Abstract
Foodborne pathogens have been a cause of a large number of diseases worldwide and more so in developing countries. This has a major economic impact. It is important to contain them, and to do so, early detection is very crucial. Detection and diagnostics relied on culture-based methods to begin with and have developed in the recent past parallel to the developments towards immunological methods such as enzyme-linked immunosorbent assays (ELISA) and molecular biology-based methods such as polymerase chain reaction (PCR). The aim has always been to find a rapid, sensitive, specific and cost-effective method. Ranging from culturing of microbes to the futuristic biosensor technology, the methods have had this common goal. This review summarizes the recent trends and brings together methods that have been developed over the years.
Keywords: Biosensors, culture based, diagnostics, ELISA, foodborne pathogens, PCR
Introduction
Microorganisms, mostly bacteria, are present in gut and skin in human body as normal flora, which are harmless and helpful in many important functions of the body. However, there are many microorganisms including bacteria, fungi and virus that are pathogenic. Gastrointestinal tract is one of the routes through which pathogens enter the human body and cause many foodborne diseases. The foodborne pathogens can enter through contaminated water or contaminated and undercooked food. Hence, it is important to detect the presence of pathogens in food and water before it enters the body to cause a serious outbreak1,2,3. Such organisms mainly include Acinetobacter spp., Bacillus subtilis, B. cereus, Campylobacter jejuni, Citrobacter koseri, C. freundii, Clostridium difficile, C. perfringens, Enterobacter sakazakii, E. cloacae, Escherichia coli O157:H7, Klebsiella oxytoca, K. pneumoniae, Listeria monocytogenes, Salmonella Enteritidis, Salmonella Typhimurium, Shigella sonnei, Staphylococcus aureus, Vibrio cholerae and Yersinia pestis4,5,6,7,8. The major requirement of detection is in public health, water and food industry, pharmaceutical industry, environment and biodefense7,9.
Shiga toxin producing E. coli O157:H7 (STEC) has always been one of the major pathogens which are responsible for foodborne outbreaks. The outbreaks can be due to different subtypes of E. coli O157:H7, termed as enterohaemorrhagic E. coli (EHEC) which has got the characteristics of both verotoxigenic E. coli and of a lesser known diarrhoeagenic enteroaggregative E. coli. Contaminated drinking water and water in the swimming pool can also be a cause for E. coli O157:H7 infection. This has been observed in Mangalore, Karnataka, India10.
China reported its first outbreak of E. coli O157:H7 in 198611. E. coli O157:H7 has been successfully isolated from humans, livestock and other animals in Fujian, Gansu, Zhejiang, Jiangsu and Anhui11,12. Powdered infant food (PIF), especially the powdered milk, is prone to pathogenic bacteria. In 2002, powdered milk produced by Wyeth was found to be contaminated with E. sakazakii7. Similar outbreak was seen in France where PIF was contaminated with Salmonella sp6. In 2010 in Trinidad a study was done with samples taken from 15 farms, and Salmonella sp. was isolated from the farms13. Germany saw one of its worst outbreaks in May 2011, when there was an unusually high number of haemolytic-uraemic syndrome (HUS) cases2. Turkey is another country that has witnessed a large number of HUS cases as the population of Turkey uses large amount of beef14. Mexico, Ireland, Belgium, England, France and Poland have also reported the presence of E. coli O157:H7 in cattle farms, carcass and faeces14. STEC detection has been reported from Canada in stool samples screened for viral gastroenteritis13. In Tanga region of Tanzania which plays a dominant role in milk marketing, various pathogens have been detected in milk as it offers a perfect medium for growth of microorganisms15. Like milk, mozzarella cheese is another consumer product that is prone to get contaminated with L. monocytogenes, E. coli and Pseudomonas fluorescens, and this was seen in Oregon State of USA16. Kefir, a fermented milk based beverage, has low percentage of alcohol and is also prone to food contamination by bacteria4. L. monocytogenes has been one of the major food pathogens which cause contamination in PIF, which was detected in the USA17. L. monocytogenes and L. ivanovii have been found to grow even at 4°C which makes it a major threat as food which is suspected of Listeria contamination has to be tested at the earliest to avoid fatal circumstances. Outbreaks due to some strains of S. aureus such as methicillin resistant S. aureus and Gram positive cocci were detected in food products in China18 and Spain19.
It has been seen that foodborne pathogens can lead to serious outbreaks irrespective of the region. This leads to the spread of disease, more so in infants and aged individuals. Hence, rapid detection becomes important to contain the spread of the pathogen before it leads to a serious outbreak. Various techniques have been evolved to detect the foodborne pathogens. The effort to improve the methods of detection has been a continuous process. The detection methods have been classified into different groups along with their principles, advantages and disadvantages, most of which are discussed in this review. Each method is supported with suitable examples for better understanding of the gradual improvement of the detection systems. The aim is to give an overall gist of the available methods for the detection of the foodborne pathogens.
Culture based methods
Culture based methods have been the oldest methods in detecting the microorganisms, even the pathogenic strains. This method gives a confirmed result regarding the presence of a particular pathogen. The success rate is found to be high, and these methods are cost-effective. However, the biggest drawback in the culture-based method is the slow growth due to which excess time is lapsed to get the final result, which can turn out to be fatal. It must be noted that all these media take up to 18-24 h to give the exact result, indicating the slow turnaround time.
One of the best known examples which shows high success rate and also shows that the method is highly cost-effective is the culture of E. coli O157:H7 on Sorbitol MacConkey agar (SMAC) which is based on the principle of fermentation of sorbitol20,21. However, the major limitation in this method is slow turnaround time and false positive results due to the emerging serotypes of sorbitol fermenting nonO157 and O157 STEC20.
The drawbacks of the SMAC agar can be overcome by the use of chromogenic medium for STEC isolation which has increased specificity and sensitivity. The major advantage of this is the easier discrimination based on colour. Due to the use of the chromogenic substance, the medium is better known as CHROMagar20,22. Though it is comparatively effective than SMAC, one notable drawback is that CHROMagar is not sensitive to all strains20. This was seen in one of the experiments where only one-fifth of the diarrhoeagenic strains were detected when compared to SMAC20.
Cefsulodin-Irgasan-Novobiocin (CIN) agar, a selective medium known for better discrimination between bacterial species, was used to differentiate Yersinia enterocolitica and non Y. enterocolitica23. Y. enterocolitica chromogenic medium is used where agar has fermentable sugar cellobiose, a chromogenic substrate and selective inhibitor which suppresses the competing bacteria. This indicates that the purple/blue colonies that are formed on the CIN agar are of Y. enterocolitica and Y. pseudotuberculosis which are important food pathogens causing yersiniosis. This method was used to study the contaminated tofu24.
Many microorganisms tend to enter starvation mode of metabolism under stress conditions. However, they will remain viable but non culturable (VBNC) which cannot be grown on conventional culture (CC) media, but can signal virulent pathways25. Detection of these pathogens is a major challenge for food safety26. Since no colonies will be formed, other methods such as fluorescent dyes are used for the detection of VBNC bacteria where different dyes are used. Binding of acridine orange to the VBNC pathogens depends on the ratio of DNA to protein in the cells. Actively reproducing cells appear green whereas slow-growing or non-reproducing cells appear orange. Another dye that is used to detect VBNC is fluorescein isothiocyanate, the principle of which is to detect the enzyme activity of living cells. If there is the presence of any living cells, violet or blue colour is seen26. Potable water, pasteurized milk and processed food are vulnerable to VBNC. Some of the foodborne pathogens that fall under VBNC category include C. jejuni, E. aerogenes, E. faecalis, E. coli (including EHEC), Pseudomonas aeruginosa, S. typhimurium, S. dysenteriae, S. sonnei and V. cholerae.
Bacteriophage-derived high-affinity binding molecules (cell wall binding domains, CBDs) have been recently introduced as tools for the detection and differentiation of Listeria in foods as conventional culture (CC) methods are hampered by lengthy enrichment and incubation steps. This when coupled with magnetic separation increases the sensitivity and speed in detection and will be more accurate when compared to the standard diagnostic methods27.
Immunoassays
Immunoassays were developed as these were easier to perform, gave faster result and were less expensive. Hence, generally before directly going into polymerase chain reaction (PCR) based methods, immunoassays are performed. Enzyme linked immunosorbent assay (ELISA) is one of the most used immunoassays to date. Antibody purity plays an important role in success of the immunoassays1. Along with purity, one more factor that affects the assay is specificity of antibody. Polyclonal antibodies have polyvalency (multiple epitopes to react with). This can affect the reaction, leading to low specificity and sensitivity. It must be noted that there are chances of false positive results. One such result was observed where there was a cross-reaction between E. coli O157:H7, Y. enterocolitica O:9 and Brucella abortus, all the samples obtained from the serum samples of infected cattle28.
The use of different substrates in ELISA has a major advantage as the substrates will bind to the respective conjugates specifically and will develop colouration which can be read in an ELISA reader in terms of wavelength. The colour change is visible to the naked eye. However, one of the disadvantages is that the binding of the chemical and conjugate is very specific, and contamination in the intermediate stages can lead to false positive result. One such substrate used is 2,2’-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) prepared in 0.05 M phosphate-citrate buffer which reacts with bovine serum albumin (BSA) solution29. Tetramethylbenzidine is another substrates that is most commonly used in ELISA. It binds to horse radish peroxidase (HRP). The colouration develops gradually. This method was used in development of sandwich ELISA for the detection of Listeria sp30. Another most commonly used substrate is p-nitrophenyl phosphate (pNPP) which binds specifically to BSA conjugated alkaline phoshatase31. In one of the experiments where detection of E. coli O157:H7 was performed, pNPP was used as the substrate32. Bispecific antibodies that recognize human red blood cell (RBC) and the foodborne pathogen L. monocytogenes were engineered. The principle behind this is an initial reduction of a mixture of anti-RBC and anti-Listeria antibodies followed by gradual reoxidation of the reduced disulphides. This facilitates association of the separated antibody chains and formation of hybrid immunoglobulins with affinity for L. monocytogenes and human RBC. The bispecific antibodies caused the agglutination of the RBCs only in the presence of L. monocytogenes cells. The agglutination process showed red coloured clumps in presence of L. monocytogenes and were readily visible to the naked eyes. This was found to be a simple approach for the rapid and highly specific screening of various pathogens in their biological niches33.
From time to time, methodologies in ELISA have been improved to suit the ever emerging new experiments. Blocking ELISA was designed with E. coli O157:H7 LPS as antigen. These were successful in detecting the pathogen in cattle and were found to be more sensitive than the normal ELISA34. Indirect ELISA has been used for detecting anti-O157 antibodies in the serum of cattle as well as humans. However, chances of the result being false positive were more due to cross reactivity34.
Sandwich ELISA is a modified form of ELISA, in which there will be two antibodies used against one antigen. The sensitivity and specificity is much higher than the existing assays. This kind of ELISA was found to be useful in detecting the Shiga-like toxin (stx) in E. coli O157 strains and also non O157 STEC strains and Listeria sp3. Polyclonal antibody was used here, with HRP as conjugate for the detection3,30. The improved version of sandwich ELISA is to detect antibody to the SEF 14 fimbrial antigen (SEF 14 – double antibody sandwich (DAS) - ELISA). This is used for the detection of chicken flocks infected with S. enteritidis. It could discriminate birds infected with S. enteritidis and those infected with Salmonella panama and S. Typhimurium14. In another novel experiment of sandwich ELISA assay, detection of stx2a was performed where the soil samples were spiked with a detection limit between 10 and 100 pg/ml and faecal samples between 100 and 500 pg/ml. When samples were tested by PCR technique, it showed 100 per cent sensitivity and specificity3.
The major advantage in reversed passive latex agglutination assay was that 6 h was sufficient for the growth of bacteria, and hence the result obtained was quicker than the culture based35. This was tried for determining the toxigenicity of diphtheria toxin of Corynebacterium diphtheriae35. Rabbit antitoxin antiserum was used to react with the antiserum with diphtheria toxin.
Monoclonal antibodies are preferred over polyclonal antibody as these have monovalency. In monoclonal antibodies, the antibody is produced against one specific antigen. While sensitivity and specificity are its major positive features, production is a laborious process and is not cost-effective. Various such experiments have been conducted to detect L. monocytogenes, S. Typhimurium, L. innocua and E. coli36.
The use of immunoglobulin G (IgG) was the beginning of a new technology, which was useful in targeting virulence in clinical microbiology37. However, gradually it is IgY, the counterpart of IgG in chicken egg yolk which has taken over. The major advantages in using IgY is that it is deposited in egg yolk in large quantities and can easily be purified by simple precipitation techniques. This property has made chicken an ideal source for specific monoclonal antibodies. It is very useful in immunotherapy and immunodiagnostics38,39. This method was proved to be successful in detecting one of the foodborne pathogens C. jejuni when present in low detection limit40. A simple and rapid gold-labelled immunosorbent assay (GLISA) has been developed which has the low detection limit of 7.3 log/cfu/g, which is found to be better than many other ELISA methods40,41. GLISA is commercially available as Singlepath Campylobacter GLISA Rapid Test40.
To overcome high detection limits, enrichment steps become important for the detection of pathogens in food products. In the enrichment step, a label-free immunoassay is used that helps in detecting the presence of the pathogen in a much simpler way. A simple and rapid detection is possible through this method with simultaneous enrichment and optical detection. The principle of this method is culture/capture/measure42.
DNAzymes are a novel class of molecular probes for detection of bacteria. DNAzymes, also called as DNA enzymes, are man-made single-stranded DNA molecules with the capability of catalyzing chemical reactions. These molecules can be isolated from vast random sequence DNA pool by a process called as ‘SELEX’ meaning systematic evolution of ligand by exponential enrichment. This process includes a DNA RNA chimeric substrate at a single ribonucleotide junction (R) that is flanked by fluorophore (F) and a quencher (Q). There will be a cleavage where the separation of fluorophore and quencher will lead to increase in fluorescence intensity which makes bacterial detection easy and rapid43. Epitope tags which confer specific properties, including affinity for resins or antibodies or detection by fluorescence microscopy, are useful for biochemical and cell biological investigations. This method has been used for the detection of Candida albicans44.
Polymerase chain reaction based methods
Kary Mullis in 1985 discovered PCR, which is considered as one of the milestone discoveries in recombinant DNA technology45. The principle behind PCR is that genes of various pathogens can be amplified and studied further46,47. Specific primers are developed for each gene. Agarose gel electrophoresis and subsequent staining with ethidium bromide are used for the identification of PCR products. Since the discovery, various types of PCR have emerged, which are given names according to the changed protocol of the original PCR. In general, the major advantages of PCR are that the process is rapid and sensitive. It is faster than the culture based methods and immunoassays. PCR has now reached new heights where the amplified product can be obtained in just 30 min, and distinguishing between the strains has become much easier as multiple primer pairs are used. The detection limits can become better with time that the DNA amplicon detection limit can be as low as femtograms (10-15g)9. This can be an alternative to tedious time consuming procedure of culturing and identification of pathogens in food safety laboratories48. However, with the advancement in PCR technology, the method has not remained cost-effective although low detection limit will remain the major criteria. PCR technique has developed as a very promising method of detection of the genes in pathogens; however, there are certain disadvantages that make it necessary to develop better methods. The difficulties include cell lysis and nucleic acid extraction, cross-contamination and failed reactions due to the presence of inhibitory substance or competing DNA from the non target cells. This can lead to inconsistent results and reduce the appeal of PCR as a reliable approach1. PCR methods are not able to differentiate between the live and dead cells. The primary disadvantage of all the PCR methods is that there are chances of generating false positive signal due to binding to non-specific double-stranded DNA sequences. Therefore, it is extremely important to have well-designed primers that do not amplify non-target sequences.
One of the initial advances in molecular cloning and recombinant DNA technology that revolutionized the detection of foodborne pathogen is the development of a PCR-based technique. In one of the methods, suitable primers were designed based on specific gene fimA of Salmonella and gene afa of pathogenic E. coli for amplification49. The size of the amplified product was 120 bp as shown by comparison with marker DNA. This is a rapid, sensitive and reliable technique for the detection of Salmonella and pathogenic E. coli49. To design loop-mediated isothermal amplification (LAMP) assays, stx1, stx2 and eae genes were chosen as targets47. LAMP employs four to six specially designed primers and a strand-displacing Bst DNA polymerase to amplify up to 109 target DNA copies under isothermal conditions (60-65°C) within an hour. The result of LAMP was compared with quantitative PCR (qPCR). The result was obtained within one hour. This method was found to be rapid, specific and sensitive for the detection of STEC strains. One more advantage is the absence of any false positive or false-negative results47. During any outbreak, it is important to detect the presence of the pathogen at the earliest. Real-time PCR allows for quantification of the target, and when combined with a rapid cycling platform, results can be generated in 30 min from the start of thermal cycling. Real-time qPCR is considered as a method of choice for the detection and quantification of microorganisms. One of its major advantages is that it is faster than CC based methods. It is also highly sensitive, specific and enables simultaneous detection of different microorganisms50. Ruggedized, advanced pathogen identification device (RAPID) system E. coli O157:H7 kit is a modified version of real time PCR which has the advantage of rapid cycle real-time PCR51. An alternate for real time PCR assay is the use of three TaqMan assay sets to detect stx1, stx2 and rfbE genes. Using multiple PCR assay sets to detect these genes allowed the very specific detection of EHEC from strains which did not possess any of these three genes. The result showed that there was horizontal transfer of stx gene between E. coli strains and in non E. coli enterobacteriaceae strains such as Citrobacter and Enterobacter52.
SYBR Green is a cyanine dye which immediately binds to all double-stranded DNA present in the sample. During PCR, DNA polymerase amplifies the target sequence which creates the PCR products. SYBR Green dye then binds to each new copy of double-stranded DNA53. As the PCR progresses, more PCR product is generated. SYBR Green dye binds to all double-stranded DNA, so the result is an increase in fluorescence intensity proportioned to the amount of PCR product produced. Real-time PCR has been combined with the dye SYBR Green and was used to detect E. coli strains. The result showed that the presence of SYBR Green increased the discriminating power between the strains54. Restriction site specific PCR was performed to detect E. coli O157:H7 which involved the amplification of DNA fragments using primers based on specific restriction enzyme recognition sequences. This method does not use endonucleases. It generates amplicons that yield ‘fingerprint’ patterns when resolved on an agarose gel55. Multiplex PCR along with SYBR Green was used to detect STEC in O157 and non O157 serotypes of E. coli in cattle faeces56. Multiplex PCR uses two sets of primers and two fluorogenic probes for simultaneous and semiautomated detection of Salmonella strains and E. coli O157:H7. This PCR assay was optimized to obtain a strong and reproducible fluorescence signal from probes labelled with two reporter dyes. This helped in immediate and specific detection in meat and faeces46,52,57. Fluorescence was combined with real-time PCR and multiplex PCR for early detection of stx1, hly and eae genes. This led to a billion fold amplification when experiment was performed under isothermal condition58,59. Large-scale multiplex (LSplex) uses 800 specific primer pairs. It can successfully amplify different pathogens whether it is Gram positive or Gram negative. It generated stronger signals with just 10 ng of DNA as compared to the ones which used 2-5 µg of DNA9. One aspect that can be improved in LSplex PCR is that its detection limit can be reduced to pico (10−12g) or to femtograms (10−15g). This will be very desirable in detection of every clinical, food or environmental samples9. Fluorescent amplification-based hybridization PCR shows good results in fluorescence intensity which is the most important aspect in detection of pathogens. It is found that fluorescent signal for E. coli O157:H7 was 6.40 while that for other related pathogens was 2.5059. It is also cost-effective. Reverse transcription PCR (RT-PCR) is another technique which uses reverse transcriptase enzyme to produce DNA from RNA followed by the normal PCR technique. This technique is used to detect virus causing dengue37. Detection of Salmonella sp. using real-time PCR is also reported in pork chop and sausage samples using SYBR Green dye in RT-PCR60,61. Real time RT-PCR has shown great potential for detecting viable pathogens such as S. enterica where mRNA is detected. In one of the studies, expression of Salmonella specific sigDE operon which encodes invasion proteins was studied and it was found that the sigDE could be a useful viable marker for the bacteria62.
The use of reporter quencher technique has been known since the early 1990s, which has developed over time63. The nucleic acid amplification technique is an indispensible tool in clinical diagnostics. Accurate and specific quantification of pathogen is very important. Hence, a new mediator probe has been developed which works on the reporter quencher methodology where release of mediator triggers signal generation of a complementary fluorogenic reporter probe. This technology was applied to detect and amplify S. aureus and E. coli64. Novel nucleic acid probes known as molecular beacons have been developed allowing for the rapid and specific detection of disease. Molecular beacons are hairpin-forming oligonucleotides labelled at one end with a quencher and at the other end with a fluorescent reporter dye65.
Markers
Conventional pathogen detection methods, such as microbiological and biochemical identification, are time-consuming and laborious while immunological or nucleic acid-based techniques require extensive sample preparation and are not amenable to miniaturization for on-site detection. Novel biological recognition elements are studied to improve the selectivity and facilitate integration on the transduction platform for sensitive detection. However, the probe that is designed has to be very specific. Bacteriophages are one such unique biological entity that show excellent host selectivity and have been actively used as recognition probes for pathogen detection66. When there is a necessity of differentiating pathogens, for example, E. coli and other enteric bacteria, gene gadAB present in E. coli strains can be obtained from the consumer food materials. However, Shigella is the only species which is gadAB-positive. To overcome the false positive results, gadAB gene can be used as a marker for just E. coli. This indicated that gadAB marker was suitable as pre-screening marker for E. coli67. This led to a large-scale genome comparison. This method is called octamer-based genome scanning68,69. The markers where gene is used are called as DNA probes. Protein probes such as green fluorescent protein (GFP) obtained from gfp gene is known for its fluorescence. This gene is obtained from the jellyfish Aequorea victoria. Expression of selectively inducible gfp gene in a plasmid transformed strain of E. coli O157:H7 was found to be a useful tool in the detection of the pathogen70. GFP protein produced by gfp gene shows a characteristic emission peak at 509 nm which indicates the presence of the organism70.
Biosensors
Biosensors are the latest among all the detection systems, some of which have better detection limits which significantly reduce and also eliminate the drawbacks associated with PCR techniques1,66,71,72,73,74,75,76. Biosensors are the devices for pathogen detection that generally consist of three elements, which are a biological capture molecule (probes and antibodies), a method for converting capture molecule – target interactions into a signal and an output data2. Despite better detection efficiencies, results derived using molecular biology methods can be affected by the various food matrices. One of such detection studies was done on Y. enterocolitica, a pathogen that can cause yersiniosis in humans and animals77. Improvements in sample preparation, data analysis and testing procedures, molecular detection techniques can simplify and increase the speed of detection. The major advantage of the biosensors is that these can detect the pathogens at low detection limits with high specificity and sensitivity, but the biosensors will require highly specific and expensive instruments, with compatible computer software, to give accurate results. Hence, these methods may not be always cost-effective.
Electrochemiluminescent assays are performed in 96-well plates and are based on electrochemical stimulation of reporter molecules such as ruthenium (II) trisbipyridal (Ru(bpy)3)2+ chloride which are attached to antibodies. The detection in this method is at a low concentration. A slightly improved version of this is called cytometric bead assay which uses a fluidic approach and have red and infrared fluorophores. These give out orange fluorescence when exposed to the electrode1. A lab-on-a-chip integrates cell pre-concentration, purification, PCR and capillary electrophoretic (CE) analysis. It is a microdevice which has a 100 nl PCR reactor and 5 cm long CE column for amplicon separation. Detection limit is 0.2 cfu/µl78. It can be used in detection of E. coli K12. Similar to lab-on-a-chip assay is cell-phone based on E. coli detection platform for screening of liquid samples. Battery powered inexpensive light emitting diodes are used. Excitation of sample is done, and the emission from the quantum dots is imaged using a phone camera unit. It was demonstrated for fat free milk mainly to detect Salmonella sp. where the detection limit of 5 to 10 cfu/ml was achieved78. Similarly, an exposure to antibody-quantum dot conjugates was used to detect E. coli and S. Typhimurium79. CdSe/ZnS quantum dots exhibited fluorescence emission shift when conjugated to antibody or DNA aptamers that are bound to bacteria. This shift in emission peak occurs when the quantum dots encounter the bacterial surface80.
A surface plasmon resonance (SPR) immunosensor was designed by means of a subtractive inhibition assay using goat polyclonal antibodies for E. coli O157:H7. The results showed that the signal was inversely correlated with the concentration of E. coli O157:H774. Fluorescence resonance energy transfer (FRET) utilizes an antibody recognizing cell surface epitopes of the target cell. It makes use of complementary oligonucleotides that are modified with fluorochromes. Fluorescence is detected using the sensors. E. coli O157:H7 and Salmonella were detected using FRET. The advantage of FRET is that it is simple, fast by giving result within five minutes. It is inexpensive and highly sensitive81.
Optical biosensors have been proven to have better detection system and separation of pathogens. These biosensors include optical fibres, planar wave guides, SPR and microarrays. Their compact design and label-free detection lead to specific and sensitive detection and this is a major advantage of optical biosensors75.
Nanobiotechnology is the latest approach for detection of pathogens. Aptamers are attracting an increasing amount of interest in the development of sensors for proteins, DNA and small molecules. An experiment design based on the combination of nucleic acid aptamer with polydiacetylene showed 98.5 per cent detection of E. coli O157:H7 (203 clinical faecal samples) when compared with the standard culture72. High affinity and specificity are found in aptamers. Gold nanoparticles (GNPs), silver nanoparticles and bioconjugated nanoparticles which give fluorescence have been used in aptamers76,82,83. GNPs have electronic, photonic and catalytic properties making their applications unique. GNPs can be used in colorimetric methods due to their optical properties76. These are non toxic and can easily conjugate to antibodies82. A bioconjugated, nanoparticle-based bioassay provides a high fluorescent signal for bioanalysis. An attempt was done using E. coli O157 cells in beef sample in a 384-well microplate format83. A new approach involves physical damage to the bacteria using a combination of pulsed laser energy and absorbing nanoparticles. When irradiated, nanoparticles absorb energy and when relaxed give out heat, which damages the cells. GNPs have been used for this method82.
Other detection methods
DNA microarray is gaining importance currently and has become a useful tool due to its rapidness, sensitivity and specificity and it allows high throughput analysis. Various studies have been conducted to detect waterborne pathogens, marine fish pathogens, which indirectly will be a threat to humans due to fish consumption6,9,15. Li et al58 reported the detection of foodborne pathogen microarrays designed to target internal transcribed spacer (ITS) sequences. In one of the studies, 10 pathogens were tested for their presence in PIF6. B. cereus, E. coli, L. monocytogenes, P. aeruginosa, S. enterica, S. aureus, V. parahaemolyticus and, C. freundii were detected using this method. E. sakazakii, K. pneumoniae, K. oxytoca, Serratia marcescens and A. baumannii are associated with contaminating PIF6. DNA microarray technique is used to detect these pathogens. ITS regions of five Bacillus sp. B. anthracis, B. cereus, B. thuringiensis, B. mycoides and B. weihenstephanensis were examined as these possess a high homology at DNA level, making it difficult to differentiate. DNA microarray was the solution to this problem6. PulseNet, a national molecular subtyping network for foodborne disease surveillance is playing a key role in detecting each of the outbreaks by the pathogens5. It mainly helps in reducing product recalls, restaurant closures and related mechanisms after the outbreak. This is done in local, State and public health and regulatory agency laboratories5.
Ultrafiltration, immunomagnetic assays (IMS), immunochromatic assay (ICA), flow cytometry (FC) and lyophilization are some of the conventional methods. Ultrafiltration has been recognized as an effective procedure for concentration and recovering microbes from large volumes of water and treated waste water52. Conventional IMS procedure uses an external source to capture magnetic particles against the side of the test tube which leads to poor results due to high background microflora84. Hence, PickPen IMS is used which increases the throughput compared to the conventional IMS. The difference is that there is an intrasolution magnetic particle transfer device in PickPen IMS which detects E. coli O157:H7, Salmonella sp. and L. monocytogenes that are prevalent in various samples. Its consistent recovery of immunobeads has high throughput and lower carryover of background microflora84,85. In one of the experiments in detecting S. Typhimurium, IMS was combined with CC, with PCR and with Fourier transform infrared spectroscopy (FTIR) as IMS-CC, IMS-PCR and IMS-FTIR where combination of IMS with FTIR was found to be the most accurate and rapid test76,86.
FC is a sensitive analytical technique which can rapidly monitor physical states of bacteria. Fluorescent probes are used to detect E. coli O157:H7, P. aeruginosa, P. syringae, S. Typhimurium and Cyclospora cayetanensis (in oocytes)75,87.
ICA has been a useful, simple, rapid, highly sensitive, specific method and does not require expensive equipment or reagents. It can be judged by naked eye in terms of cfu/ml. Immunomagnetic nanoparticles use nanopure iron as core coated with E. coli O157:H7 polyclonal antibodies in combination with ICA11. It has been found that lyophilization prior to direct DNA extraction from bovine faeces improves the quantification of C. jejuni88.
Conclusion
An ideal detection method needs to satisfy five premier requirements – high specificity (detecting only the bacterium of interest), high sensitivity (capable of detecting as low as a single live bacterial cell), short time-to-results (minutes to hours), great operational simplicity (no need for lengthy sampling procedures and use of specialized equipment) and cost effectiveness. For example, culture takes long time to give the results. On the other hand, PCR, antibody-based techniques and biosensors offer shorter waiting time, but these require the use of expensive reagents and sophisticated equipment which make the method expensive.
In this review, various methods of detection of pathogens which have been developed and improved from time to time have been discussed with the pros and cons of the respective methods (Figure). An important point that needs to be stressed here is that the search for better detection methods of pathogens cannot be stopped at one point. This will be an area of research and newer experiment will be evolving to make the detection systems rapid, sensitive, specific and cost-effective to the maximum extent.
Figure.
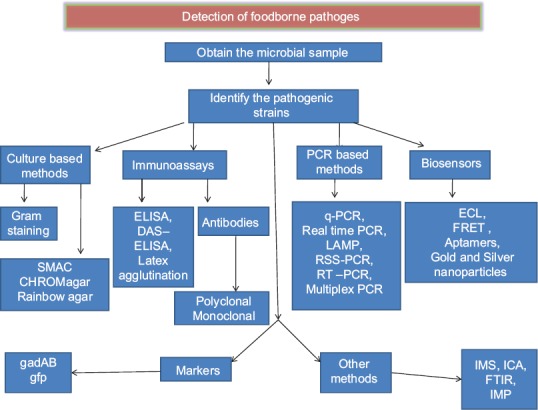
Schematic representation of the methods for the detection of pathogens. ELISA, enzyme linked immuno sorbent assay; DAS, double antibody sandwich; PCR, polymerase chain reaction; LAMP, loop mediated isothermal amplification; RSS, restriction site specific; RT, real time; ECL, electrochemilumenescence; FRET, fluorescence resonance energy transfer; IMS, immunomagnetic assay; ICA, immunochromatic assay; FTIR, fourier transform infrared spectroscopy.
Footnotes
Conflicts of Interest: None.
References
- 1.Leach KM, Stroot JM, Lim DV. Same-day detection of Escherichia coli O157: H7 from spinach by using electrochemiluminescent and cytometric bead array biosensors. Appl Environ Microbiol. 2010;76:8044–52. doi: 10.1128/AEM.01990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattaway MA, Dallman T, Okeke IN, Wain J. Enteroaggregative E. coli O104 from an outbreak of HUS in Germany 2011, could it happen again? J Infect Dev Ctries. 2011;5:425–36. doi: 10.3855/jidc.2166. [DOI] [PubMed] [Google Scholar]
- 3.He X, Patfield S, Hnasko R, Rasooly R, Mandrell RE. A polyclonal antibody based immunoassay detects seven subtypes of Shiga toxin 2 produced by Escherichia coli in human and environmental samples. PLoS One. 2013;8:e76368. doi: 10.1371/journal.pone.0076368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh AJ, O’Sullivan O, Hill C, Ross RP, Cotter PD. Sequencing-based analysis of the bacterial and fungal composition of kefir grains and milks from multiple sources. PLoS One. 2013;8:e69371. doi: 10.1371/journal.pone.0069371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxrud D, Monson T, Stiles T, Besser J. The role, challenges, and support of pulsenet laboratories in detecting foodborne disease outbreaks. Public Health Rep. 2010;125(Suppl 2):57–62. doi: 10.1177/00333549101250S207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Cao B, Gao Q, Sun Y, Liu P, Feng L, et al. Detection of Enterobacter sakazakii and other pathogens associated with infant formula powder by use of a DNA microarray. J Clin Microbiol. 2009;47:3178–84. doi: 10.1128/JCM.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan MM, Pyle BH, Camper AK. Specific and rapid enumeration of viable but nonculturable and viable-culturable Gram-negative bacteria by using flow cytometry. Appl Environ Microbiol. 2010;76:5088–96. doi: 10.1128/AEM.02932-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louie M, Louie L, Simor AE. The role of DNA amplification technology in the diagnosis of infectious diseases. CMAJ. 2000;163:301–9. doi: 10.1016/s1381-1169(00)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palka-Santini M, Cleven BE, Eichinger L, Krönke M, Krut O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhanashree B, Mallya PS. Detection of shiga-toxigenic Escherichia coli (STEC) in diarrhoeagenic stool and meat samples in Mangalore, India. Indian J Med Res. 2008;128:271–7. [PubMed] [Google Scholar]
- 11.Xu JG, Cheng BK, Jing HQ. Escherichia coli O157 H7 and Shiga-like-toxin- producing Escherichia coli in China. World J Gastroenterol. 1999;5:191–4. doi: 10.3748/wjg.v5.i3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi H, Zhong Z, Zhou HX, Deng CY, Zhu H, Li JF, et al. A rapid and highly sensitive protocol for the detection of Escherichia coli O157: H7 based on immunochromatography assay combined with the enrichment technique of immunomagnetic nanoparticles. Int J Nanomedicine. 2011;6:3033–9. doi: 10.2147/IJN.S25684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris R, Sankar K, Small JA, Suepaul R, Stewart-Johnson A, Adesiyun A. Prevalence and characteristics of enteric pathogens detected in diarrhoeic and non-diarrhoeic foals in Trinidad. Vet Med Int 2012. 2012:724959. doi: 10.1155/2012/724959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inat G, Siriken B. Detection of Escherichia coli O157 and Escherichia coli O157: H7 by the immunomagnetic separation technique and stx1 and stx2 genes by multiplex PCR in slaughtered cattle in Samsun Province, Turkey. J Vet Sci. 2010;11:321–6. doi: 10.4142/jvs.2010.11.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swai ES, Schoonman L. Microbial quality and associated health risks of raw milk marketed in the Tanga region of Tanzania. Asian Pac J Trop Biomed. 2011;1:217–22. doi: 10.1016/S2221-1691(11)60030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan J, Park SI, Daeschel MA, Zhao Y. Antimicrobial chitosan-lysozyme (CL) films and coatings for enhancing microbial safety of mozzarella cheese. J Food Sci. 2007;72:M355–62. doi: 10.1111/j.1750-3841.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- 17.Heo SA, Nannapaneni R, Story RP, Johnson MG. Characterization of new hybridoma clones producing monoclonal antibodies reactive against both live and heat-killed Listeria monocytogenes. J Food Sci. 2007;72:M008–15. doi: 10.1111/j.1750-3841.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 18.Hou Z, Meng JR, Niu C, Wang HF, Liu J, Hu BQ, et al. Restoration of antibiotic susceptibility in methicillin-resistant Staphylococcus aureus by targeting mecR1 with a phosphorothioate deoxyribozyme. Clin Exp Pharmacol Physiol. 2007;34:1160–4. doi: 10.1111/j.1440-1681.2007.04705.x. [DOI] [PubMed] [Google Scholar]
- 19.Marrero A, Mallorquí-Fernández G, Guevara T, García-Castellanos R, Gomis-Rüth FX. Unbound and acylated structures of the MecR1 extracellular antibiotic-sensor domain provide insights into the signal-transduction system that triggers methicillin resistance. J Mol Biol. 2006;361:506–21. doi: 10.1016/j.jmb.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 20.Hirvonen JJ, Siitonen A, Kaukoranta SS. Usability and performance of CHROMagar STEC medium in detection of Shiga toxin-producing Escherichia coli strains. J Clin Microbiol. 2012;50:3586–90. doi: 10.1128/JCM.01754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.March SB, Ratnam S. Sorbitol-MacConkey medium for detection of Escherichia coli O157: H7 associated with hemorrhagic colitis. J Clin Microbiol. 1986;23:869–72. doi: 10.1128/jcm.23.5.869-872.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouali M, Ruckly C, Carle I, Lejay-Collin M, Weill FX. Evaluation of CHROMagar STEC and STEC O104 chromogenic agar media for detection of Shiga Toxin-producing Escherichia coli in stool specimens. J Clin Microbiol. 2013;51:894–900. doi: 10.1128/JCM.03121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan LK, Ooi PT, Carniel E, Thong KL. Evaluation of a modified cefsulodin-irgasan-novobiocin agar for isolation of Yersinia spp. PLoS One. 2014;9:e106329. doi: 10.1371/journal.pone.0106329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weagant SD. A new chromogenic agar medium for detection of potentially virulent Yersinia enterocolitica. J Microbiol Methods. 2008;72:185–90. doi: 10.1016/j.mimet.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Ramamurthy T, Ghosh A, Pazhani GP, Shinoda S. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front Public Health. 2014;2:103. doi: 10.3389/fpubh.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fakruddin M, Mannan KS, Andrews S. Viable but nonculturable bacteria: food safety and public health perspective. ISRN Microbiol 2013. 2013:703813. doi: 10.1155/2013/703813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmelcher M, Loessner MJ. Use of bacteriophage cell wall-binding proteins for rapid diagnostics of Listeria. Methods Mol Biol. 2014;1157:141–56. doi: 10.1007/978-1-4939-0703-8_12. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen K, Smith P, Widdison J, Gall D, Kelly L, Kelly W, et al. Serological relationship between cattle exposed to Brucella abortus, Yersinia enterocolitica O: 9 and Escherichia coli O157: H7. Vet Microbiol. 2004;100:25–30. doi: 10.1016/j.vetmic.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Nakano T, Sunwoo HH, Paek BH, Chae HS, Sim JS. Effects of egg and yolk weights on yolk antibody (IgY) production in laying chickens. Poult Sci. 1998;77:266–70. doi: 10.1093/ps/77.2.266. [DOI] [PubMed] [Google Scholar]
- 30.Kim SH, Park MK, Kim JY, Chuong PD, Lee YS, Yoon BS, et al. Development of a sandwich ELISA for the detection of Listeria spp. using specific flagella antibodies. J Vet Sci. 2005;6:41–6. [PubMed] [Google Scholar]
- 31.Ko KY, Ahn DU. Preparation of immunoglobulin Y from egg yolk using ammonium sulfate precipitation and ion exchange chromatography. Poult Sci. 2007;86:400–7. doi: 10.1093/ps/86.2.400. [DOI] [PubMed] [Google Scholar]
- 32.Priyanka B, Patil RK, Dwarakanath S. Production of antibodies in chicken for Escherichia Coli O157: H7. IOSR-J EnvironSci Toxicol Food Technol. 2012;2:8–15. [Google Scholar]
- 33.Owais M, Kazmi S, Tufail S, Zubair S. An alternative chemical redox method for the production of bispecific antibodies: implication in rapid detection of food borne pathogens. PLoS One. 2014;9:e91255. doi: 10.1371/journal.pone.0091255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laegreid W, Hoffman M, Keen J, Elder R, Kwang J. Development of a blocking enzyme-linked immunosorbent assay for detection of serum antibodies to O157 antigen of Escherichia coli. Clin Diagn Lab Immunol. 1998;5:242–6. doi: 10.1128/cdli.5.2.242-246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toma C, Sisavath L, Iwanaga M. Reversed passive latex agglutination assay for detection of toxigenic Corynebacterium diphtheriae. J Clin Microbiol. 1997;35:3147–9. doi: 10.1128/jcm.35.12.3147-3149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Padova FE, Brade H, Barclay GR, Poxton IR, Liehl E, Schuetze E, et al. A broadly cross-protective monoclonal antibody binding to Escherichia coli and Salmonella lipopolysaccharides. Infect Immun. 1993;61:3863–72. doi: 10.1128/iai.61.9.3863-3872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordeiro MT, Braga-Neto U, Nogueira RMR, Marques ET., Jr Reliable classifier to differentiate primary and secondary acute dengue infection based on IgG ELISA. PLoS One. 2009;4:e4945. doi: 10.1371/journal.pone.0004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovacs-Nolan J, Mine Y. Avian egg antibodies: basic and potential applications. Avian Poult Biol Rev. 2004;15:25–46. [Google Scholar]
- 39.Shin JH, Yang M, Nam SW, Kim JT, Myung NH, Bang WG, et al. Use of egg yolk-derived immunoglobulin as an alternative to antibiotic treatment for control of Helicobacter pylori infection. Clin Diagn Lab Immunol. 2002;9:1061–6. doi: 10.1128/CDLI.9.5.1061-1066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochel I, Viochna D, Skvor J, Musil M. Development of an indirect competitive ELISA for detection of Campylobacter jejuni subsp. jejuni O: 23 in foods. Folia Microbiol (Praha) 2004;49:579–86. doi: 10.1007/BF02931537. [DOI] [PubMed] [Google Scholar]
- 41.Pölzler T, Wagner M, Slaghuis J, Schleicher C, Köfer J. Rapid monitoring of Campylobacter in high-shedding flocks for targeted disease control. J Food Prot. 2012;75:1835–8. doi: 10.4315/0362-028X.JFP-12-027. [DOI] [PubMed] [Google Scholar]
- 42.Mondani L, Roupioz Y, Delannoy S, Fach P, Livache T. Simultaneous enrichment and optical detection of low levels of stressed Escherichia coli O157: H7 in food matrices. J Appl Microbiol. 2014;117:537–46. doi: 10.1111/jam.12522. [DOI] [PubMed] [Google Scholar]
- 43.Aguirre SD, Ali MM, Kanda P, Li Y. Detection of bacteria using fluorogenic DNAzymes. J Vis Exp. 2012;pii:3961. doi: 10.3791/3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerami-Nejad M, Dulmage K, Berman J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast. 2009;26:399–406. doi: 10.1002/yea.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz TC. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp. 2012;63:e3998. doi: 10.3791/3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ooka T, Terajima J, Kusumoto M, Iguchi A, Kurokawa K, Ogura Y, et al. Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J Clin Microbiol. 2009;47:2888–94. doi: 10.1128/JCM.00792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Jiang L, Ge B. Loop-mediated isothermal amplification assays for detecting shiga toxin-producing Escherichia coli in ground beef and human stools. J Clin Microbiol. 2012;50:91–7. doi: 10.1128/JCM.05612-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radhika M, Saugata M, Murali HS, Batra HV. A novel multiplex PCR for the simultaneous detection of Salmonella enterica and Shigella species. Braz J Microbiol. 2014;45:667–76. doi: 10.1590/s1517-83822014005000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naravaneni R, Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J Med Microbiol. 2005;54:51–4. doi: 10.1099/jmm.0.45687-0. [DOI] [PubMed] [Google Scholar]
- 50.Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 2011;28:848–61. doi: 10.1016/j.fm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Bono JL, Keen JE, Miller LC, Fox JM, Chitko-McKown CG, Heaton MP, et al. Evaluation of a real-time PCR kit for detecting Escherichia coli O157 in bovine fecal samples. Appl Environ Microbiol. 2004;70:1855–7. doi: 10.1128/AEM.70.3.1855-1857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mull B, Hill VR. Recovery and detection of Escherichia coli O157: H7 in surface water, using ultrafiltration and real-time PCR. Appl Environ Microbiol. 2009;75:3593–7. doi: 10.1128/AEM.02750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kagkli DM, Folloni S, Barbau-Piednoir E, Van den Eede G, Van den Bulcke M. Towards a pathogenic Escherichia coli detection platform using multiplex SYBR®Green Real-time PCR methods and high resolution melting analysis. PLoS One. 2012;7:e39287. doi: 10.1371/journal.pone.0039287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura R, Mandrell RE, Galland JC, Hyatt D, Riley LW. Restriction-site-specific PCR as a rapid test to detect enterohemorrhagic Escherichia coli O157: H7 strains in environmental samples. Appl Environ Microbiol. 2000;66:2513–9. doi: 10.1128/aem.66.6.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conrad CC, Stanford K, McAllister TA, Thomas J, Reuter T. Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. J Microbiol Methods. 2014;105:22–30. doi: 10.1016/j.mimet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Sharma VK, Carlson SA. Simultaneous detection of Salmonella strains and Escherichia coli O157: H7 with fluorogenic PCR and single-enrichment-broth culture. Appl Environ Microbiol. 2000;66:5472–6. doi: 10.1128/aem.66.12.5472-5476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Zhao C, Zhang W, Cui S, Meng J, Wu J, et al. Use of ramification amplification assay for detection of Escherichia coli O157: H7 and other E. coli shiga toxin-producing strains. J Clin Microbiol. 2005;43:6086–90. doi: 10.1128/JCM.43.12.6086-6090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khatami F, Heidari M, Khatami M. Rapid detection of Escherichia coli O157: H7 by fluorescent amplification-based specific hybridization (FLASH) PCR. Iran Red Crescent Med J. 2012;14:594–8. [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Q, Shih WY, Shih WH. Real-Time, label-free, all-electrical detection of Salmonella typhimurium using lead zirconate titanate/gold-coated glass cantilevers at any relative humidity. Sens Actuators B Chem. 2007;125:379–88. doi: 10.1016/j.snb.2007.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Techathuvanan C, Draughon FA, D’Souza DH. Real-time reverse transcriptase PCR for the rapid and sensitive detection of Salmonella typhimurium from pork. J Food Prot. 2010;73:507–14. doi: 10.4315/0362-028x-73.3.507. [DOI] [PubMed] [Google Scholar]
- 62.Zhou M, Yang J, Zhou X, Liu B, Liu D, Yuan C, et al. Development of a sigDE-based real-time reverse-transcriptase PCR for the detection of viable Salmonella enterica. Foodborne Pathog Dis. 2014;11:537–44. doi: 10.1089/fpd.2013.1701. [DOI] [PubMed] [Google Scholar]
- 63.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. Genome Res. 1995;4:357–62. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 64.Faltin B, Wadle S, Roth G, Zengerle R, von Stetten F. Mediator probe PCR: a novel approach for detection of real-time PCR based on label-free primary probes and standardized secondary universal fluorogenic reporters. Clin Chem. 2012;58:1546–56. doi: 10.1373/clinchem.2012.186734. [DOI] [PubMed] [Google Scholar]
- 65.Tsourkas A, Bao G. Shedding light on health and disease using molecular beacons. Brief Funct Genomic Proteomic. 2003;1:372–84. doi: 10.1093/bfgp/1.4.372. [DOI] [PubMed] [Google Scholar]
- 66.Singh A, Poshtiban S, Evoy S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors (Basel) 2013;13:1763–86. doi: 10.3390/s130201763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant MA, Weagant SD, Feng P. Glutamate decarboxylase genes as a prescreening marker for detection of pathogenic Escherichia coli groups. Appl Environ Microbiol. 2001;67:3110–4. doi: 10.1128/AEM.67.7.3110-3114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steele M, Ziebell K, Zhang Y, Benson A, Konczy P, Johnson R, et al. Identification of Escherichia coli O157: H7 genomic regions conserved in strains with a genotype associated with human infection. Appl Environ Microbiol. 2007;73:22–31. doi: 10.1128/AEM.00982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harsono KD, Kaspar CW, Luchansky JB. Comparison and genomic sizing of Escherichia coli O157: H7 isolates by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1993;59:3141–4. doi: 10.1128/aem.59.9.3141-3144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma L, Zhang G, Doyle MP. Green fluorescent protein labeling of Listeria, Salmonella, and Escherichia coli O157: H7 for safety-related studies. PLoS One. 2011;6:e18083. doi: 10.1371/journal.pone.0018083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beyor N, Yi L, Seo TS, Mathies RA. Integrated capture, concentration, polymerase chain reaction, and capillary electrophoretic analysis of pathogens on a chip. Anal Chem. 2009;81:3523–8. doi: 10.1021/ac900060r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu W, Zhang J, Zheng M, Zhong Y, Yang J, Zhao Y, et al. An aptamer-based biosensor for colorimetric detection of Escherichia coli O157: H7. PLoS One. 2012;7:e48999. doi: 10.1371/journal.pone.0048999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dharmasiri U, Witek MA, Adams AA, Osiri JK, Hupert ML, Bianchi TS, et al. Enrichment and detection of Escherichia coli O157: H7 from water samples using an antibody modified microfluidic chip. Anal Chem. 2010;82:2844–9. doi: 10.1021/ac100323k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Ye Z, Si C, Ying Y. Subtractive inhibition assay for the detection of E. coli O157: H7 using surface plasmon resonance. Sensors (Basel) 2011;11:2728–39. doi: 10.3390/s110302728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narsaiah K, Jha SN, Bhardwaj R, Sharma R, Kumar R. Optical biosensors for food quality and safety assurance – A review. J Food Sci Technol. 2012;49:383–406. doi: 10.1007/s13197-011-0437-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang W, Chen C, Qian M, Zhao XS. Aptamer biosensor for protein detection using gold nanoparticles. Anal Biochem. 2008;373:213–9. doi: 10.1016/j.ab.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 77.Gui J, Patel IR. Recent advances in molecular technologies and their application in pathogen detection in foods with particular reference to Yersinia. J Pathog 2011. 2011:310135. doi: 10.4061/2011/310135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu H, Sikora U, Ozcan A. Quantum dot enabled detection of Escherichia coli using a cell-phone. Analyst. 2012;137:2541–4. doi: 10.1039/c2an35071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dwarakanatha S, Bruno JG, Athmaram TN, Bali G, Vattem D, Rao P. Antibody-quantum dot conjugates exhibit enhanced antibacterial effect vs. unconjugated quantum dots. Folia Microbiol (Praha) 2007;52:31–4. doi: 10.1007/BF02932134. [DOI] [PubMed] [Google Scholar]
- 80.Dwarakanath S, Bruno JG, Shastry A, Phillips T, John AA, Kumar A, et al. Quantum dot-antibody and aptamer conjugates shift fluorescence upon binding bacteria. Biochem Biophys Res Commun. 2004;325:739–43. doi: 10.1016/j.bbrc.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 81.Heyduk E, Heyduk T. Fluorescent homogeneous immunosensors for detecting pathogenic bacteria. Anal Biochem. 2010;396:298–303. doi: 10.1016/j.ab.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zharov VP, Mercer KE, Galitovskaya EN, Smeltzer MS. Photothermal nanotherapeutics and nanodiagnostics for selective killing of bacteria targeted with gold nanoparticles. Biophys J. 2006;90:619–27. doi: 10.1529/biophysj.105.061895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao X, Hilliard LR, Mechery SJ, Wang Y, Bagwe RP, Jin S, et al. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc Natl Acad Sci U S A. 2004;101:15027–32. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nou X, Arthur TM, Bosilevac JM, Brichta-Harhay DM, Guerini MN, Kalchayanand N, et al. Improvement of immunomagnetic separation for Escherichia coli O157: H7 detection by the PickPen magnetic particle separation device. J Food Prot. 2006;69:2870–4. doi: 10.4315/0362-028x-69.12.2870. [DOI] [PubMed] [Google Scholar]
- 85.Delibato E, Gattuso A, Minucci A, Auricchio B, De Medici D, Toti L, et al. PCR experion automated electrophoresis system to detect Listeria monocytogenes in foods. J Sep Sci. 2009;32:3817–21. doi: 10.1002/jssc.200900166. [DOI] [PubMed] [Google Scholar]
- 86.Koluman A, Celik G, Unlu T. Salmonella identification from foods in eight hours: a prototype study with Salmonella typhimurium. Iran J Microbiol. 2012;4:15–24. [PMC free article] [PubMed] [Google Scholar]
- 87.Hegde NV, Jayarao BM, DebRoy C. Rapid detection of the top six non-O157 Shiga toxin-producing Escherichia coli O groups in ground beef by flow cytometry. J Clin Microbiol. 2012;50:2137–9. doi: 10.1128/JCM.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rapp D, Waller J, Brightwell G, Muirhead RW. Lyophilization prior to direct DNA extraction from bovine feces improves the quantification of Escherichia coli O157: H7 and Campylobacter jejuni. Appl Environ Microbiol. 2010;76:1686–8. doi: 10.1128/AEM.01866-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


