myopathy (55), anorexic patients with severe protein-energy deficiency (56), and cancer patients with cachexia (57). Our finding supports the theory that plasma abundance of this protein may be expected to positively correlate with muscle mass and, thus, reflect adequacy of lean tissue. Because skeletal muscle is essential to support mechanical functions of the skeleton (58), and height-for-age and AMA were positively correlated in these study children (r = 0.46), carnosinase 1 may be a candidate biomarker for interdependent musculoskeletal growth and function in growing children.
Several proteins involved in the immune response, nutrient metabolism, and bone remodeling were specific in their associations with height-for-age. S100A12 is predominantly secreted from activated neutrophils and is considered a sensitive marker of an innate inflammatory response (42). It is not clear which mechanisms may underlie the inverse association between S100A12 and attained stature. Elevated plasma or fecal S100A12 has been found associated with chronic intestinal inflammation in children (59, 60). Thus, one plausible explanation is that it may reflect subclinically impaired gut function, which is a risk factor for child growth faltering in developing countries (61). Another correlate of height-for-age was afamin, a protein in the albumin family with emerging pleiotropic roles in glucose, lipid, and bone metabolism beyond its role and associations with vitamin E transport (17, 37). Finally, tetranectin, also solely associated with stature, is an abundant protein in bone cartilage that plays a role in bone mineralization during osteogenesis (40). In summary, proteins associated with HAZ are involved in a variety of biological functions, and their gradual differences in plasma abundance by attained height (Figure 2) suggest combined effects of multiple pathways on skeletal development in children.
Skeletal musculature of studied children, indicated by AMA, covaried with relative abundance of proteins of extracellular matrix origin, including its major structural components (collagen and proteoglycan) and homeostasis-regulating proteins. The extracellular matrix is a structural framework of connective tissue that is abundant in skeletal muscle (7). Its constant turnover occurs through interactions with myocytes that include cell-extracellular matrix adhesion, growth factor and membrane receptor signaling, and matrix proteolysis by proteinases (7, 62). These interactions are critical for muscle cell attachment, migration, proliferation, and survival (7). The observed moderate, positive correlations between extracellular matrix–related proteins (Figure 3) are likely to reflect a network of dynamics involving the extracelluar matrix that promotes skeletal muscle development.
One protein, myosin light-chain kinase (MLCK), was negatively associated with age-adjusted BMI. Although BMI does not distinguish lean from fat mass, a substantial literature supports localization of MLCK in muscle tissue where it regulates muscle contraction (49). Studies have reported an MLCK genotype to be associated with exertional muscle damage and subsequent leakage of intracellular proteins into circulation in response to high physical demand (63, 64). Thus, it is possible that children exposed to chronic stresses of malnutrition or low-grade inflammation (15) experience greater systemic breakdown of muscle tissue resulting in a higher abundance this protein in plasma (65).
Noteworthy in this study was a virtual lack of association between detectable plasma proteins and measures of adiposity. One possible explanation is that skinfolds and BMI may become less valid indicators of body fat in pediatric populations with very low adipose tissue (66), such as observed in this study in which average triceps and upper AFA measurements were <10th percentile of American children (32–34). It is also possible that fat mass–regulating or associated proteins in this lean and low-fat child phenotype were too low in plasma abundance to be detected by MS.
As a composite measurement, weight-for-age was associated with the largest and most comprehensive proteome. Because musculoskeletal mass substantially contributes to body weight, the proteome of weight included most proteins that were associated with height and arm muscle mass. In addition, positive correlates included noelin, a protein involved in nervous system development (67). Negative correlates also included proteins expressed during cellular responses to catabolic stress, such as those associated with oxidative damage (thioredoxin) (68) and vascular injury (platelet glycoprotein V). Another negative correlate, cathepsin D, is a lysosomal endopeptidase that can promote tissue degradation (69). Although body weight is nonspecific to body dimension and composition, the proteins identified in this study deserve further exploration because they may represent nutritionally regulated systems or aspects of ponderal growth that cannot be assessed by anthropometry.
Among studies that have applied omics approaches to elucidate how undernutrition may affect childhood growth and associated health outcomes (70–72), this is the first proteomics study to our knowledge that has identified protein biomarkers correlated with multiple facets of attained child growth based on extensive anthropometric data. Because study participants were sampled by a random process from a large population cohort of children residing in an area typical of the terai of Nepal and greater Gangetic flood plains region, we surmise that our findings relating the plasma proteome to child phenotype may be generalizable to similarly undernourished child populations in the region.
A limitation of this cross-sectional analysis is that we cannot assume causal inferences about relations observed between plasma proteins and anthropometric phenotype. More accurate body composition measures, such as provided by bioelectrical impedance analysis or other advanced methods, were not available in this study, which would have allowed us to explore associations between the plasma proteome and visceral or total-body fat stores. Finally, proteins detected by MS were measured on a relative scale. Because the proteomics approach has consistently provided valid direction and strength of association between detected proteins and other nutritional outcomes (14, 16, 17), the identified proteins in this study deserve further validation and replication in other populations to test plasma proteins as nutrition-sensitive biomarkers for clinical and public health practice.
This plasma proteomics study identified known growth-promoting factors and novel plasma proteins associated with anthropometric indicators in rural South Asian children. In particular, proteins were most strongly associated with lean tissue depots and may reflect pathways of lean tissue growth in these children. Because child anthropometric features are associated with immunologic and neurological functions and long-term metabolic health, proteins identified in this study may prove useful in the future as functional biomarkers of nutritional status, growth processes, and population health status.
Acknowledgments
We thank Margia Arguello, Raghothama Chaerkady, Hongie Cui, Lauren R DeVine, Jaime Johnson, Robert O’Meally, Steven C LeClerq, Ashika Nanayakkara-Bind, Hee-Sool Rho, Sudeep Shrestha, and Fredrick Van Dyke for assistance with the nutriproteomics research and Nepal field activities. We thank Ingo Ruczinski for his proteomics modeling and analytic guidance and C Conover Talbot Jr. for assistance with the Human Genome Organization gene annotation. KJS, RNC, JDY, JDG, PC, and KPW designed the research; RNC performed the research; SEL, KJS, and RNC contributed new reagents or analytic tools; SEL analyzed the data; CPS, LS-FW, SKK, RKA, PC, and KPW conducted the original field study; SEL wrote the manuscript and had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AFA, arm fat area; AMA, arm muscle area; BAZ, BMI-for-age z score; HAZ, height-for-age z score; IGF, insulin-like growth factor; IGFALS, insulin-like growth factor–binding protein, acid labile subunit; IGFBP3, insulin-like growth factor–binding protein 3; iTRAQ, isobaric Tags for Relative and Absolute Quantitation; LME, linear mixed-effects; MLCK, myosin light-chain kinase; MUAC, midupper arm circumference; S100A12, protein S100-A12; WAZ, weight-for-age z score.
References
- 1.Emery PW. Metabolic changes in malnutrition. Eye (Lond) 2005;19:1029–34. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Use and interpretation of anthropometric indicators of nutritional status. WHO Working Group. Bull World Health Organ 1986;64:929–41. [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002;1:845–67. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld RG. Endocrine control of growth. In: Cameron N, Bogin B, editors. Human growth and development. New York: Elsevier Inc.; 2012. [Google Scholar]
- 5.Shenkin A, Cederblad G, Elia M, Isaksson B. International federation of clinical chemistry. Laboratory assessment of protein-energy status. Clin Chim Acta 1996;253:S5–59. [DOI] [PubMed] [Google Scholar]
- 6.Baron J, Savendahl L, De Luca F, Dauber A, Phillip M, Wit JM, Nilsson O. Short and tall stature: a new paradigm emerges. Nat Rev Endocrinol 2015;11:735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velleman SG, Shin J, Li X, Song Y. Review: the skeletal muscle extracellular matrix: possible roles in the regulation of muscle development and growth. Can J Anim Sci 2012;92:1–10. [Google Scholar]
- 8.Raiten DJ, Raghavan R, Kraemer K. Biomarkers in growth. Ann Nutr Metab 2013;63:293–7. [DOI] [PubMed] [Google Scholar]
- 9.Schulze KJ, Christian P, Wu LS, Arguello M, Cui H, Nanayakkara-Bind A, Stewart CP, Khatry SK, LeClerq S, West KP Jr. Micronutrient deficiencies are common in 6- to 8-year-old children of rural Nepal, with prevalence estimates modestly affected by inflammation. J Nutr 2014;144:979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart CP, Christian P, LeClerq SC, West KP Jr, Khatry SK. Antenatal supplementation with folic acid + iron + zinc improves linear growth and reduces peripheral adiposity in school-age children in rural Nepal. Am J Clin Nutr 2009;90:132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart CP, Christian P, Schulze KJ, Leclerq SC, West KP Jr, Khatry SK. Antenatal micronutrient supplementation reduces metabolic syndrome in 6- to 8-year-old children in rural Nepal. J Nutr 2009;139:1575–81. [DOI] [PubMed] [Google Scholar]
- 12.Christian P, Murray-Kolb LE, Khatry SK, Katz J, Schaefer BA, Cole PM, Leclerq SC, Tielsch JM. Prenatal micronutrient supplementation and intellectual and motor function in early school-aged children in Nepal. JAMA 2010;304:2716–23. [DOI] [PubMed] [Google Scholar]
- 13.Christian P, Khatry SK, Katz J, Pradhan EK, LeClerq SC, Shrestha SR, Adhikari RK, Sommer A, West KP Jr. Effects of alternative maternal micronutrient supplements on low birth weight in rural Nepal: double blind randomised community trial. BMJ 2003;326:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole RN, Ruczinski I, Schulze K, Christian P, Herbrich S, Wu L, Devine LR, O’Meally RN, Shrestha S, Boronina TN, et al. The plasma proteome identifies expected and novel proteins correlated with micronutrient status in undernourished Nepalese children. J Nutr 2013;143:1540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SE, West KP Jr, Cole RN, Schulze KJ, Christian P, Wu LS, Yager JD, Groopman J, Ruczinski I. Plasma proteome biomarkers of inflammation in school aged children in Nepal. PLoS One 2015;10:e0144279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulze KJ, Cole RN, Chaerkady R, Wu LS, Nonyane BA, Lee SE, Yager JD, Groopman JD, Christian P, West KP Jr. Plasma selenium protein P isoform 1 (SEPP1): a predictor of selenium status in Nepalese children detected by plasma proteomics. Int J Vitam Nutr Res 2016 May 10 (Epub ahead of print; DOI: 10.1024/0300-9831/a000256). [DOI] [PubMed] [Google Scholar]
- 17.West KP Jr, Cole RN, Shrestha S, Schulze KJ, Lee SE, Betz J, Nonyane BA, Wu LS, Yager JD, Groopman JD, et al. A plasma alpha-tocopherome can be identified from proteins associated with vitamin E status in school-aged children of Nepal. J Nutr 2015;145:2646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SE, West KP Jr, Cole RN, Schulze KJ, Wu LS, Yager JD, Groopman J, Christian P. General intelligence is associated with subclinical inflammation in Nepalese children: a population-based plasma proteomics study. Brain Behav Immun 2016;56:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christian P, West KP, Khatry SK, Leclerq SC, Pradhan EK, Katz J, Shrestha SR, Sommer A. Effects of maternal micronutrient supplementation on fetal loss and infant mortality: a cluster-randomized trial in Nepal. Am J Clin Nutr 2003;78:1194–202. [DOI] [PubMed] [Google Scholar]
- 20.Zerfas AJ. The insertion tape: a new circumference tape for use in nutritional assessment. Am J Clin Nutr 1975;28:782–7. [DOI] [PubMed] [Google Scholar]
- 21.de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ 2007;85:660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbrich SM, Cole RN, West KP Jr, Schulze K, Yager JD, Groopman JD, Christian P, Wu L, O’Meally RN, May DH, et al. Statistical inference from multiple iTRAQ experiments without using common reference standards. J Proteome Res 2013;12:594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harville DA. Maximum likelihood approaches to variance component estimation and to related problems. J Am Stat Assoc 1977;72:320–38. [Google Scholar]
- 24.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B 2002;64:479–98. [Google Scholar]
- 25.Robinson GK. That BLUP is a good thing: the estimation of random effects. Stat Sci 1991;6:15–32. [Google Scholar]
- 26.Little RJA, Rubin DB. Statistical analysis with missing data. 2nd ed. New York: John Wiley; 2002. [Google Scholar]
- 27.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 2015;43:D1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2006;34:D173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fryar CD, Gu Q, Ogden CL. Anthropometric reference data for children and adults: United States, 2007–2010. Vital Health Stat 11 2012;Oct:1–48. [PubMed] [Google Scholar]
- 33.Frisancho AR. New norms of upper limb fat and muscle areas for assessment of nutritional status. Am J Clin Nutr 1981;34:2540–5. [DOI] [PubMed] [Google Scholar]
- 34.Frisancho AR, Tracer DP. Standards of arm muscle by stature for the assessment of nutritional status of children. Am J Phys Anthropol 1987;73:459–65. [DOI] [PubMed] [Google Scholar]
- 35.National Center for Biotechnology Information. Sequence identifiers: an historical note [Internet]. [cited 2016 Oct 14]. Available from: http://www.ncbi.nlm.nih.gov/Sitemap/sequenceIDs.html.
- 36.Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 1995;16:3–34. [DOI] [PubMed] [Google Scholar]
- 37.Dieplinger H, Dieplinger B. Afamin–A pleiotropic glycoprotein involved in various disease states. Clin Chim Acta 2015;446:105–10. [DOI] [PubMed] [Google Scholar]
- 38.Duchateau PN, Movsesyan I, Yamashita S, Sakai N, Hirano K, Schoenhaus SA, O’Connor-Kearns PM, Spencer SJ, Jaffe RB, Redberg RF, et al. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J Lipid Res 2000;41:1231–6. [PubMed] [Google Scholar]
- 39.Boldyrev AA, Aldini G, Derave W. Physiology and pathophysiology of carnosine. Physiol Rev 2013;93:1803–45. [DOI] [PubMed] [Google Scholar]
- 40.Wewer UM, Ibaraki K, Schjorring P, Durkin ME, Young MF, Albrechtsen R. A potential role for tetranectin in mineralization during osteogenesis. J Cell Biol 1994;127:1767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda Y, Imai Y, Kumagai H, Nosaka T, Morikawa Y, Hisaoka T, Manabe I, Maemura K, Nakaoka T, Imamura T, et al. Vasorin, a transforming growth factor beta-binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo. Proc Natl Acad Sci USA 2004;101:10732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietzsch J, Hoppmann S. Human S100A12: a novel key player in inflammation? Amino Acids 2009;36:381–9. [DOI] [PubMed] [Google Scholar]
- 43.Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer 2014;14:329–41. [DOI] [PubMed] [Google Scholar]
- 44.Byron A, Humphries JD, Humphries MJ. Defining the extracellular matrix using proteomics. Int J Exp Pathol 2013;94:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacob M, Chang L, Pure E. Fibroblast activation protein in remodeling tissues. Curr Mol Med 2012;12:1220–43. [DOI] [PubMed] [Google Scholar]
- 46.Kao YC, Jiang SJ, Pan WA, Wang KC, Chen PK, Wei HJ, Chen WS, Chang BI, Shi GY, Wu HL. The epidermal growth factor-like domain of CD93 is a potent angiogenic factor. PLoS One 2012;7:e51647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obreo J, Diez-Marques L, Lamas S, Duwell A, Eleno N, Bernabeu C, Pandiella A, Lopez-Novoa JM, Rodriguez-Barbero A. Endoglin expression regulates basal and TGF-beta1-induced extracellular matrix synthesis in cultured L6E9 myoblasts. Cell Physiol Biochem 2004;14:301–10. [DOI] [PubMed] [Google Scholar]
- 48.Reeves C, Charles-Horvath P, Kitajewski J. Studies in mice reveal a role for anthrax toxin receptors in matrix metalloproteinase function and extracellular matrix homeostasis. Toxins (Basel) 2013;5:315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil 1997;18:1–16. [DOI] [PubMed] [Google Scholar]
- 50.Kofler NM, Simons M. Angiogenesis versus arteriogenesis: neuropilin 1 modulation of VEGF signaling. F1000Prime Rep 2015;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anholt RR. Olfactomedin proteins: central players in development and disease. Front Cell Dev Biol 2014;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwa V, Fang P, Derr MA, Fiegerlova E, Rosenfeld RG. IGF-I in human growth: lessons from defects in the GH-IGF-I Axis. In: Gillman MW, Gluckman PD, Rosenfeld RG, editors. Recent advances in growth research: nutritional, molecular and endocrine perspectives. Basel (Switzerland): Nestle Nutrition Institute; 2013. [Google Scholar]
- 53.Velloso CP. Regulation of muscle mass by growth hormone and IGF-I. Br J Pharmacol 2008;154:557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hawkes CP, Grimberg A. Insulin-like growth factor-I is a marker for the nutritional state. Pediatr Endocrinol Rev 2015;13:499–511. [PMC free article] [PubMed] [Google Scholar]
- 55.Duane P, Peters TJ. Serum carnosinase activities in patients with alcoholic chronic skeletal muscle myopathy. Clin Sci (Lond) 1988;75:185–90. [DOI] [PubMed] [Google Scholar]
- 56.McLoughlin DM, Wassif WS, Morton J, Spargo E, Peters TJ, Russell GF. Metabolic abnormalities associated with skeletal myopathy in severe anorexia nervosa. Nutrition 2000;16:192–6. [DOI] [PubMed] [Google Scholar]
- 57.Arner P, Henjes F, Schwenk JM, Darmanis S, Dahlman I, Iresjo BM, Naredi P, Agustsson T, Lundholm K, Nilsson P, et al. Circulating carnosine dipeptidase 1 associates with weight loss and poor prognosis in gastrointestinal cancer. PLoS One 2015;10:e0123566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millward DJ. A protein-stat mechanism for regulation of growth and maintenance of the lean body mass. Nutr Res Rev 1995;8:93–120. [DOI] [PubMed] [Google Scholar]
- 59.de Jong NS, Leach ST, Day AS. Fecal S100A12: a novel noninvasive marker in children with Crohn’s disease. Inflamm Bowel Dis 2006;12:566–72. [DOI] [PubMed] [Google Scholar]
- 60.Leach ST, Yang Z, Messina I, Song C, Geczy CL, Cunningham AM, Day AS. Serum and mucosal S100 proteins, calprotectin (S100A8/S100A9) and S100A12, are elevated at diagnosis in children with inflammatory bowel disease. Scand J Gastroenterol 2007;42:1321–31. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe K, Petri WA Jr. Environmental enteropathy: elusive but significant subclinical abnormalities in developing countries. EBioMedicine 2016;10:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson R. The extracellular matrix: an underexplored but important proteome. Expert Rev Proteomics 2010;7:803–6. [DOI] [PubMed] [Google Scholar]
- 63.Clarkson PM, Hoffman EP, Zambraski E, Gordish-Dressman H, Kearns A, Hubal M, Harmon B, Devaney JM. ACTN3 and MLCK genotype associations with exertional muscle damage. J Appl Physiol (1985) 2005;99:564–9. [DOI] [PubMed] [Google Scholar]
- 64.Deuster PA, Contreras-Sesvold CL, O’Connor FG, Campbell WW, Kenney K, Capacchione JF, Landau ME, Muldoon SM, Rushing EJ, Heled Y. Genetic polymorphisms associated with exertional rhabdomyolysis. Eur J Appl Physiol 2013;113:1997–2004. [DOI] [PubMed] [Google Scholar]
- 65.Briend A, Khara T, Dolan C. Wasting and stunting–similarities and differences: policy and programmatic implications. Food Nutr Bull 2015; 36(1 Suppl):S15–23. [DOI] [PubMed] [Google Scholar]
- 66.Freedman DS, Sherry B. The validity of BMI as an indicator of body fatness and risk among children. Pediatrics 2009;124(Suppl 1):S23–34. [DOI] [PubMed] [Google Scholar]
- 67.Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol 2000;2:219–25. [DOI] [PubMed] [Google Scholar]
- 68.Benhar M. Nitric oxide and the thioredoxin system: a complex interplay in redox regulation. Biochim Biophys Acta 2015;1850:2476–84. [DOI] [PubMed] [Google Scholar]
- 69.Margaryan NV, Kirschmann DA, Lipavsky A, Bailey CM, Hendrix MJ, Khalkhali-Ellis Z. New insights into cathepsin D in mammary tissue development and remodeling. Cancer Biol Ther 2010;10:457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freemark M. Metabolomics in nutrition research: biomarkers predicting mortality in children with severe acute malnutrition. Food Nutr Bull 2015; 36(1 Suppl):S88–92. [DOI] [PubMed] [Google Scholar]
- 71.Semba RD, Shardell M, Sakr Ashour FA, Moaddel R, Trehan I, Maleta KM, Ordiz MI, Kraemer K, Khadeer MA, Ferrucci L, et al. Child stunting is associated with low circulating essential amino acids. EBioMedicine 2016;6:246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014;510:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]