Abstract
In 2013, an unusual increase in the number of Salmonella enterica serotype Paratyphi A (Salmonella Paratyphi A) infections was reported in patients in Phnom Penh, Cambodia, and in European, American and Japanese travellers returning from Cambodia. Epidemiological investigations did not identify a common source of exposure. To analyse the population structure and genetic diversity of these Salmonella Paratyphi A isolates, we used whole-genome sequencing on 65 isolates collected from 1999 to 2014: 55 from infections acquired in Cambodia and 10 from infections acquired in other countries in Asia, Africa and Europe. Short-read sequences from 80 published genomes from around the world and from 13 published genomes associated with an outbreak in China were also included. Pulsed-field gel electrophoresis (PFGE) was performed on a subset of isolates. Genomic analyses were found to provide much more accurate information for tracking the individual strains than PFGE. All but 2 of the 36 isolates acquired in Cambodia during 2013–2014 belonged to the same clade, C5, of lineage C. This clade has been isolated in Cambodia since at least 1999. The Chinese outbreak isolates belonged to a different clade (C4) and were resistant to nalidixic acid, whereas the Cambodian outbreak isolates displayed pan-susceptibility to antibiotics. Since 2014, the total number of cases has decreased, but there has been an increase in the frequency with which nalidixic acid-resistant C5 isolates are isolated. The frequency of these isolates should be monitored over time, because they display decreased susceptibility to ciprofloxacin, the first-choice antibiotic for treating paratyphoid fever.
Keywords: Cambodia, Salmonella Paratyphi A, whole genome sequencing, resistance
Data Summary
All supporting data, code and protocols have been provided within the article or through supplementary data files. The whole-genome sequence reads from the Salmonella Paratyphi A used in this study have been deposited in the European Nucleotide Archive. The run accession numbers and related metadata are detailed in Table S1 (available in the online Supplementary Material), which has been deposited in FigShare: DOI:10.6084/m9.figshare.346180.
Impact Statement
The bacterium Salmonella Paratyphi A can cause bloodstream infections (paratyphoid fever) and is transmitted via food or water contaminated with waste products from infected individuals. Accurate genetic information about bacterial isolates is crucial to support public health investigations, for tracing transmission and identifying the source, in particular. Classical typing methods, such as pulsed-field gel electrophoresis (PFGE), do not perform well on this genetically monomorphic bacterial population. We used whole-genome sequencing and comparative genomics for the retrospective investigation of an outbreak of Salmonella Paratyphi A infections in Cambodia. The greater discriminatory power and portability of genomic analyses, compared with PFGE, made it possible to identify the isolates associated with the outbreak unambiguously, and to assess their genetic relationships with isolates from sporadic cases, historical isolates and isolates from another outbreak in China.
Introduction
Salmonella enterica serotypes Typhi (Salmonella Typhi) and Paratyphi A (Salmonella Paratyphi A) are Gram-negative bacteria confined to human hosts. They can invade the bloodstream, causing typhoid and paratyphoid fever (known together as ‘enteric fever’). These infections have become rare in Western countries, but they continue to be responsible for a considerable burden of disease in low-resource settings (Bhan et al., 2005). Historically, enteric fever was mostly caused by Salmonella Typhi. However, the number of Salmonella Paratyphi A infections has been increasing steadily over the last 20 years, and it is now estimated that there are six million cases of paratyphoid fever annually (Newton et al., 2016). This increase is most apparent in Asia, where Salmonella Paratyphi A now accounts for 14 % (Indonesia) to 64 % (South-East China) of enteric fever cases (Ochiai et al., 2005). Furthermore, international travellers returning from countries in which Salmonella Paratyphi A is endemic are increasingly being infected with this bacterium, which accounted for 31 % of imported cases of enteric fever in the European Union in 2011 (ECDC, 2013).
During 2011–2013, an unusual increase in the number of Salmonella Paratyphi A infections was reported among patients attending the Sihanouk Hospital Centre of HOPE (SHCH), Phnom Penh, Cambodia (Vlieghe et al., 2013). This increase coincided with an observed increase in the number of cases of Salmonella Paratyphi A infection among travellers returning to Europe, the USA, New Zealand or Japan from Cambodia (Tourdjman et al., 2013; Saitoh et al., 2016; Judd et al., 2015). No similar increase in Salmonella Typhi infections was reported. Epidemiological investigations identified no common source of exposure and the route of disease transmission remained unknown. The analysis of population structure and genetic diversity among outbreak isolates is an important component of epidemiological investigations. Such information can provide clues to the origin and transmission of the bacterial pathogens. Molecular epidemiology analysis was carried out on only 22 Salmonella Paratyphi A isolates from Japanese travellers infected during 2012–2014 (Saitoh et al., 2016). PFGE with a single enzyme (XbaI) identified a predominant profile (18/22, 81.8 %) among the outbreak isolates, suggesting that the outbreak was caused by a single strain. However, PFGE, the previous gold standard for subtyping Salmonella spp., has proved unsuitable for the analysis of highly clonal serotypes like Salmonella Paratyphi A (Tien et al., 2011). In this study, our main aim was to use accurate genetic information, from whole-genome sequencing data in particular, to analyse the population structure and genetic diversity of the Salmonella Paratyphi A isolates collected during the 2013–2015 outbreak in Cambodia.
Methods
Surveillance systems – Cambodia.
The Cambodian isolates were collected at the SHCH, a 40-bed non-governmental referral hospital for adults in Phnom Penh. Since July 2007, SHCH and the Institute of Tropical Medicine in Antwerp, Belgium, have been jointly organising the surveillance of bloodstream infections at this hospital. Between 2008 and 2015, 190 Salmonella Paratyphi A and 64 Salmonella Typhi isolates (considering 1 isolate per patient) were obtained from a total of 18 917 blood cultures.
Surveillance systems – France.
In France, typhoid and paratyphoid fever have been notifiable diseases since 1903. Physicians and clinical laboratories are responsible for the mandatory reporting of cases. Clinical laboratories also send isolates to the French National Reference Centre for Escherichia coli, Shigella and Salmonella (FNRC-ESS), Institut Pasteur, Paris, on a voluntary basis. For each isolate, epidemiological data for the patient, including the date and site of isolation, sex and age, are recorded, together with the patient’s history of international travel. During the 2000s, the FNRC-ESS network included a stable number of hospital and private clinical laboratories referring about 65 % of all human Salmonella isolates identified in France to the FNRC-ESS (Le Hello et al., 2013). From 2008 to 2015, 85 430 serotyped Salmonella isolates were registered at the FNRC-ESS.
For paratyphoid fever, we merged data from both the FNRC-ESS and the notifiable diseases surveillance system, and identified duplicates. From 2008 to 2015, 311 cases of Salmonella Paratyphi A infection were reported to both the notifiable diseases surveillance system and the FNRC-ESS. These included 50 cases of Salmonella Paratyphi A infection in patients with a history of travel to Cambodia. Isolates were available from the FNRC-ESS for 47 of these cases (94 %).
For typhoid fever, only data from the FNRC-ESS were analysed, as legal constraints precluded duplicate identification. From 2008 to 2015, 1162 Salmonella Typhi isolates were received at the FNRC-ESS. Seven of these isolates were obtained from patients reporting a history of travel to Cambodia.
Bacterial isolates.
The 65 Salmonella Paratyphi A isolates sequenced for this study are listed in Table S1 (available in the Supplementary Material). They originated from the collections of the FNRC-ESS (n=31), the SHCH (n=30), and the Enteric and Leptospira Reference Laboratory at the Institute of Environmental Science and Research Limited, Wallaceville, New Zealand (n=4). The isolates were obtained from blood (n=59), stools (n=4), urine (n=1) or other sources (n=1). Conventional methods and serotyping at the FNRC-ESS, as previously described (Grimont & Weill, 2007), confirmed that all 65 isolates were Salmonella Paratyphi A.
Antibiotic-susceptibility testing.
Antibiotic susceptibility was determined by disc diffusion on Mueller–Hinton agar, in accordance with the guidelines of the Antibiogram Committee of the French Society for Microbiology (CA-SFM & EUCAST, 2014). The following 32 antimicrobial drugs (Bio-Rad) were tested: ampicillin, ticarcillin, piperacillin, piperacillin/tazobactam, cefamandole, cefoperazone, cefoxitin, cefotaxime, amoxicillin/clavulanic acid, ticarcillin/clavulanic acid, imipenem, meropenem, ertapenem, cefepime, ceftazidime, streptomycin, spectinomycin, kanamycin, amikacin, gentamicin, netilmicin, tigecycline, isepamicin, nalidixic acid, pefloxacin, ciprofloxacin, sulfonamides, trimethoprim, sulfamethoxazole/trimethoprim, chloramphenicol, tetracycline and azithromycin. Minimal inhibitory concentration (MIC) values for nalidixic acid, ciprofloxacin and azithromycin were determined by Etests (bioMérieux). E. coli CIP 76.24 (ATCC 25922) was used as a control.
Identification of gyrA mutations associated with decreased susceptibility to ciprofloxacin.
The quinolone-resistance-determining region (QRDR) of the gyrA gene encoding the DNA gyrase A subunit was sequenced in five nalidixic acid-resistant Salmonella Paratyphi A isolates obtained from travellers returning from Cambodia during 2014–2015, as previously described (Weill et al., 2006). The nucleotide and deduced amino acid sequences were analysed and compared with sequences available from the National Center for Biotechnology Information website (NCBI – http://www.ncbi.nlm.nih.gov/ – last accessed 16 May 2016).
PFGE.
PulseNet standard PFGE of XbaI-digested chromosomal DNA was carried out for a selection of 51 Salmonella Paratyphi A isolates (Ribot et al., 2006). BlnI-PFGE was also used to type a subsample of 24 isolates.
Single-nucleotide polymorphism (SNP) genotyping assay.
Three informative SNPs were selected from the 4810 SNPs identified in the comparative genomic analysis, for the reliable identification of lineage A, and clades C2 and C5 of lineage C. For lineage A, the specific SNP (A to G at position 666 468 of the Salmonella Paratyphi A ATCC 9150 reference genome, GenBank accession no. NC_006511) was within the SPA_RS02955 gene. For lineage C, the clade C2-specific SNP (G to T at position 4 294 741) was within the SPA_RS20855 gene, whereas the C5-specific SNP (G to A at position 2 381 607) was within the SPA_RS11495 gene. Three pairs of primers were designed to amplify fragments containing the three SNPs: forward primer cladeA-F (5′-TCC GAG TAG CAT TGG CTT GC-3′, ATCC 9150 co-ordinates 666 181–666 200) and reverse primer cladeA-R (5′-GAG CAG CCG CCT GAA TCA AC-3′, co-ordinates 666 755–666 736); forward primer cladeC2-F (5′-CGG AAA CTG ATG GAC TCA GC-3′, co-ordinates 4 294 490–4 294 509) and reverse primer cladeC2-R (5′-TCG ACA TAA GTC CCG TCA GC-3′, co-ordinates 4 294 992–4 294 973); and forward primer cladeC5-F (5′-CCG TTA ATC GTT GCC GTA GC-3′, co-ordinates 2 381 338–2 381 357) and reverse primer cladeC5-R (5′-CAA CGA TGC CGT TGA GTT GG-3′, co-ordinates 2 381 851–2 381 832). High-purity salt-free oligonucleotides were obtained from Eurofins Genomics. Total DNA was extracted with the InstaGene matrix kit (Bio-Rad), in accordance with the manufacturer’s recommendations. PCRs were performed in a 50 µl reaction volume containing MgCl2 (1.5 mM), deoxynucleotide triphosphates (each at 0.1 mM), GoTaq Flexi DNA polymerase (0.85 U) and its buffer (Promega), dimethyl sulfoxide (5 %), 10 pmol each primer and 2 µl template DNA. Amplification began with a first denaturation step at 94 °C; followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 57 °C for C2 and C5 and 58 °C for A, polymerisation for 1 min at 72 °C; and a final extension step at 72 °C for 10 min. Sanger DNA sequencing of the PCR products was carried out with a Big Dye Terminator V3.1 cycle sequencing kit (Applied Biosystems) and a 96-capillary 3730xl DNA Analyzer (Applied Biosystems), by Eurofins MWG Operon. The nucleotide sequences were analysed with Bionumerics v.6.6 software (Applied-Maths). The blastn program of NCBI (http://www.ncbi.nlm.nih.gov/ – last accessed 16 May 2016) was used for SNP identification.
Whole-genome sequencing.
High-throughput genome sequencing was carried out at the genomics platform of the Pasteur Institute, on the HiSeq2500 platform (Illumina) generating 138 to 151 bp paired-end reads, yielding a mean of 169-fold coverage (minimum 74-fold, maximum 407-fold).
Other genomes studied.
A selection of 93 published Salmonella Paratyphi A short-read sequences (Zhou et al., 2014; Yan et al., 2015) were downloaded from the European Nucleotide Archive (EMNL-EBI – http://www.ebi.ac.uk/ena – last accessed May 16 2016) and included in this study. These sequences were selected from high-quality short-read sequences, to cover the broadest range of time, geographical origin and genomic diversity for published Salmonella Paratyphi A isolates. We included 80 of the 149 genomes from the study by Zhou et al. (2014) and 13 of the 22 genomes from the study by Yan et al. (2015). The accession numbers of each of these genomes are listed in Table S1. The genomes of the S. enterica serotypes Choleraesuis (GenBank accession no. AE017220), Dublin (CP001144), Enteritidis (AM933172), Gallinarum (AM933173), Heidelberg (CP001120), Newport (CP001113), Agona (CP001138), Paratyphi B (CP000886), Paratyphi C (CP000857), Schwarzengrund (CP001127), Typhi (AE014613) and Typhimurium (AE006468) were used as outgroups.
Read alignment and SNP detection.
For each isolate, we aligned the paired-end reads with the Salmonella Paratyphi A ATCC 9150 reference genome (McClelland et al., 2004) (GenBank accession no. NC_006511), using Bowtie with default parameters (Langmead & Salzberg, 2012). SAMtools (Li et al., 2009) was then used to build a genome index and to identify SNPs from the Bowtie alignments. Several criteria were used to filter the resulting SNPs: a minimum coverage (number of reads mapped to the reference genome) of 20 and a minimum quality score of 25 for each SNP. The SNPs retained were concatenated to generate a multiple alignment of all SNPs by an in-house Perl script. The resulting sequences were further filtered to remove all SNPs present in insertion sequences identified by ISfinder (Siguier et al., 2006). Other repetitive regions were identified by a self-self blast analysis (Altschul et al., 1990) of the reference sequence, using the following parameters: megablast (word size 28), identity percentage >95 % and match length >400 bp. This masked 3.3 % of the reference genome. Finally, clusters of SNPs introduced via horizontal sequence transfer were detected and removed with Gubbins (Croucher et al., 2015).
Phylogenetic and temporal analyses.
SNP alignments were analysed with Bayesian evolutionary analysis sampling trees (BEAST) (Drummond & Rambaut, 2007) version 1.8.2, which was used to reconstruct temporal phylogenetic trees and to estimate divergence times. The input consisted of a continuous alignment of 4810 non-repetitive, non-recombinant SNPs from 159 strains, together with the numbers of invariant A, C, T and G nucleotides. The concatenated SNP alignments were subjected to multiple BEAST analyses with both constant-size and Bayesian skyline population size-change models, in combination with strict, log-normal or exponential relaxed clock models, to identify the best-fit model. For the BEAST analysis, the GTR+Γ substitution model was selected, and tip dates were defined as the year of isolation. For all model combinations, three independent chains of 100 million generations each were run to ensure convergence, with sampling every 1000 iterations. Convergence and effective sample size values were inspected with Tracer (Drummond & Rambaut, 2007) version 1.5. A marginal likelihood estimation was carried out, with path sampling and stepping stone sampling for each run that had converged, to compare the different combinations of clock and tree models (Baele et al., 2012, 2013). We then used marginal likelihood estimation to determine which model gave the best fit, by calculating the Bayes factor. The relaxed (uncorrelated exponential) clock model, which allows evolutionary rates to vary between the branches of the tree, and the skyline demographic model provided the best fits to the data (Table S2), as previously reported by Zhou et al. (2014).
The parameter and tree estimates of the three runs were combined with LogCombiner (Drummond & Rambaut, 2007) version 1.7.5, with the first 20 % of states in each chain removed as burn-in. Maximum clade credibility trees were generated with TreeAnnotator (Drummond & Rambaut, 2007) version 1.7.5 on the combined files, and visualised with FigTree version 1.4.2 (Drummond & Rambaut, 2007).
Furthermore, a maximum-likelihood (ML) approach with mega6 (Tamura et al., 2013) was used to support the Bayesian phylogeny. For the ML analysis, mega6 was run with the general time-reversible model and a gamma distribution, to model site-specific rate variation (i.e. the GTR+Γ substitution model). One hundred bootstrap replicate analyses were performed to assess the ML phylogeny. The final tree was visualised in FigTree version 1.4.2.
Genetic analyses.
The presence and type of antibiotic-resistance genes (ARGs) were determined with ResFinder version 2.1 (Zankari et al., 2012). The presence of mutations in the QRDR of the DNA gyrase and topoisomerase IV genes was assessed by the visual examination of de novo assembled sequences.
Pan-genome analysis.
Roary (Page et al., 2015) version 3.2.4 was used on SPAdes (Bankevich et al., 2012) assemblies annotated with Prokka (Seemann, 2014) to construct a pan-genome.
Results
Epidemic curves
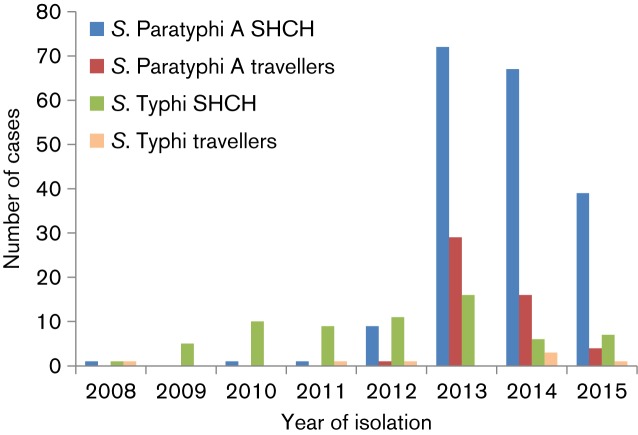
During the 2008–2012 period, enteric fever in patients attending the SHCH and in travellers returning from Cambodia to France was caused mostly by Salmonella Typhi (Fig. 1). During this period, 12 cases of Salmonella Paratyphi A infection were identified among patients attending the SHCH, and only 1 such case was reported in a traveller returning from Cambodia to France. In 2013, a sharp increase in the number of cases was observed both at the SHCH and among travellers returning from Cambodia to France, with a total of 72 and 29 cases, respectively. In 2014 and 2015, the annual number of cases decreased, but remained higher than for the period preceding 2013. During this outbreak, the number of Salmonella Typhi cases remained relatively stable.
Fig. 1.
Enteric fever cases at SHCH, Phnom Penh, Cambodia, and in travellers returning to France from Cambodia, 2008–2015. The numbers of blood culture-confirmed cases of enteric fever (only the first isolate per patient was included) at SHCH and the numbers of confirmed cases in migrants or travellers returning to France from Cambodia during the 2008–2015 period are shown.
In total, during the 2013–2015 period, 178 Salmonella Paratyphi A cases were reported among SHCH patients, and 49 cases were reported among travellers returning from Cambodia to France. The median age of the affected patients at the SHCH during this period was 26 years (interquartile range 22–31). Male and female patients were equally affected (48.9 % of the cases were male); none of these patients died. The median age of the affected travellers returning to France was 33 years (interquartile range 27–49), and 43 % were male; none of these patients died.
Phylogenetic and temporal analyses
We performed short-read genomic sequencing (Illumina) on 65 Salmonella Paratyphi A isolates (Table S1), including 55 from patients in or returning from Cambodia, obtained between 1999 and 2014. Thirty of these fifty-five Cambodian isolates were collected in Phnom Penh, between 2008 and 2013, the other 25 being collected from travellers returning to France or New Zealand from Cambodia (including seven who had also travelled to neighbouring countries, such as Vietnam, Laos, Thailand and Malaysia) or migrants from Cambodia. The other ten isolates sequenced were obtained between 2013 and 2014, from French travellers who had not visited Cambodia but had travelled to other countries in South-East Asia, Southern Asia or Africa, and from a case where the patient had not travelled abroad (laboratory contamination). Short-read sequences from 80 published Salmonella Paratyphi A genomes (Zhou et al., 2014) were selected and included in our study, to provide the population structure framework of this serotype, with its seven lineages, A to G. Short-read sequences from 13 published Salmonella Paratyphi A genomes associated with a large-scale community outbreak in Southern China during the 2010–2011 period were also included (Yan et al., 2015). The assembled genome of ATCC 9150 – a laboratory strain of unknown origin (McClelland et al., 2004) – was used as the reference genome. The genomic analysis was carried out on a final set of 159 Salmonella Paratyphi A genomes, including 63 linked to Cambodia during the 1958–2014 period.
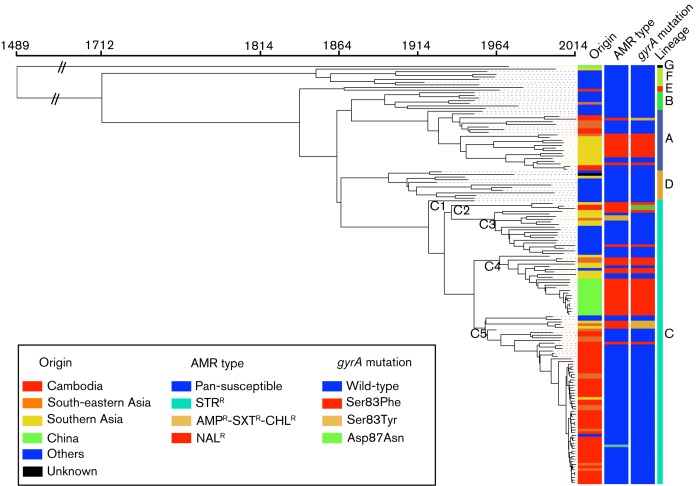
ML (Fig. S1) and BEAST (Fig. 2) phylogenetic analyses were performed on 4810 chromosomal SNPs from the non-repetitive non-recombinant core genome of 159 Salmonella Paratyphi A genomes (Table S3). Both approaches confirmed the presence of the seven lineages, A to G, described by Zhou et al. (2014). Three lineages, A (n=7, 1958–2014), E (n=1, 1965) and C (n=55, 1999–2014) were observed among the 63 genomes (55 from our study and 8 from that of Zhou et al., 2014) acquired in Cambodia between 1958 and 2014. Lineage C could be subdivided into five clades, C1 to C5, with all but 2 of the 55 Cambodian lineage C isolates belonging to C5. The two remaining isolates belonged to C2.
Fig. 2.
Timed phylogeny for 4810 SNPs in 159 Salmonella Paratyphi A genomes. A maximum clade credibility tree generated by BEAST 1.8 (exponential clock rate and Bayesian skyline population models) is shown, providing information about the geographical origin, antimicrobial resistance phenotype (AMR type), gyrA mutation encoding resistance to nalidixic acid and lineages of the genomes. The different clades (C1 to C5) of lineage C are also indicated. The tips of the tree are coloured according to the geographical origin of the genomes. The abbreviations used for the antibiotics are as follows: AMP, ampicillin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim/sulfamethoxazole. AMPR indicates resistance to AMP. NALR indicates resistance to NAL, which was associated with decreased susceptibility to ciprofloxacin (MIC in the range 0.125–1 mg l−1).
ML and BEAST analyses revealed that all but 3 of the 47 isolates collected in Cambodia or obtained from travellers returning from Cambodia between 2010 anin clade C5 (Table 1 and S1, Figs 2 and S1). During the 2013–2014 outbreak period, all but 2 of the 36 isolates acquired in Cambodia belonged to clade C5. The two remaining isolates belonged to genetic groups A and C2. Certain SNPs were identified exclusively within clade C5. Thirty-two of these specific SNPs were found in more than 70 % of the sixty-four C5 isolates, including three found in all the C5 isolates (Table S4). The mean intra-clade pairwise SNP variation within C5 isolates was 23 (minimum 1–maximum 82), whereas it was 7 (1–29) if only the C5 isolates collected between 2010 and 2014 were taken into account. No particular geographical or temporal clustering was observed for the patients hospitalised at the SHCH and infected with such C5 isolates. This outbreak strain was derived from a strain that had been circulating locally and regionally, as C5 isolates had already been isolated in Cambodia from 1999, and in Vietnam from 1963 to 2005 (Table S1). The C5 strain responsible for the outbreak was also isolated from travellers returning from Nepal (one case), Thailand (one case) and Vietnam (one case) in 2013 (Table S1, Fig. S1). However, the Cambodian outbreak strain was different from that responsible for the Chinese outbreak in 2010–2011 (Yan et al., 2015), which belonged to clade C4 (Table 2, Table S1, and Fig. 2). The C4 Chinese isolates differed from the C5 isolates by 77 SNPs on average (48–91), and the mean intra-clade pairwise SNP variation within C4 isolates was 11 (3–33).
Table 1. Characteristics of the 63 Salmonella Paratyphi A genomes linked to Cambodia, 1958–2014.
The number of isolates (n) is indicated when n>1; it includes seven isolates from patients who had also travelled to neighbouring countries, such as Vietnam, Laos, Thailand and Malaysia. AMR type, Antimicrobial resistance type; NAL, nalidixic acid; STR, streptomycin.
| Period | AMR type (n) | Lineage (n) | No. of genomes |
|---|---|---|---|
| 1958–1998 | Pan-susceptible (3) | A (2), E | 3 |
| 1999–2009 | Pan-susceptible (10) | C5 (8), A (2) | 13 |
| NAL (3) | A*, C2†, C5* | ||
| 2010–2014 | Pan-susceptible (44) | C5 (43), A | 47 |
| NAL (2) | A‡, C2† | ||
| STR | C5 |
*Ser83Phe gyrA mutation encoding NAL resistance.
†Asp87Asn gyrA mutation encoding NAL resistance.
‡Ser83Tyr gyrA mutation encoding NAL resistance.
Table 2. Characteristics of the 55 Asian Salmonella Paratyphi A genomes not linked to Cambodia, 1943–2014.
The number of isolates (n) is indicated when n>1. AMR type, Antimicrobial resistance type; MDR, multi-drug resistant (to ampicillin, streptomycin, sulfamethoxazole-trimethoprim, chloramphenicol and tetracycline); NAL, nalidixic acid; STR, streptomycin.
| Period | Southern Asia (n=28) | South-East Asia (n=12) | Eastern Asia (n=15) | |||
|---|---|---|---|---|---|---|
| AMR type (n) | Lineage (n) | AMR type (n) | Lineage (n) | AMR type (n) | Lineage (n) | |
| 1943–1998 | Pan-susceptible (5) | C4 (2), C3 (2), D | Pan-susceptible (3) | A, B, C51 | Pan-susceptible | G |
| NAL | A* | |||||
| 1999–2009 | Pan-susceptible (5) | A (2), C3, C4, F | Pan-susceptible (3) | A (2), C5 | NAL (3) | C4* (3) |
| NAL (13) | A* (8), C5† (2), C2*, C3*, C4* | NAL (2) | C4*, C5† | |||
| MDR | C3 | MDR | C3 | |||
| 2010–2014 | Pan-susceptible (2) | C4, C5 | Pan-susceptible (2) | C5 (2) | NAL (11) | C4* (11) |
| NAL | C4* | NAL | C4* | |||
Oldest C5 genome (Vietnam, 1963).
*Ser83Phe gyrA mutation encoding NAL resistance.
†Ser83Tyr gyrA mutation encoding NAL resistance.
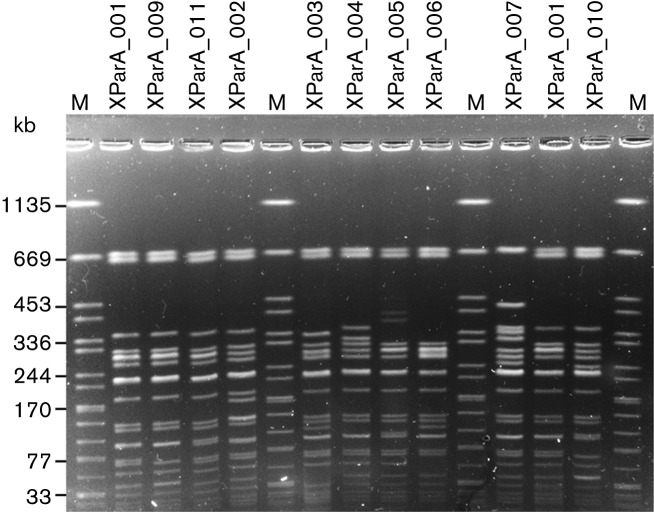
We also assessed the genetic diversity of the Salmonella Paratyphi A isolates by PulseNet standardized PFGE. A subsample of 51 Salmonella Paratyphi A isolates was first analysed by XbaI-PFGE (Table 3 and Fig. 3). A predominant profile, XParA_001 was observed in 35/41 (85.4 %) C5 isolates analysed. However, three other XbaI-PFGE profiles were seen in the remaining C5 isolates. The XParA_001 profile was also found in 75 % (3/4) of the C2 and C3 isolates analysed, but not in C4 isolates, which were phylogenetically closer to C5 (Fig. 2). Similar findings were obtained for analyses with a second enzyme, BlnI, on 24 of the 51 isolates (Fig. S2). A predominant profile, BParA_001, was observed in 14/15 C5 isolates analysed. However, another profile was seen in the remaining Cambodian outbreak C5 isolate. The BParA_001 profile was also found in all other C2 and C3 isolates tested, but not in C4 isolates.
Table 3. Correlation between PFGE data and genomic sequences from 51 Salmonella Paratyphi A isolates.
| Lineage | Combined XbaI-/BlnI-PFGE profiles (n)* | No. of genomes |
|---|---|---|
| A | XParA_003/BParA_003, XParA_006/BParA_006 | 2 |
| C2 | XParA_001/BParA_001, XParA_001/nd | 2 |
| C3 | XParA_001/BParA_001, XParA_009/BParA_001 | 2 |
| C4 | XParA_004/BParA_004 (2), XParA_002/BParA_004, XParA_007/BParA_007 | 4 |
| C5 | XParA_001/nd (24), XParA_001/BParA_001 (11), XParA_011/BParA_001 (2), XParA_011/nd (2), XParA_005/BParA_001, XParA_010/BParA_002 | 41 |
nd, Not determined.
*The number of isolates (n) is indicated when n >1.
Fig. 3.
Representative XbaI-PFGE profiles of Salmonella Paratyphi A isolates. The 10 different XbaI–PFGE profiles obtained from the analysis of 51 Salmonella Paratyphi A isolates are shown. Lanes 1, 6, 11 and 15, S. enterica serotype Braenderup H9812 used as a molecular size marker (M) (band sizes in kb); lane 2, C5 isolate 4416 (XParaA_001); lane 3, C3 isolate 201300701 (XParaA_009); lane 4, C5 isolate 201404185 (XParaA_011); lane 5, C4 isolate 201304008 (XParaA_002); lane 7, A isolate 03–2557 (XParaA_003); lane 8, C4 isolate 201301308 (XParaA_004); lane 9, C5 isolate 201311858 (XParaA_005); lane 10, A isolate 201400552 (XParaA_006); lane 12, C4 isolate 201403926 (XParaA_007); lane 13, C5 isolate 99–7427 (XParaA_001); and lane 14, C5 isolate 5288/46 (XParaA_010).
Antibiotic-susceptibility testing and determination of resistance mechanisms
The Cambodian outbreak strain was pan-susceptible to antibiotics, with the exception of one isolate that was resistant only to streptomycin. This outbreak strain was susceptible to quinolones (i.e. nalidixic acid) and fluoroquinolones (i.e. ciprofloxacin), in particular. By contrast, the two non-C5 isolates collected during the outbreak in Cambodia were resistant to nalidixic acid (MIC>256 mg l−1) with decreased susceptibility to ciprofloxacin (MIC 0.09–0.38 mg l−1). This resistance was mediated principally by mutations in the QRDR of gyrA, the chromosomal DNA gyrase A gene. We therefore checked for such mutations within this QRDR. We identified mutations leading to a serine to tyrosine substitution at codon 83 (Ser83Tyr) in the lineage A isolate from 2014, and to an aspartic acid to asparagine substitution at codon 87 (Asp87Arg) in the clade C2 isolate from 2013. Quinolone-resistant Salmonella Paratyphi A bacterial populations were also present in Cambodia before the outbreak period, as we identified three nalidixic acid-resistant strains isolated there during the 2000–2006 period. They belonged to lineage A, and clades C2 and C5 of lineage C, and they carried the gyrA mutations Ser83Phe, Asp87Asn and Ser83Phe, respectively (Table 1). The C5 nalidixic-acid resistant isolate was collected in 2000, 11 years before the outbreak. The Chinese outbreak was also caused by a strain resistant to nalidixic acid with decreased susceptibility to ciprofloxacin (Yan et al., 2015). Within the genomic sequences of the Chinese isolates, we identified the very common gyrA mutation Ser83Phe (Table 2, Table S1). Overall, various gyrA mutations were acquired at least 10 times in the 159 global Salmonella Paratyphi A genomes studied, but only in lineages A and C (Fig. 2). The gyrA mutation leading to Ser83Phe substitution was the most frequently observed.
Pan-genome analysis
The pan-genome analysis (Fig. S3) carried out by Roary identified a total of 7009 genes, including 3342 for the core genome, in the 159 genomes studied. We detected no gains of particular accessory genes nor losses of common genes in the Cambodian outbreak strain (Table S5).
Recent evolution in Cambodia
The epidemic curve based on FNRC-ESS data showed a sharp decrease in the number of Salmonella Paratyphi A infections among travellers returning to France from Cambodia between 2013 (n=25) and 2015 (n=5). However, the number of nalidixic acid-resistant isolates increased from 0 % (0/25) in 2013 to 12.5 % (2/16) in 2014 and 80 % (4/5) in 2015. The only nalidixic acid-resistant isolate sequenced here, 201400552, belonged to lineage A and had the gyrA mutation leading to a Ser83Tyr substitution. The question was, therefore, whether the other five recent nalidixic acid-resistant isolates belonged to lineage A or to the outbreak C5 clade, with the acquisition of a mutation affecting the QRDR of gyrA. As these five isolates were collected very recently (after the whole-genome study), we developed a SNP genotyping assay to determine whether these isolates belonged to lineage A or clades C2 and C5 of lineage C, which are known to contain nalidixic acid-resistant isolates. In this SNP assay, the five recent nalidixic acid-resistant isolates were found to belong to the C5 clade and to have the gyrA mutation leading to the Ser83Phe substitution.
Discussion
The analysis of whole-genome sequencing data from isolates unambiguously identified a single predominant strain (C5 clade) among the isolates collected during the recent outbreak of paratyphoid fever in Cambodia, and made it possible to assess the phylogenetic relationships between this strain and other strains circulating in Cambodia or elsewhere in Asia at or before this time. The inclusion of many other publicly available genomes, including, in particular, 80 representative genomes from a global collection of 149 Salmonella Paratyphi A isolates obtained between 1917 and 2009, increased the discriminatory power of the genomic epidemiology analysis, making it possible to define and trace back the outbreak strain.
The genomic analysis showed that the C5 clade was not confined to Cambodia, but was also found in travellers returning from Nepal, Thailand and Vietnam in 2013, suggesting that the outbreak strain may have been circulating beyond the borders of Cambodia. However, these countries did not experience similar outbreaks of Salmonella Paratyphi A infections during the same period. An outbreak of Salmonella Paratyphi A infections was reported in China in 2010–2011, but we found that this outbreak was caused by a strain from a different clade, C4. However, the C4 and C5 clades, separated by a mean of 77 SNPs, shared a common ancestor around 1950.
By contrast, PFGE, the widely-used gold standard method for subtyping Salmonella spp. to the strain level, did not attain the same discriminatory power, despite the presence of a predominant PFGE profile among the outbreak isolates. The presence of other PFGE profiles in the C5 outbreak isolates made the epidemiological analysis harder to interpret. For example, the 11 C5 isolates collected at the SHCH in Phnom Penh during 2012–2013 presented three different PFGE profiles (the main profile, XParA_001, in seven isolates, XParA_011 in three isolates and XParA_010 in one isolate). Furthermore, the predominant XbaI- and BlnI-PFGE profiles, XParA_001 and BParA_001, were also observed in C3 isolates from West Africa. In addition to the weak discriminatory power of this low-throughput fingerprinting method, the PFGE data had low portability. Despite using the same standardized protocol, the figures showing PFGE data in the papers by Saitoh et al. (2016) and Yan et al. (2015) do not contain a molecular size ladder, precluding any comparative analysis between these isolates and our isolates. By contrast, the increasing availability in public databases of short-read sequences from published genomes or of sequences generated during the routine surveillance of Salmonella infections (Ashton et al., 2016) has strongly improved the identification and investigation of outbreaks, particularly those of an international nature.
Unlike the strain responsible for the Chinese outbreak in 2010–2011, the Cambodian outbreak strain was pan-susceptible to the antibiotics tested. This antibiotic susceptibility is of particular importance, as a clear trend towards the acquisition of antibiotic resistance in Salmonella Paratyphi A isolates from Asia has been observed, despite the lack of data from South-East Asia (Gu et al., 2015; Teh et al., 2014). The susceptibility of the outbreak strain to quinolones, and the isolation of a nalidixic acid-resistant C5 strain in Cambodia 11 years before the current outbreak, suggest that neither quinolone resistance nor resistance to any other antibiotic was an important bacterial factor in this outbreak, at least before 2014. Despite a decreasing number of cases between 2013 and 2015, both at the SHCH and in travellers returning from Cambodia to France (Fig. 1), the increase in the isolation of nalidixic acid-resistant C5 isolates from 2014 is worrying, as these isolates also had decreased susceptibility to ciprofloxacin, the first-line treatment in adults. We now need to monitor Salmonella Paratyphi A infections and their susceptibility to antibiotics to determine whether there is a real trend towards an increase in nalidixic acid-resistant strains over time. These recent C5 isolates are also being investigated by whole-genome sequencing, to determine whether they form a tight cluster or are distributed evenly throughout the C5 clade, and whether they are derived from the 2013–2014 outbreak strain or from more ancestral C5 populations.
In their comprehensive comparative genomics analysis of a global collection representative of the population structure of this serotype, Zhou and co-workers concluded that the crucial genomic elements of Salmonella Paratyphi A responsible for causing paratyphoid fever were already present in the most common recent ancestor dating from the mid-15th century. Furthermore, they found that almost all genetic changes (gene acquisitions or losses and mutations) since that time had been transient (i.e. removed by purifying selection) (Zhou et al., 2014). These findings are indicative that environmental and/or human behavioural factors enhancing transmission to naive hosts are more likely to explain outbreaks of paratyphoid fever than the recent emergence of a particularly virulent Salmonella Paratyphi A strain. Such environmental and/or human behavioural risk factors for paratyphoid fever have been investigated in case–control studies in Asia. In Nepal, different routes of infection were identified for Salmonella Typhi and Salmonella Paratyphi (Karkey et al., 2013). The significant identified risk factors for Salmonella Paratyphi A infections were the consumption of street food during the 2 weeks preceding the onset of illness and residence in the capital city, Kathmandu, for less than 2 years. In Indonesia, Salmonella Typhi and Salmonella Paratyphi A were also found to follow different routes of infection (Vollaard et al., 2004). The risk factors associated with a Salmonella Paratyphi A infection were identified as the consumption of food from street vendors and flooding. In South China, during a community outbreak of 600 cases, the main risk factor was the consumption of uncooked vegetables from street stalls, local markets or small restaurants (Yan et al., 2015). The primary source was a vegetable patch irrigated with untreated hospital wastewater.
These findings highlight the need for solid public-health investigations and interventions, together with food-safety regulations, in Cambodia. Significant steps in terms of food safety were taken in Cambodia in 2015, when a draft of the country’s first food law was released.
For future public health investigations, the use of whole-genome sequencing data in real-time, together with classical epidemiological methods, would be advantageous, as demonstrated here by the unequalled discriminatory power of genomic data for tracking individual strains (i.e. distinguishing outbreak-associated isolates from those of contemporary sporadic cases and assessing the true genetic relationships between isolates) from this genetically monomorphic bacterial population. The collection of epidemiological and molecular data should be a collaborative effort, to maximise the gain from combining the two types of data. Efforts should also be made, during initial investigations, to isolate strains from suspected water and/or food sources and from identified carriers. This should make it possible to optimise the use of high resolution provided by whole-genome sequencing and to demonstrate a clear link with the suspected source, as demonstrated by Yan et al. (2015) during the outbreak in China in 2010–2011.
Acknowledgements
The surveillance of bloodstream infections at the SHCH is supported by the Belgian Directorate of Development Cooperation (DGD), through project 2.08 of the Third Framework Agreement between the Belgian DGD (Ministry of Development Cooperation) and the Institute of Tropical Medicine. We would like to thank Barbara Barbé from the Institute of Tropical Medicine, Antwerp, Belgium, Laurence Ma and Magali Tichit from the Institut Pasteur, Paris, France for technical assistance, and all colleagues involved in blood culture surveillance at SHCH. We thank all the corresponding laboratories of the FNRC-ESS. This study was supported by the Institut Pasteur and the Institut Pasteur International Network, Santé Publique France, the French government’s Investissement d’Avenir programme, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant number ANR-10-LABX-62-IBEID), and the Fondation Le Roch-Les Mousquetaires. The Institut Pasteur Genomics Platform is a member of ‘France Génomique’ consortium (ANR10-INBS-09-08). L. M.F.K. is supported by the Flemish Ministry of Sciences (EWI, SOFI project IDIS).
Supplementary Data
Abbreviations:
- BEAST
Bayesian evolutionary analysis sampling trees
- CA-SFM
Antibiogram Committee of the French Society for Microbiology
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- FNRC-ESS
French National Reference Centre for E. coli, Shigella and Salmonella
- ML
maximum likelihood
- MIC
minimal inhibitory concentration
- PFGE
pulsed-field gel electrophoresis
- QRDR
quinolone-resistance-determining region
- SHCH
Sihanouk Hospital Centre of HOPE
- SNP
single-nucleotide polymorphism
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J.(1990). Basic local alignment search tool. J Mol Biol 215403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Ashton P. M., Nair S., Peters T. M., Bale J. A., Powell D. G., Painset A., Tewolde R., Schaefer U., Jenkins C., et al. (2016). Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4,e1752. 10.7717/peerj.1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., Alekseyenko A. V.(2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 292157–2167. 10.1093/molbev/mss084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Li W. L., Drummond A. J., Suchard M. A., Lemey P.(2013). Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30239–243. 10.1093/molbev/mss243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan M. K., Bahl R., Bhatnagar S.(2005). Typhoid and paratyphoid fever. Lancet 366749–762. 10.1016/S0140-6736(05)67181-4 [DOI] [PubMed] [Google Scholar]
- CA-SFM & EUCAST (2014). Comité de l’Antibiogramme de la Société Française de Microbiologie Recommandations 2014. http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM_EUCAST_2014_V2_0.pdf.
- Croucher N. J., Page A. J., Connor T. R., Delaney A. J., Keane J. A., Bentley S. D., Parkhill J., Harris S. R.(2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43e15. 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A.(2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7,214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC (2013). Annual Epidemiological Report: Reporting on 2011 Surveillance Data and 2012 Epidemic Intelligence Data. Stockholm: European Centre for Disease Prevention and Control. http://ecdc.europa.eu/en/publications/Publications/annual-epidemiological-report-2013.pdf
- Grimont P. A., Weill F., X(2007). Antigenic formulae of the Salmonella serotypes, 9th edn. WHO Collaborating Centre for Reference and Research on Salmonella – Institut Pasteur, Paris, France. https://www.pasteur.fr/sites/www.pasteur.fr/files/wklm_en.pdf.
- Gu W., Yang Z., Chen Y., Yin J., Yang J., Li C., Zhou Y., Yin J., Xu W., et al. (2015). Molecular characteristics of Salmonella enterica Paratyphi A in Yunnan province, Southwest China. Infect Genet Evol 30181–185. 10.1016/j.meegid.2014.12.028 [DOI] [PubMed] [Google Scholar]
- Judd M. C., Grass J. E., Mintz E. D., Bicknese A., Mahon B. E.(2015). Salmonella enterica Paratyphi A infections in travelers returning from Cambodia, United States. Emerg Infect Dis 211089–1091. 10.3201/eid2106.150088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkey A., Thompson C. N., Tran Vu Thieu N., Dongol S., Le Thi Phuong T., Voong Vinh P., Arjyal A., Martin L. B., Rondini S., et al. (2013). Differential epidemiology of Salmonella Typhi and Paratyphi A in Kathmandu, Nepal: a matched case control investigation in a highly endemic enteric fever setting. PLoS Negl Trop Dis 7e2391. 10.1371/journal.pntd.0002391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L.(2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hello S., Harrois D., Bouchrif B., Sontag L., Elhani D., Guibert V., Zerouali K., Weill F. X.(2013). Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect Dis 13672–679. 10.1016/S1473-3099(13)70124-5 [DOI] [PubMed] [Google Scholar]
- Leekitcharoenphon P., Nielsen E. M., Kaas R. S., Lund O., Aarestrup F. M.(2014). Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9e87991. 10.1371/journal.pone.0087991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 252078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Clifton S. W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., et al. (2004). Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 361268–1274. 10.1038/ng1470 [DOI] [PubMed] [Google Scholar]
- Newton A. E., Routh J. A., Mahon B. E.(2016). Infectious diseases related to travel: typhoid & paratyphoid fever. CDC Health Information for International Travel. Edited by Brunette G. W.New York: Oxford University Press. [Google Scholar]
- Ochiai R. L., Wang X., von Seidlein L., Yang J., Bhutta Z. A., Bhattacharya S. K., Agtini M., Deen J. L., Wain J., et al. (2005). Salmonella Paratyphi A rates, Asia. Emerg Infect Dis 111764–1766. 10.3201/eid1111.050168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T., Fookes M., Falush D., Keane J. A., Parkhill J.(2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 313691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot E. M., Fair M. A., Gautom R., Cameron D. N., Hunter S. B., Swaminathan B., Barrett T. J.(2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for pulsenet. Foodborne Pathog Dis 359–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Morita M., Shimada T., Izumiya H., Kanayama A., Oishi K., Ohnishi M., Sunagawa T.(2016). Increase in paratyphoid fever cases in Japanese travellers returning from Cambodia in 2013. Epidemiol Infect 144602–606. 10.1017/S0950268815001648 [DOI] [PubMed] [Google Scholar]
- Seemann T.(2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 302068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M.(2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh C. S., Chua K. H., Thong K. L.(2014). Paratyphoid fever: splicing the global analyses. Int J Med Sci 11732–741. 10.7150/ijms.7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien Y. Y., Wang Y. W., Tung S. K., Liang S. Y., Chiou C. S.(2011). Comparison of multilocus variable-number tandem repeat analysis and pulsed-field gel electrophoresis in molecular subtyping of Salmonella enterica serovars Paratyphi A. Diagn Microbiol Infect Dis 69 1–6. 10.1016/j.diagmicrobio.2010.08.012 [DOI] [PubMed] [Google Scholar]
- Tourdjman M., Le Hello S., Gossner C., Delmas G., Tubiana S., Fabre L., Kerléguer A., Tarantola A., Fruth A., et al. (2013). Unusual increase in reported cases of paratyphoid A fever among travellers returning from Cambodia, January to September 2013. Euro Surveill 18,20594. 10.2807/1560-7917.ES2013.18.39.20594 [DOI] [PubMed] [Google Scholar]
- Vlieghe E., Phe T., De Smet B., Veng C. H., Kham C., Sar D., van Griensven J., Lim K., Thai S., et al. (2013). Increase in Salmonella enterica serovar Paratyphi A infections in Phnom Penh, Cambodia, January 2011 to August 2013. Euro Surveill 18,20592. 10.2807/1560-7917.ES2013.18.39.20592 [DOI] [PubMed] [Google Scholar]
- Vollaard A. M., Ali S., van Asten H. A., Widjaja S., Visser L. G., Surjadi C., van Dissel J. T.(2004). Risk factors for typhoid and paratyphoid fever in Jakarta, Indonesia. JAMA 2912607–2615. 10.1001/jama.291.21.2607 [DOI] [PubMed] [Google Scholar]
- Weill F., X, Bertrand S., Guesnier F., Baucheron S., Cloeckaert A., Grimont P. A.(2006). Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg Infect Dis 121611–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Yang B., Wang Z., Wang S., Zhang X., Zhou Y., Pang B., Diao B., Yang R., et al. (2015). A large-scale community-based outbreak of paratyphoid fever caused by hospital-derived transmission in Southern China. PLoS Negl Trop Dis 9e0003859. 10.1371/journal.pntd.0003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F. M., Larsen M. V.(2012). Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 672640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., McCann A., Weill F. X., Blin C., Nair S., Wain J., Dougan G., Achtman M.(2014). Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A 11112199–12204. 10.1073/pnas.1411012111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1.Salmonella enterica subsp. enterica serovar Choleraesuis strain SC-B67 complete genome: GenBank accession number AE017220.
- 2.Salmonella enterica subsp. enterica serovar Dublin strain CT_02021853 complete genome: GenBank accession number CP001144.
- 3.Salmonella enterica subsp. enterica serovar Enteritidis strain P125109 complete genome: GenBank accession number AM933172.
- 4.Salmonella enterica subsp. enterica serovar Gallinarum strain 287/91 complete genome: GenBank accession number AM933173.
- 5.Salmonella enterica subsp. enterica serovar Heidelberg strain SL476 complete genome: GenBank accession number CP001120.
- 6.Salmonella enterica subsp. enterica serovar Newport strain SL254 complete genome: GenBank accession number CP001113.
- 7.Salmonella enterica subsp. enterica serovar Agona strain SL483 complete genome: GenBank accession number CP000886.
- 8.Salmonella enterica subsp. enterica serovar Paratyphi B strain SPB7 complete genome: GenBank accession number CP001120.
- 9.Salmonella enterica subsp. enterica serovar Paratyphi C strain RKS4594 complete genome: GenBank accession number CP000857.
- 10.Salmonella enterica subsp. enterica serovar Schwarzengrund strain CVM19633 complete genome: GenBank accession number CP001127.
- 11.Salmonella enterica subsp. enterica serovar Typhi strain Ty2 complete genome: GenBank accession number AE014613.
- 12.Salmonella enterica subsp. enterica serovar Typhimurium strain LT2 complete genome: GenBank accession number AE006468.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.