Abstract
Background & objectives:
Polycystic ovary syndrome (PCOS) is the most common reproductive endocrine disorder of premenopausal women. Given the phenotypic overlap between PCOS and type 2 diabetes mellitus (T2DM), this study was carried out to investigate whether genes implicated in T2DM were also involved in the susceptibility to PCOS among women from southern India.
Methods:
A total of 248 women with PCOS and 210 healthy women as controls were genotyped for a panel of 15 single nucleotide polymorphisms (SNPs) from the nine T2DM genes, such as TCF7L2, IGF2BP2, SLC30A8, HHEX, CDKAL1, CDKN2A, IRS1, CAPN10 and PPARG, on Sequenom MassARRAY platform.
Results:
None of the 15 SNPs were found to be significantly associated with PCOS after Bonferroni correction for multiple testing, either in the univariate or multivariate context. The cumulative effect of risk alleles observed with reference to T2DM was also not seen with reference to PCOS.
Interpretation & conclusions:
The nine T2DM genes considered in this exploratory study might not be the primary susceptibility factors for PCOS among Indian women. Our results supplement the lack of evidence of the association of T2DM genes with PCOS among the Chinese and Caucasians hinting at the possible universality of this pattern. Specifically designed comprehensive studies that include women with T2DM and PCOS are required to explore the precise role of the diabetes genes.
Keywords: Polycystic ovary syndrome, single-nucleotide polymorphisms, type 2 diabetes mellitus, T2DM genes
Polycystic ovary syndrome (PCOS) has multiple components, such as reproductive, metabolic and cardiovascular, with long-term health implications. It is the leading cause of anovulatory infertility with an underlying complex genetic aetiology1. In genome-wide association studies (GWAS) about 70 genes have been identified by candidate gene approach and 11 loci identified as associated with PCOS1,2. However, significant GWAS variants of PCOS are not associated with risk phenotypes such as diabetes and obesity2. In addition to the reproductive defects, PCOS is hallmarked by insulin resistance and hyperinsulinaemia3. Therefore, numerous genes involved in insulin secretion and action have been explored as candidate genes in the PCOS pathology. A PCOS GWAS identified a novel polymorphism in the INSR gene related to insulin pathway4.
Genetic studies have revealed that PCOS and type 2 diabetes mellitus (T2DM) could share genetic susceptibility factors associated with both pathologies. Using this hypothesis, several studies have suggested that genes related to T2DM may also play a role in PCOS pathogenesis5,6,7, which is evident by various meta-analyses8,9,10. However, with respect to the GWAS-identified T2DM genes such as KCNJ11, TCF7L2, FTO, HHEX, CDKAL1 and SLC30A8, significant association with PCOS was not observed11,12,13. Insulin resistance is a common metabolic feature of both PCOS and T2DM. There is also strong evidence that Indians are genetically predisposed to develop T2DM and have high insulin resistance compared to other ethnic groups14,15. In a study conducted in southern India, it was observed that women with the reproductive abnormalities of PCOS had high insulin resistance16. Although some studies have investigated the role of different candidate and GWAS genes in the T2DM pathophysiology17,18,19,20, there is a paucity of such studies from India with reference to PCOS. In the Indian context, most of the PCOS studies were confined to the clinical dimensions, and only a few genetic studies were undertaken21,22,23,24. Given the high prevalence of T2DM, particularly in Hyderabad15 and because of its shared pathophysiology with PCOS, it is imperative to understand the aetiology of PCOS from the perspective of T2DM genes. Therefore, this study was aimed to examine the association of a panel of 15 single-nucleotide polymorphisms (SNPs) from nine different T2DM candidate/GWAS genes with PCOS among the women residing in Hyderabad, India. Despite a large number of SNPs that were found associated with T2DM in different populations, due to resource constraint, our choice of genotyping was restricted to 15 SNPs that were most significantly and consistently associated with T2DM, among both Indian and non-Indian populations17,20.
Material & Methods
Sampling criteria: Case and control groups: As part of the larger project entitled, “Identification of susceptibility genes associated with PCOS among Indian women” undertaken by Molecular Anthropology Group of the Biological Anthropology Unit, Indian Statistical Institute, Hyderabad, India, 250 women with PCOS and 299 normal healthy controls were enrolled in the study during 2008-2009. Due to resource constraints, only 210 of the 299 controls and 248 of 250 cases were genotyped for the present study. The patients were recruited consecutively from the Gynecology Clinic of the Osmania General Hospital and Anu Infertility Clinic in Hyderabad as per the Rotterdam criteria25. According to these criteria any two of the following three conditions need to be fulfilled for the inclusion: (i) presence of clinical and/or biochemical signs of hyperandrogenism, (ii) infrequent periods with intermenstrual interval of more than 35 days, and (iii) polycystic ovaries [an ovary with the ultrasound appearance of more than 10 subcapsular follicles (<10 mm in diameter) in the presence of prominent ovarian stroma was considered polycystic]. Patients with hyperprolactinaemia, thyroid and adrenal diseases, 21-hydroxylase deficiency and androgen-secreting tumours were excluded from the study. The ethnically matched normal controls with no history of treatment for fertility, with normal menstrual cycles every 25-32 days and with no signs of clinical hyperandrogenism (hirsutism, acne and alopecia) were recruited from the family planning centre of the Osmania General Hospital, Hyderabad and from the general population in the same city, (covering different residential complexes, government offices, academic institutions and software companies). Both cases and controls represented similar ethnic and linguistic backgrounds.
Intravenous blood samples (5 ml) were collected from both patients and controls. Informed written consent was obtained from each participant before their enrolment in the study. The Indian Statistical Institute Review Committee for Protection of Research Risks to Humans specifically approved this project and the study protocol.
Genotyping of T2DM genes: DNA was extracted using phenol–chloroform method26. Fifteen well-replicated SNPs from nine T2DM genes identified through candidate and GWASs were selected. The SNPs included were rs7903146, rs11196205 and rs12255372 (TCF7L2), rs4402960 and rs1470579 (IGF2BP2), rs13266634 (SLC30A8), rs1111875 and rs7923837 (HHEX), rs7754840 and rs7756992 (CDKAL1), rs10811661 (CDKN2A/B), rs1801278 (IRS1), rs3792267 and rs5030952 (CAPN10) and rs1801282 (PPARG). Genotyping of the 15 SNPs was performed on Sequenom MassARRAY platform (Sequenom Inc., San Diego, CA, USA) at the Centre for Genomics Application (TCGA), Delhi, India. The raw data files generated by MassARRAY Sequenom were analyzed for the intensity peaks of calibrant to ascertain the quality of the data. An overall call rate of >90 per cent was maintained. For every 96 samples (a quadrant of Sequenom chip), four samples were duplicated and the call rates were checked for concordance. The calls in the negative control (no DNA) were also monitored in all the runs. The genotype call rate for individual SNPs ranged from 96-99 per cent. Of the 248 cases, only one sample was removed with no call for all the SNPs while in the controls all the samples worked. For the interaction analysis, only samples with complete genotype information were considered for all the nine SNPs.
Statistical analysis: Allele and genotype frequencies were calculated using SPSS Statistical Software (SPSS Inc., version 20.0, SPSS, Chicago, USA). Logistic regression analysis of the alleles and genotypes was carried out for all the SNPs individually. Bonferroni correction for multiple testing was carried out by setting P = 0.003 as threshold for significance. Linkage disequilibrium (LD) plots were estimated using Haploview software (version 4.1, Broad Institute, Cambridge, MA), and THESIAS software (version 3.1, INSERM U525, Paris, France) was used to generate haplotype frequencies and logistic regression analysis of the haplotypes. Univariate and multivariate logistic regression analyses and gene–environment interaction were carried out using EpiCalc package of R program version 3.0 (R Foundation, http://www.r-project.org/), Gene–Gene interaction was carried out using multifactor dimensionality reduction (MDR) software and permutation tool (University of Pennsylvania, Philadelphia, USA). The power of the study was calculated for individual SNPs using G*Power software (version 3.1, Universitat Dusseldorf, Germany).
Results
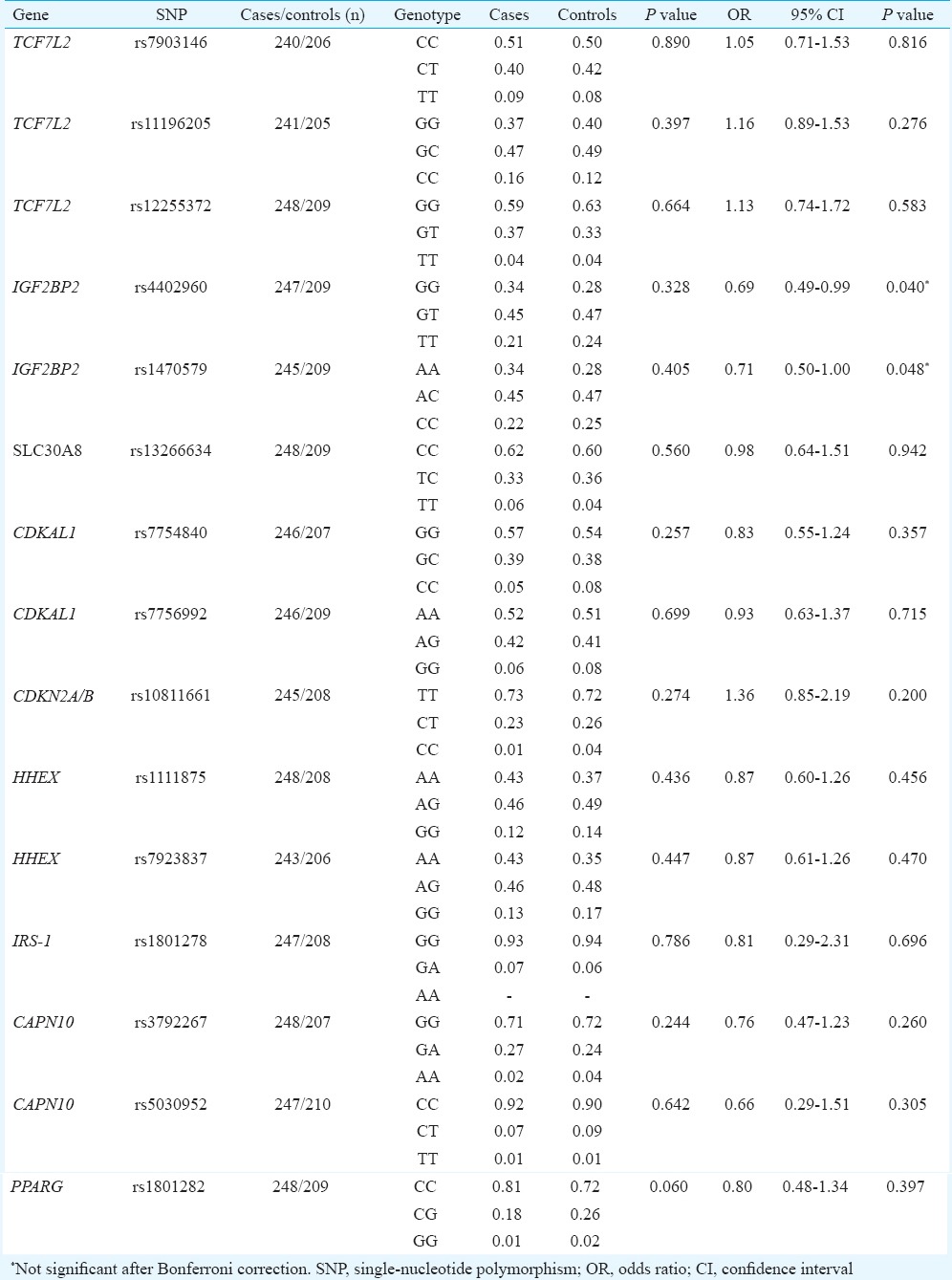
The clinical characteristics of the participants have been described elsewhere24. However, given the relatively young patients with PCOS who were encountered at the clinics in Hyderabad (ranging from 19 to 35 years), none of the PCOS cases were found to be diabetic. All the 15 SNPs followed Hardy–Weinberg equilibrium except rs5030952 of CAPN10 (P = 0.021), which also did not reach the specified threshold of significance under multiple testing (P = 0.003) to merit its exclusion from further analysis. None of the 15 SNPs were found to be significantly associated with PCOS after Bonferroni correction for multiple testing although PPARG (rs1801282) was associated with the disease (P < 0.02, Tables I and II) but in a protective role against developing PCOS. On the contrary, the two SNPs of IGF2BP2 (rs4402960 and rs1470579) showed marginally significant association (P = 0.040 and 0.048, respectively), only after adjusting for body mass index (BMI), probably indicating BMI's role as possible confounding factor in masking the protective role of this gene against the manifestation of PCOS. The pattern of distribution of genotypes in the non-obese cases and controls (P = 0.067; P = 0.060, respectively, for the two SNPs) provided empirical indication for this.
Table I.
Allele frequency distribution (in %) of the single-nucleotide polymorphisms (SNPs) of T2DM susceptibility genes in polycystic ovary syndrome (PCOS) cases and controls
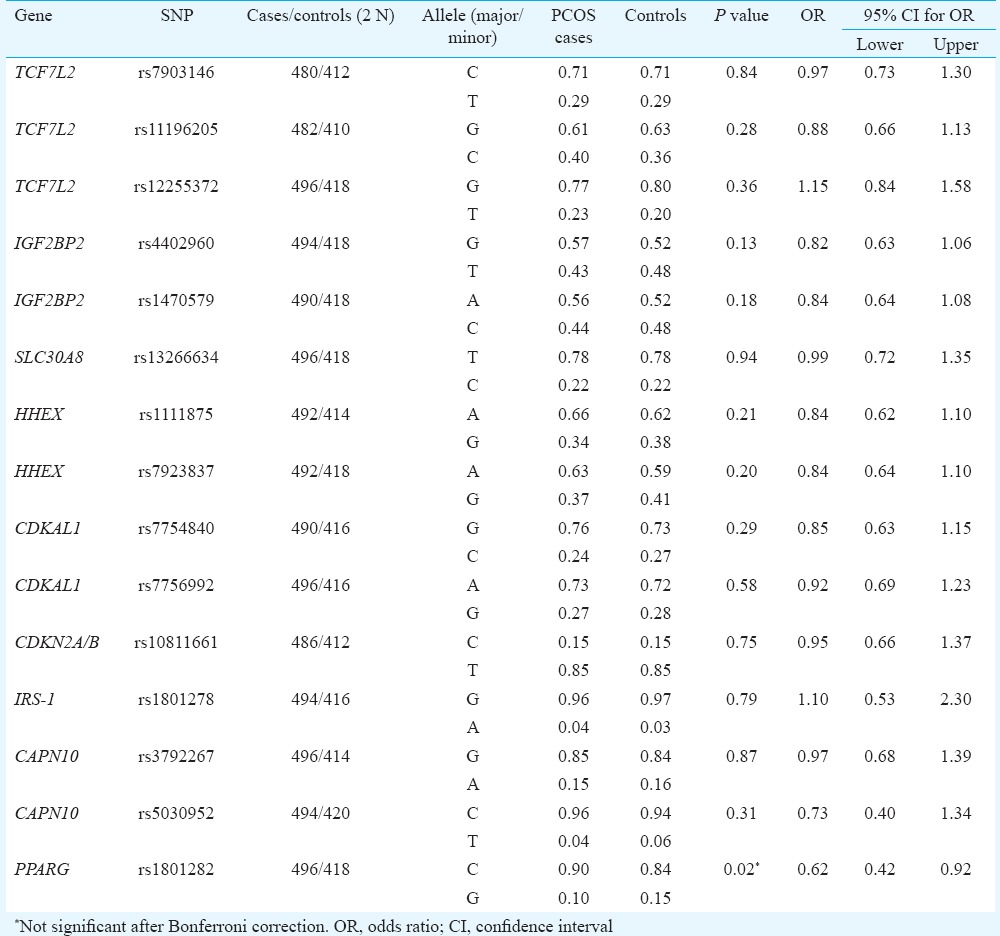
Table II.
Genotype frequency distribution (in %) and logistic regression under log-additive model and using body mass index as covariate
A strong LD was observed among the SNPs of the genes on the same chromosome, namely, TCF7L2, IGF2BP2, CDKAL1, HHEX and CAPN10, while there was no LD, as expected, between the genes located on the same chromosome (Fig. 1). Based on the observed LD pattern, only nine SNPs from the nine genes were considered for the multivariate and interaction analysis, i.e. (TCF7L2 (rs7903146), IGF2BP2 (rs1470579), SLC30A8 (rs13266634), CDKAL1 (rs7756992), CDKN2A/B (rs10811661), HHEX (rs1111875), IRS-1 (rs1801278), CAPN10 (rs3792267) and PPARG (rs1801282).
Fig. 1.
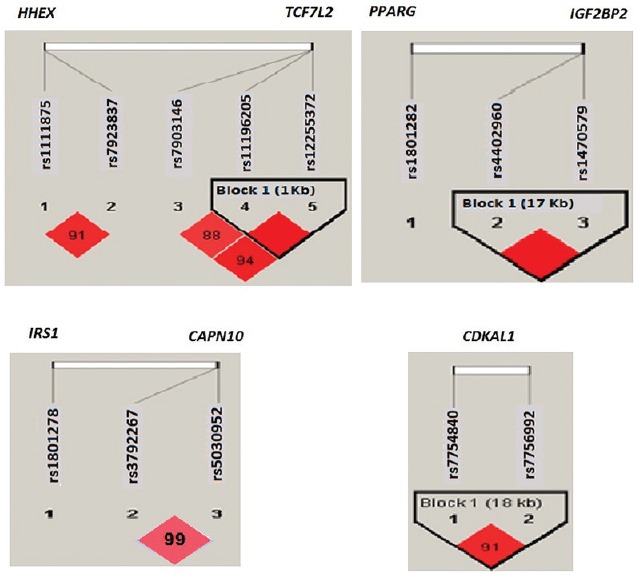
Linkage disequilibrium (LD) plots of single-nucleotide polymorphisms (SNPs) of genes in the same chromosome, i.e. TCF7L2, HHEX; IGF2BP2, PPARG; IRS1, CAPN10; CDKAL1. *D’ values mentioned in the linkage disequilibrium plot, except in two cases where the linkage disequilibrium was complete (100%). The pairs of SNPs with D’ value ≥ 80 per cent are considered to be in significant LD. Linkage disequilibrium is seen only between SNPs of the same gene, not across the genes, even on the same chromosome. The r2 values for the above linkage disequilibrium plot ranged from 0.44 to 0.98, but in case of CAPN10, it was very low (r2 = 0).
In the multivariate logistic regression analysis with covariates (results not presented), none of the nine SNPs showed significant association with PCOS after correction for multiple testing; while PPARG SNP was significant in the allele-wise analysis (P = 0.036) and disappeared after adjusting for BMI, only CAPN10 genotype (GA) showed marginally significant (P = 0.049) association with PCOS after adjusting for BMI. SNP–SNP interaction analysis using multifactor dimensionality reduction (MDR) suggested PPARG in one, IGF2BP2 and CDKAL1 in two, TCF7L2, IGF2BP2 and CDKAL1 in three and TCF7L2, IGF2BP2, CDKAL1 and HHEX in four loci combinations as the best interactions (Table III). Further, IGF2BP2 and CDKAL1 were found to be common among all the three multi locus combinations. This was evident in the dendogram (Fig. 2) which suggests synergistic interaction between IGF2BP2 and CDKAL1. However, none of the combinations turn out to be significant when permutated for 1000 iterations.
Table III.
Summary results of SNP-SNP interactions using multifactor dimensionality reduction
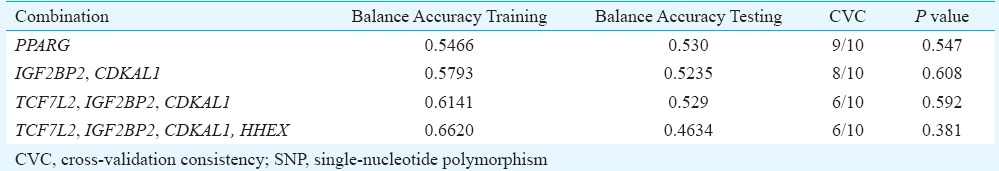
Fig. 2.
Dendogram obtained from the multifactor dimensionality reduction interaction analysis of the nine T2DM genes suggesting possible synergistic interactions. Colour indicates type of interaction: Red Synergistic interaction; Yellow - Independence; Blue - Redundancy or correlation (www.epistasis.org).
The gene–environment interaction analysis was carried out using R program, in which logistic regression analysis (assuming log-additive genetic model) was performed to gauge the possible interaction of each of the individual SNP genotypes with BMI (environmental factor) and the PCOS phenotype as the binary response variable (presence or absence of PCOS). None of the genes showed significant interaction with BMI (results not presented); CDKAL1*BMI and CAPN10*BMI interactions were significant only at 10 per cent despite BMI showing highly significant association with the phenotype (P < 0.001).
In an earlier study of T2DM based on the same 15 SNPs20, positive association was observed between the T2DM and number of risk alleles carried by an individual. Therefore, risk scores were calculated assuming that the T2DM risk alleles would be the same for PCOS as none of these SNP variants were found to be significantly risk prone to PCOS. Fig. 3 shows frequency distribution of PCOS patients and controls according to the number of risk alleles carried by them, suggesting no apparent trend of increasing risk of PCOS with increasing number of T2DM risk alleles.
Fig. 3.
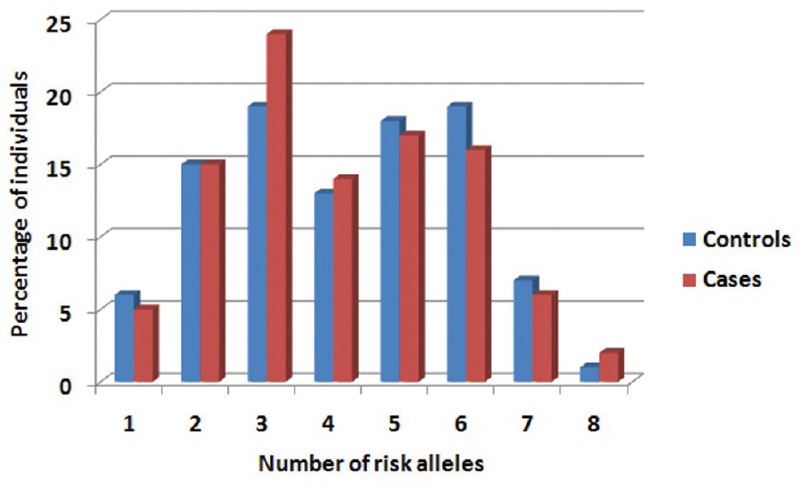
Histogram representing the frequency distribution of polycystic ovary syndrome patients and controls according to the number of risk alleles.
Discussion
Most previous studies that explored genetic susceptibility to PCOS have investigated only the effects of a single gene and/or a single polymorphism in the pathways mostly related to hyperandrogenism and ovarian dysfunction. As T2DM and PCOS share a common medical condition through insulin resistance, it has been shown that genes related to adipocyte differentiation and insulin pathways may contribute to pathogenesis of PCOS. However, the most prominent candidate genes as well as GWAS-identified T2DM genes were found to be associated with the insulin-resistant trait of PCOS, not with the overall phenotype27,28. We have earlier explored the pattern of association of T2DM genes with PCOS among the Indian women24.
Overall, our results showed that none of the 15 SNPs of the nine T2DM genes played a direct/primary role in the manifestation of PCOS phenotype. Despite the strong and universal association of TCF7L2 with T2DM, its association with PCOS could not be traced in our study population as well as in the couple of non-Indian populations studied earlier11,29. Our results were consistent with a study among Korean women with PCOS, which concluded that six T2DM-associated genes (KCNJ11, TCF7L2, SLC30A8, HHEX, FTO and CDKAL1) identified through GWAS were not associated with PCOS11. Further, none of the gene–gene or gene–environment interactions were significant. While our study had sufficiently high power [(1-β error probability) >0.90] to detect a minimal effect size of 0.1 at the five per cent significance level for individual SNPs, the power to detect interactions accurately among multiple genes would be probably low, which could be a major limitation of our study.
Insulin resistance plays a crucial role in the pathophysiology of complex traits such as T2DM and PCOS. The nine genes considered in our study were reported to play a prominent role in the manifestation of T2DM among populations of different ethnic backgrounds17, including in the population of Hyderabad20. These genes are known to be implicated in different pathways such as insulin secretion and action, pancreatic beta-cell function and glucose homeostasis, and the cross-talk between these pathways is, in turn, linked to insulin signalling cascade. Therefore, given the phenotypic overlap of T2DM and PCOS through insulin resistance, the above genes are expected to show similar pattern of genetic association, but none of these showed significant association with PCOS. In contrast, epidemiological studies conducted on premenopausal women with T2DM observed approximately one of every four to have PCOS30, and a long-term prospective study of 255 Italian women with PCOS suggested age-standardized prevalence rates of diabetes to increase from 2.2 per cent at the baseline to 39.3 per cent towards the end of follow up (average age of 45 yr)31. There can be two plausible conjectures for the lack of association: (i) there may be other genes in the diabetes pathway that might confer relatively greater effect and more specifically responsible for PCOS than the ones considered in this study, and (ii) due to the lack of significant proportion of T2DM affected women in the PCOS group.
Though many PCOS women are at increased risk for insulin resistance, pancreatic β-cell dysfunction and impaired glucose tolerance resulting in greater risk for T2DM in later life, the underlying mechanism of insulin resistance may be quite distinct in case of PCOS and T2DM13. The compensatory hyperinsulinaemia alters the steroid hormone metabolism, resulting in increased production of ovarian androgens which, in turn, leads to the manifestation of PCOS. In case of T2DM, insulin resistance results in beta-cell dysfunction leading to impaired glucose tolerance and hyperglycaemia13. Molecular evidence in support of this could be drawn from the observation of a decrease in tyrosine kinase activity accompanied by higher serine kinase activity in the fibroblasts of women with PCOS, whereas insulin resistance in T2DM could result from any defect in other intracellular insulin receptor downstream signaling to the final substrates of insulin action involved in metabolic and mitogenic aspects of cellular functions32. Further, the T2DM genes considered here constitute only the metabolic component of PCOS and may not have direct implication to its reproductive component, which is a key to the pathophysiology of PCOS33. Therefore, it is also essential to explore the interactions between set of genes in the metabolic and reproductive pathways to better appreciate the underlying genetic mechanisms that may confer risk of developing PCOS. Future studies should also focus on the larger group of women with PCOS and diabetes to extensively screen the established T2DM susceptibility loci to understand the overlapping metabolic characteristics and ascertain the genotype–phenotype correlation. This would probably enable to identify genetic variants or functional regions of the gene, and its impact on specific PCOS phenotypes, if existent and probably help unequivocally infer the universality of the lack of association of T2DM genes with PCOS.
Acknowledgment
This study was funded by the Indian Statistical Institute (ISI), India. The authors thank the Director, ISI for logistic support, Dr Neelaveni (Endocrinologist, Department of Endocrinology, Osmania Medical College, Hyderabad, India) and Dr Anuradha (Gynaecologist - Anu Test Tube Baby Centre, Somajiguda, Hyderabad, India) for help in recruiting the participants and Ms. Sireesha for assistance in blood sample collection and DNA isolation.
Footnotes
Conflicts of Interest: None.
References
- 1.Dasgupta S, Reddy BM. Present status of understanding on the genetic etiology of polycystic ovary syndrome. J Postgrad Med. 2008;54:115–25. doi: 10.4103/0022-3859.40778. [DOI] [PubMed] [Google Scholar]
- 2.Welt CK, Duran JM. Genetics of polycystic ovary syndrome. Semin Reprod Med. 2014;32:177–82. doi: 10.1055/s-0034-1371089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaikh N, Roshan D, Mukherjee S. Genetic markers of polycystic ovary syndrome: emphasis on insulin resistance. Int J Med Genet 2014. 2014:10. [Google Scholar]
- 4.Goodarzi MO, Louwers YV, Taylor KD, Jones MR, Cui J, Kwon S, et al. Replication of association of a novel insulin receptor gene polymorphism with polycystic ovary syndrome. Fertil Steril. 2011;95:1736–41.e1-11. doi: 10.1016/j.fertnstert.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks S, Gharani N, McCarthy M. Candidate genes in polycystic ovary syndrome. Hum Reprod Update. 2001;7:405–10. doi: 10.1093/humupd/7.4.405. [DOI] [PubMed] [Google Scholar]
- 6.El Mkadem SA, Lautier C, Macari F, Molinari N, Lefèbvre P, Renard E, et al. Role of allelic variants Gly972Arg of IRS-1 and Gly1057Asp of IRS-2 in moderate-to-severe insulin resistance of women with polycystic ovary syndrome. Diabetes. 2001;50:2164–8. doi: 10.2337/diabetes.50.9.2164. [DOI] [PubMed] [Google Scholar]
- 7.Haddad L, Evans JC, Gharani N, Robertson C, Rush K, Wiltshire S, et al. Variation within the type 2 diabetes susceptibility gene calpain-10 and polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:2606–10. doi: 10.1210/jcem.87.6.8608. [DOI] [PubMed] [Google Scholar]
- 8.Ruan Y, Ma J, Xie X. Association of IRS-1 and IRS-2 genes polymorphisms with polycystic ovary syndrome: a meta-analysis. Endocr J. 2012;59:601–9. doi: 10.1507/endocrj.ej11-0387. [DOI] [PubMed] [Google Scholar]
- 9.Shen W, Li T, Hu Y, Liu H, Song M. Calpain-10 genetic polymorphisms and polycystic ovary syndrome risk: a meta-analysis and meta-regression. Gene. 2013;531:426–34. doi: 10.1016/j.gene.2013.08.072. [DOI] [PubMed] [Google Scholar]
- 10.San-Millán JL, Escobar-Morreale HF. The role of genetic variation in peroxisome proliferator-activated receptors in the polycystic ovary syndrome (PCOS): an original case-control study followed by systematic review and meta-analysis of existing evidence. Clin Endocrinol (Oxf) 2010;72:383–92. doi: 10.1111/j.1365-2265.2009.03679.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim JJ, Choi YM, Cho YM, Hong MA, Chae SJ, Hwang KR, et al. Polycystic ovary syndrome is not associated with polymorphisms of the TCF7L2, CDKAL1, HHEX, KCNJ11, FTO and SLC30A8 genes. Clin Endocrinol (Oxf) 2012;77:439–45. doi: 10.1111/j.1365-2265.2012.04389.x. [DOI] [PubMed] [Google Scholar]
- 12.Saxena R, Welt CK. Polycystic ovary syndrome is not associated with genetic variants that mark risk of type 2 diabetes. Acta Diabetol. 2013;50:451–7. doi: 10.1007/s00592-012-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewens KG, Jones MR, Ankener W, Stewart DR, Urbanek M, Dunaif A, et al. Type 2 diabetes susceptibility single-nucleotide polymorphisms are not associated with polycystic ovary syndrome. Fertil Steril. 2011;95:2538–41.e1-6. doi: 10.1016/j.fertnstert.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abate N, Chandalia M. Ethnicity, type 2 diabetes and migrant Asian Indians. Indian J Med Res. 2007;125:251–8. [PubMed] [Google Scholar]
- 15.Mohan V, Sandeep S, Deepa R, Shah B, Varghese C. Epidemiology of type 2 diabetes: Indian scenario. Indian J Med Res. 2007;125:217–30. [PubMed] [Google Scholar]
- 16.Sundararaman PG, Manomani R, Sridhar GR, Sridhar V, Sundaravalli A, Umachander M. Risk of atherosclerosis in women with polycystic ovary syndrome: a study from South India. Metab Syndr Relat Disord. 2003;1:271–5. doi: 10.1089/1540419031361435. [DOI] [PubMed] [Google Scholar]
- 17.Kommoju UJ, Reddy BM. Genetic etiology of type 2 diabetes mellitus: a review. Int J Diabetes Dev Ctries. 2011;31:51–64. [Google Scholar]
- 18.Tabassum R, Chauhan G, Dwivedi OP, Mahajan A, Jaiswal A, Kaur I, et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62:977–86. doi: 10.2337/db12-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, Bjonnes A, et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes. 2013;62:1746–55. doi: 10.2337/db12-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uma Jyothi K, Reddy BM. Gene-gene and gene-environment interactions in the etiology of type 2 diabetes mellitus in the population of Hyderabad, India. Meta Gene. 2015;5:9–20. doi: 10.1016/j.mgene.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta S, Sirisha P, Neelaveni K, Anuradha K, Sudhakar G, Reddy BM. Polymorphisms in the IRS-1 and PPAR-γ genes and their association with polycystic ovary syndrome among South Indian women. Gene. 2012;503:140–6. doi: 10.1016/j.gene.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 22.Dasgupta S, Sirisha PV, Neelaveni K, Anuradha K, Reddy BM. Association of CAPN10 SNPs and haplotypes with polycystic ovary syndrome among South Indian Women. PLoS One. 2012;7:e32192. doi: 10.1371/journal.pone.0032192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaikh N, Mukherjee A, Shah N, Meherji P, Mukherjee S. Peroxisome proliferator activated receptor gamma gene variants influence susceptibility and insulin related traits in Indian women with polycystic ovary syndrome. J Assist Reprod Genet. 2013;30:913–21. doi: 10.1007/s10815-013-0025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta S, Reddy BM. The role of epistasis in the etiology of polycystic ovary syndrome among Indian women: SNP-SNP and SNP-environment interactions. Ann Hum Genet. 2013;77:288–98. doi: 10.1111/ahg.12020. [DOI] [PubMed] [Google Scholar]
- 25.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritschi EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Biyasheva A, Legro RS, Dunaif A, Urbanek M. Evidence for association between polycystic ovary syndrome (PCOS) and TCF7L2 and glucose intolerance in women with PCOS and TCF7L2. J Clin Endocrinol Metab. 2009;94:2617–25. doi: 10.1210/jc.2008-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewens KG, Jones MR, Ankener W, Stewart DR, Urbanek M, Dunaif A, et al. FTO and MC4R gene variants are associated with obesity in polycystic ovary syndrome. PLoS One. 2011;6:e16390. doi: 10.1371/journal.pone.0016390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Che Y, Cao Y, Wu X, Sun H, Liang F, et al. Polymorphisms of TCF7L2 and HHEX genes in Chinese women with polycystic ovary syndrome. J Assist Reprod Genet. 2010;27:23–8. doi: 10.1007/s10815-009-9377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peppard HR, Marfori J, Iuorno MJ, Nestler JE. Prevalence of polycystic ovary syndrome among premenopausal women with type 2 diabetes. Diabetes Care. 2001;24:1050–2. doi: 10.2337/diacare.24.6.1050. [DOI] [PubMed] [Google Scholar]
- 31.Gambineri A, Patton L, Altieri P, Pagotto U, Pizzi C, Manzoli L, et al. Polycystic ovary syndrome is a risk factor for type 2 diabetes: results from a long-term prospective study. Diabetes. 2012;61:2369–74. doi: 10.2337/db11-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rojas J, Chávez M, Olivar L, Rojas M, Morillo J, Mejías J, et al. Polycystic ovary syndrome, insulin resistance, and obesity: navigating the pathophysiologic labyrinth. Int J Reprod Med 2014. 2014:719050. doi: 10.1155/2014/719050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barber TM, Franks S. The link between polycystic ovary syndrome and both type 1 and type 2 diabetes mellitus: what do we know today? Womens Health (Lond) 2012;8:147–54. doi: 10.2217/whe.11.94. [DOI] [PubMed] [Google Scholar]



