Abstract
Shigella sonnei is a major contributor to the global burden of diarrhoeal disease, generally associated with dysenteric diarrhoea in developed countries but also emerging in developing countries. The reason for the recent success of S. sonnei is unknown, but is likely catalysed by its ability to acquire resistance against multiple antimicrobials. Between 2011 and 2013, S. sonnei exhibiting resistance to fluoroquinolones, the first-line treatment recommended for shigellosis, emerged in Bhutan. Aiming to reconstruct the introduction and establishment of fluoroquinolone-resistant S. sonnei populations in Bhutan, we performed whole-genome sequencing on 71 S. sonnei samples isolated in Bhutan between 2011 and 2013.We found that these strains represented an expansion of a clade within the previously described lineage III, found specifically in Central Asia. Temporal phylogenetic reconstruction demonstrated that all of the sequenced Bhutanese S. sonnei diverged from a single ancestor that was introduced into Bhutan around 2006. Our data additionally predicted that fluoroquinolone resistance, conferred by mutations in gyrA and parC, arose prior to the introduction of the founder strain into Bhutan. Once established in Bhutan, these S. sonnei had access to a broad gene pool, as indicated by the acquisition of extended-spectrum β-lactamase-encoding plasmids and genes encoding type IV pili. The data presented here outline a model for the introduction and maintenance of fluoroquinolone-resistant S. sonnei in a new setting. Given the current circulation of fluoroquinolone-resistant S. sonnei in Asia, we speculate that this pattern of introduction is being recapitulated across the region and beyond.
Keywords: Bhutan, fluoroquinolone, resistance, Shigella sonnei
Data Summary
1. Whole-genome sequence reads of Shigella sonnei used in this study have been deposited to the European Nucleotide Archive. The run accession numbers and related metadata are detailed in Table S1 which has been deposited in FigShare: http://figshare.com/articles/The_introduction_and_establishment_of_fluoroquinolone_resistant_Shigella_sonnei_into_Bhutan/1610693
Impact Statement
The bacterium Shigella sonnei is a major global cause of diarrhoeal disease. This organism has generally been associated with infections in developed countries, but is currently dramatically expanding into industrializing countries. The mechanisms underpinning the spread of S. sonnei are not precisely understood, but we hypothesize that their spread is partly facilitated by resistance to antimicrobials, including the current therapeutic choice to combat Shigella, the fluoroquinolones. To understand how the fluoroquinolone-resistant S. sonnei may become established in a new location, we decoded the total DNA sequences of 71 S. sonnei samples isolated in Bhutan in South Asia between 2011 and 2013. We found that these strains were likely descended from a single isolate that was introduced into Bhutan around 2006. Furthermore, we additionally found that fluoroquinolone resistance emerged prior to the introduction of this founder S. sonnei into Bhutan and these strains then became established, gaining additional genes and resistance to other antimicrobials. Our data describe how drug-resistant S. sonnei may become resident in new locations, and we predict that a similar situation is occurring currently with the epidemic of fluoroquinolone-resistant S. sonnei in Asia and beyond.
Introduction
It has recently been estimated that the global burden of diarrhoeal disease in children < 5 years of age is ∼1.7 billion new infections with 0.7 million deaths per year (Walker et al., 2013). These figures make diarrhoeal diseases, after respiratory tract infections, the second most common cause of mortality in young children in low-income countries (Liu et al., 2012; WHO, 2014). Further contemporary data from the Global Enteric Multicentre Study, the largest prospective case–control study on paediatric diarrhoeal illnesses in sub-Saharan Africa and South Asia ever conducted, identified Shigella to be amongst the most common diarrhoeal pathogens in these high-incidence settings (Kotloff et al., 2012, 2013).
The members of the genus Shigella are Gram-negative pathogens that are closely related to Escherichia coli (Pupo et al., 2000) and cause dysenteric diarrhoea (stools containing blood and/or mucus) via a cascade of virulence factors encoded principally on a large signature virulence plasmid (Sansonetti et al., 1982). The genus contains four species: Shigella sonnei, Shigella flexneri, Shigella boydii and Shigella dysenteriae, with S. sonnei and S. flexneri dominating in developed and developing countries, respectively. However, this pattern is changing, as S. sonnei is currently replacing S. flexneri in countries undergoing economic transition (Vinh et al., 2009; Thompson et al., 2015). This species replacement has positive and negative ramifications. By replacing S. flexneri, which is very diverse with multiple serotypes with respect to the single serotype S. sonnei (Connor et al., 2015), as the leading cause of dysentery means that it may be possible to control disease through vaccination (Levine et al., 2007). However, S. sonnei is also highly adept at acquiring antimicrobial resistance genes and resistance-associated mutations (Huang et al., 2005; Seol et al., 2006; Nguyen et al., 2010), limiting treatment options and making control through antimicrobial treatment alone improbable.
Recent advances in whole-genome sequencing (WGS) and analytical methodologies have enhanced our understanding of the epidemiology, international spread and antimicrobial susceptibility trends of S. sonnei (Holt et al., 2012, 2013). We now know that S. sonnei is formed of three main lineages (I, II and III), which are all related to a common ancestor that arose in Europe ∼400 years ago (Holt et al., 2012). Lineage III, which is multidrug-resistant (MDR), now dominates globally (Holt et al., 2012). We have also previously shown that once introduced into a new geographical region, into an apparently naive human population, S. sonnei can establish new isolated clonal populations which evolve locally (Holt et al., 2013). Antimicrobial resistance is a key driver of the recent S. sonnei global expansions marked by the acquisition of antimicrobial resistance genes and/or mutations in genes encoding the target proteins for antimicrobials (Pu et al., 2015).
Bhutan is a lower middle-income country in South Asia, sandwiched between China and the northwest Indian states of Assam and West Bengal. In close proximity to Nepal ( < 70 km of Indian territory separates the two countries), Bhutan is a Himalayan nation with an estimated population of 750 000 people, of which the majority (69 %) live in remote rural locations (Office of the Census Commissioner – Royal Government of Bhutan, 2006). Like other low-income countries in the region, diarrhoeal disease is a major public health problem in Bhutan, and both S. flexneri and S. sonnei are endemic (Marfin et al., 1994; Ruekit et al., 2014). Between March 2011 and October 2013, a diarrhoeal disease surveillance study was conducted in the Bhutanese capital, Thimphu. During this study, there was an increased number of S. sonnei isolated between June and July. Antimicrobial susceptibility testing, PFGE and plasmid replicon typing of 29 of these S. sonnei isolates showed that all were identical by PFGE pattern and antimicrobial susceptibility (Ruekit et al., 2014). All these S. sonnei isolates were resistant to ciprofloxacin (MIC ≥ 3 μg μl− 1), which was infrequently observed in strains circulating globally during the same period (Ruekit et al., 2014). This study suggested that these S. sonnei isolates were closely related and may have been a recent introduction into Bhutan.
To test the hypothesis that these fluoroquinolone-resistant S. sonnei had recently been introduced to Bhutan and to further characterize these Bhutanese isolates in the context of global S. sonnei, we performed WGS on 71 S. sonnei isolated in Bhutan between 2011 and 2013.
Methods
Sample collection
The Research Ethics Board of Health in Bhutan and the Institutional Review Board of the Walter Reed Army Institute of Research provided ethical approval for the study. Stool samples were collected as described previously (Ruekit et al., 2014). Stool samples were collected from cases and controls, and cultured on MacConkey and Hektoen agar, and incubated overnight at 37 °C. Non-lactose-fermenting colonies were confirmed to be Shigella by standard biochemical testing; serotype was identified by O-antigen agglutination (Denka Seiken).
Stool samples were collected during a diarrhoea surveillance study conducted at the Jigme Dorji Wangchuk National Referral Hospital (JDWNRH) in Thimphu, Bhutan, between March 2011 and October 2013, as described previously (Ruekit et al., 2014). Briefly, children aged between 3 months and 5 years presenting to JDWNRH with acute diarrhoea (three or more loose stools in the previous 24 h, with symptoms lasting ≤ 72 h at time of recruitment) and those with no diarrhoeal symptoms in the previous 2 weeks were eligible for recruitment as cases and controls, respectively (informed consent permitting).
All Shigella isolates were subjected to antimicrobial susceptibility testing using the disk diffusion method as recommended by Clinical and Laboratory Standards Institute guidelines (CLSI, 2012). The following antimicrobials were selected for susceptibility testing: azithromycin, nalidixic acid, ciprofloxacin, ampicillin, trimethoprim/sulfamethoxazole, ceftriaxone, streptomycin and tetracycline.
WGS
During the surveillance study a total of 112 Shigella were isolated (74 S. sonnei, 30 S. flexneri, seven S. boydii and one S. dysenteriae); the 74 S. sonnei were selected for WGS. The majority (64/74) of the selected S. sonnei were isolated from symptomatic diarrhoeal stool samples with the remaining strains isolated from asymptomatic infections.
Genomic DNA was extracted from all S. sonnei isolates using a Wizard Genomic DNA Extraction kit (Promega) according to the manufacturer's recommendations. For each sample, 2 μg genomic DNA was subjected to WGS on an Illumina HiSeq2000 platform, following the manufacturer's protocols to generate 100 bp paired-end reads (Quail et al., 2008). All reads were mapped to the reference sequence of S. sonnei Ss046 (GenBank accession number NC_007384) using smalt (version 0.7.4). Candidate SNPs were called against the reference sequence and filtered using SAMtools (Li et al., 2009). Low-quality SNPs were removed according to the following criteria: consensus quality < 50, mapping quality < 30, ratio of SNPs to reads at a position < 75 %, read depth < 4, number of reads per strand < 2, strand bias < 0.001, mapping bias < 0.001 or tail bias < 0.001. Three samples failed due to insufficient read depth, resulting in a final sample set including 71 S. sonnei whole-genome sequences from Bhutan.
Phylogenetic analysis, temporal analysis and Bayesian phylogenetic inference
In order to place the Bhutanese isolates in the context of global diversity, we aligned the Bhutanese S. sonnei sequences with publicly available sequences utilized in a previous global analysis (Holt et al., 2012). An initial phylogeny was inferred with these data using RAxML under the GTRGAMMA model. To further assess the relationship of the Bhutanese isolates within Central Asia, the previous alignment was then subsampled to include only the 71 Bhutanese sequences along with 20 other closely related Central Asia sequences. Previously described mobile genetic elements (Holt et al., 2012) and potential recombinant regions were removed from the alignment using Gubbins (Croucher et al., 2014). Further removal of gaps, indeterminate bases, invariant sites and 17 Bhutanese sequences that were identical to others in the dataset resulted in a final alignment of 996 SNPs across 74 taxa. A maximum-likelihood phylogeny was inferred from these sequences using garli (Zwickl, 2006) with 1000 bootstrap replicates under the TVM (transversion) model of nucleotide substitution, which was determined to be the best-fit model using jModelTest (Posada, 2008).
In order to thoroughly examine the population and temporal structure of Bhutanese S. sonnei and the population from which they emerged, all sequences from Bhutan, including identical sequences removed in the previous phylogeny, were aligned with the six most closely related isolates from other countries and trimmed down to include only the 544 SNPs present in the alignment of 77 taxa. These data were then utilized for maximum-likelihood reconstruction using PhyML under the TVM+Γ4 substitution model (Guindon et al., 2010). In order to investigate whether temporal signal was embedded in the resulting maximum-likelihood phylogeny, we applied Path-O-Gen (version 1.4) to assess the linear relationship between root-to-tip divergence and date of isolation (in day, month, year).
Bayesian MCMC (Markov Chain Monte Carlo) deployed in beast (version 1.8.0) (Drummond & Rambaut, 2007) was then used to infer the evolutionary dynamics of the aforementioned dataset, with a particular focus on Bhutanese S. sonnei. To determine the best-fit priors to the data at hand, multiple beast runs were conducted using the TVM or GTR nucleotide substitution model with constant, exponential growth or Bayesian skyline demographic history priors, in combination with either a strict or a relaxed molecular clock (uncorrelated lognormal distribution) (Drummond & Rambaut, 2007). For each beast analysis, marginal likelihood estimation was conducted using both path sampling and stepping-stone sampling approaches to facilitate model selection (Baele et al., 2012, 2013). Bayes factor comparison indicated the TVM+Γ4 substitution model with a relaxed log-normal molecular clock and a piecewise Bayesian skyline demographic model as the best-fit model to the data. Analyses using these priors were then performed in triplicate using a continuous 100 million generation MCMC chain with samples taken every 10 000 generations and parameter convergence (indicated by effective sample size values >200) was assessed in Tracer (version 1.5). LogCombiner (version 1.8) was used to combine triplicate runs, with removal of 20 % burn-in.
Gene content analysis
De novo assembly for each read set was performed using the assembler Velvet (version 1.2.03) and VelvetOptimizer, with each read set mapped back to each assembly (Zerbino & Birney, 2008). Contigs < 300 bp were discarded. Assemblies were annotated using the rapid prokaryotic genome annotation tool Prokka (Seemann, 2014). To identify contigs associated with the accessory genome, contigs were first-ordered with S. sonnei Ss046 and its large virulence plasmid pSS046 as references using abacas. The remaining contigs were visualized and examined in Artemis. Presence of specific accessory genetic determinants was confirmed by blast and visualized using act (Carver et al., 2008).
For each isolate, the entire resistance gene content (resistome) was identified by mapping the assembly to a manually curated resistome database as described previously (Chung The et al., 2015). MLST was performed for all Shigella isolates by profiling the seven MLST genes (adk, fumC, gyrB, icd, mdh, purA, recA). Mutations in specific genes were manually assessed by retrieving their alignments from the annotations and the effects of non-synonymous mutations were predicted using the sift web server (http://sift.jcvi.org/) (Sim et al., 2012). An in silico plasmid incompatibility-type PCR was performed using an in-house script and bespoke primer set.
Results
Bhutanese S. sonnei lies within lineage III (Central Asia clade)
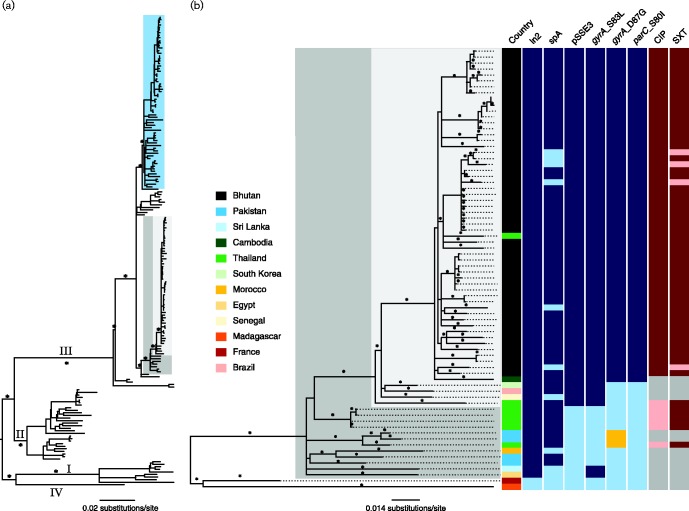
All S. sonnei (n = 74) isolated during this study were subjected to WGS; 71 isolates produced sufficient sequence for analysis. To contextualize the Bhutanese S. sonnei within the known global S. sonnei population structure, we constructed a phylogeny that included the Bhutanese sequences and those from a previous global S. sonnei sequencing project (Holt et al., 2012); within this phylogeny, all Bhutanese S. sonnei fell into a single clade within lineage III (Central Asia clade), which is a sister clade to the intercontinental expanding Global III clade (Fig. 1A). To investigate the phylogenetic relationships within the Central Asia clade, we concatenated 74 of the non-duplicate whole-genome sequences, including 54 from Bhutan and 20 selected from other countries. A maximum-likelihood phylogeny reconstructed from this alignment showed a close clonal relationship between the Bhutanese S. sonnei. Showing a similarly close relationship to the Bhutanese isolates, and interspersed within this clade, were also two isolates from Thailand and Cambodia, likely representing importations following regional travel (Fig. 1B).
Fig. 1.
The phylogenetic structure of Bhutanese S. sonnei in the context of the global phylogeny and Central Asia clade. (A) Midpoint rooted maximum-likelihood phylogenetic tree of 183 S. sonnei strains (135 from global collection and 48 from Bhutan) reconstructed using 5393 SNPs; asterisks indicate bootstrap support values ≥ 98 % on major branches. Numbers above major branches represent ones leading to major lineages (I, II, III and IV). The light blue box highlights the Global III clade; the dark grey box overlaid on the tree highlights strains belonging to the Central Asia clade; the smaller, light grey box highlights the primarily Bhutanese Central Asia clade. (B) Magnified view of the maximum-likelihood phylogenetic tree of the Central Asia clade, including 74 S. sonnei strains (54 from Bhutan and 20 others for phylogenetic context), reconstructed using 996 SNPs; asterisks indicate bootstrap support values ≥ 80 %. The tree is midpoint rooted for purposes of clarity. Columns to the right of the phylogenetic tree show isolate metadata including: country of isolation (colours indicated in the key); the presence (dark blue) or absence (light blue) of specific genetic elements and mutations [ln2, spA, pSSE3, gyrA (S83L), gyrA (D87G), parC(S80I)], or the presence of an alternative mutation (yellow) [gyrA (D87Y)]; and resistance profiles against ciprofloxacin (CIP) and trimethoprim/sulfamethoxazole (SXT), where resistance is indicated in dark red, susceptibility in light red and missing data in grey.
Temporal phylogenetic reconstruction of S. sonnei in Bhutan
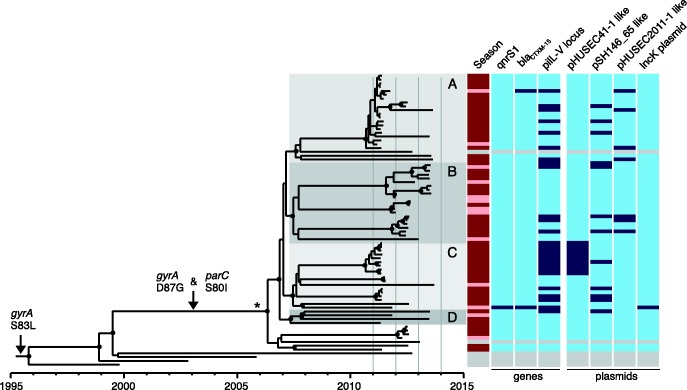
As there was such a strong temporal signature for the number of observed mutations in the global S. sonnei population (Holt et al., 2013), we explored the temporal structure of the S. sonnei isolated in Bhutan using Bayesian phylogenetic methods. To do this, the nucleotide substitution rate of the primarily Bhutanese lineage of Central Asia was determined to be 7.34 × 10− 7 substitutions per site per year [95 % highest posterior density (HPD): 5.87 × 10− 7 to 8.83 × 10− 7], which is comparable to the previously calculated substitution rate of S. sonnei lineage III (Holt et al., 2013). Using these data, we estimate that the most recent common ancestor (MRCA) of the Bhutanese S. sonnei existed around 2006 (95 % HPD 2005–2007), with the majority diverging from a hypothetical MRCA around 2007 (95 % HPD 2005–2008) (Fig. 2). The results of this analysis were further supported by the linear relationship between root-to-tip branch length and date of isolation (R2 = 0.8412). Since its projected introduction in 2006, this S. sonnei lineage diverged into at least four sublineages (labelled A, B, C and D in Fig. 2), which have been transmitted endemically within the Bhutanese capital, Thimphu: isolates taken from cases in June/July 2011 (25/71 isolates) were found to belong to two subpopulations (A and C). Sublineage B, which was not represented in June/July 2011, dominated in the subsequent years (Fig. 2).
Fig. 2.
Temporal phylogenetic reconstruction of Bhutanese Shigella sonnei between 2011 and 2013.Image shows a maximum clade credibility phylogenetic reconstruction of S. sonnei isolated primarily in Bhutan over the study period. Distinct subpopulations of Bhutanese S. sonnei are highlighted and indicated by letters. Black circles indicate posterior probability support ≥ 80 % on internal nodes. The asterisk indicates the branch leading to the Bhutanese S. sonnei clade, which represents 22 lineage-defining SNPs. Arrows denote branches characterized by select substitution events. Columns to the right of the phylogeny show metadata including: season (dark red, monsoon season: June–September; light red, other), and presence (dark blue) or absence (light blue) of mobile genetic elements, including specific genes (qnrS1, blaCTXM-15, pilL–V locus) and plasmids (pHUSEC41-1-like, pSH146_65-like, pHUSEC2011-1-like, IncK). Grey bars in metadata columns indicate sequences from S. sonnei isolated outside of Bhutan.
Mutations specific to Bhutanese S. sonnei
To identify potential genetic mechanisms associated with diversification of the Bhutanese S. sonnei, we compared their content of unique SNPs in comparison with the three most closely related outgroup isolates, taken from global isolates belonging to the same global lineage but collected outside of Bhutan (Fig. 2). The Bhutanese isolates were distinguished from these by the presence of 22 lineage-specific consensus SNPs. Of these 22 SNPs, five fell in intergenic regions and 17 mapped within predicted coding sequences; 11 represented non-synonymous and six were synonymous mutations. The genes bearing non-synonymous mutations are summarized in Table 1 together with their predicted impact on function. These include mutations in gyrA and parC associated with antimicrobial resistance, as well as those located in ycgB, mppA, hisC and rbsK, as determined by in silico sift (see Methods) analysis. With the exception of ycgB encoding a protein with unknown function, these genes all encode proteins predicted to be involved in the metabolism of histidine, ribose and formate, as well as transport of murein tripeptide (Table 1).
Table 1. Non-synonymous mutations separating the MRCA of all Bhutanese S. sonnei from other Central Asia clade strains.
| Gene | Mutation | Function | sift score*/effect |
|---|---|---|---|
| cusS | G343S | Sensory histidine kinase of CusSR, regulating the expression of the CusCFBA operon to confer resistance to high copper and silver concentrations | 0.77 |
| ycgB | Y125H | Unknown function | 0 |
| mppA | G344R | Periplasmic binding component of murein tripeptide in oligopeptide transport | 0 |
| hisC | P98S | Histidinol-phosphate aminotransferase, catalysing the biosynthesis of histidine | 0.03 |
| eutL | A22S | Carboxysomal shell protein, structural component of proteinaceous microcompartment for ethanolamine ammonia lyase | 0.5 |
| hycC | G538D | Component of HycBCDEFG, hydrogenase component (hydrogenase 3) of formate hydrogenlyase; HycC encodes an extremely hydrophobic protein with homology to NADH : ubiquinone oxidoreductase | 0 |
| sdaB | A407S | l-Serine deaminase II, catalysing the conversion of serine into pyruvate and ammonia | 0.33 |
| metC | A2T | Cystathionine B-lyase and cysteine desulfhydrase, catalysing the conversion cystathionine to homocysteine | 0.7 |
| rbsK | A50T | Ribokinase responsible for metabolism of d-ribose | 0 |
| gyrA | D87G | DNA gyrase, subunit A | Known effect: resistance to fluoroquinolones |
| parC | S80I | DNA topoisomerase IV subunit A | Known effect: resistance to fluoroquinolones |
sift scores ≤ 0.05 indicate a significant effect on protein function based on sift analysis.
Accessory genome and impact of antimicrobial resistance genes in Bhutanese S. sonnei
The composition of the accessory genome can be used to indicate the capacity of the host species to sample the local gene pool and also to highlight themes in the patterns of acquisition that may be under selection. We performed a comparative gene content analysis of Bhutanese and non-Bhutanese S. sonnei belonging to the Central Asia clade. These data revealed events that appear to have been pivotal in evolution and spread of the Central Asia clade. Notably, all the isolates of this clade possessed a specific class 2 integron In2 (dfrA1, sat1), conferring resistance to trimethoprim and streptothricin. These strains additionally gained the S. sonnei spA plasmid carrying sul2, strAB and tetRA genes, which further expanded the spectrum of antimicrobial resistance to incorporate sulphonamides, streptomycin and tetracycline resistance, respectively (Fig. 1B). This resistance gene profile concurs with the observation made in the previous global S. sonnei study (Holt et al., 2012). It is of note that the class 2 integron of the Central Asia clade differs from that of Global III in the absence of aadA1, which confers resistance to aminoglycosides. Moreover, it was clear that isolates within this clade have, over time, accumulated chromosomal mutations conferring fluoroquinolone resistance (Fig. 1B). From the lineage-specific phylogeny, we predicted that the first mutation in gyrA (S83L) occurred before 1996, and supplementary mutations in parC (S80I) and secondary mutations in gyrA (D87G), leading to high-level fluoroquinolone resistance, were fixed in the Bhutanese clade prior to its expansion and diversification in 2006 (Fig. 2).
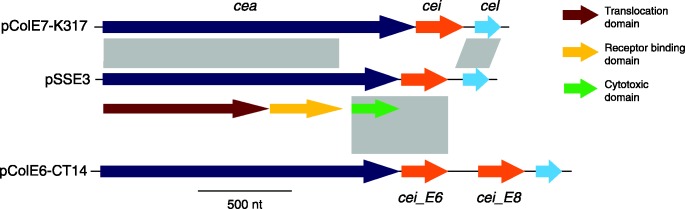
A further pivotal event in the emergence of this Central Asia clade prior to its introduction into Bhutan was the fixation of the previously described E-type colicin plasmid pSSE3 (with 100 % identity in DNA sequence). This E-type colicin has been shown to be antagonistic against a variety of diarrhoeogenic E. coli, but not against commensal E. coli (Calcuttawala et al., 2015). Plasmid pSSE3, which was found in all Bhutanese isolates (Fig. 1B), also carries an immunity function that protects the host cell against the activity of the corresponding colicin. Although it was previously characterized as a ColE3 plasmid by PCR, sequence analysis of pSSE3 indicated that the colicin biosynthesis cluster was a novel recombinant hybrid with an undetermined spectrum of activity (Fig. 3). On closer examination, the first and second domains (translocation and receptor-binding) of the colicin structural gene (cea) and the lysis gene (cel) exhibited substantial amino acid sequence identity (98 %) to that of colicin E7 (ColE7). The third domain (cytotoxic) encoded by cea was most closely related to that of ColE6 (94 % protein sequence identity), predicting that the colicin's target is actually 16S rRNA rather than DNA, which is the target of the ColE7 counterpart (Kleanthous, 2010). However, the product of the colicin immunity gene (cei) exhibited only 84 % protein sequence identity to that of colicin E6.
Fig. 3.
Novel recombinant hybrid colicin biosynthesis cluster encoded on pSSE3.Schematic representation of the colicin biosynthesis cluster on plasmid pSSE3 in comparison with colicin gene clusters of pColE7-K317 and pColE6-CT14. Grey vertical bars show homology in DNA sequences (with identity >80 %). The colicin structural gene (cea), immunity gene (cei) and lysis gene (cel) are annotated as shown. The three domains of the colicin structural gene are shown the key.
Genetic material that was acquired sporadically across the Central Asia clade within the Bhutanese isolates included the extended-spectrum β-lactamase (ESBL) gene blaCTXM-15, which was independently acquired by two S. sonnei on different incompatibility plasmids (IncK and IncI1), conferring resistance to third-generation cephalosporins (Fig. 2). The plasmid replication and transfer region of the IncK plasmid shared substantial sequence homology with that of the enteroaggregative E. coli IncI1 plasmid pSERB1. This plasmid in the Bhutanese S. sonnei incorporated the type IV pilus biosynthesis cluster pilL–V (Dudley et al., 2006). For the S. sonnei containing the IncK plasmid, the blaCTXM-15 gene was encoded along with the fluoroquinolone resistance enhancement gene qnrS1 on an IS26 transposon. The S. sonnei IncI1 plasmid (denoted pHUSEC2011-1-like) was almost identical ( ≥ 99 % DNA sequence identity) to the replication, conjugation and type IV pilus biosynthesis cluster pilL–V in pHUSEC2011-1. The pHUSEC2011-1-like plasmid was also independently acquired by six additional strains, although in these cases the blaCTXM-15 gene was not co-transferred (Fig. 2).
Lastly, nine S. sonnei strains isolated from cases occurring in June/July 2011 acquired an IncB-O plasmid, sharing homology with the MDR plasmid pHUSEC41-1 originally identified in an older O104 : H4 enteroaggregative Shiga toxin-producing E. coli (HUSEC41), which included a type IV pilus biosynthesis cluster pilL–V identical to that found on plasmid pSERB1 (Künne et al., 2012). We further identified 11 S. sonnei strains harbouring a plasmid with homology to Salmonella enterica plasmid pSH146_65, additionally carrying a distinct type IV pilus cluster pilL–V (Han et al., 2012).
Discussion
Here, we have used WGS to define a population of S. sonnei causing disease in the South Asian land-locked nation of Bhutan. These strains belong to an expansion of lineage III, which we have named the Central Asia clade, which was introduced into Bhutan around 2006. To the best of our knowledge, this is the first time that WGS has been used on this scale to study a population of S. sonnei in South Asia, a region with a substantial burden of diarrhoeal disease. In contrast to recent observations from Vietnam, the sequenced S. sonnei in Bhutan demonstrated no evidence of a single clonal expansion. This is perhaps due to the short sampling time frame or other factors such as the lack of a definitive selective sweep event. However, the evolution history of Central Asia clade S. sonnei resembles the two initial sweep events observed in Vietnam [associated with fixation of a colicin plasmid and mutation(s) in gyrA] (Holt et al., 2013), showing that the two geographically and phylogenically unrelated S. sonnei populations followed a similar evolutionary trajectory. After its introduction to Bhutan, our data predict that these S. sonnei had access to a substantial gene pool, as indicated by the sporadic acquisition of ESBL plasmids and type IV pili. The gains of ESBL plasmids, although infrequent, could potentially become the next selective sweep for the Bhutan's S. sonnei population as observed in Vietnam, given the selection path toward resistance is maintained. The characteristically thin type IV pilus has been shown to improve the efficiency and specificity of conjugation in liquid environments (Kim & Komano, 1997), and also facilitates adherence, interbacterial contact and twitching motility (Berry & Pelicic, 2015). Although this structure is known to be widespread in prokaryotes, its impact on the lifestyle of Shigella is not well described and requires further experimental investigation (Holt et al., 2013; Rohmer et al., 2014).
In addition, the Bhutanese S. sonnei population is characterized by the presence of numerous non-synonymous mutations, most of which are found in metabolic pathways. The genus Shigella is well known for its adaptive intracellular lifestyle, enabling its shedding of redundant or unnecessary functions during the course of evolution (Prosseda et al., 2012). Our in silico analysis predicted that some mutations could affect metabolic activities, which could lead to degradation. In particular, the affected d-ribose utilization and formate fermentation appear to be redundant as energy production in Shigella predominantly occurs via the uptake and metabolism of pyruvate into acetate (Kentner et al., 2014). Biosynthesis of histidine is also deemed inessential as the bacterium could readily transport it from the host's cytosol. However, the true impacts of the aforementioned mutations should be confirmed by functional analyses in vitro in future studies.
We were able to use WGS to identify the major events during the evolution of this lineage prior to, and after, its introduction into a new setting. Our data imply that the acquisition of a class 2 integron and a spA plasmid, together with the rapid accumulation of mutations associated with fluoroquinolone resistance, rendered the Bhutanese S. sonnei MDR by 2006. Furthermore, a fluoroquinolone-resistant subclade within the Central Asia clade was predicted to have arisen between 1999 and 2006. In addition, the fixation of a novel recombinant colicin E plasmid (pSSE3) may have offered this group of pathogens a competitive advantage over other enteric bacteria, similar to the colicin-associated selective sweep observed in S. sonnei in Vietnam (Holt et al., 2013). These data suggest that whilst antimicrobial resistance is clearly important, it is not sufficient in itself for driving the spread of new clones and anticompetitive functions appear to be key themes in the spread of MDR S. sonnei.
Our data additionally suggests that international travel may have resulted in the dissemination of Central Asia clade S. sonnei beyond Bhutan and the remainder of the Indian subcontinent. S. sonnei isolates with similar PFGE patterns to the strains in this analysis have been recovered from cases of travel-associated shigellosis in Ireland (De Lappe et al., 2015) and in a Shigella outbreak amongst ‘men who have sex with men’ in Canada (Gaudreau et al., 2011). The S. sonnei isolated from both instances were found to be ciprofloxacin-resistant. Studies conducted in Japan have established a strong association between this pulsotype and travel to India, and increasing prevalence of nalidixic acid-resistant S. sonnei has been observed since 2001 (Uchimura et al., 2001; Izumiya et al., 2009). Shigellosis surveys in Iran conducted from 2008 to 2012 have repeatedly shown that this characteristic Asian pulsotype is prevalent and endemic, and that these strains also possess a similar class 2 integron (dfrA1, sat1), but exhibit variability in their ciprofloxacin susceptibility phenotype (Tajbakhsh et al., 2012; Eftekhari et al., 2013; Alizadeh-Hesar et al., 2015). The first case of fluoroquinolone-resistant S. sonnei was identified in Japan in 1993 (Horiuchi et al., 1993). Since then, the incidence of ciprofloxacin-resistant Shigella in Asia and Africa has risen dramatically, from 3.9 % in 2001–2003 to 29.1 % in 2007–2009 (Gu et al., 2012). The potential emergence and dissemination of fluoroquinolone-resistant S. sonnei poses a substantial threat to public health, including low shigellosis burden areas in developed nations, as again substantiated by the recent fluoroquinolone-resistant S. sonnei outbreak in the USA (Bowen et al., 2015). Taken together, these data strongly suggest that the Central Asia clade of S. sonnei is widely present across South Asia and beyond, and may represent an emerging threat to public health.
Acknowledgements
We wish to acknowledge the staff of the Department of Enteric Disease, AFRIMS, Bangkok, Thailand for their work in isolating and maintaining the strains in this study. S. B. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and Royal Society (100087/Z/12/Z). N. R. T. is supported the Wellcome Trust grant 098051 to Wellcome Trust Sanger Institute. The views expressed in this article are those of the authors and do not represent the official policy or position of the US Department of the Army, Department of Defense or US Government.
Abbreviations:
- ESBL
extended-spectrum β-lactamase
- HPD
highest posterior density
- JDWNRH
Jigme Dorji Wangchuk National Referral Hospital
- MCMC
Markov Chain Monte Carlo
- MDR
multidrug-resistant
- MRCA
most recent common ancestor
- WGS
whole-genome sequencing
References
- Alizadeh-Hesar M., Bakhshi B., Najar-Peerayeh S. (2015). Clonal dissemination of a single Shigella sonnei strain among Iranian children during Fall 2012 in Tehran, I.R. Iran Infect Genet Evol 34260–266. [DOI] [PubMed] [Google Scholar]
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., Alekseyenko A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty Mol Biol Evol 292157–2167 10.1093/molbev/mss084 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Li W. L., Drummond A. J., Suchard M. A., Lemey P. (2013). Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics Mol Biol Evol 30239–243 10.1093/molbev/mss243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. S., Dallman T. J., Ashton P. M., Day M., Hughes G., Crook P. D., Gilbart V. L., Zittermann S., Allen V. G. (2015). Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study Lancet Infect Dis 15913–921 10.1016/S1473-3099(15)00002-X . [DOI] [PubMed] [Google Scholar]
- Berry J.-L., Pelicic V. (2015). Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives FEMS Microbiol Rev 39134–154 10.1093/femsre/fuu001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongpin M. (2015). Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin – United States, May 2014–February 2015 MMWR Morb Mortal Wkly Rep 64318–320. [PMC free article] [PubMed] [Google Scholar]
- Calcuttawala F., Hariharan C., Pazhani G. P., Ghosh S., Ramamurthy T. (2015). Activity spectrum of colicins produced by Shigella sonnei and genetic mechanism of colicin resistance in conspecific S. sonnei strains and Escherichia coli Antimicrob Agents Chemother 59152–158 10.1128/AAC.04122-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T., Berriman M., Tivey A., Patel C., Böhme U., Barrell B. G., Parkhill J., Rajandream M. A. (2008). Artemis and act: viewing, annotating and comparing sequences stored in a relational database Bioinformatics 242672–2676 10.1093/bioinformatics/btn529 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing 20th Informational SupplementWayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Connor T. R., Barker C. R., Baker K. S., Weill F. X., Talukder K. A., Smith A. M., Baker S., Gouali M., Pham Thanh D. (2015). Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri eLife 41–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Page A. J., Connor T. R., Delaney A. J., Keane J. A., Bentley S. D., Parkhill J., Harris S. R. (2014). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins Nucleic Acids Res 43e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung The H., Karkey A., Pham Thanh D., Boinett C. J., Cain A. K., Ellington M., Baker K. S., Dongol S., Thompson C. (2015). A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae EMBO Mol Med 7227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lappe N., O'Connor J., Garvey P., McKeown P., Cormican M. (2015). Ciprofloxacin-resistant Shigella sonnei associated with travel to India Emerg Infect Dis 21894–896 10.3201/eid2105.141184 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees BMC Evol Biol 7214. 10.1186/1471-2148-7-214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E. G., Abe C., Ghigo J. M., Latour-Lambert P., Hormazabal J. C., Nataro J. P. (2006). An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces Infect Immun 742102–2114 10.1128/IAI.74.4.2102-2114.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekhari N., Bakhshi B., Pourshafie M. R., Zarbakhsh B., Rahbar M., Hajia M., Ghazvini K. (2013). Genetic diversity of Shigella spp. and their integron content Foodborne Pathog Dis 10237–242 10.1089/fpd.2012.1250 . [DOI] [PubMed] [Google Scholar]
- Gaudreau C., Ratnayake R., Pilon P. A., Gagnon S., Roger M., Lévesque S. (2011). Ciprofloxacin-resistant Shigella sonnei among men who have sex with men, Canada, 2010 Emerg Infect Dis 171747–1750 10.3201/eid1709.102034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Cao Y., Pan S., Zhuang L., Yu R., Peng Z., Qian H., Wei Y., Zhao L. (2012). Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009 Int J Antimicrob Agents 409–17 10.1016/j.ijantimicag.2012.02.005 . [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0 Syst Biol 59307–321 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- Han J., Lynne A. M., David D. E., Tang H., Xu J., Nayak R., Kaldhone P., Logue C. M., Foley S. L. (2012). DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates PLoS One 7e51160. 10.1371/journal.pone.0051160 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Baker S., Weill F. X., Holmes E. C., Kitchen A., Yu J., Sangal V., Brown D. J., Coia J. E. (2012). Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe Nat Genet 441056–1059 10.1038/ng.2369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Thieu Nga T. V., Thanh D. P., Vinh H., Kim D. W., Vu Tra M. P., Campbell J. I., Hoang N. V., Vinh N. T. (2013). Tracking the establishment of local endemic populations of an emergent enteric pathogen Proc Natl Acad Sci U S A 11017522–17527 10.1073/pnas.1308632110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S., Inagaki Y., Yamamoto N., Okamura N., Imagawa Y., Nakaya R. (1993). Reduced susceptibilities of Shigella sonnei strains isolated from patients with dysentery to fluoroquinolones Antimicrob Agents Chemother 372486–2489 10.1128/AAC.37.11.2486 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I. F., Chiu C. H., Wang M. H., Wu C. Y., Hsieh K. S., Chiou C. C. (2005). Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei J Clin Microbiol 432608–2612 10.1128/JCM.43.6.2608-2612.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya H., Tada Y., Ito K., Morita-Ishihara T., Ohnishi M., Terajima J., Watanabe H. (2009). Characterization of Shigella sonnei isolates from travel-associated cases in Japan J Med Microbiol 581486–1491 10.1099/jmm.0.011809-0 . [DOI] [PubMed] [Google Scholar]
- Kentner D., Martano G., Callon M., Chiquet P., Brodmann M., Burton O., Wahlander A., Nanni P., Delmotte N. (2014). Shigella reroutes host cell central metabolism to obtain high-flux nutrient supply for vigorous intracellular growth Proc Natl Acad Sci U S A 1119929–9934 10.1073/pnas.1406694111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Komano T. (1997). The plasmid R64 thin pilus identified as a type IV pilus J Bacteriol 1793594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C. (2010). Swimming against the tide: progress and challenges in our understanding of colicin translocation Nat Rev Microbiol 8843–848 10.1038/nrmicro2454 . [DOI] [PubMed] [Google Scholar]
- Kotloff K. L., Blackwelder W. C., Nasrin D., Nataro J. P., Farag T. H., van Eijk A., Adegbola R. A., Alonso P. L., Breiman R. F. (2012). The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study Clin Infect Dis 55 (Suppl 4), S232–S245 10.1093/cid/cis753 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., Wu Y., Sow S. O., Sur D. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study Lancet 382209–222 10.1016/S0140-6736(13)60844-2 . [DOI] [PubMed] [Google Scholar]
- Künne C., Billion A., Mshana S. E., Schmiedel J., Domann E., Hossain H., Hain T., Imirzalioglu C., Chakraborty T. (2012). Complete sequences of plasmids from the hemolytic-uremic syndrome-associated Escherichia coli strain HUSEC41 J Bacteriol 194532–533 10.1128/JB.06368-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. M., Kotloff K. L., Barry E. M., Pasetti M. F., Sztein M. B. (2007). Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road Nat Rev Microbiol 5540–553 10.1038/nrmicro1662 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009). 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools Bioinformatics 252078–2079 10.1093/bioinformatics/btp352 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Johnson H. L., Cousens S., Perin J., Scott S., J.E., Rudan I., Campbell H., Cibulskis R. (2012). Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000 Lancet 3792151–2161 10.1016/S0140-6736(12)60560-1 . [DOI] [PubMed] [Google Scholar]
- Marfin A. A., Moore J., Collins C., Biellik R., Kattel U., Toole M. J., Moore P. S. (1994). Infectious disease surveillance during emergency relief to Bhutanese refugees in Nepal JAMA 272377–381 10.1001/jama.272.5.377 . [DOI] [PubMed] [Google Scholar]
- Nguyen N.T.K., Ha V., Tran N. V., Stabler R., Pham D. T., Le T. M., van Doorn H. R., Cerdeño-Tárraga A., Thomson N. (2010). The sudden dominance of blaCTX-M harbouring plasmids in Shigella spp., Circulating in Southern Vietnam PLoS Negl Trop Dis 4e702. 10.1371/journal.pntd.0000702 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: phylogenetic model averaging Mol Biol Evol 251253–1256 10.1093/molbev/msn083 . [DOI] [PubMed] [Google Scholar]
- Prosseda G., Di Martino M. L., Campilongo R., Fioravanti R., Micheli G., Casalino M., Colonna B. (2012). Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model Res Microbiol 163399–406 10.1016/j.resmic.2012.07.004 . [DOI] [PubMed] [Google Scholar]
- Pu X. Y., Pan J. C., Zhang W., Zheng W., Wang H. Q., Gu Y. M. (2015). Quinolone resistance-determining region mutations and the plasmid-mediated quinolone resistance gene qnrS played important roles in decreased susceptibility to fluoroquinolones among Shigella isolates in southeast China between 1998 and 2013 Int J Antimicrob Agents 45438–439 10.1016/j.ijantimicag.2014.12.004 . [DOI] [PubMed] [Google Scholar]
- Pupo G. M., Lan R., Reeves P. R. (2000). Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics Proc Natl Acad Sci U S A 9710567–10572 10.1073/pnas.180094797 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M. A., Kozarewa I., Smith F., Scally A., Stephens P. J., Durbin R., Swerdlow H., Turner D. J. (2008). A large genome center's improvements to the Illumina sequencing system Nat Methods 51005–1010 10.1038/nmeth.1270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Jacobs M. A., Brittnacher M. J., Fong C., Hayden H. S., Hocquet D., Weiss E. J., Radey M., Germani Y. (2014). Genomic analysis of the emergence of 20th century epidemic dysentery BMC Genomics 15355. 10.1186/1471-2164-15-355 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruekit S., Wangchuk S., Dorji T., Tshering K. P., Pootong P., Nobthai P., Serichantalergs O., Poramathikul K., Bodhidatta L., Mason C. J. (2014). Molecular characterization and PCR-based replicon typing of multidrug resistant Shigella sonnei isolates from an outbreak in Thimphu, Bhutan BMC Res Notes 795. 10.1186/1756-0500-7-95 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P. J., Kopecko D. J., Formal S. B. (1982). Involvement of a plasmid in the invasive ability of Shigella flexneri Infect Immun 35852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation Bioinformatics 302068–2069 10.1093/bioinformatics/btu153 . [DOI] [PubMed] [Google Scholar]
- Seol S. Y., Kim Y. T., Jeong Y. S., Oh J. Y., Kang H. Y., Moon D. C., Kim J., Lee Y. C., Cho D. T., Lee J. C. (2006). Molecular characterization of antimicrobial resistance in Shigella sonnei isolates in Korea J Med Microbiol 55871–877 10.1099/jmm.0.46441-0 . [DOI] [PubMed] [Google Scholar]
- Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P. C. (2012). sift web server: predicting effects of amino acid substitutions on proteins Nucleic Acids Res 40 (W1), W452–W457 10.1093/nar/gks539 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh M., García Migura L., Rahbar M., Svendsen C. A., Mohammadzadeh M., Zali M. R., Aarestrup F. M., Hendriksen R. S. (2012). Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? J Antimicrob Chemother 671128–1133 10.1093/jac/dks023 . [DOI] [PubMed] [Google Scholar]
- Thompson C. N., Duy P. T., Baker S. (2015). The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery PLoS Negl Trop Dis 9e0003708. 10.1371/journal.pntd.0003708 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura M., Kishida K., Koiwai K. (2001). [Increasing incidence and the mechanism of resistance of nalidixic acid resistant Shigella sonnei] Kansenshogaku Zasshi 75923–930(in Japanese). [DOI] [PubMed] [Google Scholar]
- Vinh H., Nhu N. T., Nga T. V., Duy P. T., Campbell J. I., Hoang N. V., Boni M. F., My P. V., Parry C. (2009). A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation BMC Infect Dis 9204. 10.1186/1471-2334-9-204 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. L., Rudan I., Liu L., Nair H., Theodoratou E., Bhutta Z. A., O'Brien K. L., Campbell H., Black R. E. (2013). Global burden of childhood pneumonia and diarrhoea Lancet 3811405–1416 10.1016/S0140-6736(13)60222-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014). World Health Statistics. http://apps.who.int/iris/bitstream/10665/112738/1/9789240692671_eng.pdf.
- Zerbino D. R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs Genome Res 18821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion PhD thesis, University of Texas at Austin, Austin, TX, USA. [Google Scholar]
Data Bibliography
- 1. Shigella sonnei Ss046 complete genome: GenBank accession number NC_007384.
- 2. Shigella sonnei strain IDH01791 plasmid pSSE3 complete sequence: GenBank accession number KP970685.
- 3. Escherichia coli plasmid pHUSEC41-1 complete sequence: GenBank accession number NC_018995.
- 4. Escherichia coli HUSEC2011 plasmid pHUSEC2011-1 complete sequence: GenBank accession number NC_022742.
- 5. Escherichia coli plasmid pSERB1 partial sequence: GenBank accession number NG_035985.
- 6. Salmonella enterica subsp. enterica serovar Heidelberg plasmid pSH146_65 complete sequence: GenBank accession number NC_019115.
- 7. Table S1. http://figshare.com/articles/The_introduction_and_establishment_of_fluoroquinolone_resistant_Shigella_sonnei_into_Bhutan/1610693.