SUMMARY
Dystonia is a brain disorder causing involuntary, often painful movements. Apart from a role for dopamine deficiency in some forms, the cellular mechanisms underlying most dystonias are currently unknown. Here, we discover a role for deficient eIF2α signaling in DYT1 dystonia, a rare inherited generalized form, through a genome-wide RNAi screen. Subsequent experiments including patient-derived cells and a mouse model support both a pathogenic role and therapeutic potential for eIF2α pathway perturbations. We further find genetic and functional evidence supporting similar pathway impairment in patients with sporadic cervical dystonia, due to rare coding variation in the eIF2α effector ATF4/CREB2. Considering also that another dystonia, DYT16, involves a gene upstream of the eIF2α pathway, these results mechanistically link multiple forms of dystonia and put forth a new overall cellular mechanism for dystonia pathogenesis – impairment of eIF2α signaling, a pathway known for its roles in cellular stress responses and synaptic plasticity.
INTRODUCTION
Dystonia is a centrally driven movement disorder characterized by sustained involuntary postures and/or slow twisting movements that lead to motor disability and pain. Clinical presentations range from focal dystonias that affect a single body part to generalized forms that involve most of the body and can severely limit the ability to be independent in activities of daily living (Waugh and Sharma, 2013). Collectively, dystonia is the 3rd most common movement disorder, and can arise in many contexts – limb trauma, traumatic brain injury, stroke, neurodegenerative diseases, antipsychotic medication use, rare inherited syndromes, and “sporadically” (Breakefield et al., 2008; Defazio, 2010). Presently, at least 24 genetic loci have been associated with isolated or combined heritable dystonias (Balint and Bhatia, 2015). While some cellular processes are implicated by those dystonia-associated genes for which there are known functions, the cellular mechanisms for dystonia remain largely unknown (Bragg et al., 2011; Lohmann and Klein, 2013). In addition, because there are both diverse causes and diverse clinical presentations of dystonia, it is further unknown whether distinct forms of dystonia may share common mechanisms. To date, dopamine deficiency is the only cellular pathomechanism for which there is support across multiple forms of dystonia (Lohmann and Klein, 2013; Waugh and Sharma, 2013).
As the need to understand disease mechanisms extends beyond Mendelian disorders with strong effect sizes and as genome-wide datasets become readily accessible, a promise and challenge of functional genomics is to integrate functional and genetic data to advance our understanding and treatment of disease. Here we applied results from a high-throughput phenotypic screen based on a rare Mendelian genetic disease to whole exomic sequence (WES) data from a small cohort of patients with sporadic disease and uncovered a cellular mechanism for dystonia pathogenesis that spans multiple forms of the disorder. Functional studies including patient-derived cell lines and genetic mouse models combined with consideration of the putative activity of PRKRA mutations found in another inherited generalized dystonia, DYT16, support a consistent directionality of reduced eIF2α pathway signaling as a mechanism for dystonia. The eIF2α pathway is a ubiquitous cellular pathway widely known as the integrated stress response, or ISR (Harding et al., 2003; Wek et al., 2006), and is also required in the brain for synaptic plasticity (Trinh and Klann, 2013).
DYT1 dystonia is a rare, early-onset, generalized form of dystonia. DYT1 is caused by an in-frame trinucleotide deletion in the TOR1A gene, leading to loss of a glutamic acid residue (ΔE) from the AAA+ ATPase Torsin1a (Ozelius et al., 1997). Both the normal function of Torsin1a and significance of the mutant protein for disease pathogenesis have been intensively studied and at least 5 cellular processes have been suggested, including roles in nuclear transport, synaptic vesicle cycling, lipid metabolism, and endoplasmic reticulum (ER) stress (Burdette et al., 2010; Chen et al., 2010; Goodchild and Dauer, 2005; Granata et al., 2011, 2008; Grillet et al., 2016; Jokhi et al., 2013; Nery et al., 2011). Although the precise cellular function(s) of Torsin1a and the pathogenic mechanism of ΔE Torsin1a remain uncertain, multiple independent laboratories have observed that the ΔE mutation has a dramatic effect on Torsin1a subcellular localization (Bragg et al., 2004; Calakos et al., 2010; Giles et al., 2008; Gonzalez-Alegre and Paulson, 2004; Goodchild and Dauer, 2005, 2004; Goodchild et al., 2005; Hewett et al., 2000, 2008; Kustedjo et al., 2000; Liang et al., 2014; Vander Heyden et al., 2009; Vulinovic et al., 2014).
Normally, wild-type (WT) Torsin1a cycles between the outer nuclear envelope (NE) and endoplasmic reticulum lumen (ER) in an ATP-dependent fashion, with the bulk of the protein detected in the ER (Goodchild and Dauer, 2004; Naismith et al., 2004). In contrast, when ΔE Torsin1a is the major species, as in overexpression experiments or homozygous knockin mouse models, it predominantly colocalizes with nuclear envelope markers and disrupts the normal subcellular NE membrane structure in a manner that is suggestive of a membrane-trafficking defect (Goodchild and Dauer, 2004; Jokhi et al., 2013; Naismith et al., 2004). At the light microscopic level, ΔE Torsin1a distribution appears as an abnormal punctate pattern (Figure 1A).
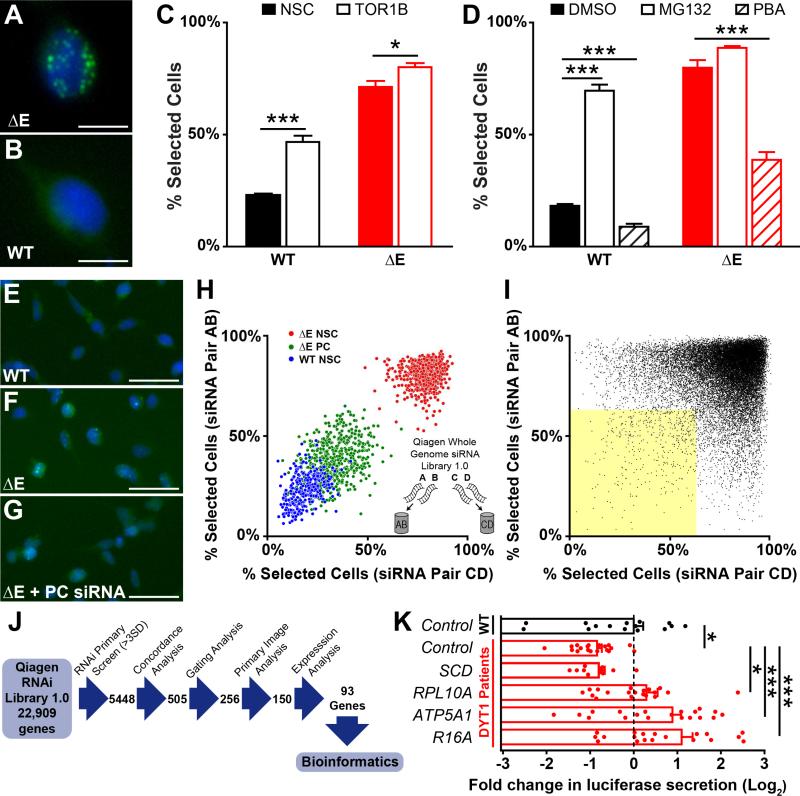
Figure 1. Torsin1a mislocalization assay and genome-wide siRNA screen.
(A and B) Flp-In T-REx 293T stable cell lines expressing ΔE (A) and WT (B) EGFP-hTorsin1a following 72 h tetracycline induction. Green – EGFP-hTorsin1a. Blue – Hoescht nuclear stain. Scale bars = 10 μm.
(C) Percent of cells with ≥ 1 EGFP puncta (“Percent Selected Cells” – see Figure S2) after siRNA silencing of Torsin1b. n = 16 wells for control siRNA and 32 wells for TOR1B siRNAs.
(D) Percent of cells with ≥ 1 EGFP puncta after treatment with the protesome inhibitor MG132 (10 μM) or the chemical chaperone phenylbutyric acid (PBA, 20 mM). n = 4 DMSO-treated wells and 8 MG132/PBA-treated wells.
(E-G) Cell lines under high-throughput screening conditions after transfection with non-silencing control siRNA (E and F) or positive control siRNA (PC, panel G). Scale bars = 50 μm.
(H) Whole genome siRNA (WGS) screen controls and siRNA pooling strategy. Inset – Four independent siRNAs targeting each gene were split into two unique pools of two siRNAs.
(I) Results of WGS screen. Dots represent data for individual gene targets, with results from independent siRNA pools plotted on orthogonal axes. Yellow shaded area – genes with concordant >3SD normalizing effects.
(J) Schematic depicting WGS workflow for analyzing primary hits.
(K) Luciferase secretion in DYT1 patient-derived fibroblasts after lentiviral delivery of shRNAs targeting 4 top WGS hits. n = 4 DYT1 lines and 3 control lines, 5 independent replicates each (except SCD – 3 replicates). *, p < 0.05; ***, p < 0.0005 by unpaired t test. All data are presented as means ± S.E.M.
In this study, we took advantage of the robust and proximal phenotype of ΔE Torsin1a mislocalization to develop a high-throughput assay to identify cellular signaling pathways involved in remediating mutant Torsin1a mislocalization. We hypothesized that pathway perturbations which correct ΔE Torsin1a mislocalization could serve as novel therapeutic targets for DYT1 dystonia. We then applied the assay to conduct a genome-wide RNAi screen, and used the results to generate unbiased hypotheses for mechanisms of dystonia.
RESULTS
Development and validation of ΔE Torsin1a localization assay
We generated two human HEK293T cell lines that stably expressed either EGFP-tagged human WT or ΔE Torsin1a from a cDNA integrated at a defined genomic site under inducible control by tetracycline (see Supplemental Experimental Procedures and Figure S1). Using this expression system, WT and ΔE Torsin1a lines consistently reproduced their respective subcellular localization phenotypes (Figures 1A and 1B). The mutant phenotype was measured as the percentage of cells with one or more EGFP puncta using automated high-content imaging analysis (“% selected cells”, see Figure S2).
We first tested the predictive validity of the automated readout in corroborating existing observations regarding Torsin1a biology. Compensation by the homologous protein, Torsin1b, is hypothesized to underlie the selective vulnerability of the brain where Torsin1b levels are low (Jungwirth et al., 2010; Kim et al., 2010; Tanabe et al., 2016). Consistent with this model, knockdown of Torsin1b significantly increased Torsin1a mislocalization in both cell lines (Figure 1C). Second, Torsin1a protein levels have also been hypothesized to contribute to DYT1 dystonia pathogenesis (Bragg et al., 2004; Goodchild and Dauer, 2004; Grundmann et al., 2007). Inhibiting protein clearance with the proteasome inhibitor MG132 significantly increased the percentage of cells with punctate EGFP-Torsin1a signal in both cell lines (Figure 1D). Lastly, ΔE Torsin1a has been associated with activation of the unfolded protein response (UPR) (Bragg et al., 2011; Chen et al., 2010; Hewett et al., 2007; Nery et al., 2011) and in DYT1 patient-derived fibroblasts, the chemical chaperone, phenylbutyric acid (PBA) reduces indicators of UPR activation (Cao et al., 2010). In our assay cell lines, we found that PBA significantly reduced punctate pathology (Figure 1D). Thus, although the biological significance of ΔE Torsin1a mislocalization in DYT1 dystonia pathogenesis is unknown, Torsin1a localization phenotypes predict known Torsin1a biology and suggests this phenotypic screen may be a useful tool to advance our understanding of the mechanisms by which ΔE Torsin1a disrupts cell function.
Genome-wide siRNA screen to correct ΔE Torsin1a mislocalization
A pilot screen of 960 genes was performed to establish proof-of-principle that gene knockdown using siRNA could normalize ΔE Torsin1a distribution (Figures 1E, 1F, and 1G). We then performed a whole genome screen (WGS) targeting 22,909 human genes (Qiagen Whole Genome siRNA Knock-Down Library 1.0). Four siRNAs targeting each gene were tested in a paired-screen design of two independent pools of two siRNAs each (Figure 1H) (Barrows et al., 2010). Hits were defined as those genes whose siRNAs concordantly improved ΔE Torsin1a localization by at least 3 standard deviations in both of the two unique siRNA pools (Figure 1I, yellow). No microRNA seed sequences were significantly enriched among screen hits (Sigoillot et al., 2012) (www.flyrnai.org/gess). In addition, hits that were cytotoxic, significantly decreased EGFP-Torsin1a expression, or were not expressed in the assay cell line (as determined by microarray analysis, see Supplemental Data File 1) were discarded (see Supplemental Experimental Procedures for detail). The primary images were then examined by blinded scorers and ranked to identify those hits that resulted in typical wild-type Torsin1a distribution. The resulting group was used for pathway analysis and consisted of 93 high-stringency and high-quality hits (Figure 1J and Table S1).
Hit validation and bioinformatic analysis
To cross-validate WGS hits in a system that did not rely upon exogenous expression of Torsin1a or a fusion protein, we tested four of the highest-quality hits in an orthogonal counter screen using patient-derived fibroblasts. We tested each shRNA for its ability to rescue a previously described defect in luciferase secretion (Cao et al., 2010; Hewett et al., 2007; Nery et al., 2011), and found that three of the four hits tested also improved this DYT1-related phenotype (Figure 1K). We then conducted bioinformatic analysis, which identified eleven significantly over-represented signaling pathways (Table 1).
Table 1.
Bioinformatic analysis of whole genome RNAi screen hits.
| Ingenuity Canonical Pathwaysa | p-value | |
|---|---|---|
| 1 | EIF2 Signaling (6) | 0.000098 |
| 2 | Assembly of RNA Polymerase II Complex (3) | 0.0011 |
| 3 | Cell Cycle: G1/S Checkpoint Regulation (3) | 0.0019 |
| 4 | Glucocorticoid Receptor Signaling (5) | 0.0041 |
| 5 | Nucleotide Excision Repair Pathway (2) | 0.0087 |
| 6 | Notch Signaling (2) | 0.010 |
| 7 | Coenzyme A Biosynthesis (1) | 0.012 |
| 8 | Estrogen Receptor Signaling (3) | 0.015 |
| 9 | Lysine Degradation II (1) | 0.020 |
| 10 | Ceramide Biosynthesis (1) | 0.024 |
| 11 | Oleate Biosynthesis II (Animals) (1) | 0.048 |
Numbers in parentheses indicate numbers of RNAi screen hits in each pathway
The most enriched pathway among WGS hits – as well as among hits derived from a less stringent analysis (≥2SD effect size, see Table S2) – was eukaryotic initiation factor 2α (eIF2α) signaling, also known as the ISR (Figure 2A) (Harding et al., 2003; Wek et al., 2006). Briefly, in the ISR, eIF2α, the rate-limiting regulatory subunit of the eIF2 complex which mediates the binding of methionyl-tRNA to the ribosome to begin protein translation (Hinnebusch and Lorsch, 2012), is phosphorylated by any of four upstream stress-sensitive kinases. Phosphorylation of eIF2α has two main effects: a general decrease in protein translation and an increased translation of a subset of transcripts containing upstream open reading frames (uORFs)(Jackson et al., 2010; Vattem and Wek, 2004). Principal among the transcripts whose translation is upregulated is ATF4, a transcription factor which stimulates the expression of stress response proteins (Harding et al., 2000; Vattem and Wek, 2004). Eukaryotic initiation factor 2α signaling also leads to expression of protein phosphatase 1 regulatory subunits CReP and GADD34, which dephosphorylate eIF2α and terminate ISR activation (Jousse et al., 2003; Novoa et al., 2001).
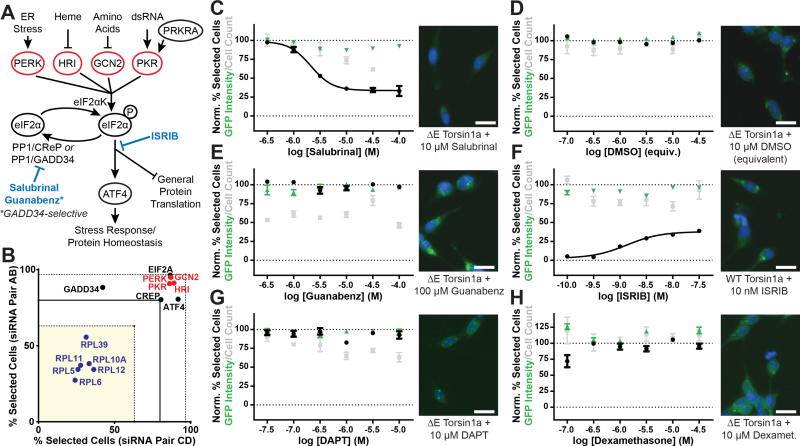
Figure 2. Enhancing eIF2α signaling corrects ΔE Torsin1a mislocalization.
(A) eIF2α signaling pathway diagram. Actions of compounds tested in panels (C) through (F) are indicated in blue.
(B) WGS results relevant to the eIF2α pathway. Blue – gene hits bioinformatically implicating the eIF2α pathway. Red – eIF2α kinases. Dashed lines indicate +/−3SD from mean.
(C-H) Left – Effects of the indicated compounds on Torsin1a localization (black), cell count (grey), and EGFP-Torsin1a expression (green) in the ΔE (C-E, G, H) or WT (F) assay cell lines. Percent Selected Cells was normalized such that vehicle-treated ΔE cells = 100 and vehicle-treated WT cells = 0. GFP Intensity and Cell Count were normalized such that vehicle-treated ΔE cells = 100. Right – Representative images from the respective treatments. Scale bars = 20 μm. n = 4 per compound dose for dose response data or 24 for untreated control data (used for normalization). All data are presented as means ± S.E.M.
Pharmacological manipulation of eIF2α signaling bidirectionally regulatesTorsin1a localization
Although our screen implicated eIF2α signaling, the hits that yielded the bioinformatics result (Figure 2B, blue) did not reveal the directionality of the signaling change normalizing ΔE Torsin1a localization. However, knockdown of each of the four eIF2α kinases significantly worsened ΔE Torsin1a localization (Figures 2A and 2B; see also Supplemental Data File 2), indicating that decreased eIF2α signaling has a deleterious effect. These results also suggest then that promoting eIF2α pathway signaling would be beneficial.
In order to test this model directly, we treated the ΔE Torsin1a cell line with compounds targeting the eIF2α signaling pathway (Figure 2A, blue). Salubrinal, a drug that prolongs ISR activation by inhibiting eIF2α dephosphorylation (Boyce et al., 2005), caused a robust dose-dependent normalization of ΔE Torsin1a localization, without modifying steady-state EGFP-Torsin1a levels or causing toxicity at effective concentrations (Figure 2C). As a control, vehicle treatment (DMSO) at concentrations present in these experiments had no effect on cell pathology or number (Figures 2D and S3). None of the compounds tested significantly altered steady-state levels of EGFP-Torsin1a (Figure 2, green). Interestingly, another eIF2α phosphatase inhibitor, guanabenz, had no effect on ΔE Torsin1a localization (Figure 2E). Guanabenz is a more selective inhibitor of GADD34-containing than CReP-containing eIF2α phosphatase complexes (Tsaytler et al., 2011). Sephin 1, another GADD34-specific inhibitor that lacks guanabenz's off-target adrenergic activity, also had no effect on ΔE Torsin1a localization (data not shown). We next examined the efficacy of a direct activator of the eIF2α kinase PERK, CCT020312 (Stockwell et al., 2012). CCT020312 also caused a dose-dependent reduction in the number of cells with Torsin1a inclusions (Figure S4), but treatment was highly toxic at effective concentrations, as indicated by cell count and morphology (Figure S4). Thus, collectively these experiments suggest that positive modulation – as opposed to direct activation – of eIF2α signaling may be more easily tolerated and therefore more likely to be of therapeutic value.
Upon finding that prolonged signaling through eIF2α phosphorylation normalizes ΔE Torsin1a distribution (as in Figure 2C), we next asked whether preventing such signaling would alter the normal distribution of Torsin1a. To do this, we tested effects of ISRIB, a compound which prevents the downstream signaling consequences of eIF2α phosphorylation (Sidrauski et al., 2015, 2013) on the WT Torsin1a assay cell line. ISRIB caused a dose-dependent increase in cells with punctate Torsin1a localization without altering Torsin1a levels (Figure 2F; for DMSO control in WT Torsin1a cell line see Figure S3). These findings indicate a dynamic role for the ISR pathway in bidirectionally regulating Torsin1a localization.
Finally, we tested compounds targeting alternative signaling pathways which could potentially mediate the effects upon Torsin1a localization, including two additional pathways enriched among the WGS hits, notch signaling and glucocorticoid signaling. Treatment with the γ-secretase inhibitor, DAPT, which blocks notch signaling, had no effect on ΔE Torsin1a localization (Figure 2G), nor did the anticonvulsant, valproic acid, which, among other activities, activates notch signaling (Figure S5). Similarly, neither the glucocorticoid receptor agonist dexamethasone (Figure 2H), or a small panel of additional compounds modulating glucocorticoid/mineralocorticoid receptor signaling (Figure S5) significantly modified ΔE Torsin1a localization. Next, because PERK-mediated eIF2α signaling is one of three branches of the unfolded protein response (Patil and Walter, 2001), we tested modulators that are available for another branch, which requires ATF6 signaling. We found that compounds targeting this branch primarily caused cytotoxicity (Figure S6). Furthermore, knockdown of ATF6 did not significantly normalize or worsen ΔE Torsin1a localization in the WGS screen. Finally, since salubrinal has been shown to inhibit NF-κB signaling in addition to its effects on the eIF2α pathway (Nakajima et al., 2015), we tested compounds that both activate and inhibit the NF-κB pathway. As with ATF6 signaling, we found that compounds targeting the NF-κB pathway were principally cytotoxic (Figure S6). Together, these results identify augmentation of signaling through the eIF2α pathway as a specific, efficacious and non-toxic target to normalize ΔE Torsin1a mislocalization.
Expression of ATF4 is sufficient to normalize ΔE Torsin1a localization
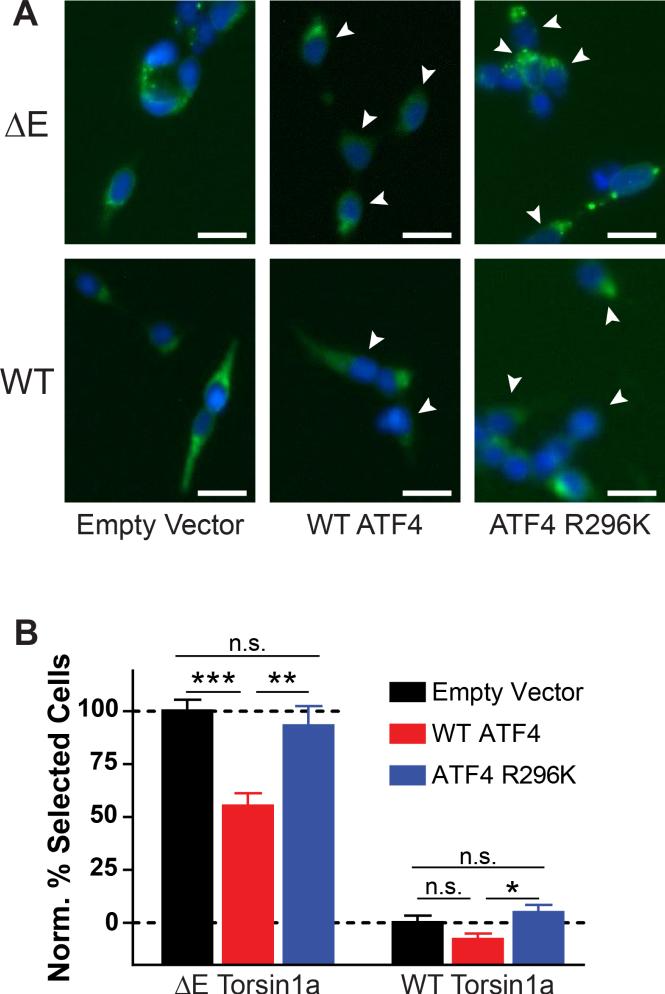
We next considered the mechanisms downstream of eIF2α phosphorylation that might mediate normalization of ΔE Torsin1a localization. Phosphorylation of eIF2α has two downstream consequences: a general decrease in translation and an increase in translation of a subset of transcripts containing uORFs, the most well-characterized of which is the transcription factor ATF4/CREB2 (Figure 2A) (Vattem and Wek, 2004). Therefore, to determine if increased ATF4 expression alone was sufficient to normalize ΔE Torsin1a localization, we overexpressed ATF4 in the ΔE Torsin1a assay cell line. ATF4 overexpression significantly improved ΔE Torsin1a localization, with negligible effects on WT Torsin1a localization (Figure 3A). Furthermore, overexpression of an ATF4 point mutant which cannot bind DNA, R296K (Aukerman et al., 1991), had no effect (Figures 3A and 3B, right).
Figure 3. ATF4 overexpression is sufficient to correct ΔE Torsin1a mislocalization.
(A) Torsin1a localization in the ΔE and WT assay cell lines after transfection with the indicated FLAG-tagged ATF4 constructs. White arrows – FLAG-positive cells. Scale bars = 20 μm.
(B) Quantification of conditions shown in (A). Percent Selected Cells was normalized such that empty vector-transfected ΔE cells = 100 and vector-transfected WT cells = 0. *, p < 0.05; **, p < 0.005; ***, p < 0.0005 by unpaired t test. n = 40 (empty vector), 28 (WT ATF4), and 12 (ATF4 R296K). All data are presented as means ± S.E.M.
eIF2α signaling is required for LTD at corticostriatal synapses and enhancing eIF2α signaling rescues deficient corticostriatal LTD in DYT1 knockin mice
We next examined whether there was evidence of eIF2α pathway dysfunction in a setting that did not involve exogenously expressed Torsin1a, mimicked the human genotype (heterozygosity), and involved the brain. Dystonia is a brain disorder and although a disturbed ER stress response is a plausible mechanism for CNS disease, eIF2α phosphorylation is also known to have brain-specific roles in long-term synaptic plasticity (Costa-Mattioli and Sonenberg, 2006; Di Prisco et al., 2014; Trinh and Klann, 2013). Coincidentally, disrupted synaptic plasticity in basal ganglia circuitry has long been hypothesized as a disease mechanism for dystonia (reviewed in Peterson et al., 2010). Furthermore, DYT1 mouse models have disruptions of long-term synaptic plasticity in the striatum – absence of type 5 metabotropic glutamate receptor (mGluR5)-dependent long-term synaptic depression (LTD) and increased long-term potentiation (LTP) magnitude (Martella et al., 2014, 2009). In the hippocampus, eIF2α phosphorylation is required for mGluR-dependent LTD (Di Prisco et al., 2014) and shifts that reduce the amount of phosphorylated protein have been previously hypothesized to lower the threshold for LTP (Costa-Mattioli et al., 2007). Thus, known brain-specific eIF2α roles directly correlate with previously well-described alterations in DYT1 mouse model synaptic plasticity.
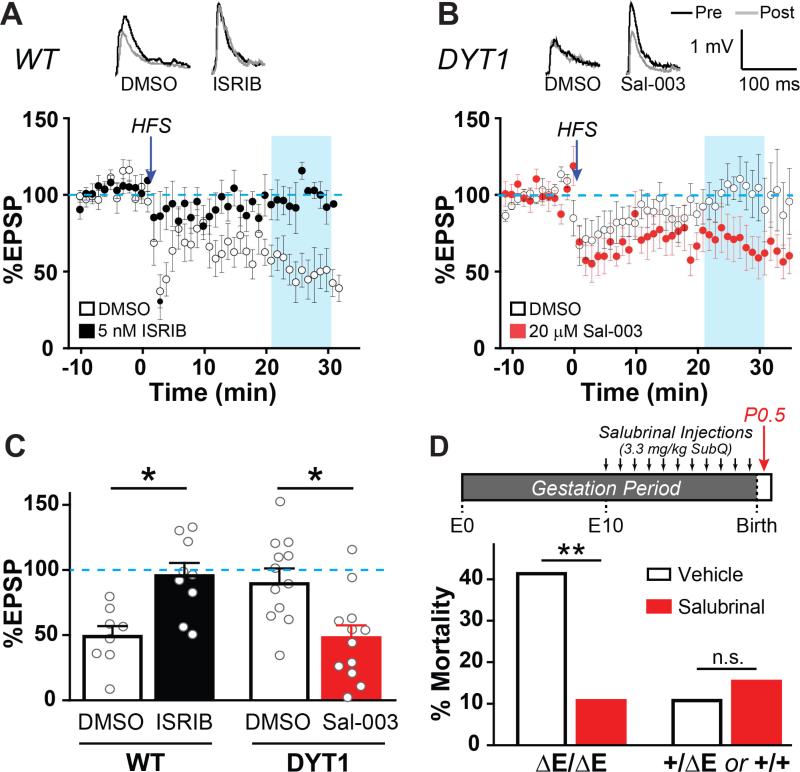
To directly test these predictions, we performed whole-cell electrophysiology of striatal projection neurons in acute brain slices to induce long-term depression in the presence of eIF2α pathway-modulating drugs. We first assessed whether eIF2α signaling normally plays a role in striatal mGluR5 LTD. We found that pre-incubation with ISRIB (5 nM), a blocker of eIF2α pathway signaling, robustly inhibited LTD in WT brain slices (Figure 4A). Additionally, it was notable that while control vehicle-treated slices display a range of responses, none were greater than the baseline amplitude; however in the presence of ISRIB, potentiating responses were now observed.
Figure 4. Enhancing eIF2α signaling rescues deficient corticostriatal LTD in heterozygous DYT1 knockin mice and improves neonatal survival of homozygous DYT1 mice.
(A) Long-term depression (LTD) of corticostriatal synapses from WT mice in the presence of 5 nM ISRIB (black dots) or vehicle control (white dots). LTD was induced by 4 trains of 100 Hz stimulation in cortex layer V (HFS, see Experimental Procedures). n = 8 slices/4 mice (vehicle) or 9 slices/4 mice (ISRIB). Traces above graph – Representative excitatory postsynaptic potentials (EPSPs) recorded before HFS (black) or 21 min post-HFS (gray).
(B) LTD of corticostriatal synapses from DYT1 mice in the presence of 20 μM Sal-003 (red dots) or vehicle control (white dots). n = 12 slices/7 mice (vehicle) or 12 slices/8 mice (Sal-003).
(C) Mean magnitude of LTD in (A) and (B). Data in blue shaded boxes in panels (A) and (B) (minutes 21-31) were averaged. *, p < 0.01 by two-tailed Mann-Whitney U-test. Data are presented as means ± S.E.M.
(D) Top – Schematic depicting experimental workflow. Bottom – Perinatal survival after in utero salubrinal exposure. n = 29 vehicle- and 28 salubrinal-treated homozygous pups and 65 vehicle- and 91 salubrinal-treated pooled heterozygous/WT pups. No effect on mortality was observed for WT or heterozygous genotypes, so results were combined to simplify presentation.
**, p < 0.01 by Chi-square test.
To test whether augmenting eIF2α signaling in DYT1 model mice would normalize synaptic plasticity, we similarly treated brain slices from heterozygous DYT1 knockin mice (delGAG) with 20 μM Sal-003, an analogue of salubrinal. In comparison to vehicle, Sal-003 restored LTD (Figure 4B) and to levels that once again approximate the normal magnitude (Figures 4B and 4C). These observations demonstrate that eIF2α signaling normally plays a role in brain processes that are disrupted in DYT1 model mice and that acute enhancement of eIF2α phosphorylation is sufficient to restore normal brain plasticity to disease model mice.
Enhancing eIF2α signaling improves neonatal survival of homozygous DYT1 mice
The findings thus far provide strong support for roles of eIF2α signaling pathway in normalizing in vitro cellular (Figures 1-3) and ex vivo brain phenotypes related to the DYT1 genotype (Figure 4). We next sought to determine whether targeting eIF2α signaling would significantly affect the in vivo deleterious consequences of the DYT1 TOR1A mutation. Although etiological mouse models of DYT1 dystonia have not had robust dystonic phenotypes (Dang et al., 2005; Tanabe et al., 2012) (a finding we believe may be related to the low genetic penetrance of this disorder among other factors), we took advantage of another deleterious genotype-dependent effect of the mutation to examine this question – neonatal lethality of homozygous delGAG knockin mice (Goodchild et al., 2005; Tanabe et al., 2012). To this end, we administered salubrinal or vehicle daily to pregnant dams (in Tor1adelGAG/+ × Tor1adelGAG/+ breedings) from approximately embryonic day 10 through delivery and tested its effects on neonatal survival. In comparison to litters from vehicle-treated dams, salubrinal dramatically reduced ΔE/ΔE pup mortality (Figure 4D). Additionally, there was no evidence that salubrinal had a non-specific mechanism in improving neonatal survival, as no mortality differences were observed between treatment and control groups in the remaining pups of either wildtype or heterozygote genotypes (Figure 4D). Although in this study design there was no postnatal salubrinal treatment except possibly through lacteal transfer (though homozygous ΔE/ΔE pups are known to not nurse (Goodchild et al., 2005)), we anecdotally observed that two salubrinal-treated homozygous ΔE Torsin1a pups survived through the second postnatal day. No vehicle-treated homozygous pups survived through even the first postnatal day.
Stress-induced eIF2α signaling is impaired in human DYT1 patient fibroblasts
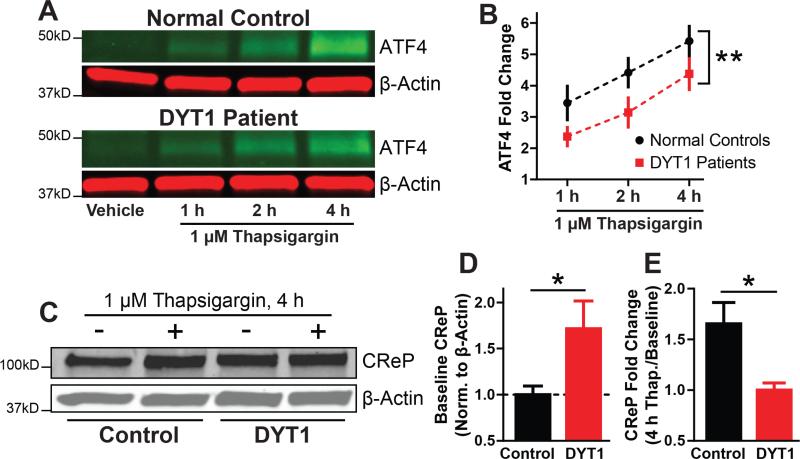
Having established the ability of eIF2α modulators to both normalize and reproduce a number of DYT1–genotype related phenotypes, a key remaining question is whether the eIF2α signaling pathway is actually disrupted in DYT1 dystonia. To address this, we examined the integrity of the eIF2α signaling pathway in fibroblasts derived from human DYT1 patients. We measured the stress-induced increase in ATF4 expression as a readout of pathway activation. In line with our predictions of impaired signaling in DYT1, we found that ATF4 upregulation in response to the potent ER stressor thapsigargin was attenuated in DYT1 patient-derived fibroblasts (Figures 5A and 5B).
Figure 5. Cells from human DYT1 dystonia patients show eIF2α pathway dysfunction.
(A) ATF4 expression in control and DYT1 patient fibroblasts after treatment with 1 μM thapsigargin. Green – anti-ATF4. Red – anti-β-actin.
(B) Quantification of conditions shown in (A). ATF4 expression was normalized to β-actin expression. n = 3 control cell lines and 4 DYT1 cell lines, 3 replicates each.
(C) CReP expression with and without 4 hours exposure to 1 μM thapsigargin.
(D and E) Quantification of conditions shown in (C). CReP expression was normalized to β-actin expression. n = 3 control cells lines and 4 DYT1 cell lines, 3 replicates each. *, p < 0.05; **, p < 0.005 by unpaired t test. All data are presented as means ± S.E.M.
DYT1 patient cells have increased basal levels of a negative feedback regulator of the eIF2α pathway
Because we the stress-induced response of ATF4 translation was still present but attenuated in DYT1 patient cells, we considered whether DYT1 may be associated with an increase in negative feedback mechanisms that attenuate phospho-eIF2α signaling. The principal negative feedback proteins in the eIF2α pathway are the eIF2α phosphatase subunits, CReP and GADD34. A role for CReP was suggested by our pharmacological studies in which salubrinal (which inhibits both CReP and GADD34) but not guanabenz (a more specific inhibitor of GADD34) corrected ΔE Torsin1a mislocalization. We therefore measured levels of CReP under basal and ER-stressed conditions. We found two striking differences in CReP levels between fibroblasts from DYT1 patients and healthy controls. First, basal CReP levels were significantly higher in fibroblasts from DYT1 patients (Figures 4C and 4D), supporting the negative feedback hypothesis. Second, whereas CReP levels in normal fibroblasts robustly increased after treatment with thapsigargin as expected (Figure 4E), CReP levels in DYT1 patient fibroblasts were unresponsive to the treatment. As the increase in CReP levels in response to stress is regulated by phospho-eIF2α-dependent translation (Andreev et al., 2015), the failure of CReP levels to rise provides additional evidence that eIF2α pathway signaling is deficient in DYT1. Altogether, these results provide evidence from human patient cell lines that eIF2α signaling is not only a novel therapeutic inroad, but is also disrupted in DYT1 dystonia. The specific disruptions of steady state and induced CReP levels further suggest a specific pathway impairment mechanism in DYT1 dystonia whereby basally increased levels of negative feedback proteins attenuate acute phasic eIF2α signaling.
Identification of a loss-of-function ATF4 mutation in sporadic cervical dystonia patients
Interestingly, the gene associated with another rare inherited dystonia supports our findings indicating a role for deficient eIF2α signaling in DYT1 dystonia pathogenesis. Mutations in the PRKRA gene, which encodes an upstream eIF2α kinase activator (Figure 2A), cause DYT16 dystonia (Camargos et al., 2008). Moreover, recent functional evidence indicates that the most common PRKRA mutation delays and reduces eIF2α phosphorylation (see Figure 5 in Vaughn et al., 2015). Considering the intersection of DYT16 and DYT1 dystonia at the eIF2α pathway, we hypothesized that eIF2α pathway dysfunction might also contribute to other forms of dystonia.
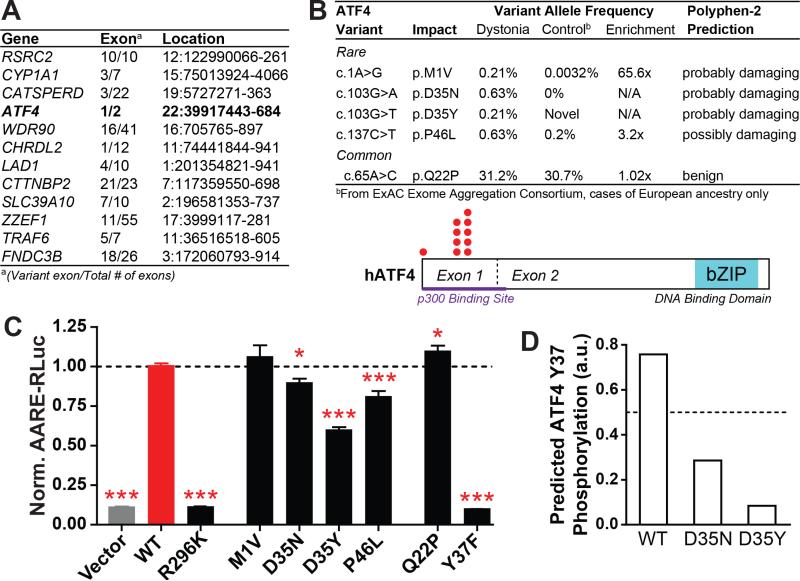
To investigate this possibility, we first reviewed results from a pilot study we had performed on whole exomic sequences (WES) obtained from 20 individuals of European ethnicity with sporadic, isolated dystonia (mean age of onset 47.4 +/− 9.6 years; Table S3). In these exomes, we performed an unbiased, exome-wide analysis to identify exons where rare (ExAC European ethnicity MAF<0.5%) coding variants were found in at least two cases and no controls (we utilized 571 controls of European ethnicity sequenced in-house using the same capture kit as the cases). We found that exome-wide, only 12 exons met these criteria (Figure 6A).
Figure 6. Identification of loss-of-function ATF4 mutations in sporadic dystonia patients.
(A) Exons bearing missense coding mutations in at least two of 20 unrelated sporadic dystonia patients and none of 571 matched controls, as determined by whole exome sequencing.
(B) Top – Rare and common variants in ATF4 exon 1 from 239 additional sporadic cervical dystonia patients; frequency, enrichment and predicted mutation severity shown at right (see also Table S5). Bottom – ATF4 protein schematic showing rare variant locations (red dots).
(C) Transcriptional activation activity of mutant ATF4 constructs, as measured by a luciferase reporter under transcriptional control of an ATF4-sensitive response element (AARE-RLuc). Data was normalized such that luciferase activity after WT ATF4 transfection = 1. *, p < 0.05; ***, p < 0.0005 vs WT ATF4 condition by unpaired t test. n = 28 (vector, WT, Q22P, P46L), 18 (R269K, D35N, D35Y), 12 (M1V), and 6 (Y37F) independent experiments. The P46L condition was measured in separate groups of 10 and 18 experiments, each of which independently reached p < 0.005 significance. All data are presented as means ± S.E.M.
(D) Homology-based prediction of the likelihood of Y37 phosphorylation in WT, D35N, and D35Y ATF4 (www.cbs.dtu.dk/services/NetPhos).
Notably, one of these was exons was in the gene for the phospho-eIF2α effector, ATF4 It is notable that the implicated exon, exon 1, encodes the amino terminal region of ATF4 which appears to have roles in recruitment of transcriptional co-factors (Chérasse et al., 2007), whereas the other exon in this gene, exon 2, encodes the canonical basic leucine-zipper DNA binding domains. We found that the same exon 1 rare variant was present in two cases (n.22:39917587C/T (GRCh37/hg19), rs111719524, p.P46L). Additionally, both P46L subjects had the same form of dystonia - cervical dystonia. Neither subject knew of any family history of dystonia.
Enrichment of rare ATF4 coding mutations in sporadic cervical dystonia
To prospectively test whether enrichment of rare coding variants in ATF4/CREB2 could be replicated in a large cohort, we Sanger sequenced ATF4 exon 1 in an additional 239 sporadic cervical dystonia cases. In total, we found that 3.3% of cases (8 of 239) had rare functional variants, compared to 0.5% of controls of European ethnicity in the ExAC database (OR 6.8, p = 4.1 × 10−5,; Figure 6B). Of the new cases, 223 self-identified as Caucasian with non-Hispanic/Latino ethnicity (see Table S4). Of the 16 other cases, one Latino case had a qualifying variant, which was P46L. Interestingly, P46L is found at a frequency of 0.2% in those with European ethnicity in the ExAC database, but at an even lower frequency in all other ethnicities in ExAC, for example, a minor allele frequency (MAF) of 0.02% in Latinos and 0.12% overall.
In addition to the three new P46L cases, another four cases were identified with qualifying variants affecting the same amino acid (D35), with two different mutations affecting this site (Figure 6B). D35 was mutated in only one sample in ExAC (of Latino descent) for an overall incidence of less than 0.0008%. While the D35N variant is classified as “probably damaging” by Polyphen2, it is notable that this residue is highly evolutionarily conserved (Figure S7) and both variants are strongly predicted to alter phosphorylation of a nearby tyrosine, Y37 (Figure 6D). Finally, a single case had a variant involving the start methionine, a location clearly suggesting a deleterious effect. Interestingly, although the functions of the N-terminal portion of ATF4 are not fully understood, the histone acetyltransferase p300 – a transcriptional co-activator – binds ATF4 within the first 85 amino acids (Lassot et al., 2005), a region which contains all of the rare variants we identified (Figure 6B, bottom, purple bar and red dots). Lastly, it is notable that the common ATF4 polymorphism Q22P, present in approximately one third of the human population (http://exac.broadinstitute.org), is not predicted to have a deleterious effect on gene function and was not enriched in our cervical dystonia patients (Figure 6B). Thus, this enrichment appears specific to rare ATF4 variations that are likely gene disrupting.
Functional characterization of rare ATF4/CREB2 variants in sporadic cervical dystonia
To evaluate the functional significance of the identified rare variants, we tested their transcriptional activation activity using a luciferase reporter driven by the ATF4-sensitive amino acid response element (AARE). As controls, we tested the activity of wildtype ATF4 (WT) and the DNA binding-dead ATF4 R296K (Aukerman et al., 1991). As expected, overexpression of WT ATF4 caused robust expression of the luciferase reporter, while expression of the DNA binding-dead ATF4 R296K had no significant effect (Figure 6C, left; R296K vs. empty vector, p = 0.89). Surprisingly, we found that the M1V variant of ATF did not significantly alter function. However, in this experimental setting, the presence of an N-terminal epitope tag (also M1V mutated) and the absence of the endogenous upstream regulatory elements for ATF4 translation confounds interpretation of the significance of this finding. It is notable that ATF4 does contain methionine residues at positions 4 and 17 which might serve as alternative start sites. Apart from M1V, the remaining coding mutations identified in sporadic cervical dystonia cases (P46L, D35N, D35Y) all showed significantly reduced activity, consistent with the direction of eIF2α pathway impairment that would be predicted by the effects described in DYT1 here and DYT16 (Vaughn et al., 2015). We also tested the most common ATF4 variant in the general population, Q22P, and found that rather than impair, it showed modestly enhanced activity relative to WT.
We were struck by the relative effects of the D35N and D35Y substitutions because they mirrored bioinformatic predictions of phosphorylation likelihood at a nearby tyrosine (Y37). Since ATF4 is known to be regulated by other posttranslational modifications but not by tyrosine phosphorylation (Pakos-Zebrucka et al., 2016), we made a conservative amino acid substitution, Y37F, to directly test the functional significance. Strikingly, severe functional effects were observed that approximated the negative control conditions (n.s. versus empty vector and R296K). These results suggest the intriguing possibility that Y37 may be an important previously unrecognized site for ATF4 regulation by phosphorylation and that impaired activity of D35 variants found in cervical dystonia may act through modulating Y37 phosphorylation. In summary, these functional observations, combined with the human genetic association validated in a second independent cohort, provide strong evidence that deficient eIF2α signaling contributes not only to rare inherited dystonias, but also to sporadic forms of adult-onset, cervical dystonia.
DISCUSSION
Using an unbiased genome-wide RNAi screen employing a novel high-throughput, DYT1 dystonia assay for Torsin1a mislocalization as an entry point, we have discovered a role for deficient eIF2α pathway signaling in dystonia pathogenesis that provides a common mechanism bridging multiple distinct forms of the disease. Although knowledge of genetic causes for isolated dystonia is rapidly increasing, the identification of specific pathomechanisms for dystonia has been far more challenging (Lohmann and Klein, 2013). To date, dopamine deficiency is the only known common cellular mechanism with strong support across multiple forms of dystonia (Lohmann and Klein, 2013; Waugh and Sharma, 2013).
eIF2α Pathway Hypothesis for Dystonia
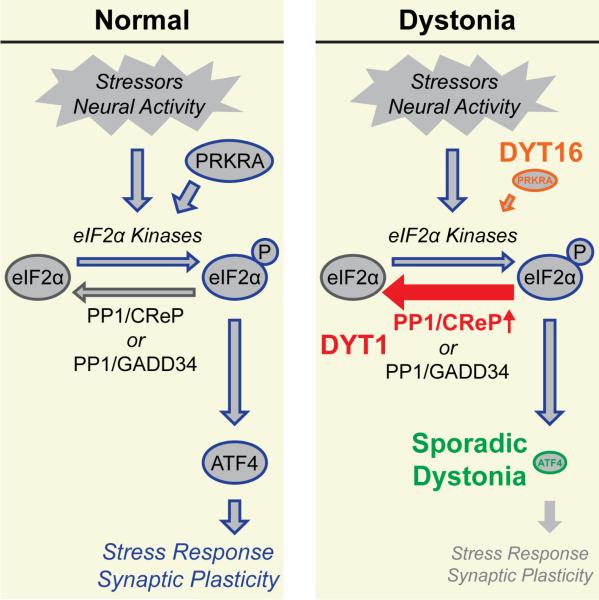
Although the specific disruptions in the eIF2α pathway are distinct among these three dystonias, they converge upon a common consequence of reduced eIF2α pathway signaling (Figure 7). In DYT1 patient-derived fibroblasts, we find an attenuated ISR alongside high basal levels of a negative feedback regulator of eIF2α phosphorylation, the CReP/PPP1R15B phosphatase (Figures 5 and 7, red). In sporadic cervical dystonia patients, we find significant enrichment of rare ATF4/CREB2 missense variants that reduce transcriptional activity (Figures 6 and 7, green). Transcriptional regulation by ATF4 is a major downstream effector of the ISR (Pakos-Zebrucka et al., 2016). In DYT16, it is noteworthy that the most common PRKRA mutation is associated with both delayed and decreased stress-induced phosphorylation of eIF2α (see Figure 5 in Vaughn et al., 2015) (Figure 7, orange) among other effects such as promoting apoptosis which has been suggested by Vaughn and colleagues as a pathomechanism. Lastly, while PRKRA's function as an activator of eIF2α kinases is well-described, additional functions have also been reported (Lee et al., 2006; Yong et al., 2015). Thus, our results in DYT1 models and the genetics of sporadic cervical dystonia provide supportive evidence that PRKRA's role in eIF2α signaling is likely relevant for DYT16 pathogenesis. Taken together these observations create a specific mechanistic model to test the hypothesis that impaired eIF2α signaling is a central mechanism for dystonia.
Figure 7.
Model depicting shared cellular mechanism of eIF2α pathway dysfunction across multiple forms of dystonia.
The failure of etiological dystonia models to reproduce dystonia symptomatology has been a major impediment to studying dystonia mechanisms and the reasons for this barrier are not yet clear. Our implication of a stress response pathway provides one possible explanation - the need for a second hit that activates this pathway. While the convergence of human genetic associations at PRKRA and ATF4/CREB2 provide genetic support that pathway dysfunction can lead to dystonia in humans, a limitation of the present study is that we were unable to examine the effects of targeting this pathway on dystonia in DYT1 mouse models with construct validity because they lack dystonic movements. Rather we find that eIF2α pathway dysfunction is present in DYT1 patient fibroblasts and leads to abnormal synaptic plasticity in DYT1 model mice, and that enhancing eIF2α signaling mitigates homozygous lethality due to the DYT1 mutation. Thus, an important future direction will be to demonstrate whether and when targeting eIF2α signaling can improve dystonia. An important related question will be to determine exactly where in the brain selective vulnerability to altered eIF2α signaling occurs and how it leads to dystonia. Both the relevant brain regions and specific cell types involved in dystonia are open areas of debate. Based on the eIF2α hypothesis, future identification of cell subtypes and brain regions with eIF2α pathway disruption considered in conjunction with other lines of evidence implicating specific cells and circuits may help accelerate this effort and enable conditional genetic tests.
eIF2α Mechanisms for Brain Dysfunction in Dystonia
ER stress and altered proteostasis, as could arise from eIF2α signaling perturbations, are widely considered potential mechanisms for selective brain vulnerability (Zhao and Ackerman, 2006). Besides having commonalities as being components of the ISR, it is notable that in the brain, both eIF2α phosphorylation and ATF4/CREB2 are also linked to expression of long-term forms of synaptic plasticity (Bartsch et al., 1995; Costa-Mattioli et al., 2007; Di Prisco et al., 2014; Trinh and Klann, 2013). In dystonia, a role for pathological plasticity has long been suggested by both clinical features, such as task-specific dystonias that occur only while performing a specific learned task and the relatively delayed time course of response to deep brain stimulation (Peterson et al., 2010), and findings of altered synaptic plasticity in dystonia animal models (Köhling et al., 2004; Martella et al., 2009).
In this study, we find evidence that the significance of eIF2α pathway dysfunction for dystonia could be related to its brain-specific role in regulating synaptic plasticity. Lack of corticostriatal mGluR5-LTD is a well-described phenotype in DYT1 mouse models (Martella et al., 2009). Previously, eIF2α phosphorylation had been shown to be necessary for hippocampal mGluR-LTD (Di Prisco et al., 2014). We have expanded this role to include mGluR5-LTD at cortical synapses onto projection neurons (Figure 4A). In addition to reproducing the DYT1 phenotype with signaling blockade in WT mice, we also find that augmenting eIF2α phosphorylation rescued plasticity in DYT1 model mice (Figure 4B). However, since it remains to be established that abnormal striatal plasticity per se is a causative feature for dystonia pathogenesis, it is important to recognize that other as yet unidentified functions of eIF2α signaling in the brain may instead be the most significant for its role in dystonia pathogenesis.
ΔE Torsin1a and eIF2α Pathway Dysfunction
An in-frame deletion of a glutamic acid residue (ΔE) is the basis for the autosomal dominant TOR1A mutation that causes DYT1 dystonia (Ozelius et al., 1997). Among various cellular effects ascribed to Torsin1a or the mutant protein, several groups have found alterations in ER stress response indicators though not specifically in the eIF2α pathway, and typically in the setting of overexpression paradigms much like our screening assay (Chen et al., 2010; Nery et al., 2011). In this study, once eIF2α signaling was suggested by the genome-wide RNAi screen (Table 1), its role was supported in 2 settings with endogenous, heterozygous delGAG TOR1A gene expression – patient fibroblast cell lines (Figures 1K and 5) and DYT1 knockin mice (Figure 4). A principal observation made in DYT1 patient fibroblasts was the finding of elevated steady-state levels of the eIF2α phosphatase, CReP (Figure 5D) and its lack of upregulation upon pathway activation as is normally seen (Figure 5E). Determining exactly how mutant Torsin1a leads to this pathway perturbation is an important future direction.
ATF4/CREB2 Contributions to Sporadic Focal Dystonia
As with many other diseases with both sporadic and inherited forms, the genetic contributions to sporadic dystonia are the least well understood. The large majority of dystonia cases are sporadic and of unknown cause, with only 10% of cases reporting an affected family member (LeDoux, 2012). In this study, we found that 3% of patients with one of the most common sporadic dystonias, cervical dystonia, had functionally significant, rare missense variants in the ATF4/CREB2 gene. The focus on this gene arose as a result of an unbiased exome analysis in which 2 of 20 cases shared a mutation (Figure 6A). While independent confirmation of this association in a second cohort establishes the enrichment, our findings raise two important future questions. The first is to determine whether other genes in this pathway are involved and in which dystonia populations. The second is to understand the mechanism of the genetic contribution to dystonia - as a risk factor or causative allele? And whether there is a need for a second hit to lead to dystonia. Certainly a two-hit model is suggested by the presence of reduced penetrance in many of the monogenic isolated dystonias (Lohmann and Klein, 2013). Attenuated activity of a major cellular stress-response pathway, the ISR, might be one mechanism for this piece of the clinical puzzle.
The eIF2α Pathway as a Diagnostic and Therapeutic Target
In all, our findings point to the eIF2α pathway as a promising biological target to aid in the development of diagnostic tools and disease-modifying treatments for dystonia. Genetic and functional phenotyping for impairment of eIF2α pathway signaling could prove useful for identifying cohorts of patients with similar underlying mechanisms/risk factors. Encouragingly, we found that pharmacologically enhancing eIF2α signaling rescued synaptic plasticity and delayed perinatal mortality (Figure 4). However, since the relationship of these two phenotypes to dystonia is unclear, further studies are warranted. Interestingly, the results of our study, by showing distinct insults to a common pathway across different dystonias (Figure 7), suggest that it may be necessary to develop therapies targeting multiple portions of the eIF2α pathway.
EXPERIMENTAL PROCEDURES
For additional experimental details and tables listing reagents used in this study, please see Supplemental Experimental Procedures.
Torsin1a localization assay
Flp-In T-REx 293 cells inducibly expressing either EGFP-WT or −ΔE Torsin1a were plated, transfected, induced, fixed, and imaged in 384-well glass-bottom plates (Corning). Transfection of siRNA pairs from the Qiagen Human Genome siRNA Library v1.0 in two unique pools per gene were performed as earlier described (Barrows et al., 2010). Fixed and Hoechst-stained cells were imaged on a Cellomics ArrayScan automated high-content imaging system. Images were analyzed using a modification of the Cellomics ArrayScan V CompartmentAnalysisV2 protocol. Similar protocols were used for pharmacological and ATF4 overexpression experiments.
Animals
ΔE Torsin1a knockin (courtesy of Dr. W. Dauer, University of Michigan) (Goodchild et al., 2005) and Drd1a-tdTomato line 6 (Ade et al., 2011) mice on C57Bl/6 background were bred in standard housing conditions with food and water provided ad libitum. All procedures were approved by the Duke University Institutional Animal Care and Use Committee (IACUC).
Electrophysiological recordings
Acute horizontal brain slices from the progeny of heterozygous ΔE Torsin1a knockin x homozygous Drd1a-tdTomato mice (age P15-21) were prepared largely as described in (Trusel et al., 2015). LTD was induced as in (Trusel et al., 2015) with modifications noted in Supplemental Experimental Procedures.
Human dystonia patients
Whole-exome sequencing subjects
Genomic DNA samples from 20 subjects with sporadic dystonia were exome sequenced. Results were compared to sequence from 571 control samples that were sequenced as part of other studies at Duke University. All studies were reviewed and approved by the Duke University Medical Center IRB, and all subjects gave written informed consent. Dystonia subjects were recruited at the Movement Disorders Center at Duke University Medical Center, Durham, NC.
ATF4 exon 1 sequencing subjects
Genomic DNA samples were acquired from subjects recruited at Movement Disorder Centers in New York (Mount Sinai Beth Israel) and Boston (Massachusetts General Hospital), and from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (ccr.coriell.org/ninds). The local institutional review boards approved the studies and all participating individuals gave informed consent prior to participation. NINDS Repository sample numbers corresponding to the samples used are listed in Supplemental Experimental Procedures.
ATF4 activity assay
HEK293T cells were transfected with pAARE-RLuc, pCMV-CLuc2, and empty vector or FLAG-tagged WT/mutant ATF4 as appropriate. The following day conditioned media was harvested and the assay cells were lysed. Conditioned media and cell lysates were assayed for CLuc and RLuc using Cyperidina Luciferase (NEB) and Renilla Luciferase (Thermo Fisher) assay kits.
Supplementary Material
Highlights.
Genome-wide RNAi screen of a novel DYT1 dystonia assay identifies the eIF2α pathway
Enhancing eIF2α signaling restores absent corticostriatal LTD in DYT1 knockin mice
DYT1 dystonia patient-derived cells have a deficient eIF2α pathway stress response
Rare loss-of-function variants in ATF4 are enriched in sporadic dystonia patients
Acknowledgements
The authors wish to thank: Drs. Xandra Breakefield and Flavia Nery for advice on the luciferase secretion assay; Dr. William Dauer for sharing cDNA, DYT1 knockin mouse lines, and insightful discussions; Dr. Takashi Kudo for providing BiX; Dr. So Young Kim for high-content imaging expertise; Dr. David Corcoran for bioinformatics and microarray expertise; Drs. Chris Nicchitta and Dennis Thiele for insightful discussions and critical reading of the manuscript; Jessi Cruger for technical laboratory assistance; the Tyler's Hope Foundation for a Dystonia Cure, Bachmann Strauss Parkinson Dystonia Center of Excellence, and Duke University School of Medicine for critical financial support (N.C.); and foremost, the many individuals who participated in the clinical research that enabled these studies. This work was funded in part by NS079860 (N.C.), NS051156 and NS095653 (J.E.R.), GM007105 (S.M.S.), R01DC011805 and R01NS088160 (collection of dystonia patient DNA samples by K.S.), and R56AI098588, RC2NS070342, and UO1AIO67854 (collection of exome sequencing control samples).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Z.F.C. designed and conducted the whole genome RNAi screen and performed all patient cell line luciferase and Western blot assays. R. H.-M. performed the electrophysiological experiments. J.E.R. designed and conducted all other experiments. M.K.S., S.M.S., A.Y.L., C.X. and K.K.T. assisted with experiments. N.C., P.H., J.J., B.S., M.S., R.S.-P., K.S. and N.S. recruited subjects with dystonia. L.O. contributed to the design and interpretation of genetic analyses. E.T.C. performed genetic analyses of whole exome and ATF4 sequences. J.L.P. assisted in the implementation of the RNAi screen. N.C. provided input to all phases of the study. J.E.R. and N.C. wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- Ade KK, Wan Y, Chen M, Gloss B, Calakos N. An Improved BAC Transgenic Fluorescent Reporter Line for Sensitive and Specific Identification of Striatonigral Medium Spiny Neurons. Front. Syst. Neurosci. 2011;5:32. doi: 10.3389/fnsys.2011.00032. doi:10.3389/fnsys.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PBF, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. doi:10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Schmidt RJ, Burr B, Burr FA. An arginine to lysine substitution in the bZIP domain of an opaque-2 mutant in maize abolishes specific DNA binding. Genes Dev. 1991;5:310–20. doi: 10.1101/gad.5.2.310. [DOI] [PubMed] [Google Scholar]
- Balint B, Bhatia KP. Isolated and combined dystonia syndromes - an update on new genes and their phenotypes. Eur. J. Neurol. 2015;22:610–7. doi: 10.1111/ene.12650. doi:10.1111/ene.12650. [DOI] [PubMed] [Google Scholar]
- Barrows NJ, Le Sommer C, Garcia-Blanco MA, Pearson JL. Factors affecting reproducibility between genome-scale siRNA-based screens. J. Biomol. Screen. 2010;15:735–47. doi: 10.1177/1087057110374994. doi:10.1177/1087057110374994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–92. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–9. doi: 10.1126/science.1101902. doi:10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Armata IA, Nery FC, Breakefield XO, Sharma N. Molecular pathways in dystonia. Neurobiol. Dis. 2011;42:136–47. doi: 10.1016/j.nbd.2010.11.015. doi:10.1016/j.nbd.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg DC, Camp SM, Kaufman CA, Wilbur JD, Boston H, Schuback DE, Hanson PI, Sena-Esteves M, Breakefield XO. Perinuclear biogenesis of mutant torsin-A inclusions in cultured cells infected with tetracycline-regulated herpes simplex virus type 1 amplicon vectors. Neuroscience. 2004;125:651–61. doi: 10.1016/j.neuroscience.2004.01.053. doi:10.1016/j.neuroscience.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Breakefield XO, Blood AJ, Li Y, Hallett M, Hanson PI, Standaert DG. The pathophysiological basis of dystonias. Nat. Rev. Neurosci. 2008;9:222–34. doi: 10.1038/nrn2337. doi:10.1038/nrn2337. [DOI] [PubMed] [Google Scholar]
- Burdette AJ, Churchill PF, Caldwell GA, Caldwell KA. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones. 2010;15:605–17. doi: 10.1007/s12192-010-0173-2. doi:10.1007/s12192-010-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N, Patel VD, Gottron M, Wang G, Tran-Viet K-N, Brewington D, Beyer JL, Steffens DC, Krishnan RR, Züchner S. Functional evidence implicating a novel TOR1A mutation in idiopathic, late-onset focal dystonia. J. Med. Genet. 2010;47:646–50. doi: 10.1136/jmg.2009.072082. doi:10.1136/jmg.2009.072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargos S, Scholz S, Simón-Sánchez J, Paisán-Ruiz C, Lewis P, Hernandez D, Ding J, Gibbs JR, Cookson MR, Bras J, Guerreiro R, Oliveira CR, Lees A, Hardy J, Cardoso F, Singleton AB. DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet. Neurol. 2008;7:207–15. doi: 10.1016/S1474-4422(08)70022-X. doi:10.1016/S1474-4422(08)70022-X. [DOI] [PubMed] [Google Scholar]
- Cao S, Hewett JW, Yokoi F, Lu J, Buckley AC, Burdette AJ, Chen P, Nery FC, Li Y, Breakefield XO, Caldwell GA, Caldwell KA. Chemical enhancement of torsinA function in cell and animal models of torsion dystonia. Dis. Model. Mech. 2010;3:386–96. doi: 10.1242/dmm.003715. doi:10.1242/dmm.003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Burdette AJ, Porter JC, Ricketts JC, Fox SA, Nery FC, Hewett JW, Berkowitz LA, Breakefield XO, Caldwell KA, Caldwell GA. The early-onset torsion dystonia-associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum. Mol. Genet. 2010;19:3502–15. doi: 10.1093/hmg/ddq266. doi:10.1093/hmg/ddq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chérasse Y, Maurin A-C, Chaveroux C, Jousse C, Carraro V, Parry L, Deval C, Chambon C, Fafournoux P, Bruhat A. The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP. Nucleic Acids Res. 2007;35:5954–65. doi: 10.1093/nar/gkm642. doi:10.1093/nar/gkm642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, Krnjević K, Lacaille J-C, Nader K, Sonenberg N. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. doi:10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N. Translational control of long-term synaptic plasticity and memory storage by eIF2alpha. Crit. Rev. Neurobiol. 2006;18:187–95. doi: 10.1615/critrevneurobiol.v18.i1-2.190. [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, McNaught KSP, Jengelley T-A, Jackson T, Li J, Li Y. Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp. Neurol. 2005;196:452–63. doi: 10.1016/j.expneurol.2005.08.025. doi:10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Defazio G. The epidemiology of primary dystonia: current evidence and perspectives. Eur. J. Neurol. 2010;17(Suppl 1):9–14. doi: 10.1111/j.1468-1331.2010.03053.x. doi:10.1111/j.1468-1331.2010.03053.x. [DOI] [PubMed] [Google Scholar]
- Di Prisco GV, Huang W, Buffington SA, Hsu C-C, Bonnen PE, Placzek AN, Sidrauski C, Krnjević K, Kaufman RJ, Walter P, Costa-Mattioli M. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat. Neurosci. 2014;17:1073–82. doi: 10.1038/nn.3754. doi:10.1038/nn.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskow Jaunarajs KL, Bonsi P, Chesselet MF, Standaert DG, Pisani A. Striatal cholinergic dysfunction as a unifying theme in the pathophysiology of dystonia. Prog. Neurobiol. 2015;127-128:91–107. doi: 10.1016/j.pneurobio.2015.02.002. doi:10.1016/j.pneurobio.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles LM, Chen J, Li L, Chin L-S. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum. Mol. Genet. 2008;17:2712–22. doi: 10.1093/hmg/ddn173. doi:10.1093/hmg/ddn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Paulson HL. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J. Neurosci. 2004;24:2593–601. doi: 10.1523/JNEUROSCI.4461-03.2004. doi:10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 2005;168:855–62. doi: 10.1083/jcb.200411026. doi:10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:847–52. doi: 10.1073/pnas.0304375101. doi:10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–32. doi: 10.1016/j.neuron.2005.11.010. doi:10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Granata A, Koo SJ, Haucke V, Schiavo G, Warner TT. CSN complex controls the stability of selected synaptic proteins via a torsinA-dependent process. EMBO J. 2011;30:181–93. doi: 10.1038/emboj.2010.285. doi:10.1038/emboj.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata A, Watson R, Collinson LM, Schiavo G, Warner TT. The dystonia-associated protein torsinA modulates synaptic vesicle recycling. J. Biol. Chem. 2008;283:7568–79. doi: 10.1074/jbc.M704097200. doi:10.1074/jbc.M704097200. [DOI] [PubMed] [Google Scholar]
- Grillet M, Dominguez Gonzalez B, Sicart A, Pöttler M, Cascalho A, Billion K, Hernandez Diaz S, Swerts J, Naismith TV, Gounko NV, Verstreken P, Hanson PI, Goodchild RE. Torsins Are Essential Regulators of Cellular Lipid Metabolism. Dev. Cell. 2016;38:235–47. doi: 10.1016/j.devcel.2016.06.017. doi:10.1016/j.devcel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Grundmann K, Reischmann B, Vanhoutte G, Hübener J, Teismann P, Hauser T-K, Bonin M, Wilbertz J, Horn S, Nguyen HP, Kuhn M, Chanarat S, Wolburg H, Van der Linden A, Riess O. Overexpression of human wildtype torsinA and human DeltaGAG torsinA in a transgenic mouse model causes phenotypic abnormalities. Neurobiol. Dis. 2007;27:190–206. doi: 10.1016/j.nbd.2007.04.015. doi:10.1016/j.nbd.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum. Mol. Genet. 2000;9:1403–13. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- Hewett JW, Nery FC, Niland B, Ge P, Tan P, Hadwiger P, Tannous BA, Sah DWY, Breakefield XO. siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cells. Hum. Mol. Genet. 2008;17:1436–45. doi: 10.1093/hmg/ddn032. doi:10.1093/hmg/ddn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett JW, Tannous B, Niland BP, Nery FC, Zeng J, Li Y, Breakefield XO. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7271–6. doi: 10.1073/pnas.0701185104. doi:10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a011544. doi:10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. doi:10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–95. doi: 10.1016/j.celrep.2013.03.015. doi:10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, Ron D. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 2003;163:767–75. doi: 10.1083/jcb.200308075. doi:10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum. Mol. Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. doi:10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- Kim CE, Perez A, Perkins G, Ellisman MH, Dauer WT. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9861–6. doi: 10.1073/pnas.0912877107. doi:10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C. Genetics in dystonia. Parkinsonism Relat. Disord. 2014;20(Suppl 1):S137–42. doi: 10.1016/S1353-8020(13)70033-6. doi:10.1016/S1353-8020(13)70033-6. [DOI] [PubMed] [Google Scholar]
- Köhling R, Koch U-R, Hamann M, Richter A. Increased excitability in cortico-striatal synaptic pathway in a model of paroxysmal dystonia. Neurobiol. Dis. 2004;16:236–45. doi: 10.1016/j.nbd.2004.01.012. doi:10.1016/j.nbd.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J. Biol. Chem. 2000;275:27933–9. doi: 10.1074/jbc.M910025199. doi:10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 2005;280:41537–45. doi: 10.1074/jbc.M505294200. doi:10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- LeDoux MS. The genetics of dystonias. Adv. Genet. 2012;79:35–85. doi: 10.1016/B978-0-12-394395-8.00002-5. doi:10.1016/B978-0-12-394395-8.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park S-Y, Kim Y-K, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. doi:10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C-C, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest. 2014;124:3080–92. doi: 10.1172/JCI72830. doi:10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann K, Klein C. Genetics of dystonia: what's known? What's new? What's next? Mov. Disord. 2013;28:899–905. doi: 10.1002/mds.25536. doi:10.1002/mds.25536. [DOI] [PubMed] [Google Scholar]
- Martella G, Maltese M, Nisticò R, Schirinzi T, Madeo G, Sciamanna G, Ponterio G, Tassone A, Mandolesi G, Vanni V, Pignatelli M, Bonsi P, Pisani A. Regional specificity of synaptic plasticity deficits in a knock-in mouse model of DYT1 dystonia. Neurobiol. Dis. 2014;65:124–32. doi: 10.1016/j.nbd.2014.01.016. doi:10.1016/j.nbd.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Martella G, Tassone A, Sciamanna G, Platania P, Cuomo D, Viscomi MT, Bonsi P, Cacci E, Biagioni S, Usiello A, Bernardi G, Sharma N, Standaert DG, Pisani A. Impairment of bidirectional synaptic plasticity in the striatum of a mouse model of DYT1 dystonia: role of endogenous acetylcholine. Brain. 2009;132:2336–49. doi: 10.1093/brain/awp194. doi:10.1093/brain/awp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7612–7. doi: 10.1073/pnas.0308760101. doi:10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S, Chi Y, Gao K, Kono K, Yao J. eIF2α-Independent Inhibition of TNF-α-Triggered NF-κB Activation by Salubrinal. Biol. Pharm. Bull. 2015;38:1368–74. doi: 10.1248/bpb.b15-00312. doi:10.1248/bpb.b15-00312. [DOI] [PubMed] [Google Scholar]
- Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, da Hora CC, Wang Q, Tagaya M, Klein C, Tannous B, Caldwell KA, Caldwell GA, Lencer WI, Ye Y, Breakefield XO. TorsinA participates in endoplasmic reticulum-associated degradation. Nat. Commun. 2011;2:393. doi: 10.1038/ncomms1383. doi:10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001;153:1011–22. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Bressman SB. Genetic and clinical features of primary torsion dystonia. Neurobiol. Dis. 2011;42:127–35. doi: 10.1016/j.nbd.2010.12.012. doi:10.1016/j.nbd.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997;17:40–8. doi: 10.1038/ng0997-40. doi:10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016 doi: 10.15252/embr.201642195. doi:10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–55. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Sejnowski TJ, Poizner H. Convergent evidence for abnormal striatal synaptic plasticity in dystonia. Neurobiol. Dis. 2010;37:558–73. doi: 10.1016/j.nbd.2009.12.003. doi:10.1016/j.nbd.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK-H, Wilson C, Okreglak V, Ashkenazi A, Hann B, Nader K, Arkin MR, Renslo AR, Sonenberg N, Walter P. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. doi:10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015;4 doi: 10.7554/eLife.05033. doi:10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods. 2012;9:363–6. doi: 10.1038/nmeth.1898. doi:10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell SR, Platt G, Barrie SE, Zoumpoulidou G, Te Poele RH, Aherne GW, Wilson SC, Sheldrake P, McDonald E, Venet M, Soudy C, Elustondo F, Rigoreau L, Blagg J, Workman P, Garrett MD, Mittnacht S. Mechanism-based screen for G1/S checkpoint activators identifies a selective activator of EIF2AK3/PERK signalling. PLoS One. 2012;7:e28568. doi: 10.1371/journal.pone.0028568. doi:10.1371/journal.pone.0028568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, Liang C-C, Dauer WT. Neuronal Nuclear Membrane Budding Occurs during a Developmental Window Modulated by Torsin Paralogs. Cell Rep. 2016;16:3322–33. doi: 10.1016/j.celrep.2016.08.044. doi:10.1016/j.celrep.2016.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, Martin C, Dauer WT. Genetic background modulates the phenotype of a mouse model of DYT1 dystonia. PLoS One. 2012;7:e32245. doi: 10.1371/journal.pone.0032245. doi:10.1371/journal.pone.0032245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GE, Sweeney AL, Beaulieu J-M, Shashidharan P, Caron MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15650–5. doi: 10.1073/pnas.0308088101. doi:10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh MA, Klann E. Translational control by eIF2α kinases in long-lasting synaptic plasticity and long-term memory. Neurobiol. Learn. Mem. 2013;105:93–9. doi: 10.1016/j.nlm.2013.04.013. doi:10.1016/j.nlm.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusel M, Cavaccini A, Gritti M, Greco B, Saintot P-P, Nazzaro C, Cerovic M, Morella I, Brambilla R, Tonini R. Coordinated Regulation of Synaptic Plasticity at Striatopallidal and Striatonigral Neurons Orchestrates Motor Control. Cell Rep. 2015;13:1353–65. doi: 10.1016/j.celrep.2015.10.009. doi:10.1016/j.celrep.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–4. doi: 10.1126/science.1201396. doi:10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 2009;20:2661–72. doi: 10.1091/mbc.E09-01-0094. doi:10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11269–74. doi: 10.1073/pnas.0400541101. doi:10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn LS, Bragg DC, Sharma N, Camargos S, Cardoso F, Patel RC. Altered activation of protein kinase PKR and enhanced apoptosis in dystonia cells carrying a mutation in PKR activator protein PACT. J. Biol. Chem. 2015;290:22543–57. doi: 10.1074/jbc.M115.669408. doi:10.1074/jbc.M115.669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulinovic F, Lohmann K, Rakovic A, Capetian P, Alvarez-Fischer D, Schmidt A, Weißbach A, Erogullari A, Kaiser FJ, Wiegers K, Ferbert A, Rolfs A, Klein C, Seibler P. Unraveling cellular phenotypes of novel TorsinA/TOR1A mutations. Hum. Mutat. 2014;35:1114–22. doi: 10.1002/humu.22604. doi:10.1002/humu.22604. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang H-Y, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. doi:10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Xiao J, Vemula SR, LeDoux MS. Recent advances in the genetics of dystonia. Curr. Neurol. Neurosci. Rep. 2014;14:462. doi: 10.1007/s11910-014-0462-8. doi:10.1007/s11910-014-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Y, Meng Y, Ding H, Fan Z, Tang Y, Zhou C, Luo J, Ke Z-J. PACT/RAX regulates the migration of cerebellar granule neurons in the developing cerebellum. Sci. Rep. 2015;5:7961. doi: 10.1038/srep07961. doi:10.1038/srep07961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. doi:10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.