Abstract
Humans are exposed to ionizing radiation not only through background radiation but also through the ubiquitous presence of devices and sources that generate radiation. With the expanded use of radiation in day-to-day life, the chances of accidents or misuse only increase. Therefore, a thorough understanding of the dynamic effects of radiation exposure on biological entities is necessary. The biological effects of radiation exposure on human cells depend on much variability such as level of exposure, dose rate, and the physiological state of the cells. During potential scenarios of a large-scale radiological event which results in mass casualties, dose estimates are essential to assign medical attention according to individual needs. Many attempts have been made to identify biomarkers which can be used for high throughput biodosimetry screening. In this study, we compare the results of different biodosimetry methods on the same irradiated cells to assess the suitability of current biomarkers and push forward the idea of employing a multiparametric approach to achieve an accurate dose and risk estimation.
Keywords: γH2AX, dicentrics, gene signatures, micronuclei, multiparametric approach, radiation biodosimetry, translocations
Introduction
Human beings are being exposed to ionizing radiation in every walk of life because of its diverse use from medical diagnostics to industrial applications. In addition, human exposure to radiation from cosmic rays and naturally occurring radon has become unavoidable. Three different models have been proposed to describe the biological effects of ionizing radiation: (i) Linear threshold, (ii) linear no-threshold, and (iii) hormesis. Among them, the linear no-threshold model assumes that radiation is harmful irrespective of the dose and that there is no threshold for radiation-induced probabilistic or stochastic effects. Therefore, stochastic effects such as gene mutations and cancer incidence are of great concern to humans. Biological effects of ionizing radiation depend on radiation quality defined by linear energy transfer (LET), dose, and dose rate. In general, high LET radiations such as neutrons and α-particles produce far more severe biological effects than γ-rays and X-rays, which are low-LET radiations. Epidemiological studies identified radiation-induced cancer risks at doses above 50 mSv based on the cancer incidence observed among the A-bomb survivors.[1] Although radiation can cause potential health effects, radiation is still the preferred treatment modality for a majority of human cancers.[2,3] Thus, ionizing radiation appears to be a double-edged sword, which can cause cancer on the one side and control cancer progression and spreading on the other.
Occupational or accidental radiological or nuclear exposure can cause serious health effects. In addition, radiological or nuclear terrorism constitutes a potential threat to several nations, where several hundreds and thousands of people may potentially get exposed to radiation. We learn from the nuclear disasters of Chernobyl and Fukushima Daiichi that the first responders to these radiation emergencies sustained high levels of radiation exposures. These incidences clearly illustrate a critical need for suitable biomarkers for personalized radiation dose assessment, which can be useful not only for appropriate medical/clinical management but also for predicting delayed stochastic (no threshold) effects such as genomic instability and cancer.
In the past several decades, various new biodosimetry assays have been developed, and the existing ones have undergone technical improvements to better estimate absorbed radiation dose. In the absence of physical dosimeters, biomarkers or biological dosimeters can be of immense help in estimating the absorbed radiation dose in certain accidental situations that involve nonnuclear workers and the general public. Further, biodosimeters can help in risk stratification of the exposed people and assist in appropriate medical management of individuals who received high doses of radiation exposure. In case of a radiological or a nuclear mass-casualty event, several hundreds and thousands of people may get exposed. Such situation(s) may require technical improvements in existing biodosimetry assays to meet the requirements by radiological triage. In addition, biodosimetry can be helpful in guiding medical treatment of radiation accident victim if assessment is made in a timely manner.
Exposure to ionizing radiation results in a variety of biological effects that depend on physical nature, exposure duration, dose, and dose rate. Although biodosimeters can help in absorbed radiation dose estimation, there is still a big gap in our knowledge with regard to prediction of health risks from estimated radiation dose. Therefore, a thorough understanding of biological effects may help in predicting the short- and long-term effects of radiation exposure. Different biodosimetry assays that are currently used are briefly described below.
Dicentric Chromosome Assay
This method is considered as the “gold standard” for radiation dose assessment.[4,5] The method was first used by Bender and Gooch as a dosimeter to assess the human exposure to radiation.[6] Dicentric analysis is widely used for radiation dose estimation in individuals following accidental and nuclear exposures. Conventionally, G-banding and/or Giemsa staining was used in the analysis of dicentrics in lymphocytes following irradiation.[7,8,9,10,11,12] The dicentric chromosome assay (DCA) is labor intensive and time consuming, requiring 72–96 h for dose estimation. Some of the recent developments of this assay include automated image capturing as well as dicentric scoring using Metafer and telescoring of digital images of metaphase spreads by a large number of skilled and experienced scorers. Recent studies have demonstrated an increased sensitivity of dicentric chromosome detection and scoring by employing fluorescence in situ hybridization (FISH) using centromere- and telomere-specific peptide nucleic acid (PNA) probes. FISH-based DCA has not only increased the sensitivity but also considerably reduced the analysis time. We have successfully employed the PNA-FISH technique for the detection of chromosome aberrations in several of our earlier studies.[13,14,15] An example of a metaphase spread stained for telomeres and centromeres using PNA probes is shown in Figure 1a.
Figure 1.
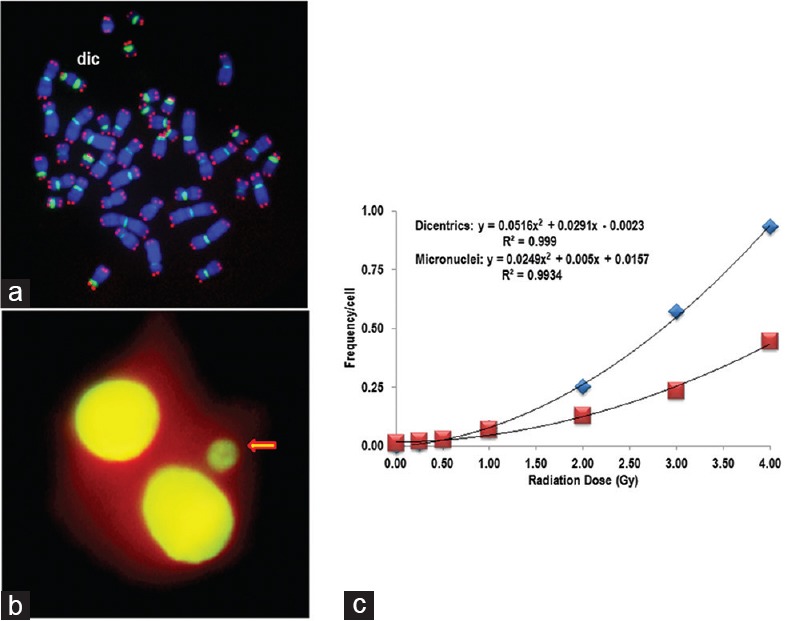
Cytogenetic damage in human lymphocytes following exposure to γ-rays analyzed by chromosome and micronuclei analyses. (a) Peptide nucleic acid-fluorescence in situ hybridization was used to detect dicentric chromosomes. Cy3-telomere (red) and FITC-centromere (green) peptide nucleic acid probes were used along with counterstain DAPI (blue). Metaphase spread shows a dicentric chromosome. (b) Acridine orange stained cytokinesis-blocked binucleated human lymphocytes. Arrow points to a micronucleus present in this cell. (c) Frequencies of dicentrics (blue rectangles) and micronuclei (red rectangles) following gamma irradiation with different doses (0, 0.25, 0.5, 1.0, 2.0, 3.0, and 4 Gy). The data were fitted with polynomial function, and the following equations were obtained: Dicentrics: y = 0.0516x2 + 0.0291x − 0.0023 (R2 =0.999); Micronuclei: y = 0.0249x2 + 0.005x + 0.0157 (R2 = 0.9934)
Using the PNA-FISH technique, we have constructed a calibration curve in our laboratory using the peripheral blood lymphocytes of three healthy human donors following ex vivo irradiation with a range of doses (0, 0.25, 0.5, 1.0, 2.0, 3.0, and 4.0 Gy) of γ rays at a dose rate of 0.85 Gy/min. The data are shown in Figure 1c. In addition to dicentric chromosomes, frequency of chromosome ends without telomeres was also estimated to determine the possibility of using this feature as a potential biomarker for radiation exposure [Figure 2a and b]. Although initial studies are promising, further experiments are required for verification and validation.
Figure 2.
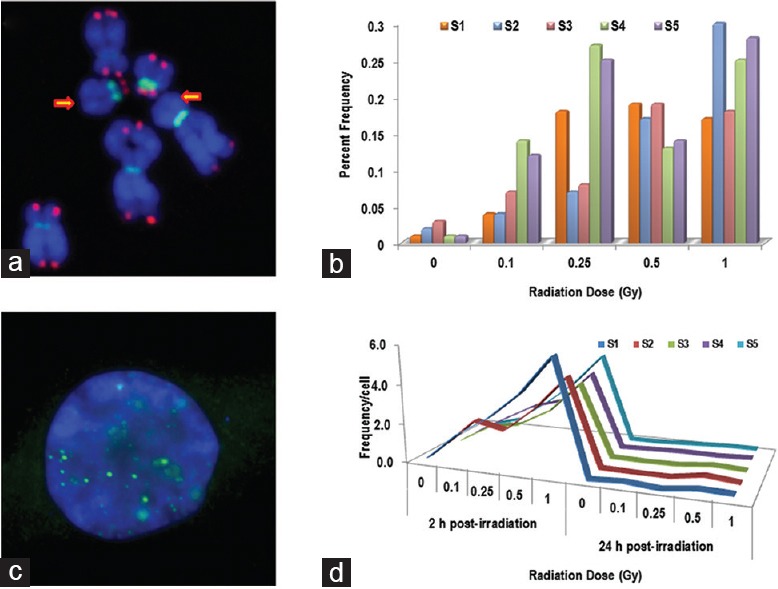
Chromosome and DNA breaks induced by radiation in human lymphocytes. (a) Chromosome ends with undetectable telomeres: Partial metaphase spread showing chromosomes and fragments with undetectable telomeres. Arrows point to the chromosome ends without any telomere signal. (b) Analysis of fragments and chromosome ends without telomere signals showed a dose-dependent response as well as heterogeneity among the different samples studied. (c and d) Induction and kinetics of γH2AX foci following exposure to gamma radiation as a measure of DNA double-strand breaks in human lymphocytes. (c) Immunofluorescence staining of nuclei with anti-γH2AX antibodies (Green) and DAPI as a counterstain (Blue). (d) At 2 h postirradiation, the γH2AX foci were induced in a dose-dependent manner and the frequency of γH2AX foci reduced to basal levels by 24 h postirradiation
Cytokinesis-block Micronucleus Assay
Cytokinesis-block micronucleus (CBMN) has gained considerable importance recently as one of the reliable indicators of radiation exposure although micronuclei formation is not entirely specific to radiation alone. This assay detects micronuclei [Figure 1b] resulting from either whole chromosomes or chromosome fragments that are excluded from mitotic spindle during cell division. Originally, the CBMN assay was developed by Fenech and Morley[16,17] using human peripheral blood lymphocytes. In the subsequent years, the CBMN technique incorporated FISH with centromeric and telomeric probes that enabled the detection of either whole chromosome or chromosome fragments in micronuclei. A recent study utilized the multicolor FISH technique to identify the chromosomes involved in micronuclei formation after radiation exposure.[18] Recently, a new high-throughput and miniaturized CBMN method was also established for rapid processing a large number of samples (in about 3 days), a critical requirement for radiological triage.[19,20] Since the CBMN assay, unlike the DCA, can be easily performed and analyzed without an extensive cytogenetic expertise, many laboratories routinely use the CBMN assay for dose estimation. Both CBMN and DCA can be used in a complementary fashion for dose estimation. Calibration curves constructed from the data of both assays performed in the lymphocytes of three human donors are given in Figure 1c.
Chromosome Translocation Assay by Multicolor Fluorescence In situ Hybridization
Multicolor FISH is very useful technique for genome-wide analysis of numerical and structural changes in humans. Using this technique, ionizing radiation-induced simple and complex chromosomal exchanges (translocations and inversions) can be easily and reliably detected [Figure 3a and b]. These stable chromosomal exchange events can be effectively used for retrospective biodosimetry because of their persistence for prolonged periods of time after radiation exposure. Using this technique, simple and complex chromosome exchanges were detected several years after exposure to plutonium in the lymphocytes of a few nuclear workers of Mayak, Russia.[21] The use of translocations in retrospective biodosimetry has been amply demonstrated in several studies. The overall assay time for translocation analysis can range from 10 to 14 days depending on the quality of metaphases and hybridization.
Figure 3.
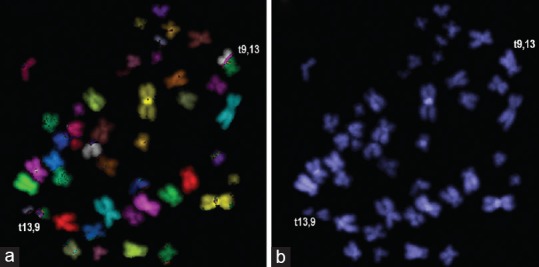
Multicolor fluorescence in situ hybridization was used to visualize the stable chromosome translocations in human lymphocytes after exposure to gamma radiation. (a) A reciprocal translocation between chromosomes 13 and 9 [t(13,9)] is shown. (b) The same metaphase as in A observed under ultraviolet filter
γ-H2ax Assay
Histone H2AX, a variant of histone H2A, is rapidly phosphorylated at serine 139 by exposure to DNA double-strand break (DSB) causing agents including ionizing radiation.[22] Phosphorylated H2AX (γ-H2AX) has been shown to accumulate at the DSB sites [Figure 2c] and is considered as a surrogate marker for DSBs.[23] Several studies have demonstrated the usefulness of γ-H2AX in dose estimation through DSB detection.[24,25,26,27] This assay, unlike DCA and CBMN, obviates the need for lymphocyte stimulation for 48–72 h making it a rapid assay for dose estimation in radiological triage.[28] However, fast kinetics of γ-H2AX foci formation [Figure 2d] and disappearance within a narrow window of 6–8 h makes it somewhat unreliable for precise dose estimation. Nevertheless, this assay can be definitely useful more as a biological indicator of radiation exposure rather than a radiation biodosimeter.
Gene Expression Profiling
It has become evident that studying a single gene response is no longer conclusive.[29] Array-based gene expression studies have yielded valuable information on ionizing radiation-specific genes. Identification of signaling pathways associated with radiation-specific genes as well as posttranslational modification of proteins encoded by these genes have given great insights for understanding the diverse biological effects induced by low and high doses of ionizing radiation with differing LET in diverse mammalian model systems including humans. Available data on tissue-specific expression of radiation-responsive genes can help in predicting the organ/tissue-specific effects. Recently, Lu et al.[30] identified changes in 29 genes which are involved in cell cycle as a biomarker for predicting low doses of radiation exposure. This method can be effectively employed to predict the postexposure impact in terms of disease outcome. One of the first studies to use microarray analysis to investigate the radiation response revealed that about 1344 genes were differentially expressed in a myeloid cancer cell line following 2 Gy irradiation at 4 h.[31]
We used Illumina microarray chip to determine gene expression changes following gamma-ray exposure in human lymphocytes. Analysis compared the expression profile between the nonirradiated and the irradiated cells from the same donor. Expression profile at two time points (2 h and 24 h) was analyzed in the study. Experimental data shown in Figure 4a represent the principle component analysis based clustering and subclustering of the various data points including donors and postirradiation time points. The data showed clear differences in the response of genes based on postirradiation time as well as individual radiosensitivity. However, from biomarker point of view, upregulated genes are preferred than the downregulated genes.
Figure 4.
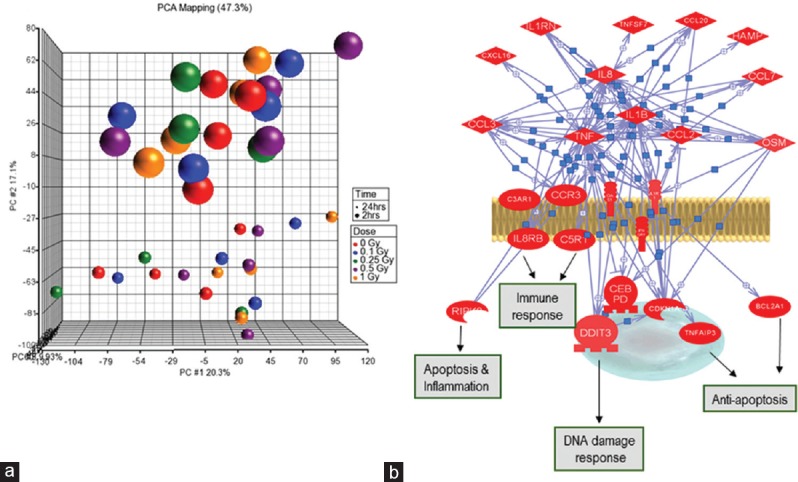
Gene expression profiling in human lymphocytes following radiation exposure. (a) Principal component analysis of transcriptomic profile of human peripheral blood lymphocytes following exposure to gamma rays. Comparison was made between donors, doses, and data points. Three-dimensional principal component analysis of expression profiles from five samples revealed a striking pattern of clustering based on the postirradiation time. This implies that postirradiation time may be the most influential factor on gene expression changes. (b) Pathway and gene network of radiation-induced changes. The gene network was constructed using Pathway Studio (Ariadne Genomics MD, USA) on the basis of biological function of the differentially expressed genes by microarrays and the knowledge obtained in the literature. Red color denotes upregulated genes. Blue lines indicate direct regulation between genes based on literature evidence
To determine whether or not the differentially regulated genes are interconnected within a larger “signaling network,” the selected genes were used to construct intracellular signaling networks based on their expression pattern. During the initial step, we searched and analyzed direct interactions between the differentially regulated genes using the information extracted from the published experimental literature and from the entire National Center for Biotechnology Information – PubMed database. This set of interactions defined the cellular potential for the assembly of protein complexes, signaling, and effector pathways as well. By positioning the expression data onto known validated physical interactions, all possible associations between the regulated genes from our microarray expression data were restricted to those that are considered to be physically possible in the cell. This approach generated condition-specific functional “signature networks,” which contained sets of functional pathways organized into a meta-network. Several well-known key components of DNA damage response are represented in the network, including DNA metabolism, DNA repair, and cell cycle regulation. Genes that were not shown to be associated with radiation-response pathways earlier were also observed such as immune and inflammation mediators responses including cytokine signaling. Recently, there is growing evidence regarding the role of immune/inflammatory response following radiation exposure which can contribute to bystander effects. Furthermore, the knowledge of radiation-induced inflammatory and immune responses would be beneficial in titrating radiation-based therapeutics. This suggests a broader genetic participation in response to radiation exposure than previously thought and highly interacting pathways previously thought to be disjoint.
Gene clusters from the pathways such as immune response, apoptosis and inflammation, and DNA damage response are clearly activated after irradiation [Figure 4b]. We have identified around 70 genes, which are differentially expressed in all the donors studied to the dose range 0.1–1.0 Gy compared to the basal levels in control cells (Data not shown). A similar number of genes were identified at 24 h postradiation exposure. Currently, we are validating these genes in our samples. Although gene expression studies have yielded valuable information, interindividual variation in transcription profile is too extensive for reliable dose estimation. In fact, no fruitful attempt has yet been made to estimate the radiation dose in exposed individuals using gene expression analysis. However, gene expression assay is an expensive and time-consuming technology; specific circulating specific proteins or microRNAs may be more functional as potential emerging biomarkers of radiation exposure. Recently, attempts have been made to analyze gene expression profiles of cancer patients undergoing radiotherapy. More robust studies are needed to verify the suitability of gene expression profiling assay for radiation dose estimation.
Perspective
A multiparametric approach involving different bioassays may have the potential for accurate exposure dose prediction. However, variations exist in radiation dose estimated by different biodosimetry assays – both current and emerging methods. Interindividual variation seems to be an important confounding factor in most of the assays. This study is an attempt to integrate several assays used in biodosimetry for the same sample to determine the best combination of biomarkers that could be used following radiation exposures. This is a study in progress, and we are consolidating the data and refining some approaches that were used in this study. In addition to the established methods, emerging proteomic approach might prove to be a promising tool for the discovery of new potential biomarkers of radiation exposure. These markers, once validated, can be used in biodosimetry in case of a radiological incident/accident. Biomarkers will complement other modes of radiation dosimetry. Apart from estimating the dose of incident radiation, biodosimetry may help in predicting health consequences of radiation exposure in a population. Biodosimetry will facilitate stratification of individuals who need immediate attention and potential medical countermeasures required to mitigate some of the radiation-associated injuries. In certain cases, it is possible to predict long-term risks associated with radiation exposures. Indeed, current refinement in the methodologies used, identification of new robust markers, and collaborations among the individual laboratories across the globe will greatly improve radiation emergency preparedness in case of unexpected radiological or nuclear mass-casualty events.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We acknowledge the support from Defence Science Organisation, Singapore and International Atomic Energy Agency, Vienna, Austria (CRP E40058), for their support.
References
- 1.Yoo SS, Jorgensen TJ, Kennedy AR, Boice JD, Jr, Shapiro A, Hu TC, et al. Mitigating the risk of radiation-induced cancers: Limitations and paradigms in drug development. J Radiol Prot. 2014;34:R25–52. doi: 10.1088/0952-4746/34/2/R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atun R, Jaffray DA, Barton MB, Bray F, Baumann M, Vikram B, et al. Expanding global access to radiotherapy. Lancet Oncol. 2015;16:1153–86. doi: 10.1016/S1470-2045(15)00222-3. [DOI] [PubMed] [Google Scholar]
- 3.Jaffray DA, Atun R, Barton M, Baumann M, Gospodarowicz M, Hoskin P, et al. Radiation therapy and the global health agenda. Clin Oncol (R Coll Radiol) 2015;27:67–9. doi: 10.1016/j.clon.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 4.IAEA. Biological Dosimetry: Chromosomal Aberrations Analysis for Dose Assesment. Vienna: International Atomic Energy Agency; 1986. [Google Scholar]
- 5.IAEA. Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies. Vienna: International Atomic Energy Agency; 2011. [Google Scholar]
- 6.Bender MA, Gooch PC. Persistent chromosome aberrations in irradiated human subjects. Radiat Res. 1962;16:44–53. [PubMed] [Google Scholar]
- 7.Natarajan AT, Vyas RC, Wiegant J, Curado MP. A cytogenetic follow-up study of the victims of a radiation accident in Goiania (Brazil) Mutat Res. 1991;247:103–11. doi: 10.1016/0027-5107(91)90038-p. [DOI] [PubMed] [Google Scholar]
- 8.Pohl-Rüling J, Haas OA, Obe G, Brøgger A, Roscher U, Daschil F, et al. The Chernobyl fallout in Salzburg/Austria and its effect on blood chromosomes. Acta Biol Hung. 1990;41:215–22. [PubMed] [Google Scholar]
- 9.Ramalho AT, Curado MP, Natarajan AT. Lifespan of human lymphocytes estimated during a six year cytogenetic follow-up of individuals accidentally exposed in the 1987 radiological accident in Brazil. Mutat Res. 1995;331:47–54. doi: 10.1016/0027-5107(95)00049-o. [DOI] [PubMed] [Google Scholar]
- 10.Ramalho AT, Nascimento AC, Littlefield LG, Natarajan AT, Sasaki MS. Frequency of chromosomal aberrations in a subject accidentally exposed to 137Cs in the Goiania (Brazil) radiation accident: Intercomparison among four laboratories. Mutat Res. 1991;252:157–60. doi: 10.1016/0165-1161(91)90016-2. [DOI] [PubMed] [Google Scholar]
- 11.Venkatesan S, Natarajan AT, Hande MP. Chromosomal instability – Mechanisms and consequences. Mutat Res Genet Toxicol Environ Mutagen. 2015;793:176–84. doi: 10.1016/j.mrgentox.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins RC, Romm H, Kao TC, Awa AA, Yoshida MA, Livingston GK, et al. Interlaboratory comparison of the dicentric chromosome assay for radiation biodosimetry in mass casualty events. Radiat Res. 2008;169:551–60. doi: 10.1667/RR1272.1. [DOI] [PubMed] [Google Scholar]
- 13.Hande MP, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low GK, Fok ED, Ting AP, Hande MP. Oxidative damage induced genotoxic effects in human fibroblasts from Xeroderma Pigmentosum group A patients. Int J Biochem Cell Biol. 2008;40:2583–95. doi: 10.1016/j.biocel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Newman JP, Banerjee B, Fang W, Poonepalli A, Balakrishnan L, Low GK, et al. Short dysfunctional telomeres impair the repair of arsenite-induced oxidative damage in mouse cells. J Cell Physiol. 2008;214:796–809. doi: 10.1002/jcp.21276. [DOI] [PubMed] [Google Scholar]
- 16.Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res. 1985;147:29–36. doi: 10.1016/0165-1161(85)90015-9. [DOI] [PubMed] [Google Scholar]
- 17.Fenech M, Morley AA. Cytokinesis-block micronucleus method in human lymphocytes: Effect of in vivo ageing and low dose X-irradiation. Mutat Res. 1986;161:193–8. doi: 10.1016/0027-5107(86)90010-2. [DOI] [PubMed] [Google Scholar]
- 18.Balajee AS, Bertucci A, Taveras M, Brenner DJ. Multicolour FISH analysis of ionising radiation induced micronucleus formation in human lymphocytes. Mutagenesis. 2014;29:447–55. doi: 10.1093/mutage/geu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertucci A, Smilenov LB, Turner HC, Amundson SA, Brenner DJ. In vitro RABiT measurement of dose rate effects on radiation induction of micronuclei in human peripheral blood lymphocytes. Radiat Environ Biophys. 2016;55:53–9. doi: 10.1007/s00411-015-0628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lue SW, Repin M, Mahnke R, Brenner DJ. Development of a high-throughput and miniaturized cytokinesis-block micronucleus assay for use as a biological dosimetry population triage tool. Radiat Res. 2015;184:134–42. doi: 10.1667/rr13991.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hande MP, Azizova TV, Geard CR, Burak LE, Mitchell CR, Khokhryakov VF, et al. Past exposure to densely ionizing radiation leaves a unique permanent signature in the genome. Am J Hum Genet. 2003;72:1162–70. doi: 10.1086/375041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothkamm K, Horn S. gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita. 2009;45:265–71. [PubMed] [Google Scholar]
- 25.Barnard S, Ainsbury EA, Al-hafidh J, Hadjidekova V, Hristova R, Lindholm C, et al. The first gamma-H2AX biodosimetry intercomparison exercise of the developing European biodosimetry network RENEB. Radiat Prot Dosimetry. 2015;164:265–70. doi: 10.1093/rpd/ncu259. [DOI] [PubMed] [Google Scholar]
- 26.Rothkamm K, Horn S, Scherthan H, Rössler U, De Amicis A, Barnard S, et al. Laboratory intercomparison on the γ-H2AX foci assay. Radiat Res. 2013;180:149–55. doi: 10.1667/RR3238.1. [DOI] [PubMed] [Google Scholar]
- 27.Rothkamm K, Beinke C, Romm H, Badie C, Balagurunathan Y, Barnard S, et al. Comparison of established and emerging biodosimetry assays. Radiat Res. 2013;180:111–9. doi: 10.1667/RR3231.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma PM, Ponnaiya B, Taveras M, Shuryak I, Turner H, Brenner DJ. High throughput measurement of γH2AX DSB repair kinetics in a healthy human population. PLoS One. 2015;10:e0121083. doi: 10.1371/journal.pone.0121083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang CM, Park KP, Song JE, Jeoung DI, Cho CK, Kim TH, et al. Possible biomarkers for ionizing radiation exposure in human peripheral blood lymphocytes. Radiat Res. 2003;159:312–9. doi: 10.1667/0033-7587(2003)159[0312:pbfire]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Lu TP, Hsu YY, Lai LC, Tsai MH, Chuang EY. Identification of gene expression biomarkers for predicting radiation exposure. Sci Rep. 2014;4:6293. doi: 10.1038/srep06293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–72. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]


