Abstract
There are many types of repeated DNA sequences in the genomes of the species of the genus Neisseria, from homopolymeric tracts to tandem repeats of hundreds of bases. Some of these have roles in the phase-variable expression of genes. When a repeat mediates phase variation, reversible switching between tract lengths occurs, which in the species of the genus Neisseria most often causes the gene to switch between on and off states through frame shifting of the open reading frame. Changes in repeat tract lengths may also influence the strength of transcription from a promoter. For phenotypes that can be readily observed, such as expression of the surface-expressed Opa proteins or pili, verification that repeats are mediating phase variation is relatively straightforward. For other genes, particularly those where the function has not been identified, gathering evidence of repeat tract changes can be more difficult. Here we present analysis of the repetitive sequences that could mediate phase variation in the Neisseria gonorrhoeae strain NCCP11945 genome sequence and compare these results with other gonococcal genome sequences. Evidence is presented for an updated phase-variable gene repertoire in this species, including a class of phase variation that causes amino acid changes at the C-terminus of the protein, not previously described in N. gonorrhoeae.
Keywords: gonococcus, phase variation, C-terminal variation, homopolymeric tract, simple sequence repeats
Data Summary
Sequence data for Neisseria gonorrhoeae strains investigated are available in GenBank under the following accession numbers: FA1090 (NC_002946.2; url - http://www.ncbi.nlm.nih.gov/nuccore/NC_002946.2); NCCP11945 (NC_011035.1; url – http://www.ncbi.nlm.nih.gov/nuccore/NC_011035.1; & CP001050.1; url - http://www.ncbi.nlm.nih.gov/nuccore/CP001050.1); MS11 (NC_022240.1; url -http://www.ncbi.nlm.nih.gov/nuccore/NC_022240.1); FA19 (NZ_CP012026.1; url – http://www.ncbi.nlm.nih.gov/nuccore/NZ_CP012026.1); FA6140 (NZ_CP012027.1; url – http://www.ncbi.nlm.nih.gov/nuccore/NZ_CP012027.1); 35/02 (NZ_CP012028.1; url – http://www.ncbi.nlm.nih.gov/nuccore/NZ_CP012028.1). These were accessed on the 15th of April 2016 for use in this study.
Sequence data were assessed via BLAST interrogation of the nr database restricted to the N. gonorrhoeae species (url – http://www.ncbi.nlm.nih.gov/).
Genome resequencing data for N. gonorrhoeae strain NCCP11945 has been deposited under BioProject PRJNA322254 (url – http://www.ncbi.nlm.nih.gov/bioproject/322254).
Genome resequencing data was assessed via Galaxy (url – usegalaxy.org).
Impact Statement
Phase variation plays a vital role in the ability of Neisseria gonorrhoeae to adapt to the various niche environments encountered. Through stochastic switching in the expression of key genes and regulatory systems, mediated by simple sequence repeats, the population of bacteria are diverse and readily able to survive in the face of selective pressures. Not all simple sequence repeats within the genome mediate phase variation. Previous investigations have sought to define the phase-variable repertoire of the species of the genus Neisseria and have identified a large number of candidates using a small number of genome sequences. With the availability of more genome sequence data and additional experimental data, we have refined the original repertoire to include those most likely to be phase-variable in N. gonorrhoeae. As these genes are important for survival, their definition as phase-variable is important for understanding pathogenesis and for potential future therapies. The advent of high-throughput sequencing has the potential to reveal additional cases of within-strain variations in repeat tracts, supporting phase-variable candidacy of genes.
Introduction
In Neisseria gonorrhoeae, the causative agent of gonorrhoea, DNA repeats are intimately linked to the biology of the organism. N. gonorrhoeae, and the closely related bacterial species Neisseria meningitidis, undergo phase-variable stochastic switching of gene expression for several surface structures, contributing to antigenic variation and immune evasion as well as niche adaptation in the course of infection (Bhat et al., 1991; Moxon et al., 2006; Carbonnelle et al., 2009; Srikhanta et al., 2009; Omer et al., 2011). Phase variation is mediated by simple sequence repeats associated with genes. In the species of the genus Neisseria the vast majority contain homopolymeric tracts within the coding sequences (Snyder et al., 2001).
Comparative sequence analysis between a single N. gonorrhoeae and several N. meningitidis genome sequences identified over 100 potentially phase-variable genes (Snyder et al., 2001), some of which have later been demonstrated to be phase-variable experimentally (Jordan et al., 2005). Transcriptional and translational phase variation have been extensively studied in the species of the genus Neisseria, however an additional class of simple sequence repeat-mediated phase variation has been described in Helicobacter canadensis following whole-genome analysis (Snyder et al., 2010). Simple sequence repeat-mediated changes in the presence or absence of C-terminal cell wall attachment motifs has also been described in Streptococcus agalactiae (Janulczyk et al., 2010). In N. meningitidis, a gene fusion between pglB2 and the downstream phosphoglycosyltransferase gene appears to be mediated by a poly-A repeat tract (Viburiene et al., 2013).
With the availability of additional gonococcal genome sequences, the gonococcal phase-variable repertoire has here been re-assessed. As a result, phase variation in which repeats at the 3′ ends of genes mediate changes in the C-terminal sequence of the proteins is described as part of a refined phase-variable gene repertoire.
Methods
Identification of phase-variable genes.
Using the previous phase-variable gene repertoires reported for N. gonorrhoeae and N. meningitidis (Snyder et al., 2001; Martin et al., 2003; Jordan et al., 2005), the homologues in N. gonorrhoeae strain NCCP11945 were sought (CP00150.1; Chung et al., 2008). In addition, pattern search in xBASE (Chaudhuri et al., 2008) was used to identify other repeats, based on previous evidence of phase variation in the species of the genus Neisseria: ≥ (G)8; ≥ (C)8; ≥ (CAAACAC)3; ≥ (CAAATAC)3; ≥ (CCCAA)3; ≥ (GCCA)3; ≥ (A)9; ≥ (T)9; ≥ (AAGC)3; ≥ (TTCC)3; and ≥ (CTTCT)3. No other repeats have been demonstrated to cause phase variation in this species. Genome sequences for N. gonorrhoeae strains NCCP11945 (NC_011035.1), FA1090 (NC_002946.2), FA19 (NZ_CP012026.1), FA6140 (NZ_CP012027.1), 35/02 (NZ_CP012028.1), and MS11 (NC_022240.1) were downloaded on 15ththApril 2016 and compared using progressive Mauve v2.3.1 (Darling et al., 2004) to identify orthologues (Table S1, available in the online Supplementary Material).
Identification of repeat variation within N. gonorrhoeae strain NCCP11945.
N. gonorrhoeae strain NCCP11945 was grown on GC agar (Oxoid) with Kellogg’s (Kellogg et al., 1963) and 5 % Fe(NO3)3 supplements at 37 °C in a candle tin for a period of 8 weeks with passages to fresh agar plates every 2 days or at 37 °C 5 % CO2 for a period of 20 weeks with passages to fresh agar plates every 2–3 days. At each passage, cells were scraped from the plate and resuspended in 1 ml of GC broth to a turbidity equivalent to a 0.5 McFarland standard before inoculation onto fresh plates using a sterile cotton swab. DNA was extracted from such resuspensions using the Puregene Yeast/Bacterial kit (Qiagen). A sample (1 µg or 100 ng) of the DNA was genome sequenced using the Ion Personal Genome Machine, Ion Express Fragment Library kit, Ion Express Template kit, and Ion Sequencing kit (Life Technologies) or using the Illumina-based methods of the MicrobesNG service (microbesng.uk). Sequence read data was interpreted using Galaxy on usegalaxy.org (Afgan et al., 2016). Briefly, the reference sequence (NC_011035.1), Ion Torrent data for eight-week passages (KU1-4, KU1-45), Ion Torrent data for 20-week passages (KU1-95, KU1-96), and Illumina data for 20-week passages (2928-NS1_1 & 2929-NS1_2 and 2929-NS2_1 & 2929-NS2_2) were uploaded to Galaxy. The Ion Torrent bam format files were converted to fastq format using BAMTools Convert (Barnett et al., 2011). FASTQ Groomer was used on all NGS data (Blankenberg et al., 2010). Bowtie2 was used to map the reads against the reference (Langmead et al., 2009; Langmead et al., 2012) before visualisation using the Integrated Genomics Viewer (Robinson et al., 2011; Thorvaldsdóttir et al., 2013).
Results and Discussion
Phase variable genes
The phase-variable gene repertoire of N. gonorrhoeae strain NCCP11945 was investigated and compared against gonococcal strains FA1090, FA19, FA6140, 35/02, and MS11 to assess the presence of similar repeat tracts across the species and variations in repeat tract lengths between strains.
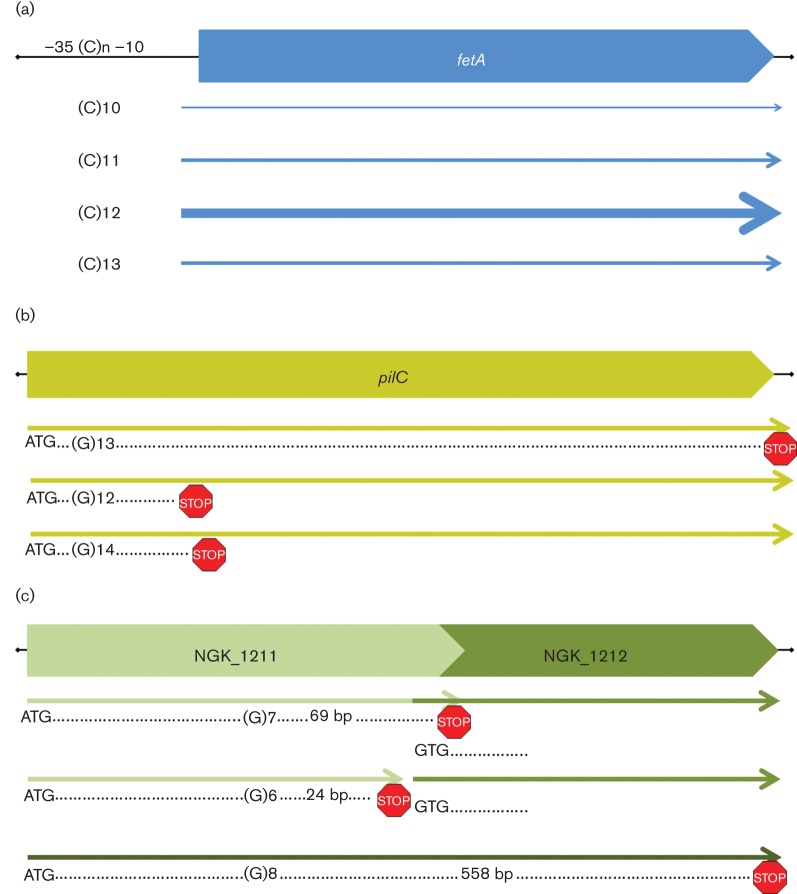
Transcriptional phase variation is mediated by repeats within or associated with the promoter region (Fig. 1a). Changes in the repeat alters the level of transcription of the gene, as in fetA (frpB; NGK_2557) where differences in the length of the poly-C homopolymeric tract between the −10 and −35 promoter regions alters expression (Carson et al., 2000). There are three transcriptional phase-variable genes in N. gonorrhoeae strain NCCP11945 (Table 1), fetA (NGK_2557), a lipoprotein (NGK_2186), and porA (NGK_0906/NGK_0907), yet in gonococci porA does not have an intact coding region. Variation in the repeats between gonococcal strains is found for all three transcriptional phase-variable genes (Table 1).
Fig. 1.
Illustrations of the types of phase variation in N. gonorrhoeae. (a): Transcriptional phase variation, in which changes in a repeat tract alter the facing and spacing of the −10 and −35 promoter elements and the level of transcription of the gene. Phase variation of fetA is used as an example, where it has been shown that differences in spacing of the −10 and −35 elements due to changes in the poly-C repeat tract alter expression levels, represented by the widths of the arrows (Carson et al., 2000). (b): Translational phase variation, in which changes in a repeat tract towards the 5′ end of the coding sequence alter the reading frame of a coding region and switch expression on and off due to frame-shift. Phase variation of pilC is used as an example, where it has been shown that changes in the poly-G repeat tract generate frame-shifts which switch protein expression on and off (Jonsson et al., 1991). (c): C-terminal phase variation, in which changes in a repeat tract towards the 3′ end of the coding sequence alter the reading frame of a coding region and switch the encoded C-terminal amino acids between the three reading frames. In the example NGK_1211, two of the reading frames result in different C-terminal ends to the protein, while the third generates a fusion with the downstream coding sequence, NGK_1212. Only some examples of C-terminal phase variation result in this type of fusion (Table 3).
Table 1. Transcriptional phase variable genes in N. gonorrhoeae.
| Gene | FA1090 locus* | Repeat in FA1090* | NCCP11945 locus† | Repeat in NCCP11945† | Repeat in FA19‡ | Repeat in FA6140§ | Repeat in 35/02|| | Repeat in MS11¶ | N. gonorrhoeae candidacy# | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| porA | NGO_04715 | (T)11C(G)6T | NGK_0906 & NGK_0907** | (T)9C(G)6T | (T)8C(G)8T | (T)9C(G)6T | (T)9C(G)6T | (T)9C(G)6TT | Known | van der Ende et al. (1995) |
| Lipoprotein | NGO2047 | (A)9 | NGK_2186†† | (A)8 | (A)8 | (A)9 | (A)9 | (A)8 | Yes | |
| fetA / frpB | NGO2093 | (C)13 | NGK_2557 | (C)14 | (C)10 | (C)11 | (C)11 | (C)11 | Known | Carson et al. (2000) |
*From the N. gonorrhoeae strain FA1090 genome sequence (NC_002946.2).
†From the N. gonorrhoeae strain NCCP11945 genome sequence (CP00150.1).
‡From the N. gonorrhoeae strain FA19 genome sequence (NZ_CP012026.1).
§From the N. gonorrhoeae strain FA6140 genome sequence (NZ_CP012027.1).
||From the N. gonorrhoeae strain 35/02 genome sequence (NZ_CP012028.1).
¶From the N. gonorrhoeae strain MS11 genome sequence (NC_022240.1).
#Gene phase variation candidacy in N. gonorrhoeae. Known, phase variation has been reported in the literature. Yes, there is evidence of repeat tract variation between strains supporting phase variation.
**This coding sequenc appears to be frame-shifted and annotated as two coding sequences.
††NGK_2186 and NGO2047 annotations are on opposite strands.
Most common in the species of the genus Neisseria is translational phase-variation where, as in pilC, the repeat is within the 5′ portion of the coding region of the gene (Fig. 1b). Changes in the repeat tract generate frame-shift mutations in two of the three open reading frames, with the gene only being translated into protein when the repeat tract length puts the gene in-frame. Whilst many phase-variable genes in the species of the genus Neisseria contain homopolymeric tracts, some experience copy number changes in repetitive sequences, such as the CTTCT repeat in opa (Muralidharan et al., 1987; Bhat et al., 1991) or the AAGC repeat in autA (Peak et al., 1999; Arenas et al., 2015). In the N. gonorrhoeae strains examined here, the AAGC repeat in virG (NGK_0804) is only present in two or three copies (Table 2), rather than several copies as in NGK_0831a and autA (NGK_2082). Although virG has low copy number for the repeat, variations between strains are observed and strains with many copies may yet be identified [there are currently none >(AAGC)3 in the NCBI nr/nt or wgs databases], therefore it is placed amongst the phase-variable genes even though this may be at low frequency or be a strain-specific effect. There are 36 translational phase-variable genes in N. gonorrhoeae based on the species examined (Table 2).
Table 2. Translational phase-variable genes in N. gonorrhoeae.
| Gene | FA1090 locus* | Repeat in FA1090* | NCCP11945 locus† | Repeat in NCCP11945† | Repeat in FA19‡ | Repeat in FA6140§ | Repeat in 35/02|| | Repeat in MS11¶ | N. gonorrhoeae candidacy# | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| pilC2 | NGO0055 | (G)13 | NGK_0074 | (G)9 | (G)10 | (G)13 | (G)9CAGG | (G)12 | Known | Jonsson et al., 1991 |
| opa | NGO0066a | (CTTCT)13CTTCG | NGK_0096 | (CTTCT)8CTTCC | (CTTCT)4CTTCC | CTT(CTTCT)10CTTCC | (CTTCT)6(CT TCC)2 | (CTTCT)8CTT CC | Known | Stern & Meyer, 1987 |
| opa | NGO0070 | (CTTCT)9CTTCG | NGK_0102 | (CTTCT)7CTTCC | (CTTCT)8CTTCC | (CTTCT)17CTTCC | (CTTCT)7CTT CC | (CTTCT)7CTT CC | Known | Stern & Meyer, 1987 |
| pglH | NGO0086 | (C)10 | np | np | no repeat | no repeat | no repeat | np | Known | Power et al., 2003 |
| pglG | NGO0087 | A(C)7 | np | np | A(C)9 | A(C)6 | A(C)6 | np | Known | Power et al., 2003 |
| pglE | NGO0207 | (CAAACAC)4 | NGK_0339 | (CAAACAC)8 (CAAATAC)3 | (CAAACAC)6CAAATACCAAACACCAAATAC | (CAAACAC)10CAAATACCAAACACCAAATACCAAACAC(CAAATAC)2 | (CAAACAC)24 CAAATAC | (CAAACAC)15(CAAATAC)3 | Known | Power et al., 2003 |
| hsdS | NGO_02155 | (G)7 | NGK_0571 | (G)7 | (G)8 | (G)9 | (G)7 | (G)7 | Known | Adamczyk-Poplawska et al., 2011 |
| Hypothetical | NGO0527 | (C)6A(C)9GC | NGK_1405 | (C)6A(C)8GC | (C)7A(C)4T(C )3GC | (C)7A(C)4T(C)3GC | (C)11A(C)8GC GC | (C)6A(C)14GC C | Yes | |
| modB | NGO0545 | (CCCAA)12 | NGK_1384 | (CCCAA)11 | (CCCAA)12 | (CCCAA)4 | (CCCAA)11 | (CCCAA)7 | Known | Srikhanta et al., 2009 |
| Replication initiation factor | NGO_06135 | (C)8TTATCTAACA(G)7 | NGK_1957 | (C)11TTATCTAACA(G)8 | (C)7TTATCTAACA(G)7 | (C)6TTATCT AACA(G)5 | (C)11TTATCT AACA(G)8 | (C)10TTATCT AACA(G)7 | Yes | |
| modA | NGO0641 | (GCCA)37 | NGK_1272 | (GCCA)18 | (GCCA)24GTCA | (GCCA)24 | (GCCA)19 | (GCCA)24 | Known | Srikhanta et al., 2009 |
| Replication initiation factor | NGO_06695 | (C)8TTATCTAACA(G)7 | NGK_1486 | (C)9TTATCTAACA(G)7 | (C)7TTATCTAACA(G)7 | (C)9TTATCT AACA(G)6 | (C)11TTATCT AACA(G)8 | (C)9TTATCTA ACA(G)7 | Yes | |
| opa | NGO0950a | (CTTCT)16CTTCC | NGK_0847 | (CTTCT)19CTTCC | (CTTCT)13CTTCC | (CTTCT)16CTTCC | (CTTCT)8CTT CC | (CTTCT)4CTT CC | Known | Stern & Meyer, 1987 |
| Hypothetical | NGO0964 | (AAGC)4 | NGK_0831a | (AAGC)8 | (AAGC)15 | (AAGC)7 | (AAGC)9 | (AAGC)7 | Yes | |
| virG | NGO0985 | (AAGC)3 | NGK_0804 | (AAGC)3 | (AAGC)2 | (AAGC)3 | (AAGC)3 | (AAGC)3 | Yes | |
| opa | NGO1040a | (CTTCT)20CTTCC | NGK_0749 | (CTTCT)20CTTCC | (CTTCT)10CTTCC | (CTTCT)7CTTCC | (CTTCT)12CT TCC | (CTTCT)13CT TCCCTTCT(C TTCC)2 | Known | Stern & Meyer, 1987 |
| opa | NGO1073a | (CTTCT)2CTTCC | NGK_0693 | (CTTCT)10CTTCC | (CTTCT)11CTTCC | (CTTCT)12CTTCC | (CTTCT)18CT TCC | (CTTCT)7CTT CC | Known | Stern & Meyer, 1987 |
| opa | NGO1277a | (CTTCT)11CTTCC | NGK_1495 | (CTTCT)7CTTCC | CTT(CTTCT)11CTTCC | (CTTCT)11CTTCC | (CTTCT)7CTT CC | (CTTCT)8CTT CC | Known | Stern & Meyer, 1987 |
| Adhesion | NGO1445 | (CAAG)20CAAA | NGK_1705 | (CAAG)12CAAA | (CAAG)12CAAA | (CAAG)6CAAA | (CAAG)9CAAA | (CAAG)6CAAA | Yes | |
| opa | NGO1463a | (CTTCT)10CTTCC | NGK_1729 | (CTTCT)7CTTCC | (CTTCT)11CTTCC | (CTTCT)12CTTCC | (CTTCT)12CT TCC | (CTTCT)10CT TCC | Known | Stern & Meyer, 1987 |
| opa | NGO1513 | (CTTCT)12CTTCG | NGK_1799 | (CTTCT)14CTTCC | CTT(CTTCT)10CTTCC | np | (CTTCT)6CTT CG | (CTTCT)7CTT CG | Known | Stern & Meyer, 1987 |
| opa | NGO1553a | (CTTCT)4CTTCC | NGK_1847 | (CTTCT)9CT TCC | (CTTCT)17CTTCC | (CTTCT)8CTTCC | (CTTCT)8CTT CC | (CTTCT)14CT TCC | Known | Stern & Meyer, 1987 |
| autA | NGO1689 | (AAGC)3 | NGK_2082 | (AAGC)3 | (AAGC)14 | (AAGC)3 | (AAGC)3 | (AAGC)3 | Known | Peak et al., 1999 ; Arenas et al., 2015 |
| pgtA | NGO1765 | (G)11 | NGK_2516 | GGGAGCGGG | (G)19 | (G)19 | GGGAGCGGG | GGGAGCGGG | Known | Banerjee et al., 2002 |
| Repetitive large surface lipoprotein | NGO_09875 & NGO_09870** | (G)8 | NGK_2422 & NGK_2423** | (G)7 | (G)7 | (G)7 | (G)7 | (G)7 | Yes | |
| opa | NGO1861a | (CTTCT)13CTTCC | NGK_2410 | (CTTCT)11CTTCC | (CTTCT)13CTTCC | (CTTCT)2CTTCC | (CTTCT)13CT TCC | (CTTCT)30CT TCC | Known | Stern & Meyer, 1987 |
| pilC1 | NGO1912 | (G)11 | NGK_2342 | (G)13 | (G)11 | GGGC(G)11 | (G)15 | (G)11 | Known | Jonsson et al., 1991 |
| Hypothetical | NGO1953 | (C)8 | NGK_2297 | (C)8 | (C)8 | (C)8 | (C)9 | (C)8 | Yes | |
| Pyrimidine 5′- nucleotidase | NGO2055 & NGO2054** | (C)6 | NGK_2176 | CAAACCCC | CAAACCCC | CAAACCCC | (C)9 | (C)10 | Yes | |
| opa | NGO2060a | (CTTCT)10CTTCG | np | np | np | (CTTCT)6CTT CC | (CTTCT)7CTT CC | Known | Stern & Meyer, 1987 | |
| opa | np | NGK_2170 | (CTTCT)14CTTCC | np | np | np | np | Known | Stern & Meyer, 1987 | |
| lgtG | NGO2072 | (C)11 | NGK_2534 & NGK_2533** | (C)12 | (C)10 | (C)10 | (C)10 | (C)10 | Known | Mackinnon et al., 2002 |
| hpuA | NGO2110 | (G)9 | NGK_2581 | (G)10 | (G)9 | (G)9 | (G)8 | (G)8 | Known | Chen et al., 1998 |
| lgtA | NGO11610 | (G)11 | NGK_2630 | (G)11 | (G)14A | (G)17A | (G)20A | (G)10A | Known | Erwin et al., 1996 |
| lgtC | NGO2156 | (G)14 | NGK_2632 | (G)13 | (G)13 | (G)10 | (G)16 | (G)8 | Known | Shafer et al., 2002 |
| lgtD | NGO2158 | A(G)14 | NGK_2634 | A(G)16 | (G)13 | A(G)12 | A(G)18 | A(G)13 | Known | Shafer et al., 2002 |
*From the N. gonorrhoeae strain FA1090 genome sequence (NC_002946.2).
†From the N. gonorrhoeae strain NCCP11945 genome sequence (CP00150.1).
‡From the N. gonorrhoeae strain FA19 genome sequence (NZ_CP012026.1).
§From the N. gonorrhoeae strain FA6140 genome sequence (NZ_CP012027.1).
||From the N. gonorrhoeae strain 35/02 genome sequence (NZ_CP012028.1).
¶From the N. gonorrhoeae strain MS11 genome sequence (NC_022240.1).
#Gene phase variation candidacy in N. gonorrhoeae. Known , phase variation has been reported in the literature. Yes, there is evidence of repeat tract variation between strains supporting phase variation.
**This coding sequence appears to be frame-shifted and annotated as two coding sequences.
np, The coding sequence is not present in this strain.
In addition, a third class of repeat-mediated phase-variable gene was identified (Snyder et al., 2010). In these C-terminal phase-variable genes, a repeat tract towards the 3′ of the coding region is able to alter the sequence at the C-terminus of the encoded protein (Fig. 1c). In N. gonorrhoeae strain NCCP11945, four of these C-terminal phase-variable genes were identified (Table 3). It is likely that in the case of the pilin sequence (NGK_2161), changes in the repeat are causing pilus protein changes, mediating antigenic variation through a phase-variable mechanism. Comparisons also show repeat tract variation in a membrane protein (NGK_1211) and mafB cassette (NGK_1624), supporting C-terminal phase variation in the species of the genus Neisseria Variations in the products of mafB cassettes are believed to contribute to competition between species within the niche (Jamet et al., 2015). Although no variation was observed in these strains in ispH (NGK_0106), (G)8 repeats are known to vary in lgtC (NGK_1632), hpuA (NGK_2581), and hsdS (NGK_0571) (Table 2), therefore it is highly likely that the repeat in ispH also has the capacity to vary.
Table 3. C-terminal phase-variable genes in N. gonorrhoeae.
| Gene | FA1090 locus* | Repeat at in FA1090* | NCCP11945 locus† | Repeat in NCCP11945† | Amino acids after the repeat in each frame of NCCP11945‡‡‡ | FA19§ | FA6140|| | 35/02 ¶ | MA11# | N. gonorrhoeae candidacy** | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ispH | NGO0072 | (G)8 | NGK_0106 | (G)8 | 26 | 50 | 61 | (G)8 | (G)8 | (G)8 | (G)8 | (Yes) |
| Menbrane protein | NGO0691 | (G)6 | NGK_1211 | (G)7 | 23 | 8 | 186^ | (G)7 | (G)6 | (G)7 | (G)7 | Yes |
| m afB cassette | WX61_RS02820 | (C)7 | NGK_1624 | (C)9 | 46 | 2 | 37 | deletion†† | (C)10 | (C)8 | (C)10 | Yes |
| Pilin cassette | WX61_RS02820 | CCGC | NGK_2161 | (C)8 | 20 | 8 | 91¶ | (C)5GCC | CCGCC | (C)5 | CCT(C)5 | Yes |
*From the N. gonorrhoeae strain FA1090 genome sequence (NC_002946.2).
†From the N. gonorrhoeae strain NCCP11945 genome sequence (CP00150.1).
‡In each column are the number of amino acids encoded 3′ of the repeat before the closest termination codon in each of the three reading frames.
§From the N. gonorrhoeae strain FA19 genome sequence (NZ_CP012026.1).
||From the N. gonorrhoeae strain FA6140 genome sequence (NZ_CP012027.1).
¶From the N. gonorrhoeae strain 35/02 genome sequence (NZ_CP012028.1).
#From the N. gonorrhoeae strain MS11 genome sequence (NC_022240.1).
**Gene phase variation candidacy in N. gonorrhoeae. Yes, there is evidence of repeat tract variation between strains supporting phase variation. (Yes), although there is no variation between strains investigated here, tracts of this length vary in other genes (Chen et al., 1998; Shafer et al., 2002; Adamczyk-Poplawska et al., 2011).
††There is a 400 bp deletion in this strain encompassing the region that would contain this repeat.
A number of previously reported candidates are not supported by evidence of phase variation, based on the absence of tract length changes between the strains (Table 4). For example, although tract variation was reported for cvaA (NGK_0168), mafA-3 (NGK_2270), and dca (NGK_1830) in N. meningitidis (Martin et al., 2003), there are no changes observed in the short (C)4 and (G)5 tracts in these genes in N. gonorrhoeae (Table 4). They are therefore unlikely to be phase-variable in this species. Likewise, neither of the dinucleotide-repeat-containing genes (NGK_1607 and NGK_2274) show variations (Table 4); dinucleotides are not likely to be phase-variable in the species of the genus Neisseria (Martin et al., 2003). All of these genes contain short repeats that do not vary or alternative nucleotide sequences in the strains investigated (Table 4).
Table 4. Genes for which there is no evidence of phase variation in N. gonorrhoeae.
| Gene | FA1090 locus* | Repeat in FA1090* | NCCP11945 locus† | Repeat in NCCP11945† | Repeat in FA19‡ | Repeat in FA6140§ | Repeat in 35/02|| | Repeat in MS11¶ | N. gonorrhoeae candidacy# |
|---|---|---|---|---|---|---|---|---|---|
| Prolyl endopeptidase | NGO0026 | GGGGCGG | NGK_0034 | GGGGCGG | GGGGCGG | GGGGCGG | GGGGCGG | GGGGCGG | No. Replacement tract |
| pill/wbpC | NGO0065 | C(G)6 | NGK_0089** | C(G)6 | C(G)6 | C(G)6 | C(G)6 | C(G)6 | No. No variation. |
| Phosphoesterase | NGO0081 | (C)7 | NGK_0 n 9 | (C)7 | (C)7 | (C)7 | (C)7 | (C)7 | No. No variation. |
| Hypothetical | NGO0121 | (A)6 | NGK_0167 | (A)6 | (A)6 | (A)6 | (A)6 | (A)6 | No. No variation. |
| cvaA | NGO0123 | (C)4 | NGK_0168 | (C)4 | (C)4 | (C)4 | (C)4 | (C)4 | No. No variation. |
| potD -2 | NGO0206 | AA(C)5 | NGK_0338 | AA(C)5 | AA(C)5 | AA(C)5 | AA(C)5 | AA(C)5 | No. No variation. |
| Hypothetical | NGO0532 | AACCGGCAAACA | NGK_1400 | AACCGGCAAACA | AACCGGCAAACA | AACCGGCAAACA | AACCGGCAAACA | AACCGGCAAACA | No. Replacement tract |
| nifS | NGO0636 | CCACACCC | NGK_1278 | CCACACCC | CCACACCC | CCACACCC | CCACACCC | CCACACCC | No. Replacement tract |
| lldD | NGO0639 | (G)7 | NGK_1275 | (G)7 | (G)7 | (G)7 | (G)7 | (G)7 | No. No variation. |
| Methylase NlalV | NGO0676 | (A)9 | NGK_1230 | (A)9 | (A)9 | (A)9 | (A)9 | (A)9 | No. No variation. |
| dnaX | NGO0743 | (C)7 | NGK_1135 | (C)7 | (C)7 | (C)7 | (C)7 | (C)7 | No. No variation. |
| mobA | NGO0754 | GGAAGG | NGK_1123 | GGAAGG | GGAAGG | GGAAGG | GGAAGG | GGAAGG | No. Replacement tract |
| ppx | NGO1041 | (C)7 | NGK_0745 | (C)7 | (C)7 | (C)7 | (C)7 | (07 | No. No variation. |
| fxP/ccoP | NGO1371 | (AT)5 | NGK_1607 | (AT)5 | (AT)5 | (AT)5 | (AT)5 | (AT)5 | No. No variation. |
| Hypothetical | NGO1384 | G(A)7 | NGK_1622 | (A)8 | (A)8 | (A)8 | (A)8 | G(A)7 | No. No variation in length. |
| pntA | NGO1470 | CCCTGCTGG | NGK_1735 | CCCTGCTGG | CCCTGCTGG | CCCTGCTGG | CCCTGCTGG | CCCTGCTGG | No. Replacement tract |
| amiC | NGO1501 | TTCGCCC | NGK_1783 | TTCGCCC | TTCGCCC | TTCGCCC | TTCGCCC | TTCGCCC | No. Replacement tract |
| dca | NGO1540 | TGTGGGGG | NGK_1830 | TGTGGGGG | TGTGGGGG | TGTGGGGG | TGTGGGGG | TGTGGGGG | No. Replacement tract |
| anmK | NGO1583 | (C)7 | NGK_1884 | (C)7 | (C)7 | (C)7 | (C)7 | (C)7 | No. No variation. |
| dinG | NGO1708 | (C)4T CC | NGK_2106 | (C)4TCC | (C)4TCC | (C)4TCC | (C)4TCC | (C)4TCC | No. Replacement tract |
| rplK | NGO1855 | (C)7 | NGK_2416 | (C)7 | (C)7 | (C)7 | (C)7 | (C)7 | No. No variation. |
| Hypothetical | NGO1970 | (TA)5 | NGK_2274 | (TA)5 | (TA)5 | (TA)5 | (TA)5 | (TA)5 | No. No variation. |
| mafA -3 | NGO1972 | (G)5 | NGK_2270 | (G)5 | (G)5 | (G)5 | (G)5 | (G)5 | No. No variation. |
| map | NGO1983 | (C)6 | NGK_2258 | (C)6 | (C)6 | (C)6 | (C)6 | (C)6 | No. No variation. |
| plsX | NGO2171 | (TTCC)3 | NGK_2652 | (TTCC)3 | (TTCC)3 | (TTCC)3 | (TTCC)3 | (TTCC)3 | No. No variation. |
| lbpA | NGO0260a | nr | NGK_0401 | GGGGGCGG | GGGGGCGG | nr | nr | TGAAACGG | No. Replacement tract |
*From the N. gonorrhoeae strain FA1090 genome sequence (NC_002946.2).
†From the N. gonorrhoeae strain NCCP11945 genome sequence (CP00150.1).
‡From the N. gonorrhoeae strain FA19 genome sequence (NZ_CP012026.1).
§From the N. gonorrhoeae strain FA6140 genome sequence (NZ_CP012027.1).
||From the N. gonorrhoeae strain 35/02 genome sequence (NZ_CP012028.1).
¶From the N. gonorrhoeae strain MS11 genome sequence (NC_022240.1).
#Gene phase variation candidacy in N. gonorrhoeae. No. Replacement tract: due to the replacement of the repeat tract with other nucleotides, this is not phase-variable. No. No variation: due to no observed variation in the repeat tract, this is not phase-variable. No. No variation in length: due to the equal length tract in all strains, this is not phase-variable.
**This coding sequence contains a point mutation, which generates a premature termination codon.
nr, The region of the coding sequence containing the repeat tract does not have homology to the aligned region in these strains.
This analysis identified 29 genes that are known to be phase variable (Tables 1, 2), either in N. gonorrhoeae or N. meningitidis including 12 paralogues of opa (11 in each strain; Muralidharan et al., 1987; Bhat et al., 1991) and 17 other known phase-variable genes (Stern et al., 1987; Jonsson et al., 1991; Van der Ende et al., 1995; Erwin et al., 1996; Chen et al., 1998; Peak et al., 1999; Carson et al., 2000; Banerjee et al., 2002; Mackinnon et al., 2002; Shafer et al., 2002; Power et al., 2003; Srikhanta et al., 2009; Adamczyk-Poplawska et al., 2011; Arenas et al., 2015). Thirteen additional genes have variations in the repeat tracts when the six N. gonorrhoeae genome sequences are compared, one transcriptional, nine translational, and three C-terminal repeats. Based on homology and presence of conserved domains, these genes are believed to encode two replication initiation factors, an adhesion protein, a pyrimidine 5′-nucleotidase, two lipoproteins, two membrane proteins, two secreted proteins, and three hypothetical proteins (Tables 1–3).
Combined with the previous data on repeat variation within and between gonococcal strains and demonstration of phase variation (Sparling et al., 1986; Yang et al., 1996; Lewis et al., 1999; Snyder et al., 2001; Power et al., 2003; Jordan et al., 2005; Srikhanta et al., 2009), a revised repertoire of 43 transcriptional (Table 1), translational (Table 2), and C-terminal (Table 3) phase-variable genes is proposed for N. gonorrhoeae as a species. This is fewer than previous predictions (76 in Jordan et al., 2005) and thus far two-thirds (67 %, 29 out of 43) have been experimentally demonstrated to be phase-variable (Tables 1, 2). The additional 14 genes, 13 of which show strain-to-strain repeat variation, require additional investigation.
Phase variable repeat copy number variation in vitro
Previously, for H. canadensis, 454 and Illumina genome sequence read data was used to support candidacy of phase-variable genes (Snyder et al., 2010). In the present study, Ion Torrent and Illumina genome sequence read data from N. gonorrhoeae strain NCCP11945 that had been passaged in the laboratory for 8 weeks or for 20 weeks was analysed for changes to phase-variable repeats for the 14 genes for which there is no within-strain evidence of phase variation (Tables 1–3). Changes were observed in known phase-variable genes pilC1, opa, and fetA, suggesting that read data can support phase variability by demonstrating within-strain variation in tracts (Table 5). Of the 14 genes, only virG (NGK_0804) and the pyrimidine 5′-nucleotidase (NGK_2176) showed no changes in repeats (Table 5). Probably, the virG (AAGC)3 copy number is too low to vary, however there may be strains with greater copy number in which it would. Likewise, the poly-C repeat in NGK_2176 has been replaced with CAAACCCC in strain NCCP11945 and therefore would not be expected to phase vary in this strain, however phase variation is likely in strain MS11, for example.
Table 5. Genes for which there is sequencing-based evidence of phase variation in N. gonorrhoeae strain NCCP11945.
| Gene | FA1090 locus* | Repeat in FA1090* | NCCP11945 locus† | Repeat in NCCP11945† | Repeat in FA19‡ | Repeat in FA6140§ | Repeat in 35/02|| | Repeat in MS11¶ | N. gonorrhoeae candidacy# | Ion Torrent* * | Illumina†† |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ispH | NGO0072 | (G)8 | NGK_0106 | (G)8 | (G)8 | (G)8 | (G)8 | (G)8 | (yes) | Repeat varies | Repeat does not vary |
| virG | NGO0985 | (AAGC)3 | NGK_0804 | (AAGC)3 | (AAGC)2 | (AAGC)3 | (AAGC)3 | (AAGC)3 | Yes | Repeat does not vary | Repeat does not vary |
| Hypothetical | NGO0964 | (AAGC)4 | NGK_0831a | (AAGC)8 | (AAGC)15 | (AAGC)7 | (AAGC)9 | (AAGC)7 | Yes | Repeat varies | Repeat varies |
| Membrane protein | NGO0691 | (G)6 | NGK_1211 | (G)7 | (G)7 | (G)6 | (G)7 | (G)7 | Yes | Repeat varies | Repeat does not vary |
| Hypothetical | NGO0527 | (C)6A(C)9GC | NGK_1405 | (C)6A(C)8GC | (C)7A(C)4T(C)3GC | (C)7A(C)4T(C)3GC | (C)11A(C)8GCGC | (C)6A(C)14GCC | Yes | Repeat varies | Repeat does not vary |
| Replication initiation factor | NGO_06695 | (C)8TTATCTAACA(G)7 | NGK1486 | (C)9TTATCTAACA(G)7 | (C)7TTATCTAACA(G)7 | (C)9TTATCTAACA(G)6 | (C)11TT ATCTAACA(G)8 | (C)9T T AT CT AACA(G)7 | Yes | Repeat varies | Repeat varies |
| mafB cassette | NGO1386 | (C)7 | NGK_1624 | (C)9 | {deletion} | (C)10 | (C)8 | (C)10 | Yes | Repeat varies | Repeat varies |
| Adhesion | NGO1445 | (CAAG)20CAAA | NGK_1705 | (CAAG)12CAAA | (CAAG)12CAAA | (CAAG)6CAAA | (CAAG)9CAAA | (CAAG)6CAA A | Yes | Repeat varies | Repeat varies |
| Replication initiation factor | NGO_06135 | (C)8TTATCTAACA(G)7 | NGK_1957 | (C)11TTATCTAACA(G)8 | (C)7TTATCTAACA(G)7 | (C)6TTATCTAACA(G)5 | (C)11TTATCTAACA(G)8 | (C)10T TAT C TAACA(G)7 | Yes | Repeat varies | Repeat varies |
| Pilin cassette | NGO_11140 | CCGC | NGK_2161 | (C)8 | (C)5GCC | CCGCC | (C)5 | CCT (C)5 | Yes | Repeat varies | Repeat varies |
| Pyrimidine 5′-nucleotidase | NGO2055 & NGO2054II | (C)6 | NGK_2176 | CAAACCCC | CAAACCCC | CAAACCCC | (C)9 | (C)10 | Yes | No repeat | No repeat |
| Lipoprotein | NGO2047 | (A)9 | NGK_2186§§ | (A)8 | (A)8 | (A)9 | (A)9 | (A)8 | Yes | Repeat varies | Repeat varies |
| Hypothetical | NGO1953 | (C)8 | NGK_2297 | (C)8 | (C)8 | (C)8 | (C)9 | (C)8 | Yes | Repeat varies | Repeat does not vary |
| Repetitive large surface lipoprotein | NGO_09875 & NGO_09870II | (G)8 | NGK_2422 & NGK_2423|| | (G)7 | (G)7 | (G)7 | (G)7 | (G)7 | Yes | Repeat varies | Repeat does not vary |
| fetA / frpB | NGO2093 | (C)13 | NGK_2557 | (C)14 | (C)10 | (C)11 | (C)11 | (C)11 | Known | Repeat varies | Repeat varies |
| pilC 1 | NGO1912 | (G)11 | NGK_2342 | (G)13 | (G)11 | GGGC(G)11 | (G)15 | (G)11 | Known | Repeat varies | Repeat varies |
| opa | NGO0950a | (CTTCT)16CTTCC | NGK_0847 | (CTTCT)19CTTCC | (CTTCT)13CTTCC | (CTTCT)16CTTCC | (CTTCT)8CTTCC | (CT T CT )4CT TCC | Known | No reads through repeat | Repeat varies |
*From the N. gonorrhoeae strain FA1090 genome sequence (NC_002946.2).
†From the N. gonorrhoeae strain NCCP11945 genome sequence (CP00150.1).
‡From the N. gonorrhoeae strain FA19 genome sequence (NZ_CP012026.1).
§From the N. gonorrhoeae strain FA6140 genome sequence (NZ_CP012027.1).
||From the N. gonorrhoeae strain 35/02 genome sequence (NZ_CP012028.1).
¶From the N. gonorrhoeae strain MS11 genome sequence (NC_022240.1).
#Gene phase variation candidacy in N. gonorrhoeae. Known, phase variation has been reported in the literature. Yes, there is evidence of repeat tract variation between strains supporting phase variation. (Yes), although there is no variation between strains investigated here, tracts of this length vary on other genes (Chen et al., 1998; Shafer et al., 2002; Adamczyk-Poplawska et al., 2011).
**Based on Ion Torrent sequencing data from cultures grown with passage for 8 weeks and from cultures grown with passage for 20 weeks (accession numbers SRR3547950, SRR3547951, SRR3547952, SRR3547953).
††Based on Illumina sequencing data from cultures grown with passage for 20 weeks (accession numbers SRR3547954, SRR3547955, SRR3547956, SRR3547957).
‡‡There is a 400 bp deletion in this strain encompassing the region that would contain this repeat.
§§NGK_2186 and NGO2047 annotations are on opposite strands.
||||This coding sequence appears to be frame-shifted and annotated as two coding sequences.
The Ion Torrent sequencing technology has been criticised for generating homopolymer-associated indels (Loman et al., 2012) and that the tracts can be incorrect at more than eight bases (Quail et al., 2012), the optimal length for phase variation. Homopolymeric tracts in Illumina data are believed to be less error prone (Schirmer et al., 2015). However, repeat sequence data from Illumina often agreed with Ion Torrent on the presence of variation (9 of 14 genes with variation in Ion Torrent, Table 5). When the Illumina data did not show repeat variation, this often corresponded to relatively low read coverage of the region compared to the Ion Torrent data (Table S2).
It is currently impossible to differentiate genuine biologically induced indels from sequencing errors (Narzisi & Schatz, 2015). We may find that what we ascribe to errors can also be subtle changes that are constantly being generated within the bacterial population. From this data, the expected biological variation supporting phase variation appears to be present in N. gonorrhoeae strain NCCP11945 for 12 as yet unexplored genes.
Conclusion
In conclusion, N. gonorrhoeae possesses three different mechanisms for phase variation: transcriptional; translational; and C-terminal. Stochastic systems obviously play important roles in the biology of the organism given the variety and number of genes involved. The functions of previously unexplored phase-variable genes, including one transcriptional phase-variable gene, nine translational phase-variable genes, and four C-terminal phase-variable genes require further investigation.
Acknowledgements
C. P. C. was supported by a Sparks project grant 11KIN01 and 13KIN01. Illumina genome sequencing was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1).
Supplementary Data
Supplementary File 1
References
- Adamczyk-Poplawska M., Lower M., Piekarowicz A.(2011). Deletion of one nucleotide within the homonucleotide tract present in the hsdS gene alters the DNA sequence specificity of type I restriction-modification system NgoAV. J Bacteriol 1936750–6759. 10.1128/JB.05672-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., Van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J., Cano S., Nijland R., Van Dongen V., Rutten L., Van der Ende A., Tommassen J.(2015). The meningococcal autotransporter AutA is implicated in autoaggregation and biofilm formation. Environ Microbiol 171321–1337. 10.1111/1462-2920.12581 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Wang R., Supernavage S. L., Ghosh S. K., Parker J., Ganesh N. F., Wang P. G., Gulati S., Rice P. A.(2002). Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J Exp Med 196147–162. 10.1084/jem.20012022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D. W., Garrison E. K., Quinlan A. R., Strömberg M. P., Marth G. T.(2011). BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 271691–1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. S., Gibbs C. P., Barrera O., Morrison S. G., Jähnig F., Stern A., Kupsch E. M., Meyer T. F., Swanson J.(1991). The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol 51889–1901. 10.1111/j.1365-2958.1991.tb00813.x [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A., Galaxy Team (2010). Manipulation of FASTQ data with Galaxy. Bioinformatics 261783–1785. 10.1093/bioinformatics/btq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D. A., Mamedov I. Z., Britanova O. V., Zvyagin I. V., Shagin D., Ustyugova S. V., Turchaninova M. A., Lukyanov S., Lebedev Y. B., Chudakov D. M.(2012). Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur J Immunol 423073–3083. 10.1002/eji.201242517 [DOI] [PubMed] [Google Scholar]
- Bragg L. M., Stone G., Butler M. K., Hugenholtz P., Tyson G. W.(2013). Shining a light on dark sequencing: characterising errors in Ion torrent PGM data. PLoS Comput Biol 9e1003031. 10.1371/journal.pcbi.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E., Hill D. J., Morand P., Griffiths N. J., Bourdoulous S., Murillo I., Nassif X., Virji M.(2009). Meningococcal interactions with the host. Vaccine 27B78–89. 10.1016/j.vaccine.2009.04.069 [DOI] [PubMed] [Google Scholar]
- Carson S. D., Stone B., Beucher M., Fu J., Sparling P. F.(2000). Phase variation of the gonococcal siderophore receptor FetA. Mol Microbiol 36585–593. 10.1046/j.1365-2958.2000.01873.x [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Loman N. J., Snyder L. A., Bailey C. M., Stekel D. J., Pallen M. J.(2008). xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res 36D543–546. 10.1093/nar/gkm928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. J., Elkins C., Sparling P. F.(1998). Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun 66987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung G. T., Yoo J. S., Oh H. B., Lee Y. S., Cha S. H., Kim S. J., Yoo C. K.(2008). Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J Bacteriol 1906035–6036. 10.1128/JB.00566-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T.(2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 141394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin A. L., Haynes P. A., Rice P. A., Gotschlich E. C.(1996). Conservation of the lipooligosaccharide synthesis locus lgt among strains of Neisseria gonorrhoeae: requirement for lgtE in synthesis of the 2C7 epitope and of the β chain of strain 15253. J Exp Med 1841233–1241. 10.1084/jem.184.4.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A., Jousset A. B., Euphrasie D., Mukorako P., Boucharlat A., Ducousso A., Charbit A., Nassif X.(2015). A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog 11e1004592. 10.1371/journal.ppat.1004592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulczyk R., Masignani V., Maione D., Tettelin H., Grandi G., Telford J. L.(2010). Simple sequence repeats and genome plasticity in Streptococcus agalactiae. J Bacteriol 1923990–4000. 10.1128/JB.01465-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson A. B., Nyberg G., Normark S.(1991). Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J 10477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. W., Snyder L. A., Saunders N. J.(2005). Strain-specific differences in Neisseria gonorrhoeae associated with the phase variable gene repertoire. BMC Microbiol 5. 10.1186/1471-2180-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D. S., Jr, Peacock W. L., Jr, Deacon W. E., Brown L., Pirkle D. I.(1963). Neisseria gonorrhoeae I. virulence genetically linked to clonal variation. J Bacteriol 851274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L.(2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L.(2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. A., Gipson M., Hartman K., Ownbey T., Vaughn J., Dyer D. W.(1999). Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol Microbiol 32977–989. 10.1046/j.1365-2958.1999.01409.x [DOI] [PubMed] [Google Scholar]
- Loman N. J., Misra R. V., Dallman T. J., Constantinidou C., Gharbia S. E., Wain J., Pallen M. J.(2012). Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30434–439. 10.1038/nbt.2198 [DOI] [PubMed] [Google Scholar]
- Mackinnon F. G., Cox A. D., Plested J. S., Tang C. M., Makepeace K., Coull P. A., Wright J. C., Chalmers R., Hood D. W., et al. (2002). Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol Microbiol 43931–943. 10.1046/j.1365-2958.2002.02754.x [DOI] [PubMed] [Google Scholar]
- Martin P., Van de Ven T., Mouchel N., Jeffries A. C., Hood D. W., Moxon E. R.(2003). Experimentally revised repertoire of putative contingency loci in Neisseria meningitidis strain MC58: evidence for a novel mechanism of phase variation. Mol Microbiol 50245–257. 10.1046/j.1365-2958.2003.03678.x [DOI] [PubMed] [Google Scholar]
- Moxon R., Bayliss C., Hood D.(2006). Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet 40307–333. 10.1146/annurev.genet.40.110405.090442 [DOI] [PubMed] [Google Scholar]
- Muralidharan K., Stern A., Meyer T. F.(1987). The control mechanism of opacity protein expression in the pathogenic Neisseriae. Antonie Van Leeuwenhoek 53435–440. 10.1007/BF00415499 [DOI] [PubMed] [Google Scholar]
- Narzisi G., Schatz M. C.(2015). The challenge of small-scale repeats for indel discovery. Front Bioeng Biotechnol 38. 10.3389/fbioe.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer H., Rose G., Jolley K. A., Frapy E., Zahar J. R., Maiden M. C., Bentley S. D., Tinsley C. R., Nassif X., Bille E.(2011). Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLoS One 6e17145. 10.1371/journal.pone.0017145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peak I. R., Jennings M. P., Hood D. W., Moxon E. R.(1999). Tetranucleotide repeats identify novel virulence determinant homologues in Neisseria meningitidis. Microb Pathog 2613–23. 10.1006/mpat.1998.0243 [DOI] [PubMed] [Google Scholar]
- Power P. M., Roddam L. F., Rutter K., Fitzpatrick S. Z., Srikhanta Y. N., Jennings M. P.(2003). Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol 49833–847. 10.1046/j.1365-2958.2003.03602.x [DOI] [PubMed] [Google Scholar]
- Quail M. A., Smith M., Coupland P., Otto T. D., Harris S. R., Connor T. R., Bertoni A., Swerdlow H. P., Gu Y.(2012). A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13341. 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P.(2011). Integrative genomics viewer. Nat Biotechnol 2924–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Ijaz U. Z., D'Amore R., Hall N., Sloan W. T., Quince C.(2015). Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res 43e37. 10.1093/nar/gku1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer W. M., Datta A., Kolli V. S., Rahman M. M., Balthazar J. T., Martin L. E., Veal W. L., Stephens D. S., Carlson R.(2002). Phase variable changes in genes lgtA and lgtC within the lgtABCDE operon of Neisseria gonorrhoeae can modulate gonococcal susceptibility to normal human serum. J Endotoxin Res 847–58. 10.1177/09680519020080010501 [DOI] [PubMed] [Google Scholar]
- Snyder L. A., Butcher S. A., Saunders N. J.(2001). Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 1472321–2332. 10.1099/00221287-147-8-2321 [DOI] [PubMed] [Google Scholar]
- Snyder L. A., Loman N. J., Linton J. D., Langdon R. R., Weinstock G. M., Wren B. W., Pallen M. J.(2010). Simple sequence repeats in Helicobacter canadensis and their role in phase variable expression and C-terminal sequence switching. BMC Genomics 11. 10.1186/1471-2164-11-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F., Cannon J. G., So M.(1986). Phase and antigenic variation of pili and outer membrane protein II of Neisseria gonorrhoeae. J Infect Dis 153196–201. 10.1093/infdis/153.2.196 [DOI] [PubMed] [Google Scholar]
- Srikhanta Y. N., Dowideit S. J., Edwards J. L., Falsetta M. L., Wu H. J., Harrison O. B., Fox K. L., Seib K. L., Maguire T. L., et al. (2009). Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog 5e1000400. 10.1371/journal.ppat.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A., Meyer T. F.(1987). Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol Microbiol 15–12. 10.1111/j.1365-2958.1987.tb00520.x [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P.(2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ende A., Hopman C. T., Zaat S., Essink B. B., Berkhout B., Dankert J.(1995). Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J Bacteriol 1772475–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viburiene R., Vik Å., Koomey M., Børud B.(2013). Allelic variation in a simple sequence repeat element of neisserial pglB2 and its consequences for protein expression and protein glycosylation. J Bacteriol 1953476–3485. 10.1128/JB.00276-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. L., Gotschlich E. C.(1996). Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med 183323–327. 10.1084/jem.183.1.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1.Zelewska, M. A., Pulijala, M., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547950 (2016).
- 2.Mahmood, H. -T. -N. A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547951 (2016).
- 3.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547952 (2016).
- 4.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547953 (2016).
- 5.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547954 (2016).
- 6.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547955 (2016).
- 7.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547956 (2016).
- 8.Churchward, C. P., Calder, A., & Snyder, L. A. S., NCBI Sequence Read Archive. Accession number SRR3547957 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1