Abstract
Background: Clinical nutrition research often lacks robust markers of compliance, complicating the interpretation of clinical trials and observational studies of free-living subjects.
Objective: We aimed to examine metabolomics profiles in response to 3 diets that differed widely in macronutrient composition during a controlled feeding protocol.
Design: Twenty-one adults with a high body mass index (in kg/m2; mean ± SD: 34.4 ± 4.9) were given hypocaloric diets to promote weight loss corresponding to 10–15% of initial body weight. They were then studied during weight stability while consuming 3 test diets, each for a 4-wk period according to a crossover design: low fat (60% carbohydrate, 20% fat, 20% protein), low glycemic index (40% carbohydrate, 40% fat, 20% protein), or very-low carbohydrate (10% carbohydrate, 60% fat, 30% protein). Plasma samples were obtained at baseline and at the end of each 4-wk period in the fasting state for metabolomics analysis by using liquid chromatography–tandem mass spectrometry. Statistical analyses included adjustment for multiple comparisons.
Results: Of 333 metabolites, we identified 152 whose concentrations differed for ≥1 diet compared with the others, including diacylglycerols and triacylglycerols, branched-chain amino acids, and markers reflecting metabolic status. Analysis of groups of related metabolites, with the use of either principal components or pathways, revealed coordinated metabolic changes affected by dietary composition, including pathways related to amino acid metabolism. We constructed a classifier using the metabolites that differed between diets and were able to correctly identify the test diet from metabolite profiles in 60 of 63 cases (>95% accuracy). Analyses also suggest differential effects by diet on numerous cardiometabolic disease risk factors.
Conclusions: Metabolomic profiling may be used to assess compliance during clinical nutrition trials and the validity of dietary assessment in observational studies. In addition, this methodology may help elucidate mechanistic pathways linking diet to chronic disease risk. This trial was registered at clinicaltrials.gov as NCT00315354.
Keywords: dietary compliance, dietary composition, low-carbohydrate diet, metabolomics, obesity, low-fat diet, glycemic index, cardiometabolic risk factors
INTRODUCTION
Among the most important methodologic challenges in behavioral studies of diet and health is the difficulty in distinguishing efficacy from effectiveness. For instance, short-term trials often demonstrate meaningful changes in body weight as a consequence of macronutrient composition, but with few exceptions, the long-term trials do not (1–3). This inconsistency might arise because differences in macronutrient composition have only a transitory effect on the biological determinants of body weight, because adherence to dietary prescriptions diminishes with time, or both. If the former, then the major scientific effort currently devoted to the study of specific diets would be better directed elsewhere. If the latter, then enhanced behavioral and environmental interventions to promote adherence to the most efficacious dietary prescriptions should yield more effective long-term obesity treatment.
The challenges to achieving and documenting compliance in long-term diet studies are substantial (4–6). Obstacles to behavioral changes include costs of purchasing food, time involved in food preparation, and willingness to comply with dietary recommendations many times a day for years. Moreover, measures of compliance tend to be biased (e.g., food recalls), logistically daunting (respiratory quotient), imprecise (nitrogen excretion), or confounded by weight loss (serum lipids). Thus, there is great need for reliable and convenient measures of compliance in nutrition research. Newly developed methods for determination of multiple serum metabolites, termed metabolomics, offer promise in this regard (7–11).
Numerous studies have used metabolomics to characterize dietary intakes, although these have ≥1 limitations, such as cross-sectional observational design subject to confounding by nondietary factors, lack of control groups, small sample size, use of a limited panel of metabolites, low resolution of profiling techniques, and data analysis based exclusively on clustering methods rather than single metabolites (12–27). The aim of the present study was to conduct high-throughput metabolomics analysis of plasma samples from a carefully controlled crossover feeding study, unconfounded by weight loss, to determine whether a molecular “fingerprint” could distinguish individuals consuming 3 diets differing in macronutrient composition. In addition, the study provided an opportunity to examine potential physiologic mechanisms through which diet affects risk for the metabolic complications of obesity.
METHODS
Study sample and overview
Plasma (EDTA) samples were obtained during a feeding study of weight-loss maintenance, reported previously (28). Supplemental Figure 1 depicts the participant flow through the study. Briefly, 21 adults with BMI (in kg/m2) ≥27, aged 18–40 y, underwent a run-in phase comprising weight monitoring for 4 wk, a hypocaloric diet made entirely from food (not formula) in a metabolic kitchen for ∼12 wk to achieve weight loss corresponding to 10–15% of initial body weight, and weight stabilization for 4 wk. During a subsequent test diet phase, participants consumed 3 diets prepared in a metabolic kitchen, each for a 4-wk period, in random order. To ensure balance and unpredictability, we prepared 30 assignments for order of diet, comprising 5 replicates of the 6 possible orders, grouped in Latin squares with random permutation within and between squares. The assignments were stored in sealed envelopes to be opened in sequence. When a participant withdrew after random assignment, we discarded the prescribed assignment. Among 21 subjects completing the study, the 6 possible orders of diet were uniformly distributed (P > 0.90 by Fisher’s exact test).
The test diets were identical in total energy content but differed in macronutrient composition. With regard to energy distribution, the low-fat diet (LF)11 had 60% carbohydrate, 20% protein, and 20% fat; the low–glycemic index diet (LGI) had 40% carbohydrate, 40% protein, and 20% fat; and the very–low carbohydrate diet (VLC) had 10% carbohydrate, 60% protein, and 30% fat. Participants consumed several meals each week under direct observation. Mean weight did not differ across the test diet periods (LF: 91.5 kg, LGI: 91.1 kg, and VLC: 91.2 kg; P = 0.80). We obtained plasma samples before weight loss and at the end of each test diet period. The study was approved by institutional review boards at Boston Children’s Hospital and Brigham and Women’s Hospital, Boston (NCT00315354).
Measurement of metabolites
We measured 4 classes of plasma metabolites using liquid chromatography–tandem mass spectrometry (LC-MS) (Q Exactive; Thermo Scientific) (29): 1) amines and polar metabolites, such as amino acids and respective metabolites, dipeptides, and other cationic metabolites; 2) central metabolites and polar metabolites, such as sugars, sugar phosphates, organic acids, purines, and pyrimidines; 3) free fatty acids and metabolites of intermediate polarity, such as bile acids and fatty acid oxidation products; and 4) polar and nonpolar lipids, such as lysophosphatidylcholines, lysophosphatidylethanolamines, phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, sphingomyelins, cholesterol esters, diacylglycerols, and triacylglycerols. We analyzed samples in random order and interspersed replicates of common pooled plasma samples as controls to allow normalization as has been done in prior studies (30–32).
Data reduction
Peaks from the LC-MS data were detected and integrated by using Progenesis CoMet software (v2.0; Nonlinear Dynamics). Identification was initially conducted by matching measured retention time and mass with known compounds (TraceFinder v3.1; Thermo Fisher). Analytic performance and data quality were assured by introducing both authentic reference standards and synthetic internal standards into each LC-MS sample during the extraction procedure. We removed from analysis those metabolites that showed a high fraction of missing values (>10%), high CV (mean ÷ SD >20%), or correlation of missing data with sample batch. We were able to proceed with analysis for 333 of the 356 metabolites for which we had data.
We log-transformed the measured metabolite concentrations to mitigate skew and adjusted the log-transformed values for baseline age, sex, measurement period, sequence of diets, and weight loss during the run-in period by analysis of covariance. We replaced any remaining missing concentrations with one-half the minimum (log-transformed, covariate-adjusted). Data are expressed in terms of change from pre–weight loss baseline for purposes of tabulating and graphically illustrating the metabolite concentrations. However, for all analyses, the statistical comparison of interest was among post–weight loss test diets. Because these test diets were administered in a randomized crossover fashion, causal inferences can be made from the analyses. (In contrast, comparisons between pre–weight loss baseline and any of the test diets are descriptive in nature and subject to confounding by weight loss and other time-varying factors.) No adjustment for change in weight during the test diet periods was made because mean weight among the test diets differed by <0.5 kg and within-subject variance in weight was minimal (0.31% of the total variance). We used SAS version 9.4 for all computations.
Differences by diet
For each metabolite and each test diet, we calculated the change from baseline in log-transformed, covariate-adjusted metabolite concentration. We performed an inverse normal transformation of these changes to reduce the influence of outliers. We used the independent-sample Student’s t test to compare the diets pairwise and compared each diet against the other 2 diets. We used a conservative Bonferroni adjustment to correct for multiple testing {P < 2.5 × 10−5 [0.05 ÷ (6 comparisons × 333 metabolites)]}.
To exploit the crossover design for maximal precision, we additionally conducted repeated-measures ANOVA on the log-transformed, covariate-adjusted metabolite concentrations from all 4 measurement periods. We used an unstructured covariance matrix to allow for arbitrary correlation between periods and applied a robust regression algorithm to detect and down-weight outliers. From parameters of the fitted model, we estimated the mean change from baseline for each diet and pairwise differences between diets. To correct for multiple comparisons, we applied the Holm step-down procedure with familywise type I error rate controlled at 5%. We performed a similar analysis on the 14 factors formed by unsupervised principal components analysis.
Correlation between metabolites
Metabolites may fall into correlated pathways. To estimate the overall correlation structure between the 333 metabolite concentrations, we calculated the Pearson correlation coefficient (r) between all pairs of metabolite concentrations. We constructed a heatmap (implemented in MetaboAnalyst v2.0) (33) that visually represents these pairwise correlations, revealing multiple clusters of highly correlated metabolites. Therefore, we used principal components analysis to construct a small number of linear combinations of metabolites that account for the overall patterns and variability seen in the complete data set. In unsupervised analysis (ignoring the diet variable), we obtained 14 combinations (factors) that together captured 80.3% of the variance in the full set of metabolites. We also conducted supervised principal components analysis as implemented in MetaboAnalyst v2.0 to identify combinations of metabolites that discriminated maximally between the 3 diets.
Metabolic pathways
We used the Human Metabolome Database (34) to manually annotate all metabolites showing association with diet composition to metabolic pathways. Further pathway-enrichment analyses were performed by using MetaboAnalyst v2.0. Fisher’s exact test was used to detect overrepresentation of metabolites among 80 human metabolic pathways from the Kyoto Encyclopedia of Genes and Genomes database (35).
Predicting diet from metabolite profiles
Using the 63 individual changes from baseline (21 participants × 3 diet groups, calculated as in method 1 above), we built Bayesian network classification models (36) to determine how accurately the diet type for an individual sample could be predicted using the metabolite measurements. We used WEKA software (v3.6.11) (37) to construct a classifier for discriminating between all 3 diets and 3 additional classifiers to discriminate the diets in a pairwise fashion. All classifiers were BayesNet models learned by using the K2 search algorithm (36) with WEKA’s default settings. To quantitate the accuracy of the approach, we omitted each data point in turn from the sample and built a classifier from the remaining data to predict the identity of the diet in the omitted sample. Accuracy was calculated as the percentage of correct predictions.
Correlating macronutrient intake from metabolite profiles
We analyzed correlation patterns between metabolite profiling and dietary intake data from 1840 individuals in the Framingham Heart Study (FHS) Offspring Cohort at the fifth examination (1991–1995). These data were obtained through the National Center for Biotechnology Information Genotypes and Phenotypes database with accession number phs000007.v19.p7. Both the study samples and the data collection procedures have been previously described in detail (30–32). Briefly, the metabolite profiling data consist of measurements for 217 metabolites generated from LC-MS profiling of fasting plasma samples. Quality control and analysis of metabolite concentrations were similar to the feeding study, with the following steps: 1) removal of any metabolites with >25% missing data, 2) log-transformation of the remaining metabolites, 3) adjustment for age, sex, and fasting duration, 4) replacement of missing values with one-half the minimum adjusted value for each metabolite, and 5) rank-based inverse normal transformation to calculate metabolite z scores.
The dietary intake data consist of daily macronutrient values (grams per day) and the glycemic index (GI) calculated from a self-reported food-frequency questionnaire asking about each participant’s dietary habits during the past year. We calculated the percentage of energy intake from each macronutrient and performed rank-based inverse normal transformation on both the macronutrient percentages and GI to generate z scores that are robust against outliers.
For each of the 103 metabolites that were measured in FHS and that differed between ≥1 pair of diets in the feeding study, we calculated the Spearman correlation between the metabolite and each of the macronutrients or GI. Directional consistency was assessed as described in Supplemental Table 4; for example, if a metabolite was higher in the VLC and the LGI than in the LF, then it should also be associated with higher protein and fat intake and lower carbohydrate intake and GI. Overall consistency of a metabolite was determined by giving priority to the macronutrient with the most significant correlation in FHS. For some metabolites, only a prediction of protein was possible, and this was used to assess directional consistency.
Disease risk
For each diet, we examined the direction of change in the concentrations of the metabolites that have been shown to predict the risk of type 2 diabetes (31, 32, 38–42). We annotated each metabolite by whether the change by diet was in the direction that increased or decreased risk of disease.
RESULTS
Differences by diet
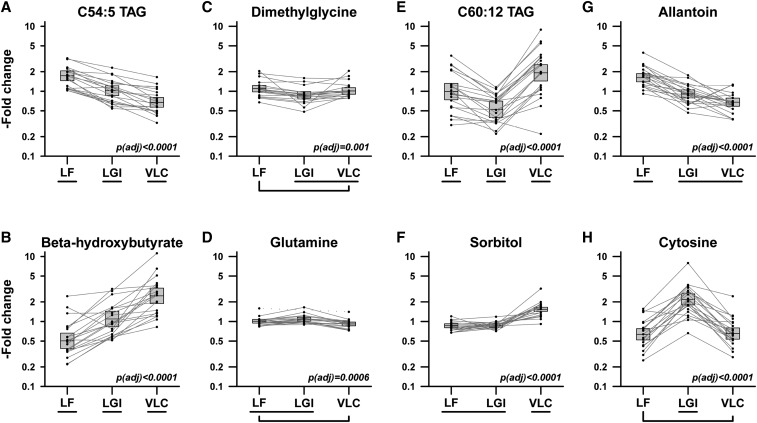
We identified 152 metabolites for which change in concentration from baseline varied significantly between diets by repeated-measures analysis; an analysis using t tests and pairwise comparisons yielded similar results (Supplemental Table 1). Metabolites displayed different characteristic profiles of variation across the 3 diets (as depicted with illustrative examples in Figure 1). Many diacylglycerols and triacylglycerols, such as C54:5 triacylglycerol (TAG), showed a linear trend across diets, with high concentrations for the LF and low concentrations for the VLC samples; others such as β-hydroxybutyrate, showed the opposite trend. In some cases, such as with dimethylglycine, glutamine or C60:12 TAG, statistically significant nonlinear patterns were observed. Some metabolites discriminated 1 diet from the other 2; for instance, certain carnitines, branched-chain amino acids, asparagine, certain cholesteryl esters, α-hydroxybutyrate, propionate, and sorbitol for the VLC; 4-pyridoxate, certain triacylglycerides, allantoin, trimethylamine-N-oxide, 2-aminoadipate, serine, and alanine for the LF; and cytosine, certain triacylglycerides, hydroxyproline, hippurate, 5-aminolevulinic acid, and pipecolic acid for the LGI. The most common pattern was that the metabolite concentration with the LGI was between the concentrations with the LF and VLC (105 of 152 metabolites), whereas the VLC least frequently had the intermediate concentration (19 of 152 metabolites). In addition to variation by diet, individual variation differed; for instance, it was higher for C60:12 TAG than C54:5 TAG.
FIGURE 1.
Patterns of metabolite response to diet. Examples of 8 metabolites demonstrating significant differential responses to 3 weight-loss maintenance diets administered to 21 young adults in a randomized crossover trial. Metabolites were selected to illustrate contrasting patterns of responses. Vertical axes show relative change from pre–weight loss metabolite concentration. Boxes indicate means and 95% CIs. Lines connect covariate-adjusted data for individual participants, omitting outliers detected by robust regression procedure. Bracketed diets did not differ significantly according to repeated-measures ANOVA on log-transformed, covariate-adjusted metabolite concentrations. P tests the hypothesis of equal mean concentration across all 3 diets, adjusted for multiple comparisons by Holm step-down procedure. Adj, adjusted; LF, low-fat diet; LGI, low–glycemic index diet; TAG, triacylglycerol; VLC, very–low carbohydrate diet.
Coordinated changes in metabolite concentrations
Analysis of the pairwise correlations between metabolites revealed groups of metabolites with similar patterns of response across diets (Supplemental Figure 2). To characterize these groupings further, we conducted unsupervised principal components analysis to generate 14 distinct factors. Essentially, these factors are different groups of metabolites that show similar patterns across the diets. For each metabolite, the factor most closely associated with that metabolite is shown in Supplemental Table 2 and Supplemental Figure 3. Ten of the factors varied significantly across diets. As with the individual metabolites, these factors exhibited different characteristic profiles of variation across diets, indicating that altered dietary composition produced characteristic coordinated changes in related classes of metabolites. For example, factor 1, comprising amino acids and their derivatives, carnitines, ceramides, and several other small molecules, increased linearly from the LF to LGI to VLC; by contrast, factor 2, comprising, phospholipids and shorter diacylglycerols and triacylglycerols, decreased from the LF to LGI to VLC.
Some coordinated changes in metabolites reflect known pathways: of 80 well-characterized human metabolic pathways described in the Kyoto Encyclopedia of Genes and Genomes database, 7 were enriched in metabolites that changed with diet (Supplemental Table 3). The majority of the enriched pathways are involved in amino acid metabolism (for example, aminoacyl–transfer RNA biosynthesis or glycine, serine, and threonine metabolism). In total, 25 unique metabolites within known pathways were highlighted by this enrichment analysis.
Predicting diet from metabolite profiles
The coordinated effects of diet on pathways and groups of metabolites suggested that combinations of metabolites might distinguish the diets from each other. To test this possibility, we withheld the data for each sample in turn and used the data from the remaining samples to construct 4 Bayesian network classifiers: 1 designed to distinguish each diet from the others (LF compared with LGI compared with VLC), and 3 designed to distinguish pairs of diets (LF compared with LGI, LF compared with VLC, and LGI compared with VLC). We then applied each classifier to the data from the withheld sample to determine whether the classifiers could correctly identify which diet the individual had consumed when the sample was obtained.
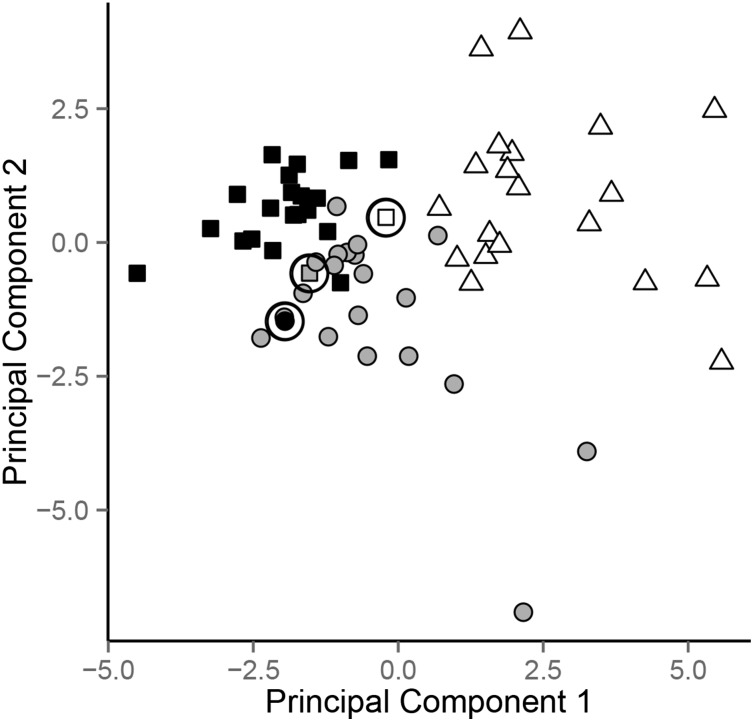
For the most difficult task, distinguishing each diet from the others, the classifier succeeded in naming the correct diet with 95% accuracy (Table 1). Consistent with this high level of accuracy, a principal component analysis showed good clustering of the samples from each diet (Figure 2, Supplemental Figure 4). The accuracy for distinguishing pairs of diets was at least as high, ranging from 95% to 100% (Table 1). We also performed 5-fold cross validation, using 80% of data to train models and 20% of data to test predictions in each fold. The resulting prediction accuracies were slightly lower, as expected with smaller training sets, but still comparable (data not shown).
TABLE 1.
Combinations of metabolites accurately identify the type of diet being consumed1
| Classifier | Accuracy, % | Misclassifications | Metabolites most strongly contributing to classifier |
| 3-way (LF vs. LGI vs. VLC) | 95 | LF misclassified as LGI (1), LGI misclassified as LF (1), VLC misclassified as LF (1) | 4-pyridoxate, C2 carnitine, C34:4 PC, C5 carnitine, C50:4 TAG, C52:5 TAG, C54:4 TAG, C56:9 TAG, C58:9 TAG, GABA, allantoin, cytidine, cytosine, gentisate, hippurate, indole-3-propionate, indoxylsulfate, pipecolic acid, sorbitol, thiamin, threonine, valine |
| LF vs. LGI | 95 | LF misclassified as LGI (2) | 11-HETE, 4-pyridoxate, C36:4 DAG, C42:0 TAG, C50:4 TAG, C56:9 TAG, GABA, alanine, allantoin, cytidine, cytosine, hippurate, hydroxyproline, thiamin |
| LF vs. VLC | 98 | VLC misclassified as LF (1) | 4-pyridoxate, C34:3 PC, C34:4 PC, C5 carnitine, C52:5 TAG, C52:6 TAG, C54:5 TAG, C54:8 TAG, C56:10 TAG, C9 carnitine, GABA, allantoin, aminoisobutyric acid, β-hydroxybutyrate, gentisate, indole-3-propionate, indoxylsulfate, leucine, propionate, sorbitol, thiamin, threonine, uracil |
| LGI vs. VLC | 100 | None | C18:2 LPE, C2 carnitine, C20:4 CE, C34:0 PC, C34:4 PC, C48:3 TAG, C5 carnitine, C54:4 TAG, C5:1 carnitine, C60:12 TAG, β-alanine, creatine, cytosine, hippurate, indole-3-propionate, indoxylsulfate, pipecolic acid, sorbitol |
Metabolites shown are those that most strongly contributed to the classifier for the comparison shown in the left-hand column. GABA, γ-aminobutyric acid; LF, low-fat diet; LGI, low–glycemic index diet; TAG, triacylglycerol; VLC, very–low carbohydrate diet.
FIGURE 2.
Accuracy of diet classification according to metabolite profile. For each sample, the data from that sample was withheld, and a Bayesian network classifier was constructed from the metabolite profile data among the remaining samples. Then, the classifier was used to predict which diet had been consumed when the withheld sample was obtained. Samples are plotted based on their position relative to the 2 principal components that capture the most variation in the metabolite profiling data for each classifier. Colors indicate the actual diet corresponding to each sample (LF, black; LGI, gray; VLC, white), and shapes indicate the classifier prediction by the classifier (LF, square; LGI, round; VLC, triangle). Misclassified samples are circled. LF, low-fat diet; LGI, low–glycemic index diet; VLC, very–low carbohydrate diet.
To assess generalizability to a population-based study involving self-reported diet, we also examined the associations between diet and metabolites in the FHS. Of the 103 diet-associated metabolites that were measured in FHS, 33 metabolites showed correlations with ≥1 macronutrient after Bonferroni’s correction for multiple testing (Supplemental Table 4). For 26 of these, the metabolite concentrations differed most between the LF and the VLC, and the direction of effect was consistent in the FHS data for 25 of the 26 metabolites. Among the other 7 metabolites, predictions related to dietary protein could be made, and these were all directionally consistent in FHS. A general pattern of directional consistency was also seen for an additional 45 metabolites that showed nominally significant (P < 0.05) correlations in FHS.
Effect of diet on metabolites that predict disease risk
Multiple metabolites that have been linked to risk of type 2 diabetes (31, 32, 38–42) showed strong or moderate associations with diet, including amino acids (isoleucine, leucine, valine, tyrosine, serine and glutamine), lipids (C18:2 lysophosphatidylethanolamine (LPE), C22:6 LPE, C56:9 TAG, C58:10 TAG, and C60:12 TAG), and others (2-aminoadipate, betaine, and C3 carnitine) as depicted in Supplemental Figures 5 and 6. The concordance between the metabolite profile that predicts type 2 diabetes risk and the diet-associated changes varied across classes of metabolites. For amino acids, concentrations with the LGI more closely resembled the protective risk profile for future type 2 diabetes risk, whereas for some other metabolites, the LF and VLC suggested protective effects.
DISCUSSION
Using a controlled, crossover feeding study as a benchmark of dietary compliance, we identified metabolomic profiles that characterized each of 3 diets differing in macronutrient composition and with high levels of confidence could distinguish which diet an individual participant had consumed. These effects were independent of body weight and time-varying confounding because the test diets were initiated after weight loss, weight was stable during the diets, and diet order was randomized.
A large number of metabolites, nearly half of the total examined, varied significantly with dietary composition (by using stringent statistical criteria and a relatively low number of participants), highlighting the broad impact of dietary composition on metabolic pathways. These metabolites fall conceptually into ≥3 categories, including those present in food, those formed through microbial action in the gastrointestinal tract, or those produced endogenously in response to diet. Some metabolites may originate directly from specific foods, such as food additives (sorbitol; propionate, an antifungal agent; and gentisate, a derivative of benzoic acid), fatty acids from high-fat foods such as butter (myristic acid), meats and plants (dietary cholines such as betaine, sarcosine, and dimethylglycine), and beverages (hippurate from tea and juices). Other substances are likely produced or transformed by gut microbiota, such as hydroxyphenylacetate, indole-3-propionate, and trimethylamine-n-oxide (43). Among endogenously generated metabolites, some suggest potential actions of diet on pathways related to diabetes (Supplemental Figures 5 and 6), inflammation (e.g., LTB4, 11-HETE and allantoin, γ-aminobutyric acid) (44–46), and energy metabolism (uridine, creatine) (47, 48). Although these associations cannot be viewed as causal, specific to all diets with similar macronutrient composition, or necessarily of biologically important magnitude, they may help elucidate the relation between dietary composition and chronic disease and stimulate additional mechanistically oriented research.
Few feeding studies, in which compliance can be reasonably ascertained, have examined metabolomic changes with macronutrient composition or GI. Barton et al. (13) compared low- and high-GI diets in a 28-d crossover trial and reported that one metabolite, kynurenate, differed statistically after adjustment for multiple comparisons. Kynurenate is an N-methyl-d-aspartate receptor antagonist with putative antiinflammatory effects. We did not find a diet effect (after adjustment for multiple comparisons) involving kynurenate in the present study, but several other potential markers or mediators of inflammation differed significantly across dietary treatments in a consistent manner, including LTB4, allantoin, and γ-aminobutyric acid. Bondia-Pons et al. (14) examined postprandial changes in metabolites following consumption of a low-insulinotropic rye bread with high-insulinotropic white wheat bread using a crossover design. Among 255 metabolites assessed, 26 showed significant differences between meal conditions, including amino acids, amino acid–derived compounds, tricarboxylic acid cycle metabolites, and organic acids. We did not observe a similar pattern of response, although differences in study design confound a direct comparison. Thus, the paucity of metabolomic feeding studies and heterogeneity between studies with regard to design and dietary treatments emphasize the need for further investigation of a generalizable fingerprint for macronutrient intake.
The primary limitation of this study, as suggested above, relates to generalizability. Here, we showed how metabolomic profiling could be used to assess compliance with studies of dietary composition focused on macronutrients, and in addition, generated hypotheses relating diet to health. But the specific results of this study would not translate directly to other research. Diets differ in many biologically important ways beyond macronutrient composition, and all eating plans described as LF, LGI, or VLC will not necessarily affect metabolomic profiles in similar ways. (This limitation would apply broadly to the outcomes of many diet studies beyond metabolomics.) In addition, metabolomic responses may differ between study populations as a consequence of age, sex, metabolic health, or other factors. Furthermore, because of the short-term nature of the intervention, we do not know how stable the observed metabolomic profiles would remain over the long term. Thus, these findings should be considered proof of principle. Even so, the consistent results obtained in an entirely independent cohort (FHS) based on self-report of people consuming self-prepared diets provide some reassurance in this regard. As additional metabolomic data from diverse studies accrue, we may be able to identify a generalizable fingerprint common to any diet that shares specific features, potentially providing a method to predict an individual’s diet from metabolomic analysis. For now, investigators using this approach would need to generate metabolomic profiles specific for their diets, with individuals representative of their study population.
In conclusion, metabolomics offers a promising approach to assess adherence in outpatient behavioral diet studies with substantial advantages over conventional dietary recall methodology and laborious or imprecise single-endpoint biomarkers. Routine use of metabolomic profiles for this purpose has potential to improve interpretation of behavioral weight loss studies with null findings (a substantial proportion of the total), by helping distinguish between lack of differential efficacy between diets compared with nonadherence. Ultimately, this distinction has direct relevance to improving the design of more effective interventions.
Acknowledgments
The authors’ responsibilities were as follows—TE, JNH, CBE, and DSL: designed the study; TE, JNH, HAF, Y-HHH, AAD, and CBC: conducted the data analyses; CBE: provided oversight of dietary interventions and sample collection; TE, JNH, and DSL: wrote the manuscript; JNH and DSL: had primary responsibility for the final content; and all authors: read and approved the final manuscript. DSL received royalties from books about obesity and nutrition. None of the other authors reported a conflict of interest related to the study.
Footnotes
Abbreviations used: FHS, Framingham Heart Study; GI, glycemic index; LC-MS, liquid chromatography–tandem mass spectrometry; LF, low-fat diet; LGI, low–glycemic index diet; VLC, very-low-carbohydrate diet.
REFERENCES
- 1.Dansinger ML, Gleason JA, Griffith JL, Selker HP, Schaefer EJ. Comparison of the Atkins, Ornish, Weight Watchers, and Zone diets for weight loss and heart disease risk reduction: a randomized trial. JAMA 2005;293:43–53. [DOI] [PubMed] [Google Scholar]
- 2.Johnston BC, Kanters S, Bandayrel K, Wu P, Naji F, Siemieniuk RA, Ball GD, Busse JW, Thorlund K, Guyatt G, et al. Comparison of weight loss among named diet programs in overweight and obese adults: a meta-analysis. JAMA 2014;312:923–33. [DOI] [PubMed] [Google Scholar]
- 3.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwig DS, Ebbeling CB, Livingston EH. Surgical vs lifestyle treatment for type 2 diabetes. JAMA 2012;308:981–2. [DOI] [PubMed] [Google Scholar]
- 5.McDonald SM, Liu J, Wilcox S, Lau EY, Archer E. Does dose matter in reducing gestational weight gain in exercise interventions? A systematic review of literature. J Sci Med Sport 2016;19:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu T, Gao X, Chen M, van Dam RM. Long-term effectiveness of diet-plus-exercise interventions vs. diet-only interventions for weight loss: a meta-analysis. Obes Rev 2009;10:313–23. [DOI] [PubMed] [Google Scholar]
- 7.Abu Bakar MH, Sarmidi MR, Cheng KK, Ali Khan A, Suan CL, Zaman Huri H, Yaakob H. Metabolomics - the complementary field in systems biology: a review on obesity and type 2 diabetes. Mol Biosyst 2015;11:1742–74. [DOI] [PubMed] [Google Scholar]
- 8.Rauschert S, Uhl O, Koletzko B, Hellmuth C. Metabolomic biomarkers for obesity in humans: a short review. Ann Nutr Metab 2014;64:314–24. [DOI] [PubMed] [Google Scholar]
- 9.Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol 2014;2:65–75. [DOI] [PubMed] [Google Scholar]
- 10.Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Draper J, Rappaport SM, van der Hooft JJ, Wishart DS. The food metabolome: a window over dietary exposure. Am J Clin Nutr 2014;99:1286–308. [DOI] [PubMed] [Google Scholar]
- 11.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation 2012;126:1110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arora T, Velagapudi V, Pournaras DJ, Welbourn R, le Roux CW, Oresic M, Backhed F. Roux-en-Y gastric bypass surgery induces early plasma metabolomic and lipidomic alterations in humans associated with diabetes remission. PLoS One 2015;10:e0126401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton S, Navarro SL, Buas MF, Schwarz Y, Gu H, Djukovic D, Raftery D, Kratz M, Neuhouser ML, Lampe JW. Targeted plasma metabolome response to variations in dietary glycemic load in a randomized, controlled, crossover feeding trial in healthy adults. Food Funct 2015;6:2949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bondia-Pons I, Nordlund E, Mattila I, Katina K, Aura AM, Kolehmainen M, Oresic M, Mykkanen H, Poutanen K. Postprandial differences in the plasma metabolome of healthy Finnish subjects after intake of a sourdough fermented endosperm rye bread versus white wheat bread. Nutr J 2011;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floegel A, von Ruesten A, Drogan D, Schulze MB, Prehn C, Adamski J, Pischon T, Boeing H. Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. Eur J Clin Nutr 2013;67:1100–8. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons H, McNulty BA, Nugent AP, Walton J, Flynn A, Gibney MJ, Brennan L. A metabolomics approach to the identification of biomarkers of sugar-sweetened beverage intake. Am J Clin Nutr 2015;101:471–7. [DOI] [PubMed] [Google Scholar]
- 17.Mondul AM, Sampson JN, Moore SC, Weinstein SJ, Evans AM, Karoly ED, Virtamo J, Albanes D. Metabolomic profile of response to supplementation with beta-carotene in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study. Am J Clin Nutr 2013;98:488–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Sullivan A, Gibney MJ, Brennan L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am J Clin Nutr 2011;93:314–21. [DOI] [PubMed] [Google Scholar]
- 19.Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, Newgard CB, Bowden DW. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab 2015;100:E463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccolo BD, Keim NL, Fiehn O, Adams SH, Van Loan MD, Newman JW. Habitual physical activity and plasma metabolomic patterns distinguish individuals with low vs. high weight loss during controlled energy restriction. J Nutr 2015;145:681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen LG, Winning H, Savorani F, Toft H, Larsen TM, Dragsted LO, Astrup A, Engelsen SB. Assessment of the effect of high or low protein diet on the human urine metabolome as measured by NMR. Nutrients 2012;4:112–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt JA, Rinaldi S, Ferrari P, Carayol M, Achaintre D, Scalbert A, Cross AJ, Gunter MJ, Fensom GK, Appleby PN, et al. Metabolic profiles of male meat eaters, fish eaters, vegetarians, and vegans from the EPIC-Oxford cohort. Am J Clin Nutr 2015;102:1518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaarhorst AA, Verhoeven A, Weller CM, Bohringer S, Goraler S, Meissner A, Deelder AM, Henneman P, Gorgels AP, van den Brandt PA, et al. A metabolomic profile is associated with the risk of incident coronary heart disease. Am Heart J 2014;168:45–52.e7. [DOI] [PubMed] [Google Scholar]
- 24.Vázquez-Fresno R, Llorach R, Urpi-Sarda M, Lupianez-Barbero A, Estruch R, Corella D, Fito M, Aros F, Ruiz-Canela M, Salas-Salvado J, et al. Metabolomic pattern analysis after mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J Proteome Res 2015;14:531–40. [DOI] [PubMed] [Google Scholar]
- 25.Wittenbecher C, Muhlenbruch K, Kroger J, Jacobs S, Kuxhaus O, Floegel A, Fritsche A, Pischon T, Prehn C, Adamski J, et al. Amino acids, lipid metabolites, and ferritin as potential mediators linking red meat consumption to type 2 diabetes. Am J Clin Nutr 2015;101:1241–50. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Yu B, Alexander D, Steffen LM, Boerwinkle E. Human metabolome associates with dietary intake habits among African Americans in the atherosclerosis risk in communities study. Am J Epidemiol 2014;179:1424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Playdon MC, Sampson JN, Cross AJ, Sinha R, Guertin KA, Moy KA, Rothman N, Irwin ML, Mayne ST, Stolzenberg-Solomon R, et al. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr 2016;104:776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebbeling CB, Swain JF, Feldman HA, Wong WW, Hachey DL, Garcia-Lago E, Ludwig DS. Effects of dietary composition on energy expenditure during weight-loss maintenance. JAMA 2012;307:2627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, Siddiqui S, Basso AS, Otterbein LE, Pardoll DM, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med 2015;21:638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 2011;121:1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 2013;123:4309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res 2012;40(Web Server issue):W127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res 2013;41:D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper G, Herskovits EA. A Bayesian method for the induction of probabilistic networks from data. Mach Learn 1992;9:309–47. [Google Scholar]
- 37.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor 2009;11:1–10. [Google Scholar]
- 38.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012;125:2222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnusson M, Lewis GD, Ericson U, Orho-Melander M, Hedblad B, Engstrom G, Ostling G, Clish C, Wang TJ, Gerszten RE, et al. A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J 2013;34:1982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stancákova A, Civelek M, Saleem NK, Soininen P, Kangas AJ, Cederberg H, Paananen J, Pihlajamaki J, Bonnycastle LL, Morken MA, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes 2012;61:1895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walford GA, Davis J, Warner AS, Ackerman RJ, Billings LK, Chamarthi B, Fanelli RR, Hernandez AM, Huang C, Khan SQ, et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism 2013;62:1772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walford GA, Ma Y, Clish C, Florez JC, Wang TJ, Gerszten RE; Diabetes Prevention Program Research Group. metabolite profiles of diabetes incidence and intervention response in the Diabetes Prevention Program. Diabetes 2016;65:1424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonso PV, Janka-Junttila M, Lee YJ, McCann CP, Oliver CM, Aamer KA, Losert W, Cicerone MT, Parent CA. LTB4 is a signal-relay molecule during neutrophil chemotaxis. Dev Cell 2012;22:1079–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhat R, Axtell R, Mitra A, Miranda M, Lock C, Tsien RW, Steinman L. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci USA 2010;107:2580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Il’yasova D, Spasojevic I, Wang F, Tolun AA, Base K, Young SP, Marcom PK, Marks J, Mixon G, DiGiulio R, et al. Urinary biomarkers of oxidative status in a clinical model of oxidative assault. Cancer Epidemiol Biomarkers Prev 2010;19:1506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joncquel-Chevalier Curt M, Voicu PM, Fontaine M, Dessein AF, Porchet N, Mention-Mulliez K, Dobbelaere D, Soto-Ares G, Cheillan D, Vamecq J. Creatine biosynthesis and transport in health and disease. Biochimie 2015;119:146–65. [DOI] [PubMed] [Google Scholar]
- 48.Scherer PE. The multifaceted roles of adipose tissue-therapeutic targets for diabetes and beyond: the 2015 Banting Lecture. Diabetes 2016;65:1452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]