Abstract
Translation in dendrites is believed to support synaptic changes during memory consolidation. Although translational control mechanisms are fundamental mediators of memory, little is known about their role in local translation. We previously found that polyribosomes accumulate in dendritic spines of the adult rat lateral amygdala (LA) during consolidation of aversive pavlovian conditioning and that this memory requires cap-dependent initiation, a primary point of translational control in eukaryotic cells. Here we used serial electron microscopy reconstructions to quantify polyribosomes in LA dendrites when consolidation was blocked by the cap-dependent initiation inhibitor 4EGI-1. We found that 4EGI-1 depleted polyribosomes in dendritic shafts and selectively prevented their upregulation in spine heads, but not bases and necks, during consolidation. Cap-independent upregulation was specific to spines with small, astrocyte-associated synapses. Our results reveal that cap-dependent initiation is involved in local translation during learning and that local translational control varies with synapse type.
SIGNIFICANCE STATEMENT Translation initiation is a central regulator of long-term memory formation. Local translation in dendrites supports memory by providing necessary proteins at synaptic sites, but it is unknown whether this requires initiation or bypasses it. We used serial electron microscopy reconstructions to examine polyribosomes in dendrites when memory formation was blocked by an inhibitor of translation initiation. This revealed two major pools of polyribosomes that were upregulated during memory formation: one pool in dendritic spine heads that was initiation dependent and another pool in the bases and necks of small spines that was initiation independent. Thus, translation regulation differs between spine types and locations, and translation that occurs closest to individual synapses during memory formation is initiation dependent.
Keywords: cap-dependent translation, local translation, pavlovian conditioning, polyribosomes, serial electron microscopy, structural plasticity
Introduction
Long-term memory consolidation requires new gene expression, culminating in protein synthesis, to support substantial biochemical and structural remodeling at synapses (Rosenberg et al., 2014; Bailey et al., 2015; Segal, 2016). It is thought that a significant amount of this protein synthesis occurs locally in dendrites (Sutton and Schuman, 2006; Bramham and Wells, 2007; Wang et al., 2010; Liu-Yesucevitz et al., 2011). Local translation can provide synapses with rapid access to new proteins, bypassing gene expression cascades and subsequent transport from the soma, and can be a means of spatiotemporal protein targeting (Martin and Ephrussi, 2009; Holt and Schuman, 2013). However, how local translation fits into the larger scheme of regulated gene expression during memory consolidation is not well understood. Protein synthesis is a universal requirement for long-term memory across species, and several translational control mechanisms are known to mediate learning and their dysregulation to underlie intellectual disabilities in humans (Davis and Squire, 1984; McGaugh, 2000; Costa-Mattioli et al., 2009; Groppo and Richter, 2009; Darnell, 2011; Gal-Ben-Ari et al., 2012; Buffington et al., 2014; Santini et al., 2014). RNA trafficking mechanisms, meanwhile, have been the main focus of research on translation at synapses (Hirokawa, 2006; Sossin and DesGroseillers, 2006; Doyle and Kiebler, 2011; Buxbaum et al., 2015). Whether protein synthesis in dendrites is subject to the translational control mechanisms that regulate learning is unknown.
Translation is a complex, highly orchestrated process, with the majority of regulatory mechanisms targeting the initiation step (Sonenberg and Hinnebusch, 2009). Initiation mechanisms are involved in synaptic plasticity and memory, and improper initiation is particularly implicated in a number of intellectual disorders (Costa-Mattioli et al., 2009; Darnell, 2011; Gal-Ben-Ari et al., 2012; Buffington et al., 2014; Santini et al., 2014). The major rate-limiting step in initiation is binding of the eukaryotic initiation factor eIF4E to eIF4G, which leads to recruitment of ribosomes to the 5′ cap of the target mRNA (Groppo and Richter, 2009). Using the eIF4E–eIF4G interaction inhibitor 4EGI-1, we previously found that initiation of cap-dependent translation in the lateral amygdala (LA) is required for consolidation of aversive pavlovian conditioning memory (Hoeffer et al., 2011). Binding of eIF4E and eIF4G was increased during consolidation; in a separate study, we found that polyribosomes were upregulated in LA dendrites as well (Ostroff et al., 2010). It is possible that initiation triggered by learning is necessary for local translation at synapses and that this, at least in part, explains why consolidation is initiation dependent. On the other hand, if dendritic translation after learning serves to provide proteins quickly ahead of transport from the soma, it could be regulated downstream of eIF4E–eIF4G binding. Once assembled, polyribosomes can be stalled and reactivated independent of initiation (Richter and Coller, 2015). This can occur in dendrites, and a substantial amount of dendritic mRNA has been proposed to be transported in the form of stalled polyribosomes (Graber et al., 2013). Cap-independent initiation can also occur; translation of several dendritic mRNAs can be initiated by ribosome binding to an internal ribosome entry site (IRES), independent of eIF4E (Pinkstaff et al., 2001; Sutton and Schuman, 2005).
To determine whether the effects of 4EGI-1 on memory consolidation are specific to translation in the soma, we used serial section transmission electron microscopy (ssTEM) to quantify polyribosomes in LA dendrites after learning. This method provides unparalleled resolution and precision, allowing us to examine localization of polyribosomes relative to synapses and to assess the relationship between polyribosomes and dendritic spine morphology. We found that administration of 4EGI-1 after training blocked some, but not all, effects on synaptic polyribosomes, and this varied by spine morphology. Our results suggest a profound but nuanced dependence of dendritic polyribosomes on cap-dependent initiation.
Materials and Methods
Subjects and surgery.
All animal procedures were approved by the New York University Animal Care and Use Committee. Subjects were adult male Sprague Dawley rats weighing ∼300 g (Hilltop Lab Animals) housed singly on a 12 h light/dark cycle with ad libitum food and water, and all procedures were conducted during the animals' light cycle. For cannula implantation, animals were anesthetized with a mixture of ketamine and xylazine (100 mg/kg and 6 mg/kg, i.p.) and secured in a stereotaxic frame (David Kopf Instruments). Bilateral craniotomies were made, and 22 g stainless steel guide cannulae (Plastics One) were placed relative to bregma (−0.8 AP, ±1.5 ML, and −1.6 mm DV) and secured with dental cement. Buprenorphine (2 mg/kg, i.p.) was used as a postoperative anesthetic. Animals were given at least 1 week to recover from surgery before experiments began and were handled briefly each day after surgery to acclimate them to having their cannulae manipulated by an experimenter.
Behavior and drug infusion.
Behavioral apparatus and paired training and box control protocols were identical to those used by Ostroff et al. (2010). Briefly, animals were habituated to the training chambers (Coulbourn Instruments) for 30 min on each of 2 d before training. The trained rats were presented with five auditory tones (30 s, 5 kHz, 80 dB) coterminating in footshocks (1 s, 0.7 mA) given at a variable interval over 32.5 min. The behavioral control rats were placed in the box but not presented with tones or shocks. Immediately after training, rats were infused with 10 μl per side of either 5 μg/μl 4EGi-1 in 50% DMSO/10% β-propylcyclodextrin or vehicle. Infusions were given with a syringe pump (Harvard Apparatus) at a rate of 1 μl/min through 28 g stainless steel injection cannulae projecting 1 mm past the tip of the guide cannulae. The cannulae were left in place for 15 min to allow the drug to diffuse, and the animals were returned to their home cages. Freezing during the tones was quantified to ensure that all animals displayed the expected behavioral response during training.
Tissue preparation.
All reagents were obtained from Electron Microscopy Sciences unless otherwise stated. Exactly 1 h after the first tone/shock pairing or the equivalent time point for the control group, animals were anesthetized with a lethal dose of chloral hydrate (1.25 g/kg, i.p.; Sigma) and perfused transcardially with saline containing 0.1 U of heparin, 0.01 m sodium cacodylate, 154 mm NaCl (Sigma), 2 mm CaCl2, and 4 mm MgCl2 at pH 7.4 to clear the bloodstream, followed by 500 ml of fixative containing 2.5% glutaraldehyde, 2% paraformaldehyde, 0.1 m sodium cacodylate, 2 mm CaCl2, and 4 mm MgCl2 at pH 7.4 at a constant rate of 75 ml/min. The brains were removed and placed in vials of the same fixative for 1 h and rinsed in the saline solution without heparin. The left hemisphere of each brain was sectioned coronally at 70 μm on a vibrating slicer (Leica Microsystems), sections were examined to confirm correct cannula placements, and the area containing the amygdala was dissected out for EM processing. Samples were postfixed in reduced osmium (1% osmium tetroxide and 1.5% potassium ferrocyanide in 0.1 m sodium cacodylate) followed by 1% osmium tetroxide in 0.1 m sodium cacodylate, dehydrated through an ascending concentration of ethanol containing 1.5% uranyl acetate (Ted Pella), and embedded and cured in LX-112 resin (Ladd Research Industries).
Sectioning and imaging.
Blocks were trimmed to the area of the LA just ventral to the lateral association fibers. Tissue was collected from four rats per experimental group, and the two rats with the best tissue preservation in each group were selected at this stage for serial sectioning. Serial sections were cut on an ultramicrotome (Leica Microsystems), picked up on pioloform-coated Synaptek slot grids (Ted Pella), and stained with aqueous uranyl acetate and Reynolds's lead citrate. Sections were imaged at 7500× on a JEOL 1230 transmission electron microscope with a 4000 × 4000 pixel digital camera (Gatan). An average of 160 sections (range, 150–202) were imaged per series. Section thickness averaged 54 nm (range, 50–62 nm) as measured by the cylindrical diameters method (Fiala and Harris, 2001).
Reconstruction and analysis.
Reconstruct software (Fiala, 2005; RRID:SCR_002716) was used to align images and perform all reconstruction and measurements, and reconstructions were rendered in 3DS Max (Autodesk). The Reconstruct software package and documentation are freely available for download at http://synapses.clm.utexas.edu/tools/reconstruct/reconstruct.stm. All analyses were performed on serial section reconstructions by experimenters blind to treatment groups. Dendritic segments that were in cross section in the central section of each series and whose protrusions were complete within the series were used for analysis, with the inclusion length taken from the first complete protrusion at the ventral end of the series up to but not including the first incomplete protrusion at the dorsal end. Microtubule number was used as a measure of dendrite caliber as it is more consistent throughout a series than diameter. Dendrites had an average of 35 microtubules (range, 13–82), and large apical dendrites (>100 microtubules) were excluded. The dataset was composed of 67 dendritic segments (8–10 per rat) with an average analyzed length of 8.7 μm (range, 5.8–13.3) with a total of 1576 protrusions. Asymmetric, presumably excitatory synapses were identified according to standard morphological criteria (Peters et al., 1991), and protrusions carrying an asymmetric synapse were defined as spines. Approximately 10% of protrusion origins gave rise to more than one spine, 8% of protrusions lacked a synapse, 2.5% of spines carried more than one synapse, and 2% carried both an asymmetric and a symmetric (inhibitory or modulatory) synapse. These features occurred equally in all groups and were not analyzed separately. Symmetric synapses were excluded from the analysis.
Statistics.
Two-way ANOVAs were used to test for interactive effects of training and drug. When significant (p < 0.05) interactions were found, simple main-effects ANOVAs were run within the drug and training groups. If no interaction was found, main-effects ANOVAs were run. For all ANOVAs, a hierarchical nested design was used with subjects nested into treatment groups to ensure that effects were not carried by single subjects.
Means and SEs for each test are shown in graphs in Figures 2–5, and corresponding F and p values are given in Tables 1–3.
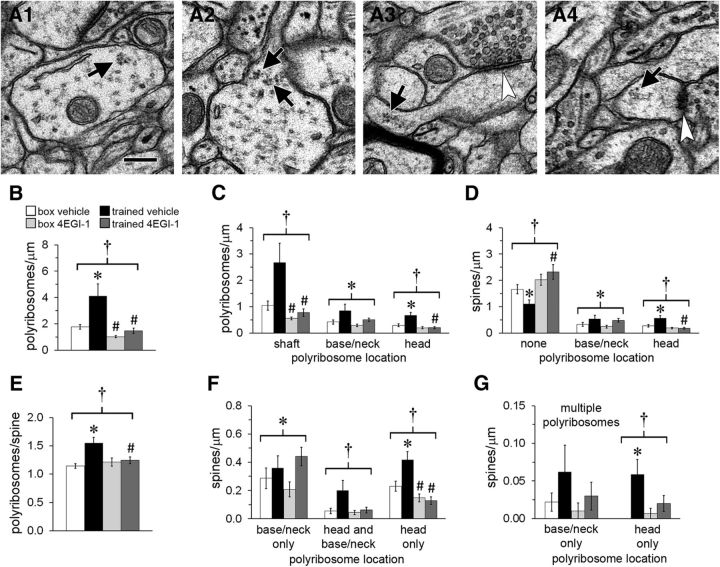
Figure 2.
Polyribosomes in dendrites. A, Electron micrographs of polyribosomes (arrows) in a dendritic shaft (A1) and in a dendritic spine base (A2), neck (A3), and head (A4). Arrowheads indicate synapses on the spines in A3 and A4. Scale bar, 250 nm. B, The overall frequency of polyribosomes in dendrites was higher in the trained-vehicle group but was reduced in both drug groups. C, The drug lowered polyribosome frequency in dendritic shafts, with a greater effect in the trained group. There was a main effect of training on polyribosomes in spine bases and necks and an increase in polyribosomes in the trained-vehicle group that was blocked by the drug. D, The drug blocked both a decrease in the frequency of spines without polyribosomes and an increase in spines with head polyribosomes in the trained-vehicle group. There was a main effect of training on spines with base or neck polyribosomes. E, In spines with polyribosomes, there were more polyribosomes per spine in the trained-vehicle group, and the drug blocked this effect. F, There was a main effect of training on spines containing polyribosomes only in the base or neck. Spines with polyribosomes in the head only were decreased by the drug in the trained and box groups and increased by training in the vehicle group. There was an interactive effect between drug and training on spines with polyribosomes in both the head and base or neck, but effects within groups did not reach statistical significance. G, Among spines with more than one polyribosome in the same location, there were more with polyribosomes in the head only in the trained-vehicle group. Shown are effects at p < 0.05 for training (*), drug (#), and training × drug (†).
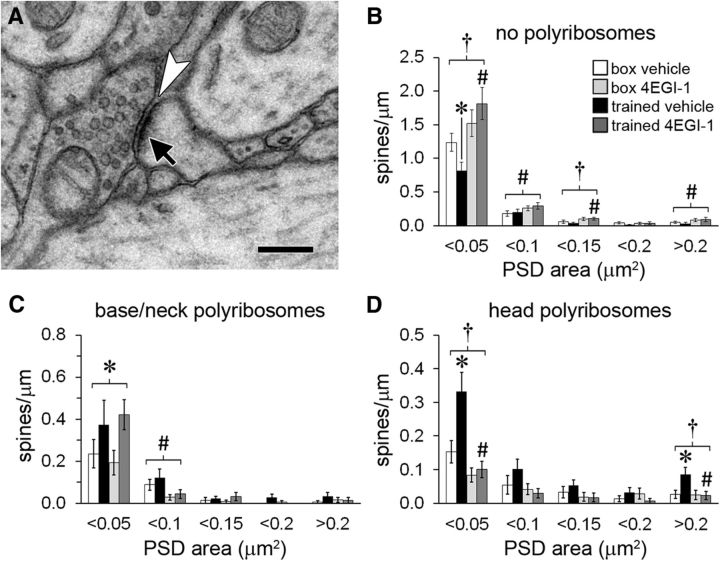
Figure 3.
Spine frequency by synapse size. A, Electron micrograph of a synapse showing the PSD (arrow) adjacent to the synaptic cleft (arrowhead). Scale bar, 250 nm. B, For spines with no polyribosomes, there was a decrease in those with the smallest synapses (PSD < 0.05 μm2) in the trained-vehicle group that was blocked by the drug, and the drug increased the frequency of larger spines. C, For spines with polyribosomes in the base or neck, there was a main effect of training on the smallest spines (PSD < 0.05 μm2) and a main effect of drug on slightly larger spines (PSD = 0.05–0.1 μm2). D, For spines with polyribosomes in the head, training induced an increase in very small (PSD < 0.05 μm2) and very large (PSD > 0.2 μm2) spines that was blocked by the drug. Shown are effects at p < 0.05 for training (*), drug (#), and training × drug (†).
Figure 4.
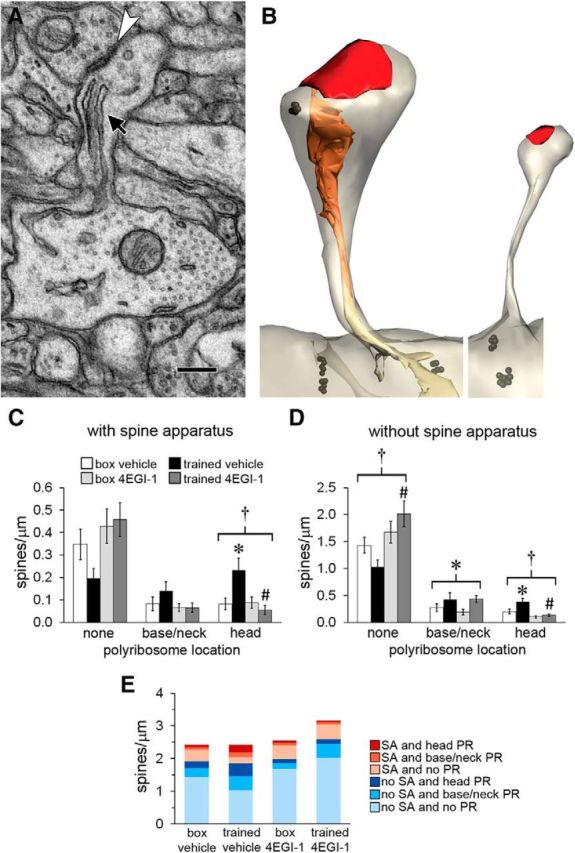
Polyribosomes and the spine apparatus. A, Electron micrograph of a spine containing a spine apparatus (arrow) and forming an asymmetric synapse (arrowhead). Scale bar, 250 nm. B, Spines reconstructed through serial sections. The spine on the left contains a spine apparatus and a polyribosome in its head, whereas the spine on the right contains neither. Black, Ribosomes; red, synapses; orange, spine apparatus; yellow, smooth ER. C, Among spines with a spine apparatus, there was an increase in spines with polyribosomes in the head in the trained-vehicle group, and this was blocked by the drug. D, Among spines without a spine apparatus, there were more spines with no polyribosomes in the trained-4EGI-1 group than in the trained-vehicle group. There was a main effect of training on spines with polyribosomes in the base or neck and an effect of training on spines with polyribosomes in the head that was blocked by the drug. E, Graph summarizing mean frequencies of spines with respect to presence of a spine apparatus (SA) and polyribosomes (PR). Shown are effects at p < 0.05 for training (*), drug (#), and training × drug (†).
Figure 5.
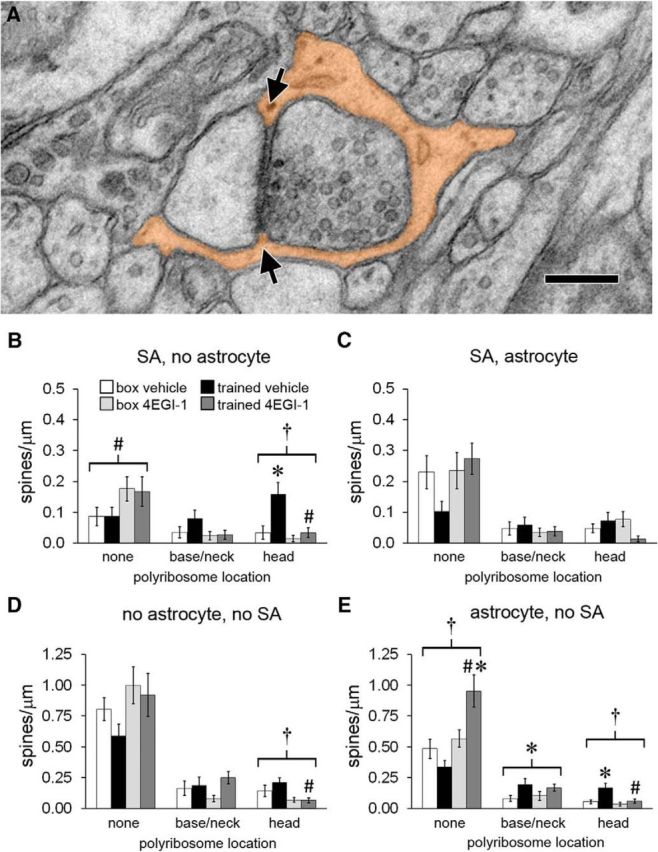
Astrocytes at synapses. A, Electron micrograph of a spine head with its synapse surrounded by an astrocytic process (orange) that makes contact with the synaptic cleft at both edges (arrows). Scale bar, 250 nm. B, For spines with a spine apparatus (SA) and no perisynaptic astrocyte, there was a main effect of drug on spines with no polyribosomes and an effect of training on spines with polyribosomes in the head that was blocked by the drug. C, There were no effects of training or drug on the frequency of spines with a spine apparatus and perisynaptic astrocyte. D, There were fewer spines with head polyribosomes but no spine apparatus or perisynaptic astrocyte in the trained-4EGI-1 group versus the trained-vehicle group. E, In spines with a perisynaptic astrocyte but no spine apparatus, there were more spines without polyribosomes in the trained-4EGI-1 group versus the box-4EGI-1 group and the trained-vehicle group. There was a main effect of training on spines with base or neck polyribosomes and an effect of training on spines with head polyribosomes that was blocked by the drug. Shown are effects at p < 0.05 for training (*), drug (#), and training × drug (†).
Table 1.
Effects of training that are sensitive to 4EGI-1
| Figure | Variable (number/μm) | Direction of training effect | Training effect |
Drug effect |
Training × drug |
|||
|---|---|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |||
| Vehicle group | Trained group | |||||||
| 2B | PR | ↑ | 5.78 | 0.02235 | 7.71 | 0.00910 | 7.80 | 0.00018 |
| 2C | Head PR | ↑ | 7.98 | 0.00821 | 13.75 | 0.00079 | 10.41 | 0.00001 |
| 2D | Sp without PR | ↓ | 5.63 | 0.02406 | 16.17 | 0.00033 | 6.99 | 0.00042 |
| 2D | Sp with head PR | ↑ | 6.19 | 0.01847 | 12.15 | 0.00145 | 8.87 | 0.00006 |
| 2E | PR (number/spine) | ↑ | 8.11 | 0.00477 | 4.05 | 0.04513 | 4.64 | 0.00338 |
| 2F | Sp with only head PR | ↑ | 6.11 | 0.01914 | 16.79 | 0.00027 | 10.61 | 0.00001 |
| 2G | Sp with multiple PR, head only | ↑ | 6.33 | 0.01725 | 2.55 | 0.11980 | 4.18 | 0.00943 |
| 3B | Sp with no PR, PSD < 0.05 μm2 | ↓ | 5.20 | 0.02965 | 14.81 | 0.00054 | 6.23 | 0.00095 |
| 3D | Sp with head PR, PSD < 0.05 μm2 | ↑ | 5.81 | 0.02210 | 11.20 | 0.00210 | 8.60 | 0.00008 |
| 3D | Sp with head PR, PSD > 0.2 μm2 | ↑ | 5.03 | 0.03220 | 6.05 | 0.01951 | 3.93 | 0.01261 |
| 4C | Sp with SA, head PR | ↑ | 4.58 | 0.04034 | 7.50 | 0.01000 | 4.85 | 0.00441 |
| 4D | Sp without SA, head PR | ↑ | 4.58 | 0.04034 | 11.13 | 0.00216 | 8.36 | 0.00010 |
| 5B | Sp with SA, no astro, head PR | ↑ | 5.62 | 0.02417 | 6.28 | 0.01750 | 5.83 | 0.00147 |
| 5E | Sp without SA, with astro, head PR | ↑ | 5.63 | 0.02403 | 5.15 | 0.03012 | 5.37 | 0.00244 |
| Drug group | Box group | |||||||
| 5E | Sp without SA, with astro, no PR | ↑ | 6.94 | 0.01360 | 0.64 | 0.43049 | 9.80 | 0.00002 |
Shown is a summary of measures that had significant (p < 0.05) training × drug interactions with training effects in only one of the drug conditions. Nonsignificant effects are shown in italics. PR, Polyribosomes; Sp, spine; astro, perisynaptic astroglia.
Table 2.
Effects of training that are insensitive to 4EGI-1
| Figure | Variable (number/μm) | Direction of training effect | Training effect |
Drug effect |
Training × drug |
|||
|---|---|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |||
| Main effect | Main effect | |||||||
| 2C | Base/neck PR | ↑ | 4.20 | 0.04494 | 2.11 | 0.15129 | 2.48 | 0.06995 |
| 2D | Sp with base/neck PR | ↑ | 4.90 | 0.03069 | 0.46 | 0.50183 | 1.87 | 0.14521 |
| 2F | Sp with only base/neck PR | ↑ | 4.69 | 0.03448 | 0.00 | 0.96378 | 1.92 | 0.13694 |
| 3C | Sp with base/neck PR, PSD < 0.05 μm2 | ↑ | 5.60 | 0.02126 | 0.05 | 0.81832 | 2.03 | 0.11916 |
| 4D | Sp without SA, base/neck PR | ↑ | 4.71 | 0.03406 | 0.14 | 0.71387 | 1.71 | 0.17422 |
| 5E | Sp without SA, with astro, base/neck PR | ↑ | 5.99 | 0.01742 | 0.00 | 0.97479 | 2.20 | 0.09704 |
Shown is a summary of measures that did not have significant (p < 0.05) training × drug interactions but showed a main effect of drug. Nonsignificant effects are shown in italics. PR, Polyribosomes; Sp, spine; astro, perisynaptic astroglia.
Table 3.
General effects of 4EGI-1
| Figure | Variable (number/μm) | Direction of drug effect | Training effect |
Drug effect |
Training × drug |
|||
|---|---|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |||
| Main effect | Main effect | |||||||
| 3B | Sp with no PR, PSD < 0.1 μm2 | ↑ | 0.36 | 0.54842 | 4.21 | 0.04460 | 1.49 | 0.22608 |
| 3B | Sp with no PR, PSD > 0.2 μm2 | ↑ | 0.05 | 0.81632 | 4.47 | 0.03878 | 1.67 | 0.18377 |
| 3C | Sp with base/neck PR, PSD < 0.1 μm2 | ↓ | 0.70 | 0.40561 | 4.91 | 0.03056 | 2.06 | 0.11464 |
| 5B | Sp with SA, no astro, no PR | ↑ | 0.00 | 0.94731 | 5.43 | 0.02329 | 1.83 | 0.15226 |
| Drug group | Box group | |||||||
| 2B | Total PR | ↓ | 3.52 | 0.07098 | 14.19 | 0.00082 | 7.80 | 0.00018 |
| 2C | Shaft PR | ↓ | 2.03 | 0.16502 | 7.12 | 0.01273 | 5.59 | 0.00191 |
| 2F | Sp with only head PR | ↓ | 0.26 | 0.61624 | 4.29 | 0.04811 | 10.61 | 0.00001 |
| Vehicle group | Trained group | |||||||
| 2C | Shaft PR | ↓ | 3.98 | 0.05483 | 5.86 | 0.02133 | 5.59 | 0.00191 |
| 3B | Sp with no PR, PSD < 0.15 μm2 | ↑ | 0.74 | 0.39566 | 8.68 | 0.00597 | 2.79 | 0.04841 |
| 4D | Sp without SA, no PR | ↑ | 3.95 | 0.05586 | 14.01 | 0.00072 | 5.30 | 0.00266 |
| 5D | Sp without SA, no astro, head PR | ↓ | 1.43 | 0.24144 | 10.58 | 0.00270 | 5.32 | 0.00260 |
| 5E | Sp without SA, with astro, no PR | ↑ | 2.40 | 0.13117 | 22.53 | 0.00004 | 9.80 | 0.00002 |
Shown are measures of main effects of drug, an effect of drug in the box condition, or an effect of drug in the trained condition in the absence of a training effect. Nonsignificant effects are shown in italics. PR, Polyribosomes; Sp, spine; astro, perisynaptic astroglia.
Results
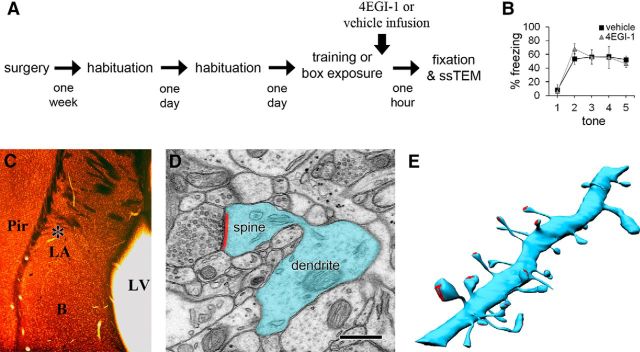
We presented adult rats with either an aversive pavlovian training protocol or equivalent time in the training box, followed immediately by infusion of either vehicle (trained-vehicle and box-vehicle groups) or 4EGI-1 (trained-4EGI-1 and box-4EGI-1 groups), and perfused them with fixative 1 h after the first tone–shock pairing or the equivalent time point for the box control groups (Fig. 1A). Our previous work found that a conditioning paradigm consisting of five tone–shock pairings resulted in a substantial upregulation of polyribosomes in LA dendrites (Ostroff et al., 2010), so we used this paradigm to investigate a possible role for translation initiation. As in the previous study, acquisition of the tone–shock association reached asymptote after the second pairing as measured by the percentage of time the animals spent freezing during the subsequent tone (Fig. 1B). Although we used only three pairings in our previous study showing that 4EGI-1 impairs consolidation (Hoeffer et al., 2011), freezing behavior reached a similar level after the second pairing, and long-term memory results were similar in the two studies. Paired training induced changes in polyribosomes relative to unpaired presentations of tone and shock or exposure to the training box alone (Ostroff et al., 2010). Unpaired training did not alter local translation relative to box exposure but did produce conditioned inhibition learning and associated morphological changes. We therefore chose to use box exposure as a control for learning induced by the paired protocol.
Figure 1.
Experimental procedures. A, Time line of behavioral training, drug infusion, and tissue collection. B, Graph of the percentage of time animals spent freezing during each of the five tones presented during training, showing equivalent acquisition between the vehicle and drug groups. Error bars indicate SEM. C, Tissue embedded for electron microscopy showing sampling location (asterisk) for serial sections. B, Basal amygdala; LV, lateral ventricle; Pir, piriform cortex. D, Electron micrograph (EM) of a dendrite (blue) with a spine forming a synapse (red). Scale bar, 500 nm. E, Three-dimensional rendering of a 10 μm segment of dendrite (blue) and its synapses (red) reconstructed through serial EM sections.
Like other forms of associative memory, pavlovian conditioning consists of a short-term, translation-independent phase that is consolidated into a persistent long-term phase during the first several hours after training. Consolidation requires new protein synthesis during a critical period immediately after training; if translation is blocked during this early period, the memory will decay within hours (Davis and Squire, 1984; Schafe and LeDoux, 2000; Maren et al., 2003; Johansen et al., 2011; Rosenberg et al., 2014). Like traditional translation inhibitors, 4EGI-1 impairs long-term, but not short-term, memory when given immediately after learning, and similarly blocks consolidation of hippocampal long-term potentiation (Hoeffer et al., 2011, 2013). Our previous ssTEM study found upregulated polyribosomes 1 h after training, which is within the window of the drug's action on consolidation processes, so we chose to use that time point in the present study as well.
4EGI-1 interferes specifically with eIF4E–eIF4G interactions, allowing us to pinpoint the effects of cap-dependent initiation, and unlike traditional protein synthesis inhibitors, it does not activate stress kinases (Hoeffer et al., 2011; Sekiyama et al., 2015). In addition, the drug has no effects on baseline synaptic transmission, short-term plasticity, or standard measures of membrane properties or neural excitability in hippocampal slices (Hoeffer et al., 2013). In our previous work with 4EGI-1, we reported that post-training infusion of the drug into the LA impairs consolidation of aversive pavlovian conditioning (Hoeffer et al., 2011).
Because the damage caused by cannula implantation adversely affects preservation of the surrounding tissue for ultrastructural work, we infused the drug into the adjacent lateral ventricle. This also impairs consolidation (C. Hoeffer, K. K. Cowansage, and E. Klann, unpublished observations) and inhibits eIF4E–eIF4G interactions and protein synthesis in the LA (Hoeffer et al., 2011). Tissue from the dorsal tip of the LA was prepared for ssTEM, and spiny dendrites were reconstructed and analyzed in three dimensions (Fig. 1C–E).
4EGI-1 reduces baseline polyribosomes in dendrites and prevents their upregulation after learning. Although translation is primarily cap dependent in eukaryotic cells, both IRES-mediated initiation and reactivation of stalled polyribosomes can occur in dendrites (Sutton and Schuman, 2005; Richter and Coller, 2015). It is conceivable that cap-independent translation predominates in dendrites during consolidation, or even under baseline conditions. If this is the case, 4EGI-1 should have no effect on polyribosomes in LA dendrites, and we therefore began by quantifying them. As in previous studies, polyribosomes were identified morphologically as clusters of at least three dense, irregularly shaped puncta 15–25 nm in diameter, often in rosette, circle, or staggered line formations (Steward and Levy, 1982; Ostroff et al., 2002, 2010). Polyribosomes were found in the shafts of dendrites, as well as in the bases, necks, and heads of dendritic spines (Fig. 2A). As in our previous study (Ostroff et al., 2010), we found more polyribosomes in the trained-vehicle group than in the box-vehicle group (Fig. 2B). This effect did not occur in the 4EGI-1 groups, indicating that the increase was not attributable to transport of preexisting polyribosomes. There were fewer polyribosomes in the box-4EGI-1 group as well as the trained-4EGI-1 group, indicating that ongoing cap-dependent initiation is necessary to maintain baseline numbers.
Polyribosomes in spine heads and dendritic shafts are specifically affected by 4EGI-1
We analyzed polyribosomes by location and found that the effects of training and 4EGI-1 were not equally distributed within dendrites. In dendritic shafts, there were fewer polyribosomes in the 4EGI-1 groups than in the vehicle groups, regardless of training (Fig. 2C). There were more polyribosomes in spine heads in the trained-vehicle group versus the box-vehicle group with no differences in the 4EGI-1 groups, yet polyribosomes in spine bases and necks were increased in both trained groups versus the control groups. Thus, ongoing initiation is necessary to sustain polyribosome levels in dendritic shafts, but not spines. Furthermore, upregulation of polyribosomes in spine heads during consolidation requires new initiation, whereas upregulation in spine bases and necks does not. The effects of training and 4EGI-1 on spine frequency reflected the effects on polyribosome distribution, and there were fewer spines without polyribosomes in the trained-vehicle group versus the box-vehicle group (Fig. 2D).
Although most spines with polyribosomes contained only one, the number of polyribosomes per spine was higher in the trained-vehicle group (Fig. 2E). To determine whether this reflected a tendency for spines to accumulate polyribosomes in multiple locations, we separated spines with polyribosomes in only the head or only in the base or neck from spines with polyribosomes in both locations (Fig. 2F). There were more spines with polyribosomes restricted to either the base or the neck with training regardless of 4EGI-1. This effect was not observed in spines that also had polyribosomes in the head. More spines had polyribosomes only in the head in the trained-vehicle group versus the box-vehicle group, whereas there were fewer of these spines in the box-4EGI-1 group versus the box-vehicle group. Thus, in the control groups, 4EGI-1 depleted polyribosomes in spine heads, except in spines that also had them in the base or neck. In contrast, during consolidation 4EGI-1 did not prevent polyribosomes from accumulating in spine bases and necks, except in spines that also had them in the head. Very few spines had multiple polyribosomes in a single location, and among these there were more in the head in the trained-vehicle group (Fig. 2G).
Polyribosomes in the bases and necks of the smallest spines are insensitive to 4EGI-1
Many neurological and intellectual disorders, including several that involve dysregulated translation, are associated with abnormal spine morphology (Fiala et al., 2002; Blanpied and Ehlers, 2004; Penzes et al., 2011; Lai and Ip, 2013). Excessive eIF4E–eIF4G interactions caused by eIF4E overexpression lead to proliferation of small spines in prefrontal cortex, suggesting that cap-dependent translation regulates spine size (Santini et al., 2013). Measures of spine morphology such as volume, head diameter, and length are correlated with synapse size, and synapse enlargement is in turn associated with synapse strengthening, stabilization, and memory formation (Harris and Stevens, 1989; Ostroff et al., 2010; Bailey et al., 2015; Segal, 2016). Synapse size is also correlated with glutamate receptor content, making it a more direct proxy for synapse function (Nicholson and Geinisman, 2009; Nusser, 2000). We used the two-dimensional area of the postsynaptic density (PSD) as a measure of synapse size and, by extension, spine size to investigate the relationship between synapse enlargement and cap-dependent translation (Fig. 3A). PSD area was quantified by multiplying the length of the PSD profile on each section by the section thickness for synapses cut in cross section, or by tracing the area of synapses cut obliquely.
Synapse size is not normally distributed, so for a more meaningful analysis we binned spine frequency by synapse size rather than comparing means. For spines without polyribosomes, there were fewer of the very smallest spines with training in the trained-vehicle group versus the box-vehicle group, whereas there were larger spines in the drug groups regardless of training (Fig. 3B). Among spines with polyribosomes in the base or neck, there were more of the smallest spines in the trained groups regardless of drug, whereas there were fewer slightly larger spines in the drug groups (Fig. 3C). There were more very small and very large spines with head polyribosomes in the trained-vehicle group versus the box-vehicle group, and no effects of 4EGI-1 on these spines (Fig. 3D). Thus, the increase in polyribosomes in spine heads occurred in both very small and very large spines, whereas the increase in spine bases and necks was restricted to very small spines. The accumulation of polyribosomes in spine heads was cap dependent regardless of synapse size. The loss of spines with polyribosomes in the drug groups was balanced by increased numbers of spines without polyribosomes, meaning that maintenance of spines is cap independent.
The very largest spines in the LA contain a spine apparatus (Fig. 4A,B), which is a membranous organelle that connects the synapse to the smooth endoplasmic reticulum in the dendrite (Westrum et al., 1980; Spacek, 1985; Ostroff et al., 2010). Its function is not understood, but there is evidence that it plays a role in synaptic plasticity and memory (Jedlicka et al., 2008; Ostroff et al., 2010). Although large spines specifically are thought to be sites of memory storage (Bourne and Harris, 2007; Segal, 2016), our synapse size groupings were dictated by the size distribution, and the individual bins have no particular functional significance. We therefore turned to the spine apparatus as a potential marker of functionally enhanced synapses. Consistent with the occurrence of the spine apparatus in large spines, there were more spines with a spine apparatus with head polyribosomes in the trained-vehicle versus the box-vehicle group, but no effects of training or drug on base and neck polyribosomes (Fig. 4C). Among spines without a spine apparatus, there were also more spines with head polyribosomes in the trained-vehicle group only and more spines with base or neck polyribosomes in both trained groups (Fig. 4D). Spines without polyribosomes were not differentially affected by the presence of a spine apparatus, reflecting the shift toward these spines in the drug groups across the distribution of synapse sizes (Fig. 3B). Between our analyses of synapse size and the spine apparatus, it is clear that 4EGI-1 affects polyribosome localization differentially depending on spine morphology. Figure 4E shows the means in Figure 4, C and D, in a single plot, which depicts the accumulation of polyribosomes in spines in the trained-vehicle group as well as the substantial spinogenesis that occurred in the trained-4EGI-1 group.
Inhibition of translation initiation after training induces proliferation of synapses with astrocytes. Astrocytic processes intercalate extensively throughout the neuropil, sometimes making direct contact with the synaptic cleft (Fig. 5A; Ghézali et al., 2016). In a previous study, we found that the number of spines with a spine apparatus whose synapses lacked astrocytic contacts increased during consolidation and that upregulation of polyribosomes was specific to these spines (Ostroff et al., 2014). This suggests an inverse association between astrocytes and learning-induced polyribosomes but does not address whether they are regulated independently. If synapse activation during learning induces local translation and retraction of astrocytes from synapses in parallel, there should be no effect of 4EGI-1 on synaptic astrocytes after training. To investigate this, we examined spine density with respect to astrocytes at the synapse, the spine apparatus, and polyribosome location. Of the spines with a spine apparatus in the box-vehicle group, most had astrocytes at the synapse and lacked polyribosomes (Fig. 5B). There were more spines with no astrocyte and head polyribosomes in the trained-vehicle group, consistent with our previous study. This was blocked by 4EGI-1, yet there were more spines with no astrocyte and no polyribosomes in both drug groups. There were no effects of either training or 4EGI-1 on spines with a spine apparatus and astrocytes at the synapse (Fig. 5C). The lack of any effects of training in the 4EGI-1 groups suggests that the shift away from perisynaptic astrocytes is specifically associated with polyribosomes in spine heads.
In contrast, the majority of spines without a spine apparatus in the box-vehicle group did not have astrocytes at the synapse. These spines were similarly stable in number, although there were fewer with polyribosomes in the head in the trained-4EGI-1 group versus the trained-vehicle group (Fig. 5D). All other effects in these spines involved spines that had synaptic astrocytes, including the higher frequency of spines with base or neck polyribosomes in the trained groups and of spines with head polyribosomes in the trained-vehicle group (Fig. 5E). There was also a substantial increase in spines with no polyribosomes in the trained-4EGI-1 group, the only increase in spine density specific to the combination of training and 4EGI-1. The interactions between training and 4EGI-1 suggest that for small spines perisynaptic astrocytes are regulated independently from polyribosomes in spine heads. Thus, the effects of training and 4EGI-1 on local translation are associated with synaptic astrocytes in small spines and with the lack of synaptic astrocytes in large spines, and perisynaptic astrocytes have different roles in these spines.
As a whole, our data depict distinct patterns of polyribosome localization with respect to cap-dependent initiation and spine morphology. Table 1 summarizes the effects of training that were sensitive to 4EGI-1, showing a net upregulation of polyribosomes carried primarily by their accumulation in the heads of two spine types: very large spines without perisynaptic astrocytes and very small spines with perisynaptic astrocytes. Training followed by inhibition of new cap-dependent translation did not produce these effects and instead resulted in proliferation of the same small spines. These spines also contained the only cap-independent polyribosomes, which accumulated in their bases and necks after training (Table 2). Independent of training, the drug caused a net depletion of polyribosomes (Fig. 2B). This occurred specifically in dendritic shafts and the heads of some spines, with the notable exception of those with additional polyribosomes in the base and neck (Table 3).
Discussion
Dendrites contain over 2500 mRNAs and a diverse assortment of mRNA targeting mechanisms, suggesting that dendritic translation serves multiple functions and could be regulated with a high degree of spatial specificity (Hirokawa, 2006; Poon et al., 2006; Bramham and Wells, 2007; Martin and Ephrussi, 2009; Doyle and Kiebler, 2011; Cajigas et al., 2012; Buxbaum et al., 2015). The hours after learning are characterized by a number of orchestrated regulatory cascades that transform synaptic activity into a stable memory trace, and local translation could serve to direct these events on the scale of a single synapse. We have taken advantage of the rich morphological detail provided by ssTEM to identify in vivo patterns of polyribosome distribution within and among dendritic spines with respect to cap-dependent translation initiation during learning. Our data reveal multiple pools of cap-dependent and cap-independent polyribosomes associated with different spine types, demonstrating separate functional identities of spines and polyribosomes and suggesting that translational control is exerted at the subspine level.
In our control groups, 4EGI-1 depleted polyribosomes by 40% within 30 min of infusion, indicating that substantial turnover occurs during this time period and that replenishment is cap dependent. Training more than doubled dendritic polyribosomes, and the drug's nearly complete blockade of this effect indicates that upregulation of local translation during consolidation is predominantly cap dependent. It also confirms that the residual polyribosomes are not an artifact of incomplete drug penetration but represent a discrete drug-insensitive population. Along with our observation of specific localization patterns between the two groups of polyribosomes, this suggests that they represent different mRNAs, different regulatory mechanisms, or both.
The simplest explanation for the cap-independent polyribosomes in our dataset is that they represent cap-dependent translation that was initiated before drug infusion and is progressing at a slow rate or on very long mRNAs. Although the rate of translation in LA dendrites is unknown, a study of 35S-methionine/cysteine incorporation in whole-brain lysates under initiation blockade showed nearly complete runoff of actively translating ribosomes within 30 min (Udagawa et al., 2013). If translation occurs at a comparable rate in LA dendrites in vivo, ribosomes that were actively translating during drug infusion would have been gone by the time the rats were killed.
Cap-independent polyribosomes could arise by IRES-mediated initiation, which bypasses the eIF4E–eIF4G binding step and is known to occur in several dendritic mRNAs (Pinkstaff et al., 2001; Sutton and Schuman, 2005). Some dendritic mRNAs appear to be transported from the soma in the form of stalled polyribosomes, which can remain in suspended animation until reactivated by synaptic activity or other signals (Richter and Coller, 2015). If the 4EGI-1-resistant polyribosomes in our study were stalled, we might expect them to be activated, and thus depleted, by learning. We did not observe this, suggesting that existing stalled, cap-dependent polyribosomes were not available for consolidation processes. Another possibility is that learning induces active polyribosomes to stall, which would result in accumulation of cap-independent polyribosomes. For example, FMRP, an mRNA-binding translation repressor whose dysfunction causes Fragile X syndrome, is suspected to regulate synapse function by stalling polyribosomes in response to activity (Darnell et al., 2011; Richter and Coller, 2015). The cap-independent pool could thus represent polyribosomes with various profiles: IRES initiated and actively translating, stalled in the soma before transport, or stalled in the dendrite.
One eIF4E effector that is especially important in learning is the mechanistic target of rapamycin complex 1 (mTORC1), which regulates cap-dependent initiation of certain mRNAs (Hoeffer and Klann, 2010). Two recent studies have identified mRNAs targeted by mTORC1; these include a large number of translation factors and ribosomal proteins, but none containing an IRES (Hsieh et al., 2012; Thoreen et al., 2012). Depending on how much dendritic translation machinery is downstream of mTORC1 and how quickly it turns over, this could explain some of the effects of 4EGI-1. If synthesis of dendritic ribosomes is cap dependent by way of mTORC1, the drug would affect both cap-dependent and IRES-mediated polyribosomes, but not stalled ones.
Polyribosomes in spine heads are particularly suggestive of synapse-specific translation regulation. The accumulation of polyribosomes in the heads of large spines in particular could reflect new translation directly supporting synapses that are strengthened during learning. The lack of perisynaptic astrocytes at these synapses is consistent with our previous study and may be a feature of enlarging or unstable large synapses (Ostroff et al., 2014). These spines comprised 7% of the population in the box-vehicle group and 14% in the trained-vehicle group, and although 4EGI-1 did deplete them of polyribosomes, it did not eliminate them. This suggests that, at least in the short term, local translation is not necessary to sustain spine size. The complete cap dependence of polyribosome upregulation in these spines again echoes the cap dependence of consolidation, supporting the connection between synapse enlargement, new translation, and memory.
In contrast to cap-dependent polyribosomes, cap-independent polyribosomes were upregulated specifically in the bases and necks of small spines with perisynaptic astrocytes. Imaging of cortical dendrites in vivo has shown that small spines have a much shorter half-life than large spines (Holtmaat et al., 2005). Along with the general association between astrocytes and synaptogenesis (Ullian et al., 2004), this suggests a distinct pattern of translation associated with unstable, possibly relatively young spines. These spines also accumulated cap-dependent polyribosomes in their heads after training, but when this was blocked, the spines proliferated. This is reminiscent of the shift toward small, immature spines seen in a number of intellectual disabilities, most notably Fragile X syndrome, which may result from dysregulated translation (Irwin et al., 2000; Fiala et al., 2002).
Because different plasticity processes are presumably happening in different spine types and locations, it is likely that their polyribosomes represent different mRNAs. Very few studies have examined the localization of individual mRNAs at the subspine level, but there is evidence that they can be precisely targeted. At least some dendritic mRNAs are trafficked singly, and their localization is regulated by a complex code of signaling and transport mechanisms (Krichevsky and Kosik, 2001; Hirokawa, 2006; Wang et al., 2010; Doyle and Kiebler, 2011; Mikl et al., 2011; Batish et al., 2012; Hutten et al., 2014). In cultured neurons, both CaMKIIα and β-actin mRNAs have been observed in spine heads (Tiruchinapalli et al., 2003; Kao et al., 2010), whereas Arc and dendrin mRNAs are primarily restricted to the base and proximal neck of spines (Dynes and Steward, 2012).
Arc, dendrin, and CaMKIIα are capable of IRES-mediated translation (Pinkstaff et al., 2001), which could explain the presence of cap-independent polyribosomes in spine bases. Arc is an immediate-early gene whose mRNA is transcribed and transported to dendrites in response to synaptic activity in adult hippocampus in vivo (Bramham et al., 2010) and is upregulated in the LA after aversive pavlovian conditioning (Ploski et al., 2008). In cultured hippocampal neurons, Arc protein is associated with inactive synapses and accumulates in spines that do not expand after LTP-inducing stimulation (Okuno et al., 2012). We found that cap-independent polyribosomes accumulated specifically in the bases and necks of the smallest spines, consistent with IRES-mediated translation at inactive synapses. Dynes and Steward (2012), however, found that puromycin, which disassembles active ribosomes, did not eliminate polyribosomes at spine bases in a stimulated area of dentate gyrus, supporting the presence of stalled polyribosomes at spine bases.
In our biochemical experiments, we found that infusion of 4EGI-1 into the ventricles of naive animals reduced eIF4E–eIF4G interactions by ∼50% at 30 min and protein synthesis by ∼40% at 45 min, similar to its effects on polyribosomes (Hoeffer et al., 2011). Training induced an ∼130% increase in eIF4E–eIF4G interactions that was mostly blocked by 4EGI-1, mirroring the ∼130% increase in dendritic polyribosomes that was abolished by 4EGI-1 in this study. Thus, dendritic polyribosomes depend on cap-dependent initiation to a similar extent and with similar kinetics as average levels of translation in the tissue. This could reflect an even distribution of translation rates and control mechanisms throughout the tissue, a substantial fraction of overall translation occurring in dendrites, or both. The amount of translation that occurs in dendrites relative to neuronal cell bodies, glial cells, and other cells in brain tissue remains an open question.
It is important to note that although polyribosomes have been traditionally viewed as the main, if not only, sites of translation in the cytoplasm, this is not necessarily the case. One study of cultured hippocampal neurons reported a substantial amount of translation despite the fact that nearly all of the polyribosomes in the soma and dendrites were stalled, and the authors speculated that this was attributable to translation by monosomes (Graber et al., 2013). Until recently, it was unknown whether this was possible; however, a recent study in yeast revealed that translation does occur on monosomes, with a strong bias toward low-abundance regulatory proteins (Heyer and Moore, 2016). If this is the case in neurons, our counts of polyribosomes may dramatically underestimate the amount of translation in dendrites. More work is needed to decipher the complexity of translational control at individual synapses, but our results suggest that it is function and synapse specific.
Footnotes
This work was supported by National Institutes of Health Grants MH083583 and MH094965 (L.E.O.), NS034007 and NS047384 (E.K.), and NS086933 and DA036673 (C.H.); Simons Foundation Grant SFARI 27444 (C.H.); Brain and Behavior Research Foundation Grant 21069 (C.H.); and Alzheimer's Association Grant MNIRGDP-12-258900 (C.H.). We are grateful to Elizabeth Perry and Robert Smith for expert technical assistance.
The authors declare no competing financial interests.
References
- Bailey CH, Kandel ER, Harris KM (2015) Structural components of synaptic plasticity and memory consolidation. Cold Spring Harb Perspect Biol 7:a021758. 10.1101/cshperspect.a021758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S (2012) Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A 109:4645–4650. 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Ehlers MD (2004) Microanatomy of dendritic spines: emerging principles of synaptic pathology in psychiatric and neurological disease. Biol Psychiatry 55:1121–1127. 10.1016/j.biopsych.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM (2007) Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol 17:381–386. 10.1016/j.conb.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG (2007) Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 8:776–789. 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K (2010) The Arc of synaptic memory. Exp Brain Res 200:125–140. 10.1007/s00221-009-1959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M (2014) Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 37:17–38. 10.1146/annurev-neuro-071013-014100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Yoon YJ, Singer RH, Park HY (2015) Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol 25:468–475. 10.1016/j.tcb.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74:453–466. 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61:10–26. 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC. (2011) Defects in translational regulation contributing to human cognitive and behavioral disease. Curr Opin Genet Dev 21:465–473. 10.1016/j.gde.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146:247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR (1984) Protein synthesis and memory: a review. Psychol Bull 96:518–559. 10.1037/0033-2909.96.3.518 [DOI] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA (2011) Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 30:3540–3552. 10.1038/emboj.2011.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O (2012) Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol 520:3105–3119. 10.1002/cne.23073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC. (2005) Reconstruct: a free editor for serial section microscopy. J Microsc 218:52–61. 10.1111/j.1365-2818.2005.01466.x [DOI] [PubMed] [Google Scholar]
- Fiala JC, Harris KM (2001) Cylindrical diameters method for calibrating section thickness in serial electron microscopy. J Microsc 202:468–472. 10.1046/j.1365-2818.2001.00926.x [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM (2002) Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev 39:29–54. 10.1016/S0165-0173(02)00158-3 [DOI] [PubMed] [Google Scholar]
- Gal-Ben-Ari S, Kenney JW, Ounalla-Saad H, Taha E, David O, Levitan D, Gildish I, Panja D, Pai B, Wibrand K, Simpson TI, Proud CG, Bramham CR, Armstrong JD, Rosenblum K (2012) Consolidation and translation regulation. Learn Mem 19:410–422. 10.1101/lm.026849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghézali G, Dallérac G, Rouach N (2016) Perisynaptic astroglial processes: dynamic processors of neuronal information. Brain Struct Funct 221:2427–2442. 10.1007/s00429-015-1070-3 [DOI] [PubMed] [Google Scholar]
- Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS (2013) Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci U S A 110:16205–16210. 10.1073/pnas.1307747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppo R, Richter JD (2009) Translational control from head to tail. Curr Opin Cell Biol 21:444–451. 10.1016/j.ceb.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Stevens JK (1989) Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci 9:2982–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ (2016) Redefining the translational status of 80S monosomes. Cell 164:757–769. 10.1016/j.cell.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Hirokawa N. (2006) mRNA Transport in dendrites: RNA granules, motors, and tracks. J Neurosci 26:7139–7142. 10.1523/JNEUROSCI.1821-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E (2010) mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 33:67–75. 10.1016/j.tins.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, Rodriguez R, Schmidt EK, Klosi E, Chorev M, Lloyd RE, Pierre P, Wagner G, LeDoux JE, Klann E (2011) Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc Natl Acad Sci U S A 108:3383–3388. 10.1073/pnas.1013063108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Santini E, Ma T, Arnold EC, Whelan AM, Wong H, Pierre P, Pelletier J, Klann E (2013) Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J Neurophysiol 109:68–76. 10.1152/jn.00342.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80:648–657. 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K (2005) Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45:279–291. 10.1016/j.neuron.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485:55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Sharangdhar T, Kiebler M (2014) Unmasking the messenger. RNA Biol 11:992–997. 10.4161/rna.32091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT (2000) Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex 10:1038–1044. 10.1093/cercor/10.10.1038 [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Vlachos A, Schwarzacher SW, Deller T (2008) A role for the spine apparatus in LTP and spatial learning. Behav Brain Res 192:12–19. 10.1016/j.bbr.2008.02.033 [DOI] [PubMed] [Google Scholar]
- Johansen JP, Cain CK, Ostroff LE, LeDoux JE (2011) Molecular mechanisms of fear learning and memory. Cell 147:509–524. 10.1016/j.cell.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao DI, Aldridge GM, Weiler IJ, Greenough WT (2010) Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci U S A 107:15601–15606. 10.1073/pnas.1010564107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696. 10.1016/S0896-6273(01)00508-6 [DOI] [PubMed] [Google Scholar]
- Lai K-O, Ip NY (2013) Structural plasticity of dendritic spines: the underlying mechanisms and its dysregulation in brain disorders. Biochim Biophys Acta 1832:2257–2263. 10.1016/j.bbadis.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B (2011) Local RNA translation at the synapse and in disease. J Neurosci 31:16086–16093. 10.1523/JNEUROSCI.4105-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maren S, Ferrario CR, Corcoran KA, Desmond TJ, Frey KA (2003) Protein synthesis in the amygdala, but not the auditory thalamus, is required for consolidation of Pavlovian fear conditioning in rats. Eur J Neurosci 18:3080–3088. 10.1111/j.1460-9568.2003.03063.x [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136:719–730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. (2000) Memory–a century of consolidation. Science 287:248–251. 10.1126/science.287.5451.248 [DOI] [PubMed] [Google Scholar]
- Mikl M, Vendra G, Kiebler MA (2011) Independent localization of MAP2, CaMKIIα and β-actin RNAs in low copy numbers. EMBO Rep 12:1077–1084. 10.1038/embor.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DA, Geinisman Y (2009) Axospinous synaptic subtype-specific differences in structure, size, ionotropic receptor expression, and connectivity in apical dendritic regions of rat hippocampal CA1 pyramidal neurons. J Comp Neurol 512:399–418. 10.1002/cne.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser Z. (2000) AMPA and NMDA receptors: similarities and differences in their synaptic distribution. Curr Opin Neurobiol 10:337–341. 10.1016/S0959-4388(00)00086-6 [DOI] [PubMed] [Google Scholar]
- Okuno H, Akashi K, Ishii Y, Yagishita-Kyo N, Suzuki K, Nonaka M, Kawashima T, Fujii H, Takemoto-Kimura S, Abe M, Natsume R, Chowdhury S, Sakimura K, Worley PF, Bito H (2012) Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell 149:886–898. 10.1016/j.cell.2012.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Fiala JC, Allwardt B, Harris KM (2002) Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron 35:535–545. 10.1016/S0896-6273(02)00785-7 [DOI] [PubMed] [Google Scholar]
- Ostroff LE, Cain CK, Bedont J, Monfils MH, Ledoux JE (2010) Fear and safety learning differentially affect synapse size and dendritic translation in the lateral amygdala. Proc Natl Acad Sci U S A 107:9418–9423. 10.1073/pnas.0913384107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Manzur MK, Cain CK, Ledoux JE (2014) Synapses lacking astrocyte appear in the amygdala during consolidation of pavlovian threat conditioning. J Comp Neurol 522:2152–2163. 10.1002/cne.23523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14:285–293. 10.1038/nn.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay S, Webster H (1991) The fine structure of the nervous system, Ed 3 New York: Oxford UP. [Google Scholar]
- Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA (2001) Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci U S A 98:2770–2775. 10.1073/pnas.051623398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploski JE, Pierre VJ, Smucny J, Park K, Monsey MS, Overeem KA, Schafe GE (2008) The activity-regulated cytoskeletal-associated protein (Arc/Arg3.1) is required for memory consolidation of pavlovian fear conditioning in the lateral amygdala. J Neurosci 28:12383–12395. 10.1523/JNEUROSCI.1662-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC (2006) Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci 26:13390–13399. 10.1523/JNEUROSCI.3432-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Coller J (2015) Pausing on polyribosomes: make way for elongation in translational control. Cell 163:292–300. 10.1016/j.cell.2015.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg T, Gal-Ben-Ari S, Dieterich DC, Kreutz MR, Ziv NE, Gundelfinger ED, Rosenblum K (2014) The roles of protein expression in synaptic plasticity and memory consolidation. Front Mol Neurosci 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E (2013) Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Klann E (2014) Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci 122:131–167. 10.1016/B978-0-12-420170-5.00005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, LeDoux JE (2000) Memory consolidation of auditory pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. J Neurosci 20:RC96(1–5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M. (Available June 14, 2016) Dendritic spines: morphological building blocks of memory. Neurobiol Learn Mem. Advance online publication. 10.1016/j.nlm.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Sekiyama N, Arthanari H, Papadopoulos E, Rodriguez-Mias RA, Wagner G, Léger-Abraham M (2015) Molecular mechanism of the dual activity of 4EGI-1: dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc Natl Acad Sci U S A 112:E4036–45. 10.1073/pnas.1512118112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L (2006) Intracellular trafficking of RNA in neurons. Traffic 7:1581–1589. 10.1111/j.1600-0854.2006.00500.x [DOI] [PubMed] [Google Scholar]
- Spacek J. (1985) Three-dimensional analysis of dendritic spines. II. Spine apparatus and other cytoplasmic components. Anat Embryol 171:235–243. 10.1007/BF00341418 [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 2:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2005) Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol 64:116–131. 10.1002/neu.20152 [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127:49–58. 10.1016/j.cell.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ (2003) Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, Lorenz LJ, Bassell GJ, Akbarian S, Chattarji S, Klann E, Richter JD (2013) Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med 19:1473–1477. 10.1038/nm.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Christopherson KS, Barres BA (2004) Role for glia in synaptogenesis. Glia 47:209–216. 10.1002/glia.20082 [DOI] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS (2010) Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33:173–182. 10.1016/j.tins.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrum LE, Jones DH, Gray EG, Barron J (1980) Microtubules, dendritic spines and spine apparatuses. Cell Tissue Res 208:171–181. [DOI] [PubMed] [Google Scholar]