Abstract
Gene-knockout pigs hold great promise as a solution to the shortage of organs from donor animals for xenotransplantation. Several groups have generated gene-knockout pigs via clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) and somatic cell nuclear transfer (SCNT). Herein, we adopted a simple and micromanipulator-free method, handmade cloning (HMC) instead of SCNT, to generate double gene-knockout pigs. First, we applied the CRISPR/Cas9 system to target α1,3-galactosyltransferase (GGTA1) and cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH) genes simultaneously in porcine fetal fibroblast cells (PFFs), which were derived from wild-type Chinese domestic miniature Wuzhishan pigs. Cell colonies were obtained by screening and were identified by Surveyor assay and sequencing. Next, we chose the GGTA1/CMAH double-knockout (DKO) cells for HMC to produce piglets. As a result, we obtained 11 live bi-allelic GGTA1/CMAH DKO piglets with the identical phenotype. Compared to cells from GGTA1-knockout pigs, human antibody binding and antibody-mediated complement-dependent cytotoxicity were significantly reduced in cells from GGTA1/CMAH DKO pigs, which demonstrated that our pigs would exhibit reduced humoral rejection in xenotransplantation. These data suggested that the combination of CRISPR/Cas9 and HMC technology provided an efficient and new strategy for producing pigs with multiple genetic modifications.
Keywords: CRISPR/Cas9, Double-knockout pigs, Handmade cloning, Xenotransplantation
Gene-knockout pigs have great promise for clinical xenotransplantation as sources of organs and cells [1]. Because of the lack of identifiable embryonic stem cells, the generation of gene-knockout pigs has proved difficult and slow [2].
Zinc-finger nuclease (ZFN) and transcription activator-like effector nuclease (TALEN) gene-editing technologies have made the creation of gene-knockout pigs easier and faster. To date, several gene-knockout pigs have been generated by ZFNs and TALENs [3,4,5,6,7]. However, both of these systems are expensive and complex to design. Additionally, TALENs cannot easily target some parts of the genome [2].
Recently, some problems have been solved by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system, which has been developed as a new gene-editing technology [8,9,10]. This efficient and powerful system has revolutionized genome engineering. The CRISPR/Cas9 system has been applied in various animals, such as mice [10], rats [9], sheep [11], chickens [12], and pigs [2, 13, 14].
The somatic cell nuclear transfer (SCNT) technique, which was invented in 1997, was a great scientific innovation in the animal cloning field. Based on SCNT, handmade cloning (HMC), which was invented by Vajta et al., is not only a promising and simple version of SCNT but also a hand-guided technique or zona-free cloning without the use of a micromanipulator [15, 16]. In the HMC process, the oocytes are enucleated using a microblade under a stereomicroscope. Then, a single donor cell is placed in the middle of two enucleated cytoplasts and fused by an appropriate electric shock. The constructed embryos are cultured in vitro and D6 blastocysts are transferred to uterine horns of surrogates. Finally, the cloned piglets are delivered approximately 114 days later. HMC has revolutionized the field of embryology without the use of expensive micromanipulators and skilled expertise [17, 18].
In the present study, we attempted to generate two gene-knockout pigs rapidly by combining CRISPR/Cas9 and HMC. The α1,3-galactosyltransferase (GGTA1) and cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH) genes were chosen as targets, as these are responsible for the synthesis of Galα-1,3-Gal (Gal) and N-glycolylneuraminic acid (Neu5Gc), respectively—two carbohydrate xenoantigens that are important for xenotransplantation. Deletion of the two genes together may further reduce antibody-mediated rejection after xenotransplantation. In fact, GGTA1/CMAH double-knockout (DKO) pigs have been generated by ZFNs [5], a combination of ZFNs and TALENs [6], and the CRISPR/Cas9 system [19, 20]. However, in this study, we generated the DKO pigs with a novel strategy that combined the CRISPR/Cas9 system and HMC. We disrupted these two genes simultaneously in the same somatic cell using the CRISPR/Cas9 system, followed by HMC to produce the gene DKO pigs. DKO pigs were obtained within 6 months. Our study provided a new strategy to generate pigs with more than one gene modification.
Materials and Methods
Animals care and chemicals
All the animal experiments were approved by the Institutional Review Board on Bioethics and Biosafety of Beijing Genomics Institute (BGI-IRB). All surgical procedures were performed under full anesthesia, and all efforts were made to minimize animal suffering. All chemicals were purchased from Sigma Chemical Co., except where otherwise indicated.
sgRNA design and vector construction
Single guide RNAs (sgRNAs) targeting pig GGTA1 and CMAH genes were designed following protocols described previously [21]. The GGTA1 sgRNA sequence was 5′-GCAAATACATACTTCATGGTTGG-3′, which targets exon 6, and the CMAH sgRNA sequence was 5′-GAAGCTGCCAATCTCAAGGAAGG-3′, which targets exon 1. The cas9-coding DNA fragment was synthesized and cloned into the pMD-18T vector (Takara, Dalian, Liaoning, China). A cytomegalovirus (CMV) promoter was used to drive transcription of Cas9 in the vector. The U6-sgRNA fragment was synthesized and cloned into the pMD-18T vector. Two BsaI restriction sites were introduced into the region between the U6 promoter and sgRNA tail. For sgRNA vector construction, two complementary oligo DNAs were synthesized and then annealed to a double-strand DNA, ligated to the BsaI site of the U6-sgRNA vector to form an integral sgRNA-expressing frame.
sgRNA cleavage activity validation
For sgRNA activity validation, PK15 cells were cultured in Dulbecco′s Minimum Essential Medium (DMEM; Gibco, Gaithersburg, MD, USA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco) in a 24-well plate. When the cells grew to 50–60% confluence, sgRNAs and cas9 vectors were mixed at a weight ratio of 3:1 and co-transfected into PK15 cells using the lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer′s protocols. To enhance the transfection efficiency and increase the sensitivity of the subsequent Surveyor assay, the transfection was completed twice. The second transfection was performed on the same cell well 12–24 h after the first, and all the cells were collected 48 h later. Cells transfected with the pN1-EGFP vector (Clontech, Palo Alto, CA, USA) acted as a control group.
Subsequently, the Surveyor assay was performed to identify sgRNA cleavage activity, as described previously [22]. Briefly, PCR was performed on DNA extracted from transfected cells using specific primers, which amplified exon 6 of pig GGTA1 and exon 1 of pig CMAH. For the GGTA1 gene, the forward primer was 5′-AGCCACCCTGCCAGAATCAC-3′ and the reverse primer was 5′-GGAGGATTCCCTTGAAGCACTC-3′. For the CMAH gene, the forward primer was 5′-GCATCTAAAAGCGAGGTAGTAG-3′ and the reverse primer was 5′- ATCCAAAAGAGGCTTGAGTC-3′. The PCR conditions were 94°C for 5 min, 94°C for 30 sec, 57°C for 30 sec, 72°C for 30 sec, for 35 cycles, 72°C for 5 min, and held at 16°C. Subsequently, a mixture containing 18 µl PCR product and 2 µl 10 × NEB buffer 2 was incubated at 95°C for 10 min and then incubated at room temperature for 30 min. After the annealing reaction, 0.2 µl of T7 endonuclease I (T7E1) (NEB, Ipswich, MA, USA) was added to the sample and the mixture was incubated at 37°C for 1 h. Finally, the digested product was electrophoresed on a 2% agarose gel and the electropherogram was visually analyzed using Tanon 1.1 (Tanon, Shanghai, China).
Preparation of Wuzhishan porcine fetal fibroblasts (PFFs)
PFFs used throughout this study were primary cells isolated from the Chinese Wuzhishan male fetuses at 40 days after insemination, as described previously [23]. The PFFs were cultured in Dulbecco′s Minimum Essential Medium (DMEM; Gibco) supplemented with 15% (vol/vol) FBS, 1% non-essential amino acid (NEAA; Invitrogen), and 1% l-glutamine (Invitrogen) at 38°C in 5% CO2 in air.
Selection of GGTA1 and CMAH double gene bi-allelic knockout cells
Before nucleofection, PFF cells were thawed and cultured until 70–90% confluent. Approximately 1× 106 cells were subjected to nucleofection using the Basic Nucleofector® Kit for Primary Mammalian Fibroblasts (Lonza, Allendale, NJ, USA) following the manufacturer′s protocols. Briefly, the required number of cells were suspended in 100 µl Nucleofector® Solution at room temperature. Cyclic vectors of GGTA1-sgRNA, CMAH-sgRNA, Cas9, and linearized pcDNA3.1(+) were mixed in advance at a weight ratio of 3:3:2:0.4. A total of 5 µg of DNA was mixed with the cell resuspension. Nucleofection was performed on a Nucleofector™ 2b Device (Lonza) by running the U-023 program. After 24 h recovery, the cells were selected with G418 at a concentration of 450 µg/ml for 8–10 days. Individual cell colonies were selected and genotyped.
The cell colonies were subjected to two rounds of T7E1 digestion. In the first round, PCR products were digested immediately after the annealing reaction for identification of heterozygotes. In the second round, PCR products of the candidate homozygotes and wild-types were mixed at a volume ratio of 1:1 and the T7E1 assay was performed. Positive digestion during the second round was considered indication of probable homozygotes. Then GGTA1 and CMAH loci of the positive cell colonies were genotyped by Sanger sequencing.
Oocyte maturation
Slaughterhouse-derived sow ovaries were collected in physiological saline solution and transported to the laboratory at 32°C within 4 h. Cumulus-oocyte complexes (COCs) were recovered from 3–5 mm follicles. The compact COCs were selected in groups of 50. Each group was matured in 400 µl of bicarbonate buffered TCM-199 (Gibco) supplemented with 10% (v/v) pig follicular fluid, 10% cattle serum (CS; Gibco), 5 IU/ml hCG and 10 IU/ml eCG at 38.5°C in a humidified atmosphere of 5% (v/v) CO2 for 41–43 h. Subsequently, COCs were treated with 1 mg/ml hyaluronidase dissolved in Hepes-buffered TCM-199 to remove the cumulus cells.
Handmade cloning (HMC)
For HMC, cells of passages 8–12 (total passage) were used as nuclear donors. Confluent cell monolayers were harvested by digestion with 0.05% trypsin-EDTA (Gibco) and the cell suspension was placed at room temperature for 0.5–1 h before fusion.
HMC was performed, as previously described [17]. Briefly, zona pellucida of oocytes were partially digested by 3 mg/ml pronase dissolved in T33 (T means Hepes-buffered TCM 199 medium; the number for percentage (v/v) of calf serum supplement). Then, enucleation was performed with a microblade (AB Technology, Pullman, WA, USA) under a stereo microscope. The cytoplasm was washed in T2 and T20 drops (to wash out the zona pellucida fragments), and collected in a T10 drop (for buffering before fusion with the donor cells).
Fusion was performed in two steps in which the second step included the initiation of activation. For the first step, each cytoplast was transferred to 20 µl T0 containing 1 mg/ml of phytohemagglutinin (PHA) for 1 to 3 sec and subsequently attached to a single donor cell. Then, cytoplast-fibroblast pairs were fused with a single 100 V direct current (DC) impulse of 2.0 kV/cm for 9 µs (BLS CF-150/B fusion machine) in fusion medium (0.3 M mannitol and 0.01% PVA). Approximately 1 h later, each fused pair was fused with another putative cytoplast in activation medium (0.3 M mannitol, 0.1 mM MgSO4, 0.1 mM CaCl2, and 0.01% PVA) with a single DC pulse of 0.86 KV/cm for 80 µs. Reconstructed embryos were treated with 10 mg/ml cycloheximide and 5 mg/ml cytochalasin B dissolved in PZM-3 at 38.5°C in 5% CO2 and 5% O2 for 4 to 6 h.
Embryo culture, transfer, and cloned piglet genotype assay
Zona-free embryos produced from HMC were cultured in groups of 20 to 30 in 400 µl PZM-3 supplemented with 4 mg/ml bovine serum albumin (BSA) in a modified ‘Well of the Well′ (WOW) system [24]. Day 6 (D6) blastocysts produced from HMC were surgically transferred to uterine horns of surrogates. Pregnancies were confirmed by B ultrasound scanning on day 28 and monitored every 2 weeks thereafter. The cloned piglets were delivered by natural birth. The genomic DNA extracted from an ear skin biopsy of the newborn cloned piglets was assessed for mutagenesis at the targeted site by PCR-based assays.
Preparation of peripheral blood mononuclear cells (PBMCs), red blood cells (RBCs), and artery endothelial cells (AECs)
PBMCs and RBCs from wild-type, GGTA1/CMAH DKO, and GGTA1-KO pigs (which were kindly supplied by Dr. Dengke Pan) were isolated as previously described [25, 26]. AECs were isolated, cultured, and passaged as previously described [27, 28].
Immunofluorescence staining
Staining for Gal and Neu5Gc was performed as previously described [29]. Gal was stained with fluorescein isothiocyanate (FITC)-conjugated BS-IB4 lectin (Sigma-Aldrich). Neu5Gc was stained with an anti-Neu5Gc antibody kit (Biolegend, San Diego, CA, USA, secondary antibody and tertiary antibody from Jackson ImmunoResearch, West Grove, PA, USA), following the manufacturer′s protocols. The picture was obtained using a Leica DMIL fluorescent inverted microscope (Leica Microsystems) provided with an ebq 50 ac-L lamp (Leistungselektronik Jena GmbH, Jena, Germany), and UV filter.
Expression of Gal and Neu5Gc on PBMCs and AECs
Flow cytometry analysis of Gal and Neu5Gc expression in cells was previously described [26, 28]. Briefly, for Gal staining, cells were stained with FITC-conjugated BS-IB4 lectin for 30 min at 4°C; unstained cells were used as a negative control. For Neu5Gc staining, cells were stained with an anti-Neu5Gc antibody for 30 min at 4°C; an isotype-matched antibody was used as a negative control. After washing with PBS, the cells were stained with a secondary antibody (Jackson ImmunoResearch) for 30 min at 4°C, washed again, and incubated with a tertiary antibody (Biolegend) for 30 min at 4°C in the dark. After washing, the treated cells were analyzed by flow cytometry.
Human antibody binding to RBCs and PBMCs
IgG and IgM binding assays were performed as previously described [26, 27]. In brief, serum from several healthy volunteers (n = 20, including all ABO types) were mixed and heat-inactivated at 56°C for 30 min. Cells (1 × 106 RBCs or 1 × 105 PBMCs) were incubated with 2.5% (RBCs) or 13% (PBMCs) heat-inactivated serum for 30 min (RBCs) or 120 min (PBMCs) at 4°C. After washing with PBS, 10% goat serum was used to prevent non-specific binding. Next, the cells were incubated with FITC-conjugated goat anti-human IgM or IgG antibodies (Invitrogen) for 30 min at 4°C in the dark. After washing, the treated cells were analyzed by flow cytometry. The extent of IgM and IgG binding was evaluated by relative mean fluorescence intensity (MFI), using the following formula:
Relative MFI = (actual MFI)/(MFI obtained with secondary antibodies only without serum)
Antibody-mediated complement-dependent cytotoxicity
The assay was conducted as previously described [30]. Briefly, PBMCs (2 × 106) were incubated with various diluted serum samples in 200 µl for 30 min at 37°C. PBMCs were incubated with heat-inactivated serum as a negative control. After washing twice with PBS, the cells were incubated with propidium iodide (PI, 1:1000; Invitrogen) for 15 min at 4°C. After washing, the treated cells were analyzed by flow cytometry. The PI-positive cells were used to assess cell death. The percentage of cytotoxicity was calculated by the following formula:
%cytotoxicity = (A-B)/(100-B) × 100%, where A represents the percentage of PI-positive cells incubated with serum, and B represents the percentage of PI-positive cells incubated with heat-inactivated serum.
Statistical analysis
Data are presented as the mean ± SEM. A two-tailed Student′s t test was used for analysis of the differences between the groups. All P values < 0.05 were considered statistically significant.
Results
Generation of bi-allelic GGTA1 and CMAH DKO cells
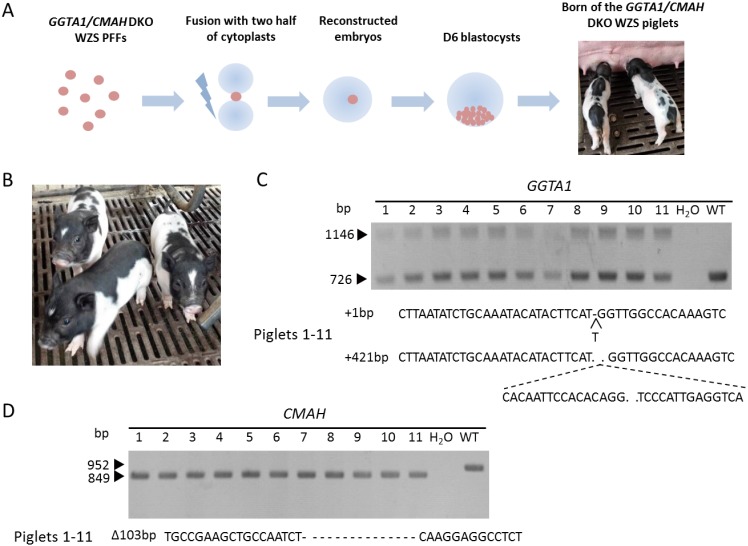
sgRNAs were designed to target exon 6 of pig GGTA1 and exon 1 of pig CMAH (Fig. 1A). We selected at least three sgRNAs of each gene locus and tested their cleavage activities in vitro via PK15 cell transfection and T7E1 Surveyor assay. One sgRNA within each gene locus exhibited cleavage activity (Fig. 1B). Then, nucleofection was performed on Wuzhishan PFFs and G418-resistant cell colonies were selected. One candidate positive cell colony (#8) was found via Surveyor assay among the 11 cell colonies selected (Fig. 2A), and the PCR and sequencing results validated that the genotype of the #8 cell colony was a bi-allelic GGTA1 and CMAH DKO cell colony (Fig. 2B–C). For the GGTA1 gene locus, one allele was inserted with 1 base-pair, which led to a frameshift mutation exactly after the CRISPR cut site in exon 6, and the another allele was inserted with 421 base-pairs, in which there was a partial DNA sequence of the pcDNA3.1 vector (Fig. 2B–C). For the CMAH gene locus, there was a 103 base-pair deletion after the CRISPR cut site in exon 1, and the genotype was homozygous by validation of the electrophoresis assay and PCR product sequencing (Fig. 2B–C).
Fig. 1.
CRISPR/Cas9 system targeting pig GGTA1 and CMAH loci. (A) Schematic of sgRNAs targeting GGTA1 and CMAH loci. The target sgRNAs were designed in the last exon of the GGTA1 locus and the first exon of the CMAH locus. sgRNA-target sequences are labeled in lowercase and the protospacer adjacent motif (PAM) is underlined. (B) Surveyor assay (T7E1) for Cas9-mediated cleavage at GGTA1 and CMAH loci in PK15 cells.
Fig. 2.
Selection of GGTA1 and CMAH gene bi-allelic DKO cell colonies. (A) Cell colonies were selected and subjected to two rounds of T7E1 Surveyor assays. The first round (left, PCR products of the candidate colonies were digested by T7E1) excluded heterozygotes, and the second round (right, PCR products of the candidate colonies and wild-type cells were mixed and then digested by T7E1) identified the candidate homozygotes. * indicates T7E1-digested fragments of the #8 cell colony at the GGTA1 and CMAH loci. (B) PCR amplicons of GGTA1 (left) and CMAH (right) loci of the #8 cell colony and wild-type cells. (C) Genotypes of the #8 cell colony at the GGTA1 and CMAH loci. PAM sequence is underlined.
Production of GGTA1/CMAH DKO pigs via HMC
The #8 colony cells were used as nuclear donors of HMC, and the GGTA1/CMAH DKO pigs were generated (Fig. 3A–B). A total of 1,307 reconstructed embryos derived from GGTA1/CMAH DKO cells were created. The overall D6 blastocyst rate was 45%, which was statistically higher than that of the average rate in our laboratory over the past 4 years (45% vs. 36%), and also higher than that of previous knockout cells (45% vs. 34%) (Table 1) [23]. Then, 517 blastocysts were transferred into eight recipients, and three of them became pregnant, indicating a pregnancy rate of 38% (Table 1). All three surrogate mothers developed to term, which led to the birth of 13 piglets, of which two piglets were stillborn and 11 piglets were live born (Table 1). Total cloning efficiency was 2.5% (Table 1). Genomic DNA extracted from ear skin biopsies of the 11 cloned piglets was assessed for mutagenesis at the targeted sites by PCR-based assays and sequencing analysis to determine the genotype of the pigs. The results indicated that all 11 cloned piglets were derived from one nuclear donor and the genotypes of the GGTA1 and CMAH loci were the same as that of the #8 cell donor (Fig. 3C–D).
Fig. 3.
Generation of GGTA1 and CMAH gene DKO piglets by HMC. (A) A schematic description of GGTA1/CMAH Wuzhishan PFFs used as nuclear donors and subjected to HMC resulting in the birth of GGTA1 and CMAH DKO piglets. (B) Photograph of three GGTA1/CMAH DKO piglets. (C) PCR products (above) and genotype analyzed by Sanger sequencing (below) of the 11 cloned piglets at the GGTA1 loci. (D) PCR products (above) and genotype analyzed by Sanger sequencing (below) of the 11 cloned piglets at the CMAH loci.
Table 1. Handmade cloning for generating GGTA1/CMAH double-knockout pigs.
| Recipients | Reconstructed embryos | D6 blastocyst (%) | Blastocyst transferred | Pregnancy | Still born | Live born | Cloning efficiency % |
| 01 | 219 | 79 (36) | 63 | No | – | – | – |
| 02 | 243 | 128 (53) | 103 | No | – | – | – |
| 03 | 239 | 96 (40) | 81 | Yes | 0 | 3 | 3.7 |
| 04 | 117 | 58 (50) | 55 | Yes | 2 | 4 | 10.9 |
| 05 | 105 | 52 (50) | 49 | No | – | – | – |
| 06 | 105 | 47 (45) | 46 | Yes | 0 | 4 | 8.7 |
| 07 | 104 | 45 (43) | 45 | No | – | – | – |
| 08 | 175 | 77 (44) | 75 | No | – | – | – |
| Total | 1307 | 582 (45) | 517 | 3 (38%) | 2 | 11 | 2.5 |
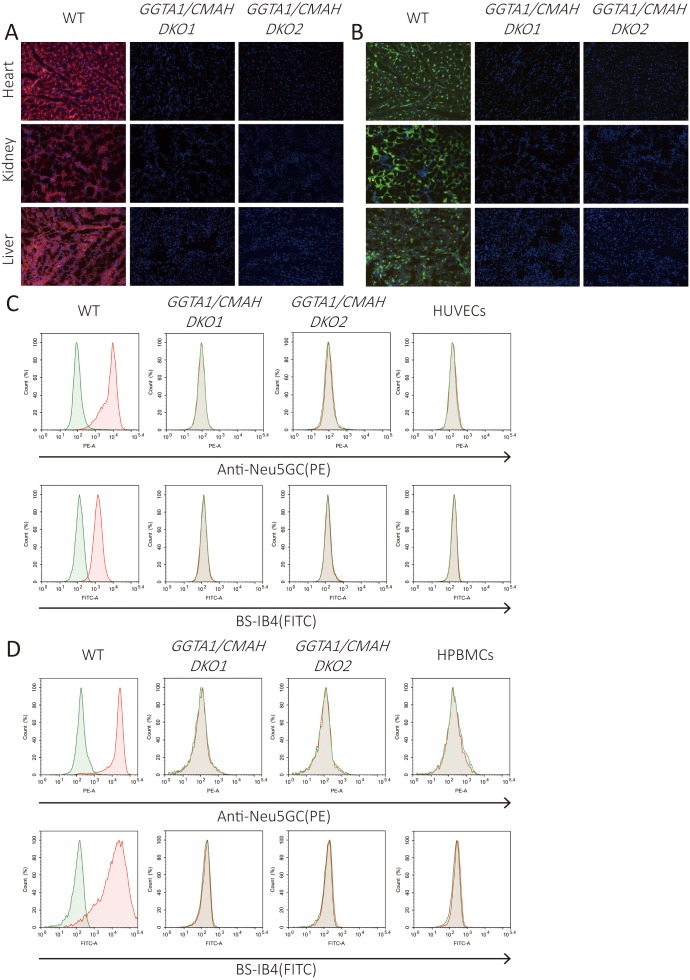
Next, we checked Neu5Gc and Gal antigen expression in the DKO pigs we generated. Histological analysis of various tissues obtained from two GGTA1/CMAH DKO pigs indicated that neither the Neu5Gc nor the Gal antigen were detected on any of the tissues from the DKO pigs, whereas antigen expression was normal in wild-type pigs (Fig. 4A–B). Flow cytometry analysis of PAECs and PBMCs from the GGTA1/CMAH DKO pigs revealed similar results (Fig. 4C–D), and demonstrated that the DKO pigs lacked both Neu5Gc and Gal antigen expression.
Fig. 4.
Immunofluorescence and flow cytometry analysis of Gal and Neu5Gc expression in GGTA1/CMAH DKO pigs. (A–B) Immunofluorescence of various tissues from wild-type or GGTA1/CMAH DKO pigs stained with anti-Neu5Gc antibody (A) and BS-IB4 lectin (B) (Magnification × 200; Neu5Gc, red; Gal, green; Nuclei, blue). (C–D) Flow cytometry analysis of AECs (C) and PBMCs (D) from the pigs indicated in A-B stained with anti-Neu5Gc antibody and BS-IB4 lectin. Human umbilical vein endothelial cells (HUVECs) and human PBMCs (HPBMCs) were negative controls, respectively. The green lines represent negative controls; the red lines represent the experimental groups.
Off-target analysis of the newborn piglets
A major concern of utilizing the CRISPR/Cas9 system for genome editing is the potential for off-target effects [31]. Several studies have shown that CRISPR/Cas9 cleaves genomic DNA with mismatches in the guide RNA strand [32,33,34]. To test whether there was an undesired off-target mutagenesis in the genome-modified cloned piglets, the predicted off-target sites (OTS) were screened and identified, as previously described [14, 35]. Genome DNA extracted from the ear tissues of one piglet was used to perform off-target analysis. Thirteen and 12 potential OTS of the GGTA1 and CMAH locus, respectively, were PCR amplified and sequenced. Primers for amplification of the OTS are listed in Supplementary Table 1 (online only). Results showed that the GGTA1-OTS6, CMAH-OTS9, and CMAH–OTS11 presented undesired DNA mutations, which were 3 bp, 4 bp, and 2 bp deletions, respectively. However, the three deletions were in non-coding regions of the genome, which were deemed to have no negative effects on functional roles of any genes. Moreover, these 11 gene-modified pigs were healthy and some of them seemed even more active than some wild-type pigs.
Human antibody binding to cells from GGTA1/CMAH DKO pigs
Having generated these DKO pigs (without expression of Neu5Gc and Gal), we determined the level of xenoantigen expression by flow cytometry. After incubation with heat-inactivated serum from several healthy human volunteers, the mean relative MFI of IgM binding to RBCs from the wild-type, GGTA1-knockout (KO), and GGTA1/CMAH DKO pigs was 134.95, 3.09, and 1.31, respectively, whereas the mean relative MFI of IgG binding to RBCs from the above pigs was 17.28, 3.87, and 1.06, respectively (Fig. 5A). Human IgM and IgG binding to RBCs from GGTA1-KO pigs was much less than to RBCs from wild-type pigs, and there was further reduction of IgM and IgG binding to RBCs from the GGTA1/CMAH DKO pigs. We also obtained similar results with PBMCs (Fig. 5B). The data revealed that the majority of human IgM and IgG was anti-Gal antibody, and the majority of non-Gal IgM and IgG was anti-Neu5Gc antibody. Anti-Neu5Gc IgM and IgG accounted for >50% of non-Gal IgM and IgG.
Fig. 5.
Human antibody binding and antibody-mediated complement-dependent cytotoxicity of cells from GGTA1/CMAH DKO pigs. (A–B) Flow cytometry analysis of human IgM and IgG binding to RBCs (A) and PBMCs (B) from wild-type (n = 3), GGTA1-KO (n = 3), and GGTA1/CMAH DKO (n = 3) pigs. (C) Flow cytometry analysis of cytotoxicity of PBMCs from GGTA1-KO (n = 3) and GGTA1/CMAH DKO (n = 3) pigs incubated with different human serum concentrations. (*P < 0.05, ** P < 0.01, *** P < 0.001).
Effect of GGTA1/CMAH DKO PBMCs on antibody-mediated complement-dependent cytotoxicity
Next, we assessed immune rejection using an antibody-mediated complement-dependent cytotoxicity assay by flow cytometry in vitro. Using 40%, 80%, and 100% serum concentrations, cytotoxicity of PBMCs from GGTA1/CMAH DKO pigs was 2.11%, 2.86%, and 3.44%, respectively, whereas cytotoxicity of PBMCs from GGTA1-KO pigs was 6.14%, 11.58%, and 14.52%, respectively (Fig. 5C). Cytotoxicity of PBMCs from GGTA1-KO pigs was much higher than that of PBMCs from GGTA1/CMAH DKO pigs. These data indicated that the Neu5Gc antigen is a major non-Gal antigen, and suggests that it will be critical for clinical xenotransplantation.
Discussion
Organ/tissue transplantation is an important therapy for end-stage organ failure and some other human diseases, but the shortage of human organs/tissues is a major limitation [36]. Pigs are considered to be the most likely animal to solve the problem, and there are several reasons to choose pigs as xenograft ‘donors′, (e.g., physiological, anatomical, economic, and ethical) [37, 38]. However, immune rejection is a major hurdle for pig-to-human organ/tissue transplantation.
Gal is a major xenoantigen, and GGTA1-KO pigs were produced in 2002, thus reducing the humoral barrier to xenotransplantation [39]. Transplantation experiments employing hearts and kidneys from GGTA1-KO pigs into baboons proved an advance [40,41,42]. Several studies revealed that Neu5Gc, which is controlled by CMAH, is a major non-Gal antigen and would be problematic for clinical xenotransplantation [43,44,45,46]. Thus, generation of GGTA1/CMAH DKO pigs became a matter of urgency. Herein, we showed that the combination of CRISPR/Cas9 and HMC is a new strategy to generate gene DKO pigs.
We generated GGTA1/CMAH DKO pigs by targeting the GGTA1 and CMAH genes together in PFFs with CRISPR/Cas9 and HMC. We knocked out these two genes simultaneously using wild-type PFFs in a single step using the CRISPR/Cas9 system. Additionally, we obtained bi-allelic GGTA1/CMAH DKO pigs with only one round of nucleofection, clone selection, and HMC. Previously, Miyagawa et al. (2015) generated GGTA1/CMAH DKO pigs using GGTA1-KO cells and a combination of ZFN and TALEN technology [6]. This group also obtained heterozygous pigs, and subsequently, homozygous pigs, by two rounds of SCNT. Lutz et al. also produced these pigs with two rounds of clone screening using ZFNs [5, 6]. In summary, the method we used is efficient and convenient in the production of gene-modified pigs.
Tector′s group generated the GGTA1/CMAH DKO pigs by CRISPR/Cas9 and SCNT [19, 20]. However, we used a different strategy. First, we obtained a single mutation (GGTA1/CMAH DKO) cell colony by G418 selection, Surveyor assay, and sequencing prior to HMC (instead of lectin selection prior to SCNT). We screened the cells dependent on a resistance gene and genome sequence. This method could be applied to any other gene selection to generate new genetic-modified pigs without the limitation of specific antibody availability for the target gene product. In addition, the 11 piglets we obtained were theoretically of identical genotype, whereas the 10 piglets obtained by Li et al. had different genotypes [20]. For the CMAH gene, three of the 10 piglets were wild-types, one was heterozygous, and six were homozygous; however, the genotypes of the homozygous piglets differed from one another [20].
We used HMC technology where there is no need for microinjection, which is associated with some technical difficulties and is not easy to master, although more oocytes were required for HMC because of the two oocytes essential for the reconstruction of a single embryo. Moreover, HMC is less costly without the expensive micromanipulator; however, it is as efficient as SCNT [15]. Using HMC, approximately 100–140 constructed embryos can be generated by two skilled technicians in 2.5–3 h and the average blastocyst rate can be 30–50% [23, 47]. Although a skilled person using SCNT can produce 100–120 cloned embryos in 1 h, this method is dependent on an expensive micromanipulator. As such, HMC is suitable for an ‘industry-scale′ cloning platform. It benefits from the micromanipulator-free system and has the potential for automation, with which we could produce cloned pigs at a large scale. Additionally, we used the PFFs from Wuzhishan miniature pigs that have potential economic benefits and comparatively lower cost. Therefore, we used different cells from a different strain of pigs with a new method, which differed from those reported previously.
Our data indicated that both GGTA1 and CMAH genes were completely disrupted by the CRISPR/Cas9 system, as Gal and Neu5Gc were not expressed in our DKO pigs. Our results also revealed that, whereas a large percentage of IgM and IgG from human serum is targeted for Gal, approximately half of non-anti-Gal IgM and IgG bound to Neu5Gc. The data suggest Neu5Gc is a major non-Gal antigen target. Antibody-mediated complement-dependent cytotoxicity using different human serum concentrations confirmed that anti-Neu5Gc antibodies are major antibodies directed toward non-Gal antigens and will be important in the success or failure of clinical xenotransplantation. As we expected, these results demonstrated that GGTA1/CMAH DKO cells are more profoundly protected from the humoral response than GGTA1-KO cells.
In summary, this study provided a new strategy, combining CRISPR/Cas9 with HMC, which can efficiently produce gene-modified pigs. All our observations support the conclusion that this technique provides a new, less costly strategy to produce pigs with multiple gene modifications. GGTA1/CMAH DKO pigs should be useful for xenotransplantation research and may be essential for clinical xenotransplantation.
Supplementary
Acknowledgments
Work in the Shenzhen Second People′s Hospital was supported by grants from the National Natural Science Foundation of China (81502410), China Postdoctoral Science Foundation (2015M570744), Shenzhen Foundation of Health and Family Planning Commission (201505006), Sanming Project of Medicine in Shenzhen, the Project of Shenzhen Engineering Center (GCZX2015043017281705), Shenzhen Foundation of Science and Technology (JCJY20160229204849975), Fund for High Level Medical Discipline Construction of Shenzhen (2016031638). Clinical Doctor-Basic Scientist Combination Foundation of Shenzhen Second People′s Hospital, and Key Laboratory Project of Shenzhen Second People′s Hospital.
References
- 1.Butler JR, Ladowski JM, Martens GR, Tector M, Tector AJ. Recent advances in genome editing and creation of genetically modified pigs. Int J Surg 2015; 23(Pt B): 217–222. [DOI] [PubMed] [Google Scholar]
- 2.DeMayo FJ, Spencer TE. Editors-in-Chief BoR. CRISPR bacon: a sizzling technique to generate genetically engineered pigs. Biol Reprod2014; 91:79. [DOI] [PubMed]
- 3.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA 2011; 108: 12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon DN, Lee K, Kang MJ, Choi YJ, Park C, Whyte JJ, Brown AN, Kim JH, Samuel M, Mao J, Park KW, Murphy CN, Prather RS, Kim JH. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep 2013; 3: 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutz AJ, Li P, Estrada JL, Sidner RA, Chihara RK, Downey SM, Burlak C, Wang ZY, Reyes LM, Ivary B, Yin F, Blankenship RL, Paris LL, Tector AJ. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose α-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation 2013; 20: 27–35. [DOI] [PubMed] [Google Scholar]
- 6.Miyagawa S, Matsunari H, Watanabe M, Nakano K, Umeyama K, Sakai R, Takayanagi S, Takeishi T, Fukuda T, Yashima S, Maeda A, Eguchi H, Okuyama H, Nagaya M, Nagashima H. Generation of α1,3-galactosyltransferase and cytidine monophospho-N-acetylneuraminic acid hydroxylase gene double-knockout pigs. J Reprod Dev 2015; 61: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y, Zou Q, Yan Q, Zeng Y, Lai L. Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS ONE 2013; 8: e84250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 2013; 31: 681–683. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Crénéguy A, Brusselle L, Anegón I, Menchaca A. Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS ONE 2015; 10: e0136690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oishi I, Yoshii K, Miyahara D, Kagami H, Tagami T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci Rep 2016; 6: 23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Wang Y, Yuan Y, Zhang W, Ren Z, Jin Y, Liu X, Xiong Q, Chen Q, Zhang M, Li X, Zhao L, Li Z, Wu Z, Zhang Y, Hu F, Huang J, Li R, Dai Y. Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics 2015; 42: 437–444. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban MA, Zeng Y, Yang H, Lai L. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 2015; 72: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma G, Arora JS, Sethi RS, Mukhopadhyay CS, Verma R. Handmade cloning: recent advances, potential and pitfalls. J Anim Sci Biotechnol 2015; 6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vajta G, Lewis IM, Hyttel P, Thouas GA, Trounson AO. Somatic cell cloning without micromanipulators. Cloning 2001; 3: 89–95. [DOI] [PubMed] [Google Scholar]
- 17.Du Y, Kragh PM, Zhang Y, Li J, Schmidt M, Bøgh IB, Zhang X, Purup S, Jørgensen AL, Pedersen AM, Villemoes K, Yang H, Bolund L, Vajta G. Piglets born from handmade cloning, an innovative cloning method without micromanipulation. Theriogenology 2007; 68: 1104–1110. [DOI] [PubMed] [Google Scholar]
- 18.Vajta G. Handmade cloning: the future way of nuclear transfer? Trends Biotechnol 2007; 25: 250–253. [DOI] [PubMed] [Google Scholar]
- 19.Estrada JL, Martens G, Li P, Adams A, Newell KA, Ford ML, Butler JR, Sidner R, Tector M, Tector J. Evaluation of human and non-human primate antibody binding to pig cells lacking GGTA1/CMAH/β4GalNT2 genes. Xenotransplantation 2015; 22: 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, Wang ZY, Paris LL, Blankenship RL, Downey SM, Tector M, Tector AJ. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation 2015; 22: 20–31. [DOI] [PubMed] [Google Scholar]
- 21.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol 2015; 1239: 197–217. [DOI] [PubMed] [Google Scholar]
- 22.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 2010; 649: 247–256. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Dou H, Xiang X, Li L, Li Y, Lin L, Pang X, Zhang Y, Chen Y, Luan J, Xu Y, Yang Z, Yang W, Liu H, Li F, Wang H, Yang H, Bolund L, Vajta G, Du Y. Factors determining the efficiency of porcine somatic cell nuclear transfer: data analysis with over 200,000 reconstructed embryos. Cell Reprogram 2015; 17: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vajta G, Peura TT, Holm P, Páldi A, Greve T, Trounson AO, Callesen H. New method for culture of zona-included or zona-free embryos: the Well of the Well (WOW) system. Mol Reprod Dev 2000; 55: 256–264. [DOI] [PubMed] [Google Scholar]
- 25.Long C, Hara H, Pawlikowski Z, Koike N, dArville T, Yeh P, Ezzelarab M, Ayares D, Yazer M, Cooper DK. Genetically engineered pig red blood cells for clinical transfusion: initial in vitro studies. Transfusion 2009; 49: 2418–2429. [DOI] [PubMed] [Google Scholar]
- 26.Lee W, Hara H, Ezzelarab MB, Iwase H, Bottino R, Long C, Ramsoondar J, Ayares D, Cooper DK. Initial in vitro studies on tissues and cells from GTKO/CD46/NeuGcKO pigs. Xenotransplantation 2016; 23: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara H, Long C, Lin YJ, Tai HC, Ezzelarab M, Ayares D, Cooper DK. In vitro investigation of pig cells for resistance to human antibody-mediated rejection. Transpl Int 2008; 21: 1163–1174. [DOI] [PubMed] [Google Scholar]
- 28.Lee W, Miyagawa Y, Long C, Ekser B, Walters E, Ramsoondar J, Ayares D, Tector AJ, Cooper DK, Hara H. Expression of NeuGc on pig corneas and its potential significance in pig corneal xenotransplantation. Cornea 2016; 35: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen D, Miyagawa Y, Mehra R, Lee W, Isse K, Long C, Ayares DL, Cooper DK, Hara H. Distribution of non-gal antigens in pig cornea: relevance to corneal xenotransplantation. Cornea 2014; 33: 390–397. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wang L, Xiang Y, Ricklin D, Lambris JD, Chen G. Using an in vitro xenoantibody-mediated complement-dependent cytotoxicity model to evaluate the complement inhibitory activity of the peptidic C3 inhibitor Cp40. Clin Immunol 2016; 162: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan N, Lai L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond) 2014; 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endo M, Mikami M, Toki S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 2015; 56: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 2015; 4: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014; 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper DK, Ekser B, Ramsoondar J, Phelps C, Ayares D. The role of genetically engineered pigs in xenotransplantation research. J Pathol 2016; 238: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lunney JK. Advances in swine biomedical model genomics. Int J Biol Sci 2007; 3: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsunari H, Nagashima H. Application of genetically modified and cloned pigs in translational research. J Reprod Dev 2009; 55: 225–230. [DOI] [PubMed] [Google Scholar]
- 39.Phelps CJ, Koike C, Vaught TD, Boone J, Wells KD, Chen SH, Ball S, Specht SM, Polejaeva IA, Monahan JA, Jobst PM, Sharma SB, Lamborn AE, Garst AS, Moore M, Demetris AJ, Rudert WA, Bottino R, Bertera S, Trucco M, Starzl TE, Dai Y, Ayares DL. Production of alpha 1,3-galactosyltransferase-deficient pigs. Science 2003; 299: 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuwaki K, Tseng YL, Dor FJ, Shimizu A, Houser SL, Sanderson TM, Lancos CJ, Prabharasuth DD, Cheng J, Moran K, Hisashi Y, Mueller N, Yamada K, Greenstein JL, Hawley RJ, Patience C, Awwad M, Fishman JA, Robson SC, Schuurman HJ, Sachs DH, Cooper DK. Heart transplantation in baboons using alpha1,3-galactosyltransferase gene-knockout pigs as donors: initial experience. Nat Med 2005; 11: 29–31. [DOI] [PubMed] [Google Scholar]
- 41.Tseng YL, Kuwaki K, Dor FJ, Shimizu A, Houser S, Hisashi Y, Yamada K, Robson SC, Awwad M, Schuurman HJ, Sachs DH, Cooper DK. alpha1,3-Galactosyltransferase gene-knockout pig heart transplantation in baboons with survival approaching 6 months. Transplantation 2005; 80: 1493–1500. [DOI] [PubMed] [Google Scholar]
- 42.Yamada K, Yazawa K, Shimizu A, Iwanaga T, Hisashi Y, Nuhn M, OMalley P, Nobori S, Vagefi PA, Patience C, Fishman J, Cooper DK, Hawley RJ, Greenstein J, Schuurman HJ, Awwad M, Sykes M, Sachs DH. Marked prolongation of porcine renal xenograft survival in baboons through the use of alpha1,3-galactosyltransferase gene-knockout donors and the cotransplantation of vascularized thymic tissue. Nat Med 2005; 11: 32–34. [DOI] [PubMed] [Google Scholar]
- 43.Zhu A, Hurst R. Anti-N-glycolylneuraminic acid antibodies identified in healthy human serum. Xenotransplantation 2002; 9: 376–381. [DOI] [PubMed] [Google Scholar]
- 44.Magnusson S, Månsson JE, Strokan V, Jussila R, Kobayashi T, Rydberg L, Romano E, Breimer ME. Release of pig leukocytes during pig kidney perfusion and characterization of pig lymphocyte carbohydrate xenoantigens. Xenotransplantation 2003; 10: 432–445. [DOI] [PubMed] [Google Scholar]
- 45.Miwa Y, Kobayashi T, Nagasaka T, Liu D, Yu M, Yokoyama I, Suzuki A, Nakao A. Are N-glycolylneuraminic acid (Hanganutziu-Deicher) antigens important in pig-to-human xenotransplantation? Xenotransplantation 2004; 11: 247–253. [DOI] [PubMed] [Google Scholar]
- 46.Padler-Karavani V, Varki A. Potential impact of the non-human sialic acid N-glycolylneuraminic acid on transplant rejection risk. Xenotransplantation 2011; 18: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt M, Kragh PM, Li J, Du Y, Lin L, Liu Y, Bøgh IB, Winther KD, Vajta G, Callesen H. Pregnancies and piglets from large white sow recipients after two transfer methods of cloned and transgenic embryos of different pig breeds. Theriogenology 2010; 74: 1233–1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.