Abstract
Background & objectives:
The Arg>Pro polymorphism in codon 72 of p53 gene is known to affect the susceptibility of cervical cancer differently in different population worldwide although information regarding its role in determining survival status and disease outcome in patients is lacking. The present study was conducted to determine the genotype frequency and prognostic role of p53 codon 72 Arg>Pro polymorphism in patients with advanced stage cervical cancer in India.
Methods:
The p53 codon 72 polymorphism was determined in tumour biopsies (n = 107) and matched blood samples (n = 19) in cervical cancer patients using polymerase chain reaction-restriction fragment length polymorphism method (PCR-RFLP). Effect of p53 genotype on the overall survival (OS) and recurrence-free survival (RFS) was analyzed. Individual Arg or Pro alleles were studied for their significance on survival as Pro carriers (Pro/Pro + Arg/Pro) versus Arg/Arg individuals or Arg carriers (Arg/Arg + Arg/Pro) versus Pro/Pro individuals.
Results:
The frequencies for Arg/Arg, Arg/Pro and Pro/Pro genotypes were 27.2, 49.5 and 23.3 per cent, respectively. There was no significant difference in the genotypes with respect to patients’ OS or RFS.
Interpretation & conclusions:
The findings of our study indicated that p53 codon 72 polymorphism might not be an independent marker in predicting clinical outcome in advanced stage cervical cancer patients. Further studies need to be done in larger samples to confirm these findings.
Keywords: Cervical cancer, overall survival, p53 codon 72 polymorphism, PCR-RFLP, recurrence-free survival
Cervical cancer is the third most common cancer among women worldwide. In India, with an estimated 1,34,000 (27%) new cases, with mortality of 72,000 in 2008, have made it the leading cause of gynaecological cancer related deaths1. Human papillomavirus (HPV) infection is established as the major aetiological cause for cervical carcinogenesis. As reported in our previous study in a group of Indian women with advanced stage cervical cancer, 95 per cent of the cases were HPV positive, with the two high-risk HPVs - HPV16 and/or HPV18 being most prevalent2. The E6 and E7 oncoproteins encoded by HPV16 and HPV18 bind and functionally inactivate the tumour suppressor proteins - p53 and Rb, respectively, thereby disrupting G1 arrest during DNA damage and possibly leading to carcinogenic transformation of the cervical epithelium following HPV infection3.
Although infection with HPV has been established as the major aetiological reason for cervical carcinogenesis, only a few of the women infected with HPV develop cancer in their lifetime4. This suggests probable involvement of genetic cofactors in the development of cancer. The p53 gene exon 4 codon 72 Arg>Pro polymorphism has long been investigated for its association with the risk of cervical carcinogenesis4,5,6. A sequence change from CGC to CCC of p53 gene at codon 72 results in either Arg72 or Pro72 protein variants, both of which act as wild type but with different primary structures and biochemical functions7,8. According to the hypothesis by Storey et al5, the p53Arg variant is seven times more susceptible to E6-mediated ubiquitin-dependent proteolysis than the p53Pro variant. It has been shown that women homozygous for arginine are at a higher risk of developing cervical cancer than the heterozygous ones6. On the contrary, there are studies that refute any association of this polymorphism with cervical cancer susceptibility7; yet others have found the p53 Pro/Pro genotype to be more hazardous than Arg/Arg for the development of the disease4. The p53Arg variant has been found to be about five times more susceptible to apoptosis than Pro variant due to its greater ability to localize in the mitochondria, thereby resulting in enhanced ubiquitination by an E3 ubiquitin-protein ligase (MDM2)8. It has been suggested that tumour cells with mutations resulting in reduced p53-dependent apoptosis are less vulnerable to chemo/radiotherapy (RT) and hence are more aggressive9. This can be correlated with the possibility that patients carrying Pro allele (which are less liable to apoptosis as compared to Arg genotype) might have a better prognosis following chemo/radiotherapy. However, there are only a few studies that support the effect of this polymorphism on the survival of patients with the disease10,11,12,13. Therefore, the aim of the present study was to analyze the frequency of p53 codon 72 polymorphism in cervical cancer patients with advanced stage (FIGO stage IIIB) of the disease. It was also proposed to determine the association of this polymorphism, if any, with the disease outcome, either independently or in combination with HPV infection.
Material & Methods
The study population included 107 Indian women with histologically proven, advanced stage cervical cancer (FIGO stage IIIB) who received radical RT or concomitant chemoradiation (RT+CT) at the Radiation Oncology Department of Tata Memorial Hospital, Mumbai, India, from August 2003 to November 2010. The HPV status of 83 of these 107 patients was known from an earlier study2. One hundred and seven pre-treatment cervical cancer biopsies and 19 paired blood samples (selected randomly) were included in the study after approval from the Hospital's Institute Ethics Committee. The tissue specimens were coded to maintain confidentiality and were frozen in liquid nitrogen and stored at −80°C until use. All the patients were followed up from the day the treatment began and the time when recurrence occurred, or till the patient's last visit to the clinic, which was taken as the termination date for follow up. Follow up was done once every three months in the first year, 4-6 months between second and fifth years and annually subsequently. The follow up evaluation consisted of pelvic examination and appropriate investigations including tissue diagnosis to establish recurrence and extent of it.
DNA extraction and p53 genotyping at codon 72: Genomic DNA was isolated from 107 tumour and 19 blood samples by standard phenol–chloroform method14. The samples were genotyped at the p53 codon 72 locus by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. For this, genomic DNA was amplified using primers as described by Xu et al14. A 279 bp segment of the p53 gene was amplified in a 25 µl reaction mixture containing 60 ng of genomic DNA, ×10 PCR buffer, 0.6 mM MgCl2, 0.2 mM deoxynucleotide triphosphates (dNTPs), 0.2 µM primers and 1.5 units Taq DNA Polymerase (Thermo Scientific, USA). PCR cycling conditions used were initial DNA denaturation at 94°C, followed by 30 cycles of DNA denaturation at 94°C for 30 sec, primer annealing at 60°C for 30 sec, primer extension at 72°C for 30 sec and a final extension at 72°C for 10 min. By comparison of the band intensity with the standard O’GeneRuler 100 bp DNA Ladder (Thermo Scientific, USA), approximately 500 ng of the PCR product was subjected to restriction digestion with 10 units of BstUI restriction enzyme (New England Biolabs, USA) for two hours at 60°C. The digested products were resolved on 2 per cent agarose gel containing ethidium bromide, and the p53 genotype at codon 72 was identified by the band pattern obtained. A single band of 279 bp indicated Pro/Pro genotype, two bands of 160 and 119 bp indicated Arg/Arg genotype and three bands corresponding to 279, 160 and 119 bp indicated Arg/Pro genotype.
Statistical analysis: Statistical analysis was done using SPSS software (IBM SPSS Statistics, version 21.0, Armonk, NY, USA). The association of p53 codon 72 polymorphism with the clinical parameters/characteristics was carried out using Chi-square test. Survival probabilities were plotted using Kaplan–Meier method and compared using log-rank test. Overall survival (OS) was defined as time from registration of the patient to the clinic till death due to any cause or the last follow up and recurrence-free survival (RFS) as time from registration till recurrence or death due to disease or the last follow up. Multivariate analysis was carried out using Cox proportional hazard model to identify the factors that predicted survival.
Results
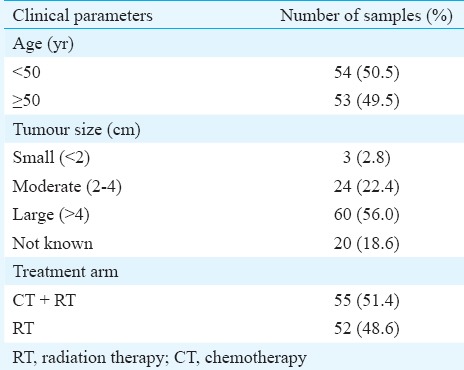
A total of 107 pre-treatment cervical tumour biopsies and 19 matched blood samples selected randomly from patients of Indian origin (median age, 49 yr; age range, 33-80 yr; IQR, 40-55 yr) were included in the study. The clinicopathological characteristics of the patients are given in Table I. Of the 107 patients, 52 received RT alone and 55 were treated with RT+CT. More than three years of follow up data (median follow up, 5.2 yr; range, 3.7-7.0 yr; IQR, 4.6-5.8 yr) were available for these patients. Sixty one patients developed recurrence of the disease while the remaining patients were disease free at the last follow up.
Table I.
Clinicopathological characteristics of cervical carcinoma patients (n=107)
P53 codon 72 genotyping: The DNA from the patients was genotyped at codon 72 locus of p53 for Arg/Arg, Arg/Pro and Pro/Pro genotype using PCR-RFLP method (Table II). The genotypes of randomly selected 18 tumour samples and 4 paired blood samples were also confirmed by Sanger sequencing (data not shown). The genotypic frequencies of p53 codon 72 locus (Table II) were in accordance with the Hardy–Weinberg equilibrium distribution (χ2 = 0.006946).
Table II.
p53 codon 72 Arg >Pro genotypes and allele frequencies in cervical carcinoma patients (n=107)
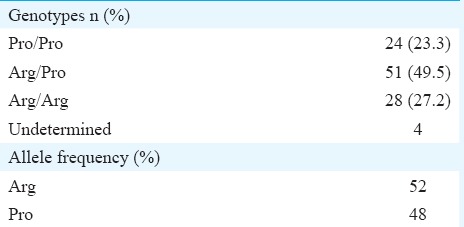
Role of p53 codon 72 polymorphism in cervical cancer prognosis: Kaplan–Meier survival curves were plotted for OS and RFS to study the effect of p53 codon 72 polymorphism on disease prognosis. The median OS and RFS of the patients were 3.2 [95% confidence interval (CI); 1.9-4.5 yr] and 3.2 years (95% CI; 1.9-4.6 yr), respectively. No significant differences were observed in the OS and RFS of the patients with the three genotypes (Fig. 1A, B and Table III). To determine the significance of individual alleles in the survival of the patients, the OS and RFS of Pro-allele carriers (Pro/Pro + Arg/Pro) were compared with those of Arg homozygous individuals and also of Arg allele carriers (Arg/Arg + Arg/Pro) with Pro homozygous individuals. In both the cases, homozygous Pro and Pro carrier women had a slightly poor prognosis than women with Arg homozygous genotype, but the difference was not significant (Fig. 2 and Table III).
Fig. 1.
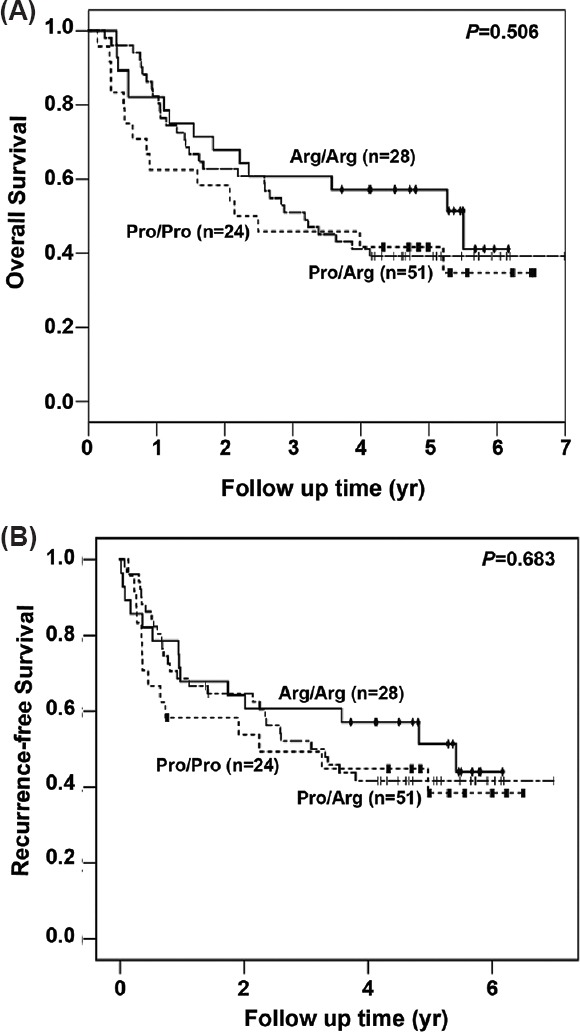
Survival curve of patients carrying Arg/Arg, Arg/Pro and Pro/Pro genotypes. Kaplan–Meier analysis of overall (A) and recurrence-free survival (B) of 103 invasive cervical cancer cases with Arg/Arg (n=28), Arg/Pro (n=51) and Pro/Pro (n=24) genotypes is depicted.
Table III.
Univariate analyses of overall and recurrence-free survival (RFS) using Cox proportional hazard model
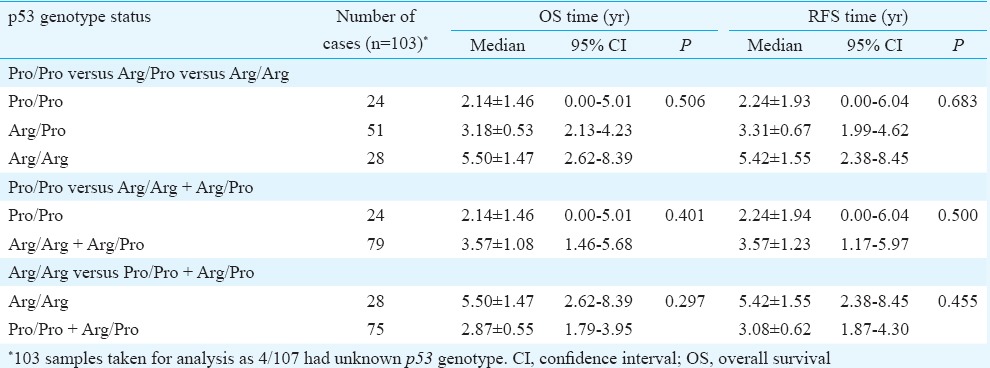
Fig. 2.
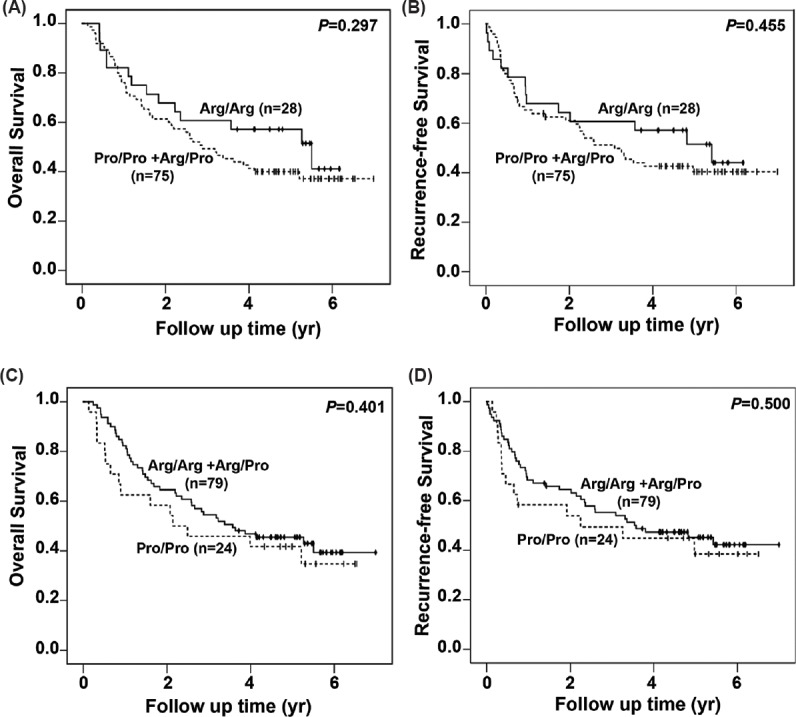
Survival curves for overall and recurrence- free survival of the cases with different combinations of the p53 genotype. Kaplan–Meier survival analysis of overall and recurrence-free survival for the genotypes Arg/Arg (n=28) versus Pro/Pro and Arg/Pro (n=75) taken together is shown (A and B). Similar analysis was done for Pro/ Pro (n=24) versus Arg/Arg + Arg/Pro (n=79) genotypes. This is represented in C and D.
HPV typing of 83 of these samples was available as part of a previous study from our laboratory2. This information was used for multivariate analysis using Cox proportional hazard method. This however, was not found to be significant (data not shown).
Discussion
The p53 codon 72 Arg>Pro polymorphism has been studied extensively for its association with the risk of cervical carcinogenesis. Reports stating the over-representation of homozygous Arg in cases as compared to controls in lung cancer15, head and neck cancer16 and breast cancer17 suggested its role in cancer susceptibility, whereas studies in other cancers such as chronic myeloid leukaemia18 and nasopharyngeal carcinoma19 reported association of Pro/Pro genotype in cancer development. Overall, this difference might be due to differences in sample size and methodologies used in these studies, or variation in the ethnic distribution of this polymorphism.
The present study was conducted to study the effect of p53 codon 72 polymorphism on the disease outcome in a group of invasive cervical cancer patients of Indian origin. The observed genotype frequencies of Pro/Pro (23.3%), Arg/Pro (49.5%) and Arg/Arg (27.2%) were in concordance with other similar studies from India4,20.
The association of p53 codon 72 polymorphism with disease prognosis differs in different cancer types. Studies in cancers such as breast14, lung15, head and neck squamous cell carcinoma21, colorectal cancer22 and multiple myeloma23 indicated Arg/Arg genotype to be beneficial for survival. A report on nasopharyngeal carcinoma indicated Pro/Pro genotype to be of prognostic value24. However, several other studies questioned the involvement of this polymorphism in disease outcomes or response to therapies25,26. A study in Brazilian population by Brenna et al10 reported Arg/Pro genotype to be associated with good clinical outcome and Arg/Arg genotype with decreased survival time in cervical cancer patients. These investigators did not find any Pro/Pro genotype in their study population. Contrary to this report, in our study, we observed no correlation between the three different p53 genotypes with OS as well as RFS in a group of advanced stage cervical cancer patients. Our observation, however, corroborated with other reports11,11,12,13,27,28 where no significant correlation was observed between codon 72 polymorphism and cervical cancer prognosis.
Though no significant differences in OS or RFS were observed in our study, the Arg allele was slightly more favourable for survival as compared to Pro allele. These results were in agreement with studies in other cancers such as breast cancer14, lung cancer15, multiple myeloma23 and colorectal cancer22. Our findings were also consistent with the finding of Dumont et al8 that apoptosis was more efficiently induced by the Arg72-p53 isoform as compared to the Pro72-p53 one. This superior activity of Arg72-p53 variant was also reflected in an in vivo study by Sullivan et al21 who showed more favourable outcomes following chemo-RT in the advanced stage head and neck cancer patients expressing Arg72 variant of the protein as compared to the Pro72.
The HPV-dependent effect of this polymorphism on patient's OS and RFS, in multivariate analysis, also revealed no significant results which was in concordance with other similar studies on cervical cancer11,13. The probable reason for the insignificant difference in the relapse time among the three genotypes, whether analyzed individually or in combination, might be the advanced FIGO (International Federation of Gynecology and Obstetrics) stage IIIB of the disease and lower sample size in each subgroup. A study29 has reported FIGO staging as important and independent prognostic factor for disease-free survival and OS of cervical cancer patients.
In conclusion, our study showed that p53 codon 72 polymorphism might not be an independent and significant predictor of survival outcome in advanced stage cervical cancer patients of Indian origin. However, this finding needs to be tested in a larger cohort.
Acknowledgment
The authors thank late Dr K. A. Dinshaw and all individuals from the Gynaecology Disease Management Group, Tata Memorial Hospital, Mumbai, who were involved in judiciously compiling the clinical history and collecting and storing the biopsies from cervical cancer patients. Financial support from Women's Cancer Initiative (IRG#634) is gratefully acknowledged.
Footnotes
Conflicts of Interest: None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Das P, Thomas A, Mahantshetty U, Shrivastava SK, Deodhar K, Mulherkar R. HPV genotyping and site of viral integration in cervical cancers in Indian women. PLoS One. 2012;7:e41012. doi: 10.1371/journal.pone.0041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya P, Duttagupta C, Sengupta S. Proline homozygosity in codon 72 of p53: a risk genotype for human papillomavirus related cervical cancer in Indian women. Cancer Lett. 2002;188:207–11. doi: 10.1016/s0304-3835(02)00430-5. [DOI] [PubMed] [Google Scholar]
- 5.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–34. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 6.Burroni E, Bisanzi S, Sani C, Puliti D, Carozzi F. Codon 72 polymorphism of p53 and HPV type 16 E6 variants as risk factors for patients with squamous epithelial lesion of the uterine cervix. J Med Virol. 2013;85:83–90. doi: 10.1002/jmv.23417. [DOI] [PubMed] [Google Scholar]
- 7.Calhoun ES, McGovern RM, Janney CA, Cerhan JR, Iturria SJ, Smith DI, et al. Host genetic polymorphism analysis in cervical cancer. Clin Chem. 2002;48:1218–24. [PubMed] [Google Scholar]
- 8.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 9.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–67. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 10.Brenna SM, Silva ID, Zeferino LC, Pereira JS, Martinez EZ, Syrjänen KJ. Prognostic value of P53 codon 72 polymorphism in invasive cervical cancer in Brazil. Gynecol Oncol. 2004;93:374–80. doi: 10.1016/j.ygyno.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Ngan HY, Liu VW, Liu SS, Cheng DK, Ng TY, Wong LC. Homozygous arginine at codon 72 of p53 has no prognostic significance in cervical cancer. Tumour Biol. 2000;21:135–8. doi: 10.1159/000030119. [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa A, Fujimoto T, Akutagawa N, Iwasaki M, Takeuchi M, Fujinaga K, et al. p53 polymorphism (codon-72) has no correlation with the development and the clinical features of cervical cancer. Int J Gynecol Cancer. 2000;10:402–7. doi: 10.1046/j.1525-1438.2000.010005402.x. [DOI] [PubMed] [Google Scholar]
- 13.Wong YF, Chung TK, Cheung TH, Nobori T, Hampton GM, Wang VW, et al. p53 polymorphism and human papillomavirus infection in Hong Kong women with cervical cancer. Gynecol Obstet Invest. 2000;50:60–3. doi: 10.1159/000010282. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, et al. p53 codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–33. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- 15.Wang YC, Chen CY, Chen SK, Chang YY, Lin P. p53 codon 72 polymorphism in Taiwanese lung cancer patients: association with lung cancer susceptibility and prognosis. Clin Cancer Res. 1999;5:129–34. [PubMed] [Google Scholar]
- 16.Cortezzi SS, Provazzi PJ, Sobrinho JS, Mann-Prado JC, Reis PM, de Freitas SE, et al. Analysis of human papillomavirus prevalence and TP53 polymorphism in head and neck squamous cell carcinomas. Cancer Genet Cytogenet. 2004;150:44–9. doi: 10.1016/j.cancergencyto.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Ohayon T, Gershoni-Baruch R, Papa MZ, Distelman Menachem T, Eisenberg Barzilai S, Friedman E. The R72P P53 mutation is associated with familial breast cancer in Jewish women. Br J Cancer. 2005;92:1144–8. doi: 10.1038/sj.bjc.6602451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergamaschi G, Merante S, Orlandi E, Galli A, Bernasconi P, Cazzola M. TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica. 2004;89:868–9. [PubMed] [Google Scholar]
- 19.Hadhri-Guiga B, Toumi N, Khabir A, Sellami-Boudawara T, Ghorbel A, Daoud J, et al. Proline homozygosity in codon 72 of TP53 is a factor of susceptibility to nasopharyngeal carcinoma in Tunisia. Cancer Genet Cytogenet. 2007;178:89–93. doi: 10.1016/j.cancergencyto.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Mitra S, Misra C, Singh RK, Panda CK, Roychoudhury S. Association of specific genotype and haplotype of p53 gene with cervical cancer in India. J Clin Pathol. 2005;58:26–31. doi: 10.1136/jcp.2004.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–37. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 22.Starinsky S, Figer A, Ben-Asher E, Geva R, Flex D, Fidder HH, et al. Genotype phenotype correlations in Israeli colorectal cancer patients. Int J Cancer. 2005;114:58–73. doi: 10.1002/ijc.20645. [DOI] [PubMed] [Google Scholar]
- 23.Hattori Y, Ikeda Y, Suzuki Y, Ichikawa D, Matsushita M. Codon 72 polymorphism of TP53 gene is a novel prognostic marker for therapy in multiple myeloma. Br J Haematol. 2014;165:728–31. doi: 10.1111/bjh.12784. [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Wang H, Jin H, Ouyang S, Zhou J, Hu J, et al. Expression of pAkt affects p53 codon 72 polymorphism-based prediction of response to radiotherapy in nasopharyngeal carcinoma. Radiat Oncol. 2013;8:117. doi: 10.1186/1748-717X-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matakidou A, El Galta R, Webb EL, Rudd MF, Bridle H, Eisen T, et al. Lack of evidence that p53 Arg72Pro influences lung cancer prognosis: an analysis of survival in 619 female patients. Lung Cancer. 2007;57:207–12. doi: 10.1016/j.lungcan.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Brant O, Hoffmann M, Kanappilly A, Görögh T, Gottschlich S. P53 codon 72 polymorphism in squamous cell carcinoma of the head and neck region. Anticancer Res. 2007;27:3301–5. [PubMed] [Google Scholar]
- 27.Hernádi Z, Szarka K, Sápy T, Krasznai Z, Veress G, Póka R. The prognostic significance of HPV-16 genome status of the lymph nodes, the integration status and p53 genotype in HPV-16 positive cervical cancer: a long term follow up. BJOG. 2003;110:205–9. [PubMed] [Google Scholar]
- 28.Szarka K, Veress G, Juhász A, Kónya J, Sápy T, Soós G, et al. Integration status of virus DNA and p53 codon 72 polymorphism in human papillomavirus type 16 positive cervical cancers. Anticancer Res. 2000;20:2161–7. [PubMed] [Google Scholar]
- 29.Stehman FB, Bundy BN, DiSaia PJ, Keys HM, Larson JE, Fowler WC. Carcinoma of the cervix treated with radiation therapy. I. A multi-variate analysis of prognostic variables in the Gynecologic Oncology Group. Cancer. 1991;67:2776–85. doi: 10.1002/1097-0142(19910601)67:11<2776::aid-cncr2820671111>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]


