Abstract
Firmicutes and Bacteroidetes are the predominant bacterial phyla colonizing the healthy human large intestine. Whilst both ferment dietary fibre, genes responsible for this important activity have been analysed only in the Bacteroidetes, with very little known about the Firmicutes. This work investigates the carbohydrate-active enzymes (CAZymes) in a group of Firmicutes, Roseburia spp. and Eubacterium rectale, which play an important role in producing butyrate from dietary carbohydrates and in health maintenance. Genome sequences of 11 strains representing E. rectale and four Roseburia spp. were analysed for carbohydrate-active genes. Following assembly into a pan-genome, core, variable and unique genes were identified. The 1840 CAZyme genes identified in the pan-genome were assigned to 538 orthologous groups, of which only 26 were present in all strains, indicating considerable inter-strain variability. This analysis was used to categorize the 11 strains into four carbohydrate utilization ecotypes (CUEs), which were shown to correspond to utilization of different carbohydrates for growth. Many glycoside hydrolase genes were found linked to genes encoding oligosaccharide transporters and regulatory elements in the genomes of Roseburia spp. and E. rectale, forming distinct polysaccharide utilization loci (PULs). Whilst PULs are also a common feature in Bacteroidetes, key differences were noted in these Firmicutes, including the absence of close homologues of Bacteroides polysaccharide utilization genes, hence we refer to Gram-positive PULs (gpPULs). Most CAZyme genes in the Roseburia/E. rectale group are organized into gpPULs. Variation in gpPULs can explain the high degree of nutritional specialization at the species level within this group.
Keywords: Carbohydrate, comparative genomics, gut microbiota, Lachnospiraceae, obligate anaerobe, Roseburia
Data Summary
The high-quality draft genomes generated in this work were deposited at the European Nucleotide Archive under the following accession numbers:
1. Eubacterium rectale T1-815; CVRQ01000001–CVRQ01000090: http://www.ebi.ac.uk/ena/data/view/PRJEB9320
2. Roseburia faecis M72/1; CVRR01000001–CVRR01000101: http://www.ebi.ac.uk/ena/data/view/PRJEB9321
3. Roseburia inulinivorans L1-83; CVRS01000001–CVRS01000151: http://www.ebi.ac.uk/ena/data/view/PRJEB9322
Impact Statement
Firmicutes and Bacteroidetes are the predominant bacterial phyla that colonize the healthy human large intestine. Whilst both phyla include species that ferment dietary fibre, genes responsible for this important activity have been analysed only in the Bacteroidetes and this paper represents the first detailed analysis for a group of human colonic Firmicutes. This paper will be of interest to those working in the fields of bacterial genomics, intestinal microbiology, human nutrition and health, and microbial polysaccharide breakdown. In particular, interest is growing rapidly in the human gut microbiota and its contribution to health and disease, including the potential for manipulating the microbiota through diet to achieve health benefits. The bacteria studied here are of special interest as they play a dominant role in producing the health-protective metabolite butyrate from dietary carbohydrates. This analysis reveals distinct polysaccharide utilization loci that comprise genes encoding degradative enzymes (glycoside hydrolases (GHs)) linked to genes encoding carbohydrate transporters and regulatory functions in the genomes of Roseburia spp. and Eubacterium rectale. Key differences are reported between these PULs and those of colonic Bacteroidetes, whilst the GH distribution allows strains to be categorized into carbohydrate-utilization ecotypes that utilize different carbohydrates for growth.
Introduction
The human large intestine supports an extremely dense and diverse microbial community that plays an important role in human health (Flint et al., 2012b; Sekirov et al., 2010). Carbohydrates derived from the diet and from the host that remain undigested by host enzymes provide the major energy sources for growth and metabolism of the colonic microbiota. In addition to interactions with the host involving microbial cells and cell components, the short-chain fatty acid products of carbohydrate fermentation by gut bacteria exert multiple effects on the host as energy sources, and as regulators of inflammation, proliferation and apoptosis (Louis et al., 2014). There is particular interest in the role played by butyrate-producing species of the gut microbiota in health maintenance, as their populations are found to be less abundant in a range of conditions that involve dysbiosis, including inflammatory bowel disease and colorectal cancer (Balamurugan et al., 2008; Wang et al., 2012; Machiels et al., 2014). The predominant butyrate-producing bacteria in the healthy human colon belong to the phylum Firmicutes (Barcenilla et al., 2000; Louis et al., 2010), and include Faecalibacterium prausnitzii (Ruminococcaceae) and Roseburia spp., Eubacterium rectale, Eubacterium hallii and Anaerostipes spp. (Lachnospiraceae) (Louis & Flint, 2009).
So far, the only group of human colonic bacteria to have been investigated in any detail with respect to polysaccharide utilization are Bacteroides spp. (Martens et al., 2011; Flint et al., 2012a). These species possess large genomes with extremely high numbers of predicted carbohydrate-active enzymes (CAZymes). These CAZyme genes are located in the genome adjacent to genes encoding regulators and carbohydrate transport functions, forming multiple polysaccharide utilization loci (PULs) whose organization is typified by the Bacteroides thetaiotaomicron starch utilization system (Sus) (Martens et al., 2011; McNulty et al., 2013; El Kaoutari et al., 2013). This, together with the far lower proportional numbers of CAZymes found in the genomes of human colonic Firmicutes, has led to the suggestion that Bacteroides spp. play the predominant role in carbohydrate degradation in the human colon (El Kaoutari et al., 2013). However, various Firmicutes have been shown to respond to changes in the major dietary carbohydrate in human volunteer studies, with relatives of Ruminococcus spp., Roseburia spp. and E. rectale increasing with diets enriched with resistant starch or wheat bran (Duncan et al., 2007; Martínez et al., 2010, 2013; Walker et al., 2011; Salonen et al., 2014; David et al., 2014). This suggests an alternative interpretation, i.e. that Firmicutes might typically be nutritionally highly specialized, whereas Bacteroides spp. may typically retain a greater plasticity for glycan utilization. Such nutritional specialization has already been noted among the ruminococci (Ze et al., 2012; Wegmann et al., 2014). Given that Firmicutes can account for ∼70 % of bacterial phylogenetic diversity in the human colon (Eckburg et al., 2005), there is an obvious need for better understanding of carbohydrate utilization in this phylum. Roseburia spp. together with E. rectale form a coherent group of butyrate-producing Firmicutes, based on 16S rRNA gene sequences and multiple shared genotypic and phenotypic traits, including butyrate pathway genes and flagellar motility (Aminov et al., 2006; Louis & Flint, 2009; Neville et al., 2013). The fact that this group of bacteria are flagellated provides an additional mechanism for interaction with the host immune system (Neville et al., 2013). The availability of genome sequence information for multiple representatives of the Roseburia and E. rectale group isolated from the human colon therefore provides an excellent opportunity to gain an understanding of polysaccharide utilization by this important group of butyrate-producing Firmicutes, which typically accounts for 5–20 % of total colonic bacteria in human adults (Hold et al., 2003; Aminov et al., 2006; Tap et al., 2009; Walker et al., 2011). Our analysis reveals for the first time the existence and organization of Gram-positive PULs (gpPULs) in this group of Lachnospiraceae. Furthermore, considerable specialization in the utilization of different dietary carbohydrates was observed at the species level that is likely to underlie species-specific responses to dietary carbohydrates observed in human volunteer studies (Salonen et al., 2014).
Methods
Genomes, bacterial strains and growth conditions
The bacterial genomes used in this work are described in Table 1. Routine culturing of bacterial strains was in anaerobic M2GSC medium (Miyazaki et al., 1997) in 7.5 ml aliquots in Hungate tubes, sealed with butyl rubber septa (Bellco Glass). Single-carbohydrate growth experiments were carried out in basal YCFA medium (Lopez-Siles et al., 2012) supplemented with 0.5 % (w/v) of the carbohydrate being examined. All carbohydrates and manufacturers are detailed in Table S1 (available in the online Supplementary Material). Cultures were inoculated using the anaerobic methods described by Bryant (1972) and incubated anaerobically without agitation at 37 °C. Growth experiments were routinely carried out in flat-bottom 96-well microtitre plates (Corning; Sigma-Aldrich) prepared in the anaerobic ConceptPlus workstation. Sample blanks containing uninoculated medium were used as controls. Substrates (10 μl of a 10 % stock) were placed directly in wells and a 190 μl aliquot of the master mix (7.5 ml basal YCFA containing 100 μl bacterial inoculum) was added. Microtitre plates were covered and tightly sealed (Bio-Rad iCycler iQ optical tape 2239444) to prevent evaporation and maintain the anaerobic atmosphere. Cells were incubated for 24 h at 37 °C in a BioTek spectrophotometer, with OD650 readings taken automatically every hour with low-speed shaking for 5 s prior to each reading. In cases where the substrate was particularly cloudy, experiments were repeated in basal YCFA (7.5 ml) Hungate tubes, containing 1 % (mucin T2 and T3), 0.5 % (inulin) or 0.2 % (β-mannan) substrate, and 100 μl inoculum.
Table 1. Genomes and bacterial strains.
All strains were isolated from human faecal samples. Sequencing and genome assembly of E. rectale T1-815, R. inulinivorans L1-83 and R. faecis M72/1 was performed by the Wellcome Trust Sanger Institute. The genomes of E. rectale A1-86, and M104/1 and R. intestinalis XB6B4 and M50/1 were sequenced by the Pathogen Genomics group at the Wellcome Trust Sanger Institute as part of the EU MetaHit project (http://www.sanger.ac.uk/resources/downloads/bacteria/metahit/). The locus tag is the strain identifier used throughout this work.
| Species | Strain | Locus tag | GenBank accession No. | No. ORFs | Size (nt) | Genome publication reference | Strain publication reference | Institute of isolation* |
| E. rectale | ATCC33656 | EUBREC_ | NC_012781.1 | 3621 | 3 449 685 | Unpublished | Unpublished | VPI |
| A1-86 | EUR_ | NC_021010.1 | 2898 | 3 344 951 | Unpublished | Barcenilla et al. (2000) | RINH | |
| M104/1 | ERE_ | NC_021044.1 | 3206 | 3 698 419 | Unpublished | Louis et al. (2004) | RINH | |
| T1-815 | T1_815_ | CVRQ01000001–CVRQ01000090 | 2896 | 3 045 135 | This work | Barcenilla et al. (2000) | RINH | |
| R. inulinivorans | A2-194 | RINU_ | ACFY01000000 | 4522 | 4 048 462 | Unpublished | Duncan et al. (2006) | RINH |
| L1-83 | L1-83_ | CVRS01000001–CVRS01000151 | 3488 | 3 781 521 | This work | Barcenilla et al. (2000) | RINH | |
| R. intestinalis | L1-82 | RINT_ | ABYJ00000000.2 | 4766 | 4 411 375 | Unpublished | Duncan et al. (2006) | RINH |
| M50/1 | ROI_ | NC_021040.1 | 3461 | 4 143 550 | Unpublished | Louis et al. (2004) | RINH | |
| XB6B4 | RO1_ | NC_021012 | 3610 | 4 286 292 | Unpublished | Chassard et al. (2007) | INRA | |
| R. hominis | A2-183 | RHOM_ | CP003040.1 | 3362 | 3 592 125 | Travis et al. (2015) | Duncan et al. (2006) | RINH |
| R. faecis | M72/1 | M72_ | CVRR01000001–CVRR01000101 | 3205 | 3 334 694 | This work | Duncan et al. (2006) | RINH |
* VPI, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA; RINH, Rowett Institute of Nutrition and Health, University of Aberdeen, Aberdeen, UK; INRA, INRA Clermont-Ferrand/Theix, France.
Gas production was measured by displacement of a syringe inserted into the butyl stopper following 48 h growth in Hungate tubes. The final pH of the media was recorded and compared with that of the starting medium. These formed additional checks to assess bacterial growth on cloudy substrates.
Substrate-agarose overlay plates were used to assess the ability of strains to degrade substrates, without necessarily being able to grow on them. Bacterial broth cultures grown overnight in Hungate tubes (M2GSC) were streaked onto YCFA agar plates containing 0.2 % glucose, soluble potato starch and cellobiose. Following overnight incubation, a hand-hot molten 0.4 % agarose overlay containing 0.2 % of the appropriate substrate prepared in 50 mM sodium phosphate buffer (pH 6.5) was carefully poured over the colonies. After a further overnight incubation, plates were stained for 30 min, washed and the formation of clear zones noted. Mucin overlays were stained with 0.1 % Amido black prepared in 3.5 M acetic acid and washed in 1.2 M acetic acid; glucagel overlays were stained with 0.1 % Congo red and washed with 1 M NaCl; β-mannan overlays were stained with either 0.1 % Congo red or with Grams Iodine Solution (Sigma).
Sequencing, assembly and automated annotation of high-quality draft genomes of E. rectale T1-815, Roseburia faecis M72/1 and Roseburia inulinivorans L1-83
Genomic DNA was sequenced on the Illumina HiSeq platform generating paired-end reads with a read length of 100 bp. A de novo assembly of the three strains was carried out using Velvet (Zerbino & Birney, 2008), and the assemblies were manually improved using a combination of Gapfiller (Boetzer & Pirovano, 2012) to close sequence gaps and iCORN (Otto et al., 2010) to correct for sequence errors. Annotation of the improved assemblies consisted of identifying coding sequences using Prodigal (Hyatt et al., 2010) and transferring functional gene annotation using closely related references in a best-hit reciprocal manner. Further annotation was then incorporated, principally using Pfam (Punta et al., 2012), Prosite (Sigrist et al., 2010) and RNAmmer (Lagesen et al., 2007) to identify protein families, functional protein sites and rRNA. These high-quality draft genomes were deposited at the European Nucleotide Archive.
Pan-genome homology and motif identification
Orthology detection was performed using QuartetS software (Yu et al., 2011). Orthologues were assigned based on the bidirectional best hit of amino acid sequences, with thresholds of 45 % sequence identity over 50 % of sequence. Additional criteria for orthologue prediction were E values < 1e− 5 and bit scores >50, and a minimum clustering number of two sequences. Sequences were then separated into the Roseburia/E. rectale group core and variable genome using a presence/absence matrix. Sequences with no orthologues in the other 10 strains were considered to be unique genes.
All protein sequences annotated as having an Enzyme Commission number of EC 3.2.1 [glycoside hydrolase (GH)] were extracted from the KEGG database (Kanehisa & Goto, 2000) to form a 24 981 amino acid sequence GH protein reference database. The proteins of the pan-genome of the Roseburia/E. rectale group were queried against this GH protein database using blastp. The results were filtered to exclude all matches with E values >1e− 10, sequence identity < 35 % or bit scores < 200. The database for carbohydrate-active enzyme annotation (dbCAN) HMM (hidden Markov model) database version 3 (http://csbl.bmb.uga.edu/dbCAN/) was downloaded locally and used to query the pan-genome for conserved domains with the programme hmmscan (a command in the hmmer 3.0 package; hmmer.org). These results were filtered by excluding E values >1e− 3 for alignments < 80 amino acids and E values >1e− 5 for alignments ≥ 80 amino acids, and using alignment coverage of >0.3 as the threshold.
Carbohydrate utilization ecotype (CUE) and gpPUL determination
Where all the members of a GH family hydrolysed the same carbohydrate type, carbohydrate sets were assigned by GH family e.g. all GH13s were assigned to the α-glucans set. Where different members of a GH family hydrolysed different carbohydrate types, carbohydrate sets were assigned by KEGG GH annotation (Table S2). As type 1 arabinogalactan could be interpreted as belonging to the carbohydrate sets ‘Xylans and Arabinans’, ‘Pectins’ or ‘Alpha- and Beta-galactosides’, endo-1,4-β-galactanases, which cleave the 1,4-β-galactan backbone of type 1 arabinogalactan, were assigned to a separate carbohydrate set termed ‘Type-1 Arabinogalactans’. GH heatmap analyses were performed using MeV software from the tm4 suite (Saeed et al., 2003). CUEs were determined by hierarchical clustering of the GH heatmap using Kendall τ distance and Spearman distance with complete linkage.
The pan-genome was queried against the KEGG and COG (http://www.ncbi.nlm.nih.gov/COG/) databases, excluding top hit matches of E values >1e–5. The putative CAZymes discovered in this work were incorporated into this annotation and GHs less than 11 genes from a putative carbohydrate transporter system were further investigated by manual curation in Artemis (Carver et al., 2008). The boundaries of gpPULs, both upstream and downstream, were determined by the presence of three adjacent genes not predicted to be involved in carbohydrate degradation. gpPULs were defined as encoding, at minimum, a polysaccharide-degrading enzyme, a transport system and a transcriptional regulator.
Additional annotation tools
Phylogenetic trees were reconstructed in mega 6 software (Kumar et al., 2008). Principal coordinate analyses were performed in the statistical software r using Kendall τ distances of the first five eigenvectors. Visual comparisons of intra-species and inter-species genome variability were performed using the blast Ring Image Generator (brig) software (Alikhan et al., 2011).
Results
In vitro utilization of carbohydrates by the Roseburia/E. rectale group
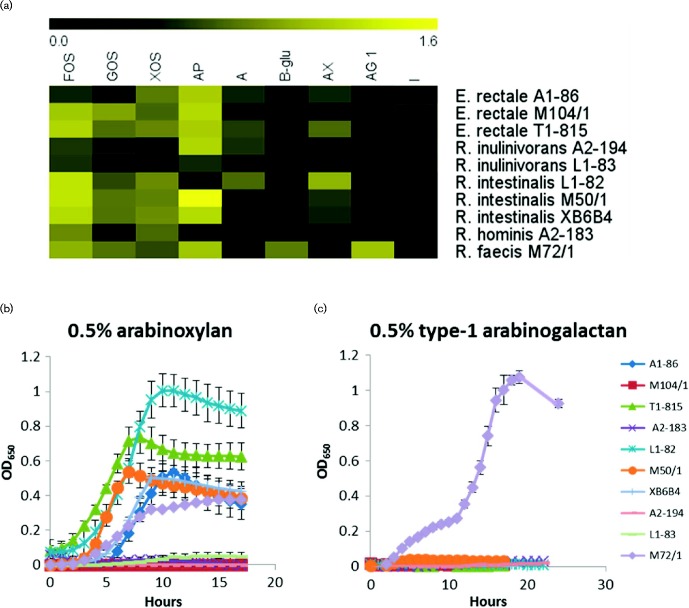
The ability of three strains of E. rectale, two of R. inulinivorans, three of Roseburia intestinalis and one each of R. faecis and Roseburia hominis to degrade and utilize a variety of carbohydrates for growth was tested by anaerobic culturing. Growth in microtitre plates revealed that all 10 strains could utilize fructo-oligosaccharides (Fig. 1a). The ability to grow on galacto-oligosaccharides and xylo-oligosaccharides was more limited with E. rectale A1-86, R. inulinivorans L1-83 and R. hominis A2-183 unable to utilize galacto-oligosaccharides, and the two R. inulinivorans strains unable to grow on xylo-oligosaccharides.
Fig. 1.
Growth of Roseburia/E. rectale strains on selected carbohydrates in microtitre plates. (a) Heatmap representing the mean maximum OD650 obtained by a strain during growth on a specific carbohydrate (OD650 0.0–1.6). Growth was observed on fructo-oligosaccharide (FOS), galacto-oligosaccharide (GOS), xylo-oligosaccharide (XOS), amylopectin (AP), amylose (A), 1,3–1,4-β-glucan (B-glu), arabinoxylan (AX), type 1 arabinogalactan (AG1) and inulin (I). No growth was observed for any the 11 strains on β-mannan, xyloglucan, type 2 arabinogalactan, mucin core type 2 or mucin core type 3. Data plotted in graphs are the mean ± sd OD650 readings of six replicates of strains grown on (b) 0.5 % arabinoxylan or (c) 0.5 % type 1 arabinogalactan. Full growth data are presented in Table S3.
Nine of the 10 strains were able to utilize amylopectin and/or amylose for growth (Fig. 1a, Table S3), the exception being R. hominis A2-183 which was not unable to grow on either type of starch. All strains of E. rectale and R. inulinivorans were capable of utilizing inulin, whereas R. intestinalis, R. hominis and R. faecis strains did not grow with inulin as the sole carbohydrate source (Fig. S1).
Growth on the plant cell wall polysaccharides arabinoxylan, xyloglucan, arabinogalactan, 1,3–1,4-β-glucan and β-mannan was also variable. Arabinoxylan could be utilized by all the three R. intestinalis strains, by E. rectale A1-86 and T1-815, and by R. faecis M72/1 (Fig. 1b). Xyloglucan could not be utilized for growth by any of the 10 strains tested, although xyloglucan overlay plates revealed that the R. intestinalis strains formed clear zones (Fig. S2).
Only R. inulinivorans L1-83 and R. faecis M72/1 were capable of utilizing 1,3–1,4-β-glucan whilst no strain utilized β-mannan (1,4-β-mannan backbone) or type 2 arabinogalactan (1,3-β-galactan backbone). Only R. faecis M72/1 was capable of utilizing type 1 arabinogalactan (1,4-β-galactan backbone) (Fig. 1c).
None of the strains was capable of using either type II or type III pig gastric mucin for growth (data not shown) and no degradation of type II or type III pig gastric mucin could be detected on overlay plates of the 10 strains (data not shown).
Pan-genome of Roseburia/E. rectale group
Eleven genomes representing E. rectale and the four Roseburia spp. were investigated (Table 1). These comprised the 10 strains whose growth characteristics were compared in Fig. 1, with the addition of E. rectale ATCC33656 (Table S4). The genomes of E. rectale A1-86, ATCC33656 and M104/1, R. intestinalis L1-82, XB6B4 and M50/1, R. inulinivorans A2-194, and R. hominis A2-183 were sequenced previously and are publicly available in the GenBank database. In addition, high-quality draft genomes of R. faecis M72/1, R. inulinivorans L1-82 and E. rectale T1-815 were sequenced, assembled and automatically annotated in this work. The draft genomes were compared with the complete genome of E. rectale ATCC33656 (Fig. S3a). This revealed a high level of genome plasticity in the Roseburia/E. rectale group; in particular, sections of the E. rectale ATCC33656 genome were not present in the other genomes despite the fact that the genomes were of similar size (Table 1).
Core, variable and unique genes were identified by assigning all ORFs in the Roseburia/E. rectale pan-genome to orthologous groups (OGs). Orthologues present in all 11 strains were considered to be core genes, with all 11 orthologues forming a core OG; sequences with orthologues present in two to 10 strains were considered to be variable genes and sequences that were found only in one strain were considered unique genes. In this way, 794 core OGs, 5513 variable OGs and 7825 unique genes in the pan-genome (pan-genome details in Fig. S4) were identified. The distribution of genes between the core (mean 22.9 %, range 16.7–27.4 %), variable (mean 57.7 %, range 51.9–64.2 %) and unique (mean 19.4 %, range 10.9–29.4 %) genomes was similar in all strains.
Detection of CAZymes
Predicted CAZyme-encoding genes in the pan-genome were identified to the protein family level in silico using HMMs representing conserved regions of all CAZyme families (Yin et al., 2012). In addition, a protein database focusing on carbohydrate metabolism was established by collecting all the protein sequences in KEGG that had been assigned EC 3.2.1 (GH) and the Roseburia/E. rectale pan-genome was queried against this database using blastp. The combined results from these analyses resulted in the identification of 1840 CAZyme genes in the Roseburia/E. rectale pan-genome, including 932 GHs (Table S5), 503 glycosyltransferases, 243 carbohydrate esterases (CEs) and one polysaccharide lyase (Table S6). Only 74 (7.9 %) of these GHs were predicted to possess signal peptides (SPs) by SignalP software (Petersen et al., 2011), indicating cell-bound or extracellular enzymes. These results are presented in Table S7. Alternative protein secretion of Roseburia/E. rectale pan-genome xylanases was also investigated using SecretomeP (Bendtsen et al., 2005), but no new predicted secreted proteins were identified. Of the 932 GHs, 148 (16 %) were predicted to possess carbohydrate-binding modules (CBMs) (Table S8). GHs with CBMs were more likely to possess SPs (23 %) than GHs without CBMs (5 %).
The distribution of CAZymes, GHs and ‘all genes’ of the Roseburia/E. rectale pan-genome between the core, variable and unique genome fractions was compared (Fig. S5). There was a higher percentage of GHs and CAZymes (75 and 74 %, respectively) in the variable genome, compared with ‘all genes’ (58 %). Strikingly, only 26 out of a total of 538 OGs representing CAZymes within the pan-genome were found in all 11 genomes. These included 13 GH enzymes, including five GH13 and two putative oligosaccharide phosphorylases (Table S9).
The majority of GH OGs were therefore species-specific or strain-specific. For example, 107 CAZyme OGs (including 70 GHs) were found only in R. intestinalis, of which 31 (including 18 GHs) were present in all three R. intestinalis strains (Table S6). Meanwhile, 85 CAZyme OGs (including 24 GHs) were found only in E. rectale, of which only four CAZyme OGs (two GHs) were conserved in all four E. rectale strains. This represents considerable inter-strain variation within these species.
Phylogenetic relationships within GH families
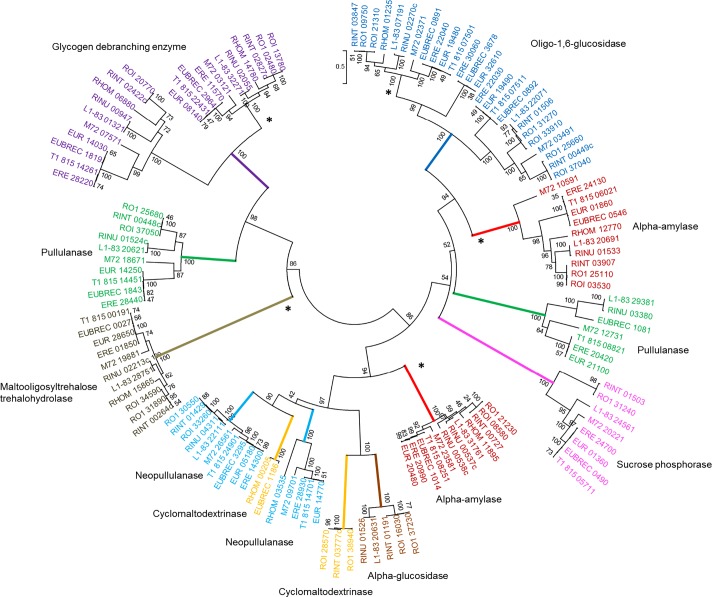
Within each genome, GH families were often represented by multiple genes, as exemplified by the 16 GH43 genes present in R. intestinalis L1-82 (Table S5). In order to investigate the sequence relationships more closely, protein sequence-based phylogenetic trees of GH13 (α-glucan degradation), GH32 (fructan degradation), GH10, GH43 and GH51 (plant cell wall polysaccharide degradation) were reconstructed. Many of the GHs clustered into strongly supported clades (bootstrap ≥ 90) that correlated largely with the annotations assigned to them by the KEGG GH database.
GH13 family enzymes and starch utilization
The phylogenetic tree of the 130 GH13 sequences (Fig. 2) revealed clades similar to previously identified subfamilies that have tentatively assigned divergent functions (Stam et al., 2006). A group of seven ‘pullulanases’ (the green clade at the right of tree, Fig. 2), which possess N-terminal SPs and (with the exception of M72_12731) putative C-terminal sortase signals, included the enzyme RINU_03380 which is responsible for the major amylase activity detected in R. inulinivorans A2-194 cell extracts (Ramsay et al., 2006). The overexpressed gene product of RINU_03380 (Amy13C) hydrolysed α-1,4-glucan linkages in starch, but not α-1,6-glucan linkages (Ramsay et al., 2006), whilst the related EUR_21100 enzyme from E. rectale has recently been shown to cleave α-1,4 linkages to release maltotetraose (Cockburn et al., 2015), indicating that these enzymes are not true type 1 pullulanases. The enzymes in this clade also possess CBMs, either CBM26 (EUR_21100, ERE_20420, T1-815_08821 and M72_12731) or CBM41 (RINU_03380, L1-83_29381 and EUBREC_1081) (this work), which have both been described as starch-binding domains (Lammerts van Bueren et al., 2004; Boraston et al., 2006). Interestingly, whilst this clade was found in all four E. rectale strains, in both strains of R. inulinivorans and in R. faecis, no representative was present in R. hominis A2-183 or in the three R. intestinalis strains. R. hominis A2-183 also lacked representatives of two other GH13 OGs (annotated as a pullulanase and a neopullanase) that were found in the other 10 strains, which presumably explains why it was the only one of the 11 strains that was unable to grow with soluble starch as substrate. The major active extracellular GH13 enzyme (RINT_03777c) detected on a zymogram in R. intestinalis strains by Ramsay et al. (2006) belongs to a different clade than EUR_21100 and RINU_03380.
Fig. 2.
Phylogenetic tree of Roseburia/E. rectale GH13s. Gene names are colour-coded based on KEGG GH annotation. Strongly supported clades (bootstrap ≥ 90) are coloured at their most proximal branch. The branch colour corresponds to the KEGG GH annotation of the genes within the clade. Colour coding is as follows: neopullulanase (EC 3.2.1.135) (light blue), cyclomaltodextrinase (EC 3.2.1.54) (orange), α-glucosidase (EC 3.2.1.3) (brown), α-amylase (EC 3.2.1.1) (red), sucrose phosphorase (EC 3.2.1.7) (pink), oligo-1,6-glucosidase (EC 3.2.1.10) (blue), pullulanase (EC 3.2.1.41) (green), glycogen-debranching enzyme (EC 3.2.1.-) (purple) and malto-oligosyltrehalose trehalohydrolase (EC 3.2.1.141) (gold). Bootstrap values, expressed as a percentage of 1000 replications, are given at the branching nodes. This tree is unrooted and reconstructed using the maximum-likelihood method. The scale bar refers to the number of amino acid differences per position. Clades of core GH13s are indicated by asterisks at their most proximal branch.
GH32 family enzymes and utilization of fructans
Sequences belonging to GH32 (Fig. S6) were divided into five strongly supported clades: three β-fructofuranosidases, one levanase clade (unique to the R. intestinalis strains) and one divergent clade with no KEGG GH annotation (Fig. S6). The β-fructofuranosidase clade indicated by the red branch in Fig. S6 includes the R. inulinivorans A2-194 gene RINU_03877c, whose expression is upregulated 25-fold during growth on inulin, compared with growth on glucose, and encodes a β-fructofuranosidase that degrades intermediate- and long-chain fructan substrates (Scott et al., 2011). This clade was found in all strains that were able to utilize inulin for growth and in only one other strain (R. faecis M72/1).
GH families for utilization of hemicelluloses
Only five GH10 genes, which typically encode xylanases, were found in the Roseburia/E. rectale group pan-genome, two in R. intestinalis L1-82, and one each in R. intestinalis XB6B4, E. rectale T1-815 and R. faecis M72/1. Four of these GH10 genes (one in each strain) belonged to the same OG, and possessed two CBM9s and a SP. Hemicellulase activities are also found in the families GH43 (Fig. S7) and GH51 (Fig. S8). In total, 93 % (64 of 69) of the GH43s could be assigned into 11 clades, four of which were unique to R. intestinalis strains. The GH51 sequences fell into five putative α-l-arabinofuranosidase clades and one putative xylan-1,4-β-xylosidase clade (Fig. S8). When the GH43 and GH51 sequences were combined (Fig. S9), four GH43 family xylan-1,4-β-xylosidases clustered into the only xylan-1,4-β-xylosidase clade in GH51, suggesting some overlap between these families.
Only the three R. intestinalis strains encoded GH74 and GH26 enzymes, which are typically involved in utilization of xyloglucan and β-mannan, respectively (Table S5). The two R. inulinivorans strains lacked any representatives of GH10, GH26, GH43, GH51, consistent with their inability to utilize xylans or xylo-oligosaccharides for growth (Fig. 1) (Duncan et al., 2006; Scott et al., 2014).
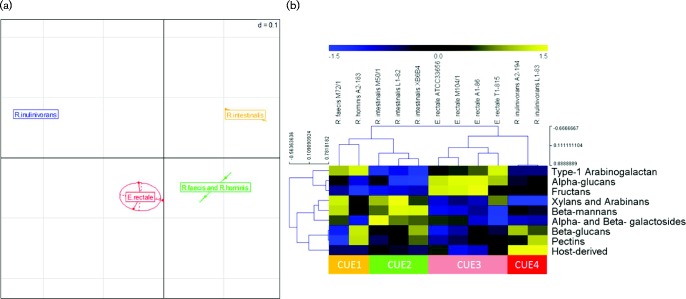
Determination of CUEs
Principal coordinate analysis revealed that the 11 strains formed four statistically significant (P < 0.001) clusters based on their GH family complement (Fig. 3a). Three of these clusters were specific to E. rectale, R. intestinalis and R. inulinivorans. The single strains of R. faecis M72/1 and R. hominis A2-183 did not fall into these three clusters. The GHs of each strain were also assigned into sets, based on their predicted activity against carbohydrate substrates (Table S2). Strain clustering was retained after these data transformations (Fig. S10) and resulted in four statistically significant carbohydrate set-based clusters that we call carbohydrate utilization ecotypes (CUEs; Fig. 3b). CUE1, which includes R. hominis A2-183 and R. faecis M72/1, was enriched in type 1 arabinogalactan-degrading genes (P = 0.03).
Fig. 3.
(a) Principal coordinate analysis of Roseburia/E. rectale strains based on complement of GH families and (b) heatmap showing CUEs. Values of a given GH family or carbohydrate set were taken as the number of these genes each genome possessed. In (a), coordinates were calculated using Kendal τ distance applied to the first five eigenvectors. R. intestinalis strains L1-82, M50/1 and XB6B4 (orange); R. inulinivorans strains A2-194 and L1-83 (blue); R. faecis M72/1 and R. hominis A2-183 (green); and E. rectale strains A1-86, T1-815, M104/1 and ATCC33656 (red) form separate clusters (P < 0.001, non-parametric multivariate ANOVA). In (b), CUEs were determined by complete linkage clustering using Kendall τ (as shown) and Spearman distance (not shown). GH53, an endo-1,4-β-galactanase that cleaves the β-1,4-d-galactosidic linkages in type I arabinogalactans, was assigned to the carbohydrate set ‘Type 1 arabinogalactan’ and is excluded from ‘Xylans and Arabinans’, ‘Pectins’ and ‘Alpha- and Beta-galactosides’. CUE1 consists of R. faecis M72/1 and R. hominis A2-183. CUE2 consists of R. intestinalis strains L1-82, M50/1 and XB6B4. CUE3 consists of E. rectale strains A1-86, T1-815, M104/1 and ATCC33656. CUE4 consists of R. inulinivorans strains A2-194 and L1-83. GH assignment to each carbohydrate set is described in Table S6.
CUE2, which includes the three R. intestinalis strains, was enriched for xylan, arabinan, pectin, β-mannan and galactose sugar degradation genes (P < 0.03). CUE3, which includes all four E. rectale strains, was enriched for fructan degradation genes (P = 0.02) and CUE4, which includes both R. inulinivorans strains, was enriched for host-derived carbohydrate degradation genes, such as mucin glycans (P = 0.04). The relationship between predicted CUE and actual growth behaviour will be discussed below.
PULs in the Roseburia/E. rectale group
PULs are an important feature of Bacteroides genomes (Martens et al., 2008; Larsbrink et al., 2014; Cuskin et al., 2015) and it was therefore decided to investigate the Roseburia/E. rectale group genomes for the presence of gene clusters dedicated to carbohydrate utilization.
R. intestinalis XB6B4 was selected for detailed analysis because it possessed the second highest number of GHs (131 GHs compared with 146 in R. intestinalis L1-82), but the draft genome had a smaller number of contigs. PULs of Bacteroidetes are defined as possessing, at minimum, a TonB-dependent transporter/SusD family lipoprotein-encoding gene pair. As Gram-positive bacteria lack outer membrane transporters, a new definition for PULs is required for these organisms. Here, we define a Gram-positive PUL (gpPUL) as being a locus encoding, at minimum, one polysaccharide-degrading enzyme, a carbohydrate transport system and a transcriptional regulator.
R. intestinalis XB6B4, which was originally selectively isolated for its high xylan-degrading activity (Chassard et al., 2007), was predicted to possess 33 gpPULs. Predicted carbohydrate transport systems were adjacent to GH genes in gpPULs. Of the 35 carbohydrate transporters identified in R. intestinalis XB6B4 gpPULs, 26 (79 %) were ATP-binding cassette (ABC) transporters, 7 (20 %) were glycoside–pentoside–hexuronide (GPH) : cation symporter family transporters, one was a major facilitator superfamily (MFS) transporter and one was a phosphotransferase system (PTS) transporter. No evidence of SusC and SusD homologues, which are the carbohydrate transporters almost universally observed in PULs from the Bacteroidetes, could be found in R. intestinalis XB6B4.
Many of the gpPULs in R. intestinalis XB6B4 encoded GH and CE enzymes that can target multiple carbohydrates, making functional predictions difficult. Despite this, the likely target substrate(s) of 10 (out of 33) gpPULs could be predicted with reasonable confidence based on the GHs and CEs present (Table 2): these included a gpPUL concerned with utilization of pectin and xylan (Ros-2), arabinoxylan (Ros-6 and Ros-7), arabinan (Ros-8), arabinogalactan (Ros-5), glycogen (Ros-4), and glucomannan/galactomannan (Ros-3). R. intestinalis XB6B4 also had gpPULs predicted to utilize O-linked mucus glycans (Ros-1) and N-linked mucus glycans (Ros-10). The remaining gpPUL in R. intestinalis XB6B4 (Ros-9) was predicted to encode genes for the utilization of arabinogalactan and glucomannan.
Table 2. gpPULs identified in R. intestinalis XB6B4 and E. rectale A1-86 for which the substrate target(s) could be confidently predicted.
The carbohydrates utilized by these gpPULs were predicted by their complement of GHs and CEs. ABC transporters, GPH : cation symporter family transporters and MFS transporters were predicted to mediate carbohydrate transport for some of the gpPULs. Transcriptional regulators were identified similar to those of the l-arabinose operon (AraC), lactose operon (LacI), arsenic resistance operon (AsrR), methyl-accepting chemotaxis sensory transducer (MCST), tetracycline resistance genes (TetR) and N-acetyl-d-galactosamine operon (NagC), histidine kinase (HK), response regulator with AraC-like DNA-binding domain and CheY-like receiver domain (RR[AraC-CheY]) and response regulator with LytTR-like DNA-binding domain (RR[LytTR]). The gpPULs of R. intestinalis XB6B4 and E. rectale A1-86 possess the prefixes ‘Ros-’ and ‘Eub-’, respectively.
| PUL | Predicted substrate | GHs and CEs | Transport system | Transcriptional regulation |
| Ros-1 | O-linked mucus glycans | GH29, GH42 | ABC | HK |
| Ros-2 | Pectin and xylan | GH28, 2 GH43, CE12 | ABC | AraC |
| Ros-3 | Gluco- and galactomannan | GH1, GH36, GH76, GH113, 2 GH130, CE2, CE3 | ABC | AraC, LacI |
| Ros-4 | Glycogen | GH13, GH77, GH78/15, | ABC | AsrR, LacI |
| Ros-5 | Xylan and arabinogalactan | GH2, GH3, GH8, GH42, GH43, GH53, GH115, | 2 × ABC | HK, RR[AraC–CheY], LacI, AraC |
| Ros-6 | Arabinoxylan | 2 GH39, GH43/51, GH43, 2 GH51, GH120, 2 CE1 | ABC | LacI, AraC |
| Ros-7 | Arabinoxylan | GH25, 2 GH43, GH51 | ABC and GPH | AraC, MCST, RR[LytTR], HK |
| Ros-8 | Arabinan | GH51, GH127 | ABC | TetR |
| Ros-9 | Arabinogalactan and glucomannan | GH2, GH5, GH53, GH130, CE1F | 2 × ABC | LacI |
| Ros-10 | N-linked mucus glycans | GH3, GH38, GH85, GH125, GH130, GH20, CE1, | ABC and MFS | 2 LacI, 2 NagC |
| Eub-1 | Starch | 2 GH31 | ABC | LacI |
| Eub-2 | Arabinogalactan | GH2, GH53 | GPH | AraC |
| Eub-3 | Fructans | GH32 | ABC | LacI |
| Eub-4 | Fructans | GH32 | ABC | LacI |
E. rectale A1-86 was also selected for detailed analysis because E. rectale is the most abundant species of the Roseburia/E. rectale group present in the colonic microbiota (Walker et al., 2011; Louis et al., 2010). Again, SusC and SusD homologues were not identified in the genome of E. rectale A1-86, but alternative carbohydrate transport systems were adjacent to GH genes in gpPULs. This strain was predicted to possess 15 gpPULs. Of the 18 carbohydrate transporters identified in E. rectale A1-86, 10 (56 %) were ABC transporters, six (33 %) were GPH : cation symporter family transporters and two (11 %) were PTS transporters. The likely carbohydrate targets of the four of these gpPULs that could be predicted with reasonable confidence were starch (Eub-1) and fructan (Eub-3 and Eub-4) (Table 2). E. rectale A1-86 also possessed a gpPUL predicted to utilize arabinogalactan (Eub-2). This gpPUL was orthologous to Ros-5, possessed by R. intestinalis XB6B4.
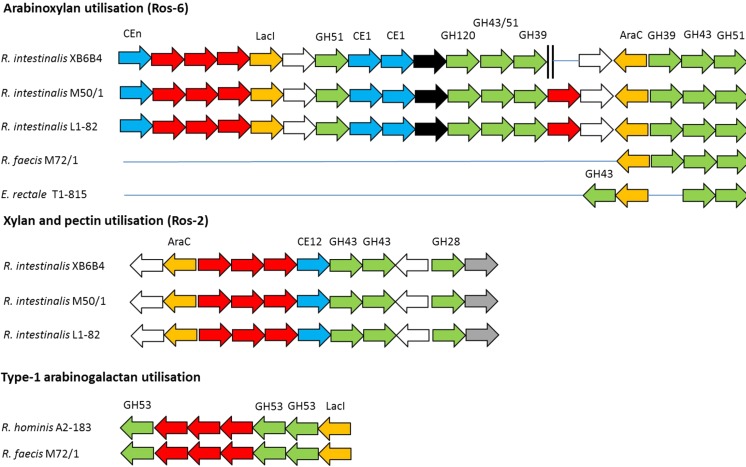
Specific gpPULs of interest were selected for comparison across strains. The predicted xylan utilization gpPUL Ros-6 showed well-conserved gene order for the three R. intestinalis strains, but R. faecis M72/1 and E. rectale T1-815 contained only a few of the genes from this gpPUL (Fig. 4a), and this gpPUL was completely absent in the other E. rectale strains, R. inulinivorans and R. hominis.
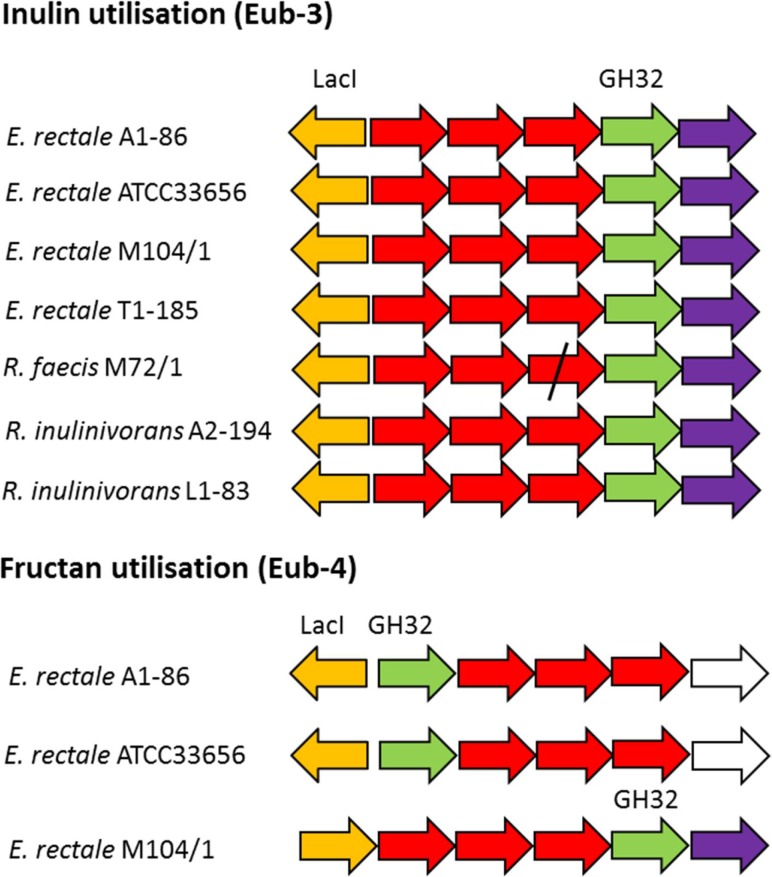
Fig. 4.
Schematic representation of gpPULs concerned with xylan and arabinogalactan utilization. Glycoside hydrolase genes are coloured green. Carbohydrate esterase genes are coloured blue. ABC-transporter system component genes are coloured red. Transcriptional regulator genes are coloured yellow. Uncharacterized transporter genes are coloured black. Hypothetical genes are coloured white. Xylose isomerase genes are coloured grey. Two parallel black bars between genes indicate sections that are separated in the genome sequence. Roseburia/E. rectale strains not represented in the diagram lack an orthologous gpPUL. Genes located vertically to each other are orthologues. Solid blue lines between genes are for easy visual comparison of the genes between species and do not represent real gaps in the genome. Locus tags of gpPULs are listed in Table S7.
Ros-2 was unique to R. intestinalis and its gene order was perfectly conserved between the three R. intestinalis strains. This gpPUL possessed two GH43 genes predicted to encode xylan-1,4-β-xylosidases (EC 3.2.1.37), a xylose isomerase gene, a CE12 gene (xylan/pectin esterases) and a GH28 gene predicted to encode a polygalacturonase (EC 3.2.1.15). Ros-2 also encoded an AraC-like transcriptional regulator and an ABC transporter system (Fig. 4a). R. hominis A2-183 and R. faecis M72/1 both possessed a predicted arabinogalactan gpPUL that was absent in the other nine strains of the Roseburia/E. rectale pan-genome (Fig. 4b, Table S10). This gpPUL consisted of three GH53 genes predicted to encode arabinogalactan endo-1,4-β-galactosidases (EC 3.2.1.89), two of which possessed a CBM61 (1,4-β-galactan binding; Cid et al., 2010) and a SP.
The predicted inulin utilization gpPUL Eub-3 was present in all E. rectale and R. inulinivorans strains, and in R. faecis M72/1, whilst E. rectale strains A1-86, ATCC33656 and M104/1 also possessed a second fructan gpPUL Eub-4 (Fig. 5, Table S10). Of the 10 strains tested for growth on inulin, all strains possessing Eub-3 were capable of utilizing inulin for growth, with the exception of R. faecis M72/1 (Fig. 1a). The R. faecis M72/1 Eub-3 contained a substitution SNP (C replaced with T) at nucleotide 381 of an ABC transporter permease, predicted to result in a truncated protein and likely explaining the inability of R. faecis M72/1 to grow on inulin. This mutation, first observed in the genome sequence, was subsequently confirmed by targeted Sanger sequencing. None of the R. intestinalis of R. hominis strains, which lack Eub-3, were capable of utilizing inulin.
Fig. 5.
Schematic representation of gpPULs concerned with fructan utilization. GH genes are coloured green. ABC transporter system component genes are coloured red. Transcriptional regulator genes are coloured yellow. Fructokinase genes are coloured purple. Hypothetical genes are coloured white. Any of the 11 Roseburia/E. rectale strains not represented in the diagram lack an orthologous gpPUL. Genes located vertically to each other are orthologues. The diagonal line through the R. faecis gene represents a frameshift mutation. Locus tags of gpPULs are listed in Table S7.
Eub-4 possesses a GH32 gene predicted to encode a β-fructofuranosidase (EC 3.2.1.26). Although the GH32 genes in this gpPUL were predicted to be orthologues, the E. rectale M104/1 gene lacked the CBM66 (binding of terminal fructose moiety of levantriose; Cuskin et al., 2012) present in the GH32 genes of E. rectale A1-86 and ATCC33656.
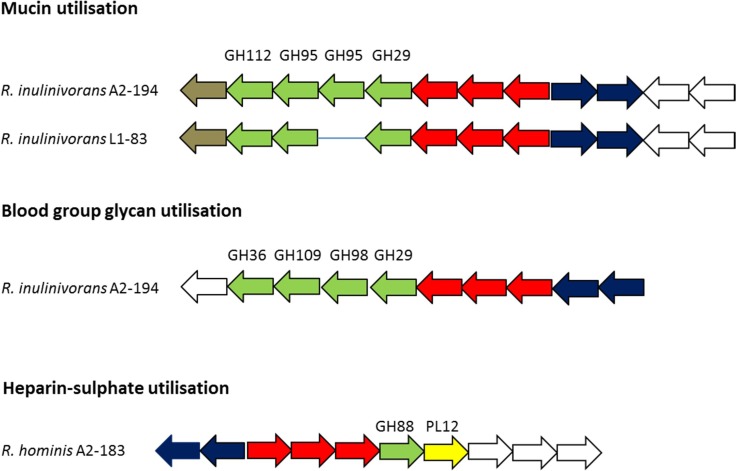
The two R. inulinivorans strains possessed a predicted mucin gpPUL that is absent in the other nine strains (Fig. 6, Table S10), which encoded a mucin desulphatase, four mucin-degrading GHs and an ABC transporter system. R. inulinivorans A2-194 also possessed a predicted blood group glycan gpPUL that was absent in the other strains (Fig. 6). This gpPUL contained four GH genes predicted to encode enzymes for the degradation of blood group glycans, including a SP possessing blood-group endo-1,4-β-galactosidase (EC 3.2.1.102) harbouring two CBM51 domains – a CBM family shown to bind blood group A/B antigens in Clostridium perfringens (Gregg et al., 2008). This gpPUL was also predicted to encode a GH109 enzyme, but particular caution should be taken when annotating members of this GH family in silico, as the sequences of true GH109 enzymes are highly similar to oxidoreductases that do not degrade carbohydrates. An additional gpPUL, dedicated to the utilization of fucose as a growth substrate, was previously identified and shown to be inducible in R. inulinivorans A2-194 during growth on fucose (Scott et al., 2006).
Fig. 6.
Schematic representation of gpPULs concerned with host-derived carbohydrates. GH genes are coloured green. ABC transporter system component genes are coloured red. The polysaccharide lyase gene is coloured bright yellow. Mucin desulphatase genes are coloured gold. Hypothetical genes are coloured white. Two-component signal transduction component genes consisting of a histidine kinase and a response regulator containing a CheY-like receiver domain and an AraC-like DNA-binding domain are coloured navy. Solid blue lines between genes are for easy visual comparison of the genes between species and do not represent real gaps in the genome. Any of the 11 Roseburia/E. rectale strains not represented in the diagram lack an orthologous gpPUL. Locus tags of gpPULs are listed in Table S7.
The only polysaccharide lyase found in the Roseburia/E. rectale pan-genome was encoded by R. hominis A2-183. This gene was part of a gpPUL predicted to utilize heparin sulphate (components of extracellular matrix and cell surface proteoglycans) (Fig. 6).
Discussion
Whilst carbohydrate utilization in the Gram-negative Bacteroidetes phylum has been investigated extensively and is well understood (D'Elia & Salyers, 1996; Reeves et al., 1997; Shipman et al., 2000; Martens et al., 2008, 2011; McNulty et al., 2013; Larsbrink et al., 2014), the present work represents the first detailed analysis of carbohydrate utilization genes and their organization within a dominant group of human colonic Firmicutes. The 11 strains of Roseburia and E. rectale (the ‘Roseburia/E. rectale group’) examined here encoded a mean number of 85 GHs per genome. This is much higher than the mean number of GHs reported per genome for Firmicutes (40 GHs), but much lower than the mean number of GHs per genome for Bacteroidetes (130 GHs) in a ‘mini-microbiome’ of human colonic bacteria (El Kaoutari et al., 2013). The possession of relatively large numbers of GH genes is in general agreement with findings from human dietary studies that illustrate the dependence of Roseburia and E. rectale populations upon dietary sources of carbohydrate (Duncan et al., 2007; Martínez et al., 2010; Walker et al., 2011; Salonen et al., 2014).
A fundamental feature of carbohydrate utilization genes in Bacteroides spp. is their clustering into genomic regions, termed PULs. Polysaccharide utilization in Bacteroidetes involves limited extracellular cleavage of polysaccharides, followed by the binding and translocation into the periplasm of the released oligosaccharides via outer membrane Sus protein homologues. In addition to GH genes, these PULs encode the Sus proteins and also transcriptional regulation systems (most frequently hybrid two-component regulators) that respond to the presence of specific carbohydrates (Martens et al., 2011).
We report here that PULs are an equally important feature of genome organization in the Roseburia/E. rectale group of Firmicutes. The genome of R. intestinalis XB6B4 was found to contain 33 gpPULs, which contained 106 of its 131 GH genes. As in Bacteroides, these gpPULs appear to be substrate-specific, and include linked transport systems and regulatory genes. ABC transport systems predominate, accounting for 79 % of transporters within gpPULs in R. intestinalis XB6B4 and 56 % in E. rectale A1-86, with cation symporters and PTS systems found in smaller numbers. No evidence was found for close homologues of the Bacteroidetes Sus proteins; binding of polysaccharides in the Roseburia group therefore seems likely to involve the CBMs present in many GH enzymes, whilst ABC transport components are assumed to mediate binding of oligosaccharides prior to transport in the majority of cases. gpPUL-encoded GHs in E. rectale and R. inulinivorans are known to be highly inducible (Scott et al., 2011; Cockburn et al., 2015), as is seen in Bacteroides (Martens et al., 2008, 2011; McNulty et al., 2013). The adjacent transcriptional regulators of the Roseburia/E. rectale group tend to be LacI- and AraC-type proteins with only a few examples of the hybrid two-component system transcriptional regulators. Hybrid two-component system transcriptional regulators and extracytoplasmic function sigma factors are, however, the most frequently observed regulators in Bacteroides PULs (Sonnenburg et al., 2006). The differences revealed here in membrane organization, SPs, transport and regulatory systems all suggest that the detailed organization and regulation of degradative enzymes differs in this group of Gram-positive bacteria from that in Bacteroides spp. It is also apparent that these features may differ substantially in a second family of Firmicutes that is highly abundant in the human colon, i.e. the Ruminococcaceae (Wegmann et al., 2014; Ben David et al., 2015; Ze et al., 2015).
Another important conclusion of the present study is that different species of the Roseburia/E. rectale group show considerable specialization in their abilities to utilize different carbohydrate substrates. Based initially on the CAZyme content of their genomes, these strains could be assigned to CUEs that consisted, in three out of four cases, entirely of members of a single species. The remaining CUE (CUE1) consists of the single available genome sequences for R. hominis and R. faecis. Our data suggest that most members of the Roseburia/E. rectale group share a core capacity to utilize starch and fructo-oligosaccharides, with only R. hominis A2-183 less capable of utilizing both. In addition, however, R. intestinalis is predicted to specialize in the degradation of plant cell wall matrix polysaccharides (e.g. arabinoxylan), R. inulinivorans in degrading host-derived carbohydrates, and R. hominis and R. faecis in type 1 arabinogalactan degradation. The correspondence between the genome-predicted ecotype and the observed growth of strains on different substrates was not always straightforward and requires some comment. The enrichment of genes associated with xylan breakdown in R. intestinalis strains corresponded well with their ability to grow on arabinoxylan and xylo-oligosaccharides. However, two E. rectale strains lacking many of these gpPULs were also able to grow on arabinoxylan and xylo-oligosaccharides. This might perhaps be explained by an as yet undiscovered xylanase or the utilization of different breakdown products, e.g. removal of arabinose substituents as opposed to cleavage of the main xylan chain. In addition, production of a GH74 enzyme by R. intestinalis strains and enzymic activity against xyloglucan did not correlate with growth on this substrate, presumably because hydrolysis products were not utilized. Furthermore, possession of hydrolases concerned with particular host glycans did not lead to growth on mucin in any of the strains, presumably because this requires a wider repertoire of enzymic specificities. Particularly in the case of mucin and plant structural polysaccharides, it should be recognized that the complexity and variability of the substrates make simple predictions from genomic data tentative. Nevertheless, in vivo evidence from human studies confirms that these species show variation with respect to dietary carbohydrate supplementation and individual microbiota composition (Louis et al., 2010; Martínez et al., 2010; Walker et al., 2011; Salonen et al., 2014). Work by Louis et al. (2010) based on amplification of the butyryl-CoA : acetate CoA transferase gene revealed striking inter-individual variation within the Roseburia/E. rectale group, with E. rectale dominant in six individuals, R. faecis in two individuals and R. inulinivorans in one individual. The nutritional specialization revealed by the present work, assuming variations in dietary intakes, provides a plausible explanation for such differences.
The percentage of Roseburia/E. rectale GHs possessing SPs was unusually low at only 7.9 %. This is in marked contrast with some other human colonic bacteria, such as Bacteroidetes, that are predicted to secrete ∼85 % of their GHs (El Kaoutari et al., 2013). El Kaoutari et al. (2013) also reported that only 19 % of Firmicutes GHs in their ‘mini-microbiome’ possessed SPs, although SPs are found in a high proportion of GHs in Ruminococcus spp. from the rumen and human colon (Rincon et al., 2010; Wegmann et al., 2014). The low percentage of SPs among GH enzymes might therefore be a feature mainly of the Lachnospiraceae – the most abundant family of Firmicutes in the human colon. It remains to be established whether GHs in the Roseburia/E. rectale group of Lachnospiraceae that lack SPs are mostly intracellular, or whether (as seems more likely) many possess alternative signal sequences enabling secretion or positioning within the cell membrane. Of the two amylases found to be upregulated by growth on starch in E. rectale, one possessed a SP and the other a hydrophobic region suggesting a possible membrane location (Cockburn et al., 2015). Previous analysis of amylopullulanases in R. inulinivorans identified an inducible multidomain enzyme involved in starch degradation that had a SP and a hydrophobic region, as well as both catalytic and carbohydrate-binding domains (Ramsay et al., 2006; Scott et al., 2011). Our work revealed the SP-possessing amylases of R. inulinivorans and E. rectale to be orthologues of each other, with R. faecis M72/1 and all strains of both R. inulinivorans and E. rectale possessing a copy of this gene.
In conclusion, understanding the impact of diet on the human gut microbiota and gut metabolism requires a far better understanding of these important but little-studied groups of Firmicutes bacteria that appear to make a highly significant contribution to the fermentation of polysaccharides. This work has shown that this can come initially from comparative genome analysis that can subsequently be used to guide functional studies (Flint et al., 2008).
Acknowledgements
The Rowett Institute of Nutrition and Health (University of Aberdeen) receives financial support from the Scottish Government Rural and Environmental Sciences and Analytical Services (RESAS). POS is a PhD student supported by the Scottish Government (RESAS) and the Science Foundation Ireland, through a centre award to the APC Microbiome Institute, Cork, Ireland.
Supplementary Data
Supplementary Data
Abbreviations:
- ABC
ATP-binding cassette
- CAZyme
carbohydrate-active enzyme
- CE
carbohydrate esterase
- CUE
carbohydrate utilization ecotype
- CBM
carbohydrate-binding module
- GH
glycoside hydrolase
- GPH
glycoside–pentoside–hexuronide
- gpPUL
Gram-positive polysaccharide utilization loci
- HMM
hidden Markov model
- MFS
major facilitator superfamily
- OG
orthologous group
- PTS
phosphotransferase system
- PUL
polysaccharide utilization loci
- SP
signal peptide
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). blast Ring Image Generator (brig): simple prokaryote genome comparisons BMC Genomics 12402. 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R. I., Walker A. W., Duncan S. H., Harmsen H. J., Welling G. W., Flint H. J. (2006). Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale Appl Environ Microbiol 726371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R., Rajendiran E., George S., Samuel G. V., Ramakrishna B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer J Gastroenterol Hepatol 231298–1303. [DOI] [PubMed] [Google Scholar]
- Barcenilla A., Pryde S. E., Martin J. C., Duncan S. H., Stewart C. S., Henderson C., Flint H. J. (2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut Appl Environ Microbiol 661654–1661 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben David Y., Dassa B., Borovok I., Lamed R., Koropatkin N. M., Martens E. C., White B. A., Bernalier-Donadille A., Duncan S. H., other authors (2015). Ruminococcal cellulosome systems from rumen to human Environ Microbiol 173407–3426 10.1111/1462-2920.12868. [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Kiemer L., Fausbøll A., Brunak S. (2005). Non-classical protein secretion in bacteria BMC Microbiol 558. 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Pirovano W. (2012). Toward almost closed genomes with GapFiller Genome Biol 13R56. 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston A. B., Healey M., Klassen J., Ficko-Blean E., Lammerts van Bueren A., Law V. (2006). A structural and functional analysis of alpha-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition J Biol Chem 281587–598 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- Bryant M. P. (1972). Commentary on the Hungate technique for culture of anaerobic bacteria Am J Clin Nutr 251324–1328,. [DOI] [PubMed] [Google Scholar]
- Chassard C., Goumy V., Leclerc M., Del'homme C., Bernalier-Donadille A. (2007). Characterization of the xylan-degrading microbial community from human faeces FEMS Microbiol Ecol 61121–131 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- Cid M., Pedersen H. L., Kaneko S., Coutinho P. M., Henrissat B., Willats W. G., Boraston A. B. (2010). Recognition of the helical structure of beta-1,4-galactan by a new family of carbohydrate-binding modules J Biol Chem 28535999–36009 10.1074/jbc.M110.166330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn D. W., Orlovsky N. I., Foley M. H., Kwiatkowski K. J., Bahr C. M., Maynard M., Demeler B., Koropatkin N. M. (2015). Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale Mol Microbiol 95209–230 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F., Flint J. E., Gloster T. M., Morland C., Baslé A., Henrissat B., Coutinho P. M., Strazzulli A., Solovyova A. S., other authors (2012). How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity Proc Natl Acad Sci U S A 10920889–20894 10.1073/pnas.1212034109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F., Lowe E. C., Temple M. J., Zhu Y., Cameron E. A., Pudlo N. A., Porter N. T., Urs K., Thompson A. J., other authors (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism Nature 517165–169 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia J. N., Salyers A. A. (1996). Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch J Bacteriol 1787173–7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., other authors (2014). Diet rapidly and reproducibly alters the human gut microbiome Nature 505559–563 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Aminov R. I., Scott K. P., Louis P., Stanton T. B., Flint H. J. (2006). Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces Int J Syst Evol Microbiol 562437–2441 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- Duncan S. H., Belenguer A., Holtrop G., Johnstone A. M., Flint H. J., Lobley G. E. (2007). Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces Appl Environ Microbiol 731073–1078 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005). Diversity of the human intestinal microbial flora Science 3081635–1638 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A., Armougom F., Gordon J. I., Raoult D., Henrissat B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota Nat Rev Microbiol 11497–504 10.1038/nrmicro3050.. [DOI] [PubMed] [Google Scholar]
- Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis Nat Rev Microbiol 6121–131 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E. (2012a). Microbial degradation of complex carbohydrates in the gut Gut Microbes 3289–306 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., Scott K. P., Louis P., Duncan S. H. (2012b). The role of the gut microbiota in nutrition and health Nat Rev Gastroenterol Hepatol 9577–589 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- Gregg K. J., Finn R., Abbott D. W., Boraston A. B. (2008). Divergent modes of glycan recognition by a new family of carbohydrate-binding modules J Biol Chem 28312604–12613 10.1074/jbc.M709865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold G. L., Schwiertz A., Aminov R. I., Blaut M., Flint H. J. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces Appl Environ Microbiol 694320–4324 10.1128/AEM.69.7.4320-4324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification BMC Bioinformatics 11119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes Nucleic Acids Res 2827–30 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). mega: a biologist-centric software for evolutionary analysis of DNA and protein sequences Brief Bioinform 9299–306 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rødland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes Nucleic Acids Res 353100–3108 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerts van Bueren A., Finn R., Ausió J. (2004). α-Glucan recognition by a new family of carbohydrate-binding modules found primarily in bacterial pathogens Biochemistry 4315633–15642 10.1021/bi048215z. [DOI] [PubMed] [Google Scholar]
- Larsbrink J., Rogers T. E., Hemsworth G. R., McKee L. S., Tauzin A. S., Spadiut O., Klinter S., Pudlo N. A., Urs K., other authors (2014). A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes Nature 506498–502 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M., Khan T. M., Duncan S. H., Harmsen H. J., Garcia-Gil L. J., Flint H. J. (2012). Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth Appl Environ Microbiol 78420–428 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Flint H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine FEMS Microbiol Lett 2941–8 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- Louis P., Duncan S. H., McCrae S. I., Millar J., Jackson M. S., Flint H. J. (2004). Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon J Bacteriol 1862099–2106 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Young P., Holtrop G., Flint H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene Environ Microbiol 12304–314 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G. L., Flint H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer Nat Rev Microbiol 12661–672 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., Ballet V., Claes K., Van Immerseel F., other authors (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis Gut 631275–1283 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- Martens E. C., Chiang H. C., Gordon J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont Cell Host Microbe 4447–457 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens E. C., Lowe E. C., Chiang H., Pudlo N. A., Wu M., McNulty N. P., Abbott D. W., Henrissat B., Gilbert H. J., other authors (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts PLoS Biol 9e1001221. 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Kim J., Duffy P. R., Schlegel V. L., Walter J. (2010). Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects PLoS One 5e15046. 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Lattimer J. M., Hubach K. L., Case J. A., Yang J., Weber C. G., Louk J. A., Rose D. J., Kyureghian G., other authors (2013). Gut microbiome composition is linked to whole grain-induced immunological improvements ISME J 7269–280 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty N. P., Wu M., Erickson A. R., Pan C., Erickson B. K., Martens E. C., Pudlo N. A., Muegge B. D., Henrissat B., other authors (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome PLoS Biol 11e1001637. 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Martin J. C., Marinsek-Logar R., Flint H. J. (1997). Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14 Anaerobe 3373–381 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- Neville B. A., Sheridan P. O., Harris H. M., Coughlan S., Flint H. J., Duncan S. H., Jeffery I. B., Claesson M. J., Ross R. P., other authors (2013). Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans PLoS One 8e68919. 10.1371/journal.pone.0068919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T. D., Sanders M., Berriman M., Newbold C. (2010). Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology Bioinformatics 261704–1707 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions Nat Methods 8785–786 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., other authors (2012). The Pfam protein families database Nucleic Acids Res 40 (D1), D290–D301 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay A. G., Scott K. P., Martin J. C., Rincon M. T., Flint H. J. (2006). Cell-associated alpha-amylases of butyrate-producing Firmicute bacteria from the human colon Microbiology 1523281–3290 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- Reeves A. R., Wang G. R., Salyers A. A. (1997). Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron J Bacteriol 179643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M. T., Dassa B., Flint H. J., Travis A. J., Jindou S., Borovok I., Lamed R., Bayer E. A., Henrissat B., other authors (2010). Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium Ruminococcus flavefaciens FD-1.PLoS One 5e12476. 10.1371/journal.pone.0012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., other authors (2003). tm4: a free, open-source system for microarray data management and analysis Biotechniques 34374–378. [DOI] [PubMed] [Google Scholar]
- Salonen A., Lahti L., Salojärvi J., Holtrop G., Korpela K., Duncan S. H., Date P., Farquharson F., Johnstone A. M., other authors (2014). Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men ISME J 82218–2230 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P., Martin J. C., Campbell G., Mayer C. D., Flint H. J. (2006). Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium Roseburia inulinivorans J Bacteriol 1884340–4349 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P., Martin J. C., Chassard C., Clerget M., Potrykus J., Campbell G., Mayer C. D., Young P., Rucklidge G., other authors (2011). Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch Proc Natl Acad Sci U S A 108 (Suppl 14672–4679 10.1073/pnas.1000091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P., Martin J. C., Duncan S. H., Flint H. J. (2014). Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro FEMS Microbiol Ecol 8730–40 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- Sekirov I., Russell S. L., Antunes L. C., Finlay B. B. (2010). Gut microbiota in health and disease Physiol Rev 90859–904 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shipman J. A., Berleman J. E., Salyers A. A. (2000). Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron J Bacteriol 1825365–5372 10.1128/JB.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J., Cerutti L., de Castro E., Langendijk-Genevaux P. S., Bulliard V., Bairoch A., Hulo N. (2010). PROSITE, a protein domain database for functional characterization and annotation Nucleic Acids Res 38D161–D166 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg E. D., Sonnenburg J. L., Manchester J. K., Hansen E. E., Chiang H. C., Gordon J. I. (2006). A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism Proc Natl Acad Sci U S A 1038834–8839 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M. R., Danchin E. G., Rancurel C., Coutinho P. M., Henrissat B. (2006). Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins Protein Eng Des Sel 19555–562 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J. P., Ugarte E., Muñoz-Tamayo R., Paslier D. L., other authors (2009). Towards the human intestinal microbiota phylogenetic core Environ Microbiol 112574–2584 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Travis A. J., Kelly D., Flint H. J., Aminov R. I. (2015). Complete genome sequence of the human gut symbiont Roseburia hominis Genome Announc 3e01286–e01215 10.1128/genomeA.01286-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. W., Ince J., Duncan S. H., Webster L. M., Holtrop G., Ze X., Brown D., Stares M. D., Scott P., other authors (2011). Dominant and diet-responsive groups of bacteria within the human colonic microbiota ISME J 5220–230 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., Jia W., Cai S., Zhao L. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers ISME J 6320–329 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U., Louis P., Goesmann A., Henrissat B., Duncan S. H., Flint H. J. (2014). Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans Environ Microbiol 162879–2890. [DOI] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation Nucleic Acids Res 40 (W1), W445–W451 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Zavaljevski N., Desai V., Reifman J. (2011). QuartetS: a fast and accurate algorithm for large-scale orthology detection Nucleic Acids Res 39e88. 10.1093/nar/gkr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X., Duncan S. H., Louis P., Flint H. J. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon ISME J 61535–1543 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X., Ben David Y., Laverde-Gomez J. A., Dassa B., Sheridan P. O., Duncan S. H., Louis P., Henrissat B., Juge N., other authors (2015). Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii MBio 6e01058–e01015 10.1128/mBio.01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs Genome Res 18821–829 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Data References
- 1. Eubacterium rectale T1-815 genome (2015); CVRQ01000001–CVRQ01000090: http://www.ebi.ac.uk/ena/data/view/PRJEB9320.
- 2. Roseburia faecis M72/1 genome (2015); CVRR01000001–CVRR01000101: http://www.ebi.ac.uk/ena/data/view/PRJEB9321.
- 3. Roseburia inulinivorans L1-83 genome (2015); CVRS01000001–CVRS01000151: http://www.ebi.ac.uk/ena/data/view/PRJEB9322.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data