Abstract
CD36 is a multifunctional membrane-type receptor glycoprotein that reacts with oxidized low-density lipoprotein and long-chain fatty acid (LCFA). However, much remains to be understood about the molecular mechanism of the cardio-myopathy observed in CD36-KO mice. In this study, we identify different genes pathways involved in response to CD36 cardio-myopathy phenotype by identifying the differences among biological processes, molecular pathways and networks of interactions that emerge from knocking CD3 and using different bioinformatics tools such as STRING, GeneMANIA and Cytoscape. We were able list all the CD36-regulated genes, their related function and their specific networks. Data analysis showed that CD36-regulated genes differentially expressed are involved in biological processes such as FA metabolism, angiogenesis/apoptosis and cell structure. These results provide the first look at mechanisms involved in CD36 deficiency and development of cardio-myopathy and the opportunity to identify new therapeutic targets.
Keywords: CD36, cardio-myopathy, genes networks, genes pathways, metabolism, angiogenesis/apoptosis
Background
Hypertrophic cardio-myopathy ("HCM") is characterized by a myocardium hypertrophy .To date the molecular mechanisms underlying this pathology remain elusive [1,2, 3,4, 5,6]. It is most well known as a leading cause of sudden cardiac death in young athletes [7]. The occurrence of hypertrophic cardio-myopathy is a significant cause of sudden unexpected cardiac death in any age group and is a cause of disabling cardiac symptoms. Younger people are likely to have a more severe form of hypertrophic cardio-myopathy. HCM is frequently asymptomatic until sudden cardiac death. As a consequence, re-curent screening has been suggested for certain populations [8].
The CD36 gene encodes a membrane glycoprotein (also known as platelet glycoprotein IV or IIIb), that is expressed in a wide variety cell types, including platelets, monocytes, erythroblasts, capillary endothelial cells and mammary epithelial cells [9,10]. In the heart CD36 is also expressed both in epithelial and muscle cells [11]. In these latter, CD36 has been shown to be a long chain fatty acids (FA) receptor/transporter [12]. In endothelial cells, it has been proposed as one of thrombospondin receptor [13]. CD36 is a multifunctional membrane-type receptor glycoprotein that reacts with thrombospondin, collagen, oxidized low-density lipoprotein and long-chain fatty acid (LCFA).1,2 LCFA is one of the major cardiac energy substrates; hence, LCFA metabolism may have an important role in cardiac diseases. We present here a patient with hereditary hypertrophic cardiomyopathy (HCM) and type I CD36 deficiency that showed no myocardial LCFA accumulation.
CD36-KO mice exhibit between 60 and 80% reduction in betamethyl- p-[123I]-iodophenyl-pentadecanoic acid (BMIPP) uptake by heart tissue [14] which is paralleled by an increase in the heart/body index and the left ventricular size [13]. We propose in this study to define the molecular defects that lead to cardiac hypertrophy and consequent of the fatty acid transporter CD36 deficiency. To define this molecular defect, we used two microarrays technologies (Affymetrix, Agilent) in order to identify genetic alterations related to the myocardial phenotype of CD36-Ko mice.
In a previous study [15], heterogeneity of the arrays data was analyzed by splitting them on three levels: genes, genes set, and network/pathway. We were able to identify the CD36-regulated candidate genes linked to Hypertrophic cardio-myopathy. At the second level we were able to cluster group of genes (gene sets) that may have similar functions [15]. In this study, we aim to ascertain whether each gene set from each subtype is significantly enriched in a list of selected phenotypes. The third level of analysis was to construct gene networks of the proposed CD36 regulated genes from three selected web-based tools: Ingenuity Pathway Analysis (IPA), Search Tool for The Retrieval of Interacting Genes (STRING), and Gene Multiple Association Network Integration Algorithm (GeneMANIA).
Materials and Methods
Dataset
To identify a comprehensive set of genes that are differentially regulated by CD36 expression in the heart, we used two microarray technologies (Affymetrix and Agilent) to compare gene expression in heart tissues from CD36 KnocK-Out (KOCD36) versus wild type (WT-CD36) mice. The obtained results using the two technologies were similar with around 35 genes differentially expressed using both technologies [15]. Absence of CD36 led to down-regulation of the expression of three groups of genes involved in pathways of FA metabolism, angiogenesis/apoptosis and structure. These data are consistent with the fact that the CD36 protein binds FA and thrombospondin 1 involved respectively in lipid metabolism and anti-angiogenic activities [15]. Summary of dataset analysis methodology used in this study is shown in Figure 1.
Figure 1.
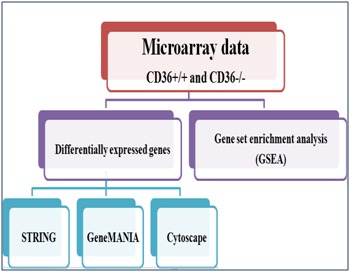
Summary of dataset analysis methodology used in this study
Gene Set Enrichment Analysis and Enrichment Map
Gene set enrichment analyses (GSEA) [16,17] are commonly used to determine the biological characterization, statistical significance, and concordant differences between an experimental gene set and a selected gene list from annotated gene sets knowledge bases to red on Molecular Signatures Database (MSigDB). GSEA can be downloaded from http://www .broadinstitute.org/gsea/downloads.jsp. The Jaccard coefficient is used to compare the similarity between two sample gene sets A and B and defined as the intersection between group A and B divided by their union. The results from GSEA are then visualized through the enrichment map [18], a Cytoscape plugin for network visualization. The ranked experimental gene list along with the enriched gene sets from GSEA is used to build the network of gene sets (nodes) where edges represent their similarity. The size of a node varies by gene set size and the thickness of the edge represents the degree of correlation between two gene sets.
Networks and Pathway Analysis
Analysis of Network Invoked by CD36-Regulated Genes
The biological knowledge of gene and protein interactions is growing rapidly and there are many tools and curated databases available on a large scale. Insightful knowledge gained from studying gene sets rather than individual genes using networkbased approaches can reveal network patterns and relevant molecular pathways from the experiment gene sets. In this study, we utilized two different freely accessible and user-friendly web tools as follows. Gene Multiple Association Network Integration Algorithm (GeneMANIA) [19,20] (http://www.genemania.org/) is a web-based tool for prediction of gene function or implemented as a Cytoscape plugin tool. Based on single gene or gene set query from 7 organisms, it shows results for interactive functional associative network according to their co-expression data from Gene Expression Omnibus (GEO), physical and genetic interaction data derived from BioGRID, predicted protein interaction data based on orthology from I2D, co-localization, shared protein domain, and GO function. Search Tool for the Retrieval of Interacting Genes (STRING) version 9.1 [21,22] (http://string-db.org/) is an online protein-protein interaction database curated from literature and predicted associations from systemic genome comparisons. The user can query using single or multiple name(s) and protein sequence(s). The protein interactions can be displayed according to their confidence, evidence, actions, or interactions.
Results and Discussion
Identification of CD36-Regulated Genes through a Refined Analysis of Data
Our initial analysis of this time series gene expression data for differentially expressed gene identification followed the same method used in a previous paper [15]. All files were processed and normalized by Robust Multiarray Average (RMA) in R as in the original study. The selected normalization method may have an effect on downstream analysis, for example, reverse engineering analyses [15]; however, investigating this effect is beyond the scope of this study. We found that combining CD36+/+ and CD36-/- data compromises the accuracy of selection of differentially expressed. Table 1 summarizes the 30 differentially expressed genes from the combined dataset.
Table 1. Gene description with corresponding reference.
| Gene Name | Synonyms / Identifier | Description | R |
| CD36 | ENSG00000135218, ENSP00000308165, ENSP00000378268, ENSP00000392298, ENSP00000396258, ENSP00000398760, ENSP00000399421, ENSP00000401863, ENSP00000407690, ENSP00000409762, ENSP00000410371, ENSP00000411411, ENSP00000415743, ENSP00000416388, ENSP00000431296, ENSP00000433659, ENSP00000435698, ENSP00000439543, ENSP00000441956, 948, CD36, NP_000063, NP_001001547, NP_001001548, NP_001120915, NP_001120916, NM_000072, NM_001001547, NM_001001548, NM_001127443, NM_001127444, GP3B, GP4, GPIV, SCARB3, CD36_HUMAN, P16671, | CD36 molecule (thrombospondin receptor) (472 aa) updated on 21-Jun-2015 | [a] |
| Dgat1 | ENSG00000185000, ENSG00000261698, ENSP00000332258, ENSP00000432795, ENSP00000454624, ENSP00000457814, 8694, DGAT1, NP_036211, NM_012079, ARGP1, DGAT, DGAT1_HUMAN, O75907, | diacylglycerol O-acyltransferase 1; Catalyzes the terminal and only committed step in triacylglycerol synthesis by using diacylglycerol and fatty acyl CoA as substrates. In contrast to DGAT2 it is not essential for survival. May be involved in VLDL (very low density lipoprotein) assembly. In liver, plays a role in esterifying exogenous fatty acids to glycerol. Functions as the major acyl-CoA retinol acyltransferase (ARAT) in the skin, where it acts to maintain retinoid homeostasis and prevent retinoid toxicity leading to skin and hair disorders (488 aa) | [b] |
| Adh4 | ENSG00000198099, ENSP00000265512, ENSP00000397939, ENSP00000423571, ENSP00000424583, ENSP00000424630, ENSP00000425416, ENSP00000426667, ENSP00000427261, ENSP00000427525, 127, ADH4, NP_000661, NM_000670, ADH-2, ADH4_HUMAN, P08319, | alcohol dehydrogenase 4 (class II), pi polypeptide (380 aa) | [c] |
| Irs3 | ENSMUSP00000060844 | insulin receptor substrate 3 | [d] |
| Irs1 | ENSG00000169047, ENSP00000304895, 3667, IRS1, NP_005535, NM_005544, HIRS-1, IRS1_HUMAN, P35568, | insulin receptor substrate 1, May mediate the control of various cellular processes by insulin. When phosphorylated by the insulin receptor binds specifically to various cellular proteins containing SH2 domains such as phosphatidylinositol 3-kinase p85 subunit or GRB2. Activates phosphatidylinositol 3-kinase when bound to the regulatory p85 subunit (By similarity) | [e] |
| IL2 | ENSG00000109471, ENSP00000226730, 3558, IL2, NP_000577, NM_000586, IL-2, TCGF, IL2_HUMAN, P60568, | interleukin 2, Produced by T-cells in response to antigenic or mitogenic stimulation, this protein is required for T-cell proliferation and other activities crucial to regulation of the immune response. Can stimulate B-cells, monocytes, lymphokine- activated killer cells, natural killer cells, and glioma cells | [f] |
| Fat4 | ENSG00000196159, ENSP00000335169, ENSP00000377862, 79633, FAT4, NP_078858, NM_024582, CDHF14, CDHR11, FAT-J, FAT4_HUMAN, Q6V0I7, | FAT tumor suppressor homolog 4 (Drosophila), May function in the regulation of planar cell polarity. Cadherins are cell-cell interaction molecules (By similarity) | [g] |
| Pdhb | ENSG00000168291, ENSP00000307241, ENSP00000373220, ENSP00000417267, ENSP00000418448, | pyruvate dehydrogenase (lipoamide) beta, The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2), and thereby links the glycolytic pathway to the tricarboxylic cycle | [h] |
| Lpl | ENSG00000175445, ENSP00000309757, ENSP00000428237, ENSP00000428496, ENSP00000428557, | lipoprotein lipase, The primary function of this lipase is the hydrolysis of triglycerides of circulating chylomicrons and very low density lipoproteins (VLDL). Binding to heparin sulfate proteogylcans at the cell surface is vital to the function. The apolipoprotein, APOC2, acts as a coactivator of LPL activity in the presence of lipids on the luminal surface of vascular endothelium (By similarity) | [i] |
| Ide | ENSG00000175445, ENSP00000309757, ENSP00000428237, ENSP00000428496, ENSP00000428557, | lipoprotein lipase, The primary function of this lipase is the hydrolysis of triglycerides of circulating chylomicrons and very low density lipoproteins (VLDL). Binding to heparin sulfate proteogylcans at the cell surface is vital to the function. The apolipoprotein, APOC2, acts as a coactivator of LPL activity in the presence of lipids on the luminal surface of vascular endothelium (By similarity) | [j] |
| Uqcrh | ENSG00000173660, ENSP00000309565, 440567, 7388, UQCRH, NP_001083060, NP_005995, NM_001089591, NM_006004, QCR6, UQCR8, P07919, QCR6_HUMAN, | ubiquinol-cytochrome c reductase hinge protein, This is a component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is part of the mitochondrial respiratory chain. This protein may mediate formation of the complex between cytochromes c and c1 (91 aa) | [k] |
| Uqcrc1 | ENSG00000010256, ENSP00000203407, ENSP00000388660, ENSP00000393696, 7384, UQCRC1, NP_003356, NM_003365, D3S3191, QCR1, UQCR1, P31930, QCR1_HUMAN, | ubiquinol-cytochrome c reductase core protein I, This is a component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is part of the mitochondrial respiratory chain. This protein may mediate formation of the complex between cytochromes c and c1 (480 aa) | [l] |
| Gyk | ENSMUSP00000119564 | glycerol kinase, Key enzyme in the regulation of glycerol uptake and metabolism (By similarity) (553 aa) | [m] |
| Thbs1 | ENSG00000137801, ENSP00000260356, ENSP00000380720, 7057, THBS1, NP_003237, NM_003246, THBS, THBS-1, TSP, TSP-1, TSP1, P07996, TSP1_HUMAN, | thrombospondin 1, Adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. Binds heparin. May play a role in dentinogenesis and/or maintenance of dentin and dental pulp (By similarity). Ligand for CD36 mediating antiangiogenic properties (1170 aa) | [n] |
| Sdc4 | ENSP00000361818 | Cell surface proteoglycan that bears heparan sulfate | [o] |
| CD9 | ENSG00000010278, ENSP00000009180, ENSP00000371955, ENSP00000371958, ENSP00000371959, ENSP00000440985, 928, CD9, NP_001760, NM_001769, BA2, MIC3, MRP-1, TSPAN29, CD9_HUMAN, P21926, | molecule, Involved in platelet activation and aggregation. Regulates paranodal junction formation. Involved in cell adhesion, cell motility and tumor metastasis. Required for sperm-egg fusion | [p] |
| Pdgfra | ENSG00000134853, ENSP00000257290, ENSP00000424218, ENSP00000425232, ENSP00000425626, ENSP00000425648, ENSP00000425902, ENSP00000426472, 5156, PDGFRA, NP_006197, NM_006206, CD140a, PDGFR2, P16234, PGFRA_HUMAN, | platelet-derived growth factor receptor, alpha polypeptide, Tyrosine-protein kinase that acts as a cell-surface receptor for PDGFA, PDGFB and PDGFC and plays an essential role in the regulation of embryonic development, cell proliferation, survival and chemotaxis. Depending on the context, promotes or inhibits cell proliferation and cell migration. Plays an important role in the differentiation of bone marrow-derived mesenchymal stem cells. Required for normal skeleton development and cephalic closure during embryonic development. Required for normal development of the mucosa lining th [...] (1089 aa) | [q] |
| Hand1 | ENSG00000113196, ENSP00000231121, 9421, HAND1, NP_004812, NM_004821, bHLHa27, eHand, Hxt, Thing1, HAND1_HUMAN, O96004, | heart and neural crest derivatives expressed 1, Transcription factor that plays an essential role in both trophoblast-giant cells differentiation and in cardiac morphogenesis. In the adult, could be required for ongoing expression of cardiac-specific genes. Binds the DNA sequence 5'- NRTCTG-3' (non-canonical E-box) (By similarity) (215 aa) | [r] |
| Arnt2 | ENSG00000172379, ENSP00000307479, ENSP00000452961, ENSP00000453651, ENSP00000453792, 9915, ARNT2, NP_055677, NM_014862, bHLHe1, KIAA0307, ARNT2_HUMAN, Q9HBZ2, | aryl-hydrocarbon receptor nuclear translocator 2, Specifically recognizes the xenobiotic response element (XRE) | [s] |
| Rag1 | ENSG00000166349, ENSP00000299440, ENSP00000434610, 5896, RAG1, NP_000439, NM_000448, MGC43321, RNF74, P15918, RAG1_HUMAN, | recombination activating gene 1, Catalytic component of the RAG complex, a multiprotein complex that mediates the DNA cleavage phase during V(D)J recombination. V(D)J recombination assembles a diverse repertoire of immunoglobulin and T-cell receptor genes in developing B and T- lymphocytes through rearrangement of different V (variable), in some cases D (diversity), and J (joining) gene segments. In the RAG complex, RAG1 mediates the DNA-binding to the conserved recombination signal sequences (RSS) and catalyzes the DNA cleavage activities by introducing a double-strand break between t [...] | [t] |
| Api5 | ENSG00000166181, ENSP00000368129, ENSP00000399341, ENSP00000402540, ENSP00000431391, ENSP00000434462, ENSP00000436189, ENSP00000436436, 8539, API5, NP_001136402, NP_001136403, NP_006586, NM_001142930, NM_001142931, NM_006595, AAC-11, AAC11, API5L1, API5_HUMAN, Q9BZZ5, | apoptosis inhibitor 5, Antiapoptotic factor that may have a role in protein assembly. Negatively regulates ACIN1. By binding to ACIN1, it suppresses ACIN1 cleavage from CASP3 and ACIN1-mediated DNA fragmentation. Also known to efficiently suppress E2F1-induced apoptosis. Its depletion enhances the cytotoxic action of the chemotherapeutic drugs | [u] |
| Map3K2 | ENSG00000166181, ENSP00000368129, ENSP00000399341, ENSP00000402540, ENSP00000431391, ENSP00000434462, ENSP00000436189, ENSP00000436436, 8539, API5, NP_001136402, NP_001136403, NP_006586, NM_001142930, NM_001142931, NM_006595, AAC-11, AAC11, API5L1, API5_HUMAN, Q9BZZ5, | apoptosis inhibitor 5, Antiapoptotic factor that may have a role in protein assembly. Negatively regulates ACIN1. By binding to ACIN1, it suppresses ACIN1 cleavage from CASP3 and ACIN1-mediated DNA fragmentation. Also known to efficiently suppress E2F1-induced apoptosis. Its depletion enhances the cytotoxic action of the chemotherapeutic drugs | [v] |
| Mertk | ENSG00000153208, ENSP00000295408, ENSP00000387277, ENSP00000389152, ENSP00000402129, ENSP00000412660, 10461, MERTK, NP_006334, NM_006343, mer, RP38, MERTK_HUMAN, Q12866, | c-mer proto-oncogene tyrosine kinase, Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to several ligands including LGALS3, TUB, TULP1 or GAS6. Regulates many physiological processes including cell survival, migration, differentiation, and phagocytosis of apoptotic cells (efferocytosis). Ligand binding at the cell surface induces autophosphorylation of MERTK on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with GRB2 or PLCG2 and induces phosph [...] (999 aa) | [w] |
| Myl4 | ENSG00000198336, ENSP00000347055, ENSP00000377096, ENSP00000442375, ENSP00000458194, ENSP00000458907, ENSP00000459035, ENSP00000460734, ENSP00000461121, ENSP00000461570, ENSP00000461747, 4635, MYL4, NP_001002841, NP_002467, NM_001002841, NM_002476, AMLC, GT1, PRO1957, MYL4_HUMAN, P12829, | myosin, light chain 4, alkali, atrial, embryonic, Regulatory light chain of myosin. Does not bind calcium (197 aa) | [x] |
| Tnnt2 | ENSG00000118194, ENSP00000236918, ENSP00000353535, ENSP00000356284, ENSP00000356286, ENSP00000356287, ENSP00000356289, ENSP00000356291, ENSP00000387874, ENSP00000395163, ENSP00000402238, ENSP00000404134, ENSP00000408731, ENSP00000414036, ENSP00000422031, 7139, TNNT2, NP_000355, NP_001001430, NP_001001431, NP_001001432, NP_001263274, NP_001263275, NP_001263276, NM_000364, NM_001001430, NM_001001431, NM_001001432, NM_001276345, NM_001276346, NM_001276347, CMH2, P45379, TNNT2_HUMAN, | troponin T type 2 (cardiac), Troponin T is the tropomyosin-binding subunit of troponin, the thin filament regulatory complex which confers calcium-sensitivity to striated muscle actomyosin ATPase activity (288 aa) | [y] |
| Pdlim3 | ENSP00000284770 | PDZ and LIM domain 3,May play a role in the organization of actin filament arrays within muscle | [z] |
| Ifng | ENSG00000111537, ENSP00000229135, 3458, IFNG, NP_000610, NM_000619, IFNG_HUMAN, P01579, | interferon, gamma, Produced by lymphocytes activated by specific antigens or mitogens. IFN-gamma, in addition to having antiviral activity, has important immunoregulatory functions. It is a potent activator of macrophages, it has antiproliferative effects on transformed cells and it can potentiate the antiviral and antitumor effects of the type I interferons (By similarity) | [aa] |
| Pdlim3 | ENSP00000284770 | PDZ and LIM domain 3,May play a role in the organization of actin filament arrays within muscle cells (By similarity) (364 aa) | [bb] |
Networks and Pathways Analysis
Network analysis can help understand the molecular and cellular interactions [23]. It can be visualized to represent entities (nodes) and their relationships (arcs). The advent of high-throughput technology has led to a large increase in publicly available information. Each data type can capture different aspect of functional roles of interested genes. In this section, we investigated functional interaction among genes and proteins in the cell using available data and knowledge bases.
We selected two different web-based network tools: GeneMANIA and STRING using the differentially expressed genes as a query gene sets. These toolsets integrates computational methods to predict the gene functions based on a collection of interaction networks. GeneMANIA is a large collection of interaction networks from several data sources which identify genes and networks that are functionally associated (protein and genetic interactions, pathways, coexpression, colocalization, and protein domain similarity) with the query gene sets. Another advantage is that the user can run this as a plugin with the Cytoscape tool allowing the user to apply other tools to analyze the networks (Figure 2). STRING relies on the phylogeny to infer the functional interaction (protein networks) with direct interaction to score nodes, while GeneMANIA uses functional genomic data with label propagation to score nodes and generate gene networks (Figure 3). STRING uses precomputed networks, while those of GeneMANIA are not precomputed, and user can upload their own networks. STRING covers a large number of organisms, while GeneMANIA only covers 7 organisms but allows the user to upload or add more networks through the plugin. In addition, users can run enrichment analysis (GO, KEGG, PFAM, INTERPRO, and protein-protein interaction) on STRING (Figure 4).
Figure 2.
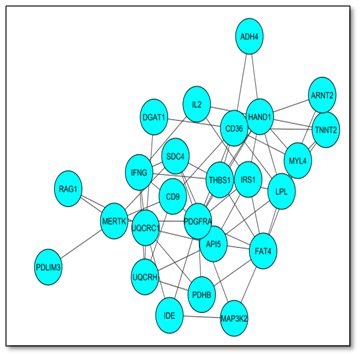
Interaction network Visualization using Cytoscape². Cytoscape and enrichment map were used for visualization of the results; only gene sets from Gene Ontology were used. Nodes represent enriched GO gene sets, whose size reflects the total number of genes in that gene set. Edge thickness represents the number of overlapping genes between gene sets calculated using Jaccard coefficient.
Figure 3.
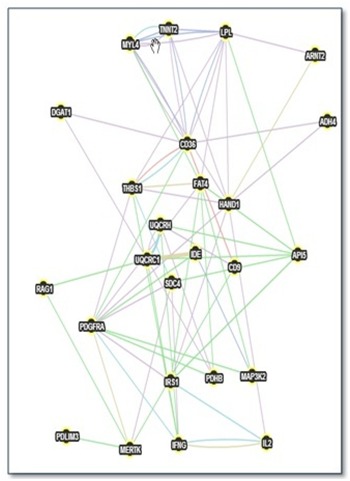
Gene network of CD36 derived from GeneMANIA. A gene network from GeneMANIA shows the relationships for genes from the list (nodes) connected (with edges) according to the functional association networks from the databases.
Figure 4.
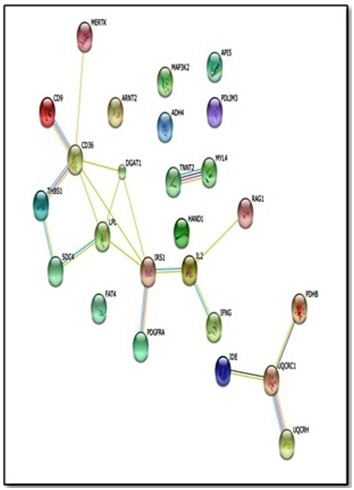
Results from STRING search of Protein interaction for CD36. The figure illustrates the protein interaction upon querying STRING protein network (evidence view) in Mus Musculus with 25 proteins. Additional information from other resources can be retrieved for each protein and interaction. Nodes represent proteins and different line colours denote the type of evidence for the interaction.
The results of genes and network that are functionally associated with the gene set from early response of CD36 are shown in Figure 3 and Figure 4.We compared the two networks from STRING and GeneMANIA based on the interactions they revealed; here we used CD36 as the centre gene in the comparison. All interactions found in STRING were found in GeneMANIA as described in more detail in Table 2 (Cf. Annex). Comparison between the results from both tools can be used to confirm the functional associations of the interested gene sets. There is evidence of overlap and uniqueness in the interactions revealed by the two web-based tools.
Table 2. Interactions for CD36 (early response) network using GeneMania and STRING.
| Interactions | GENMANIA | STRING |
| CD36 -> Dgat1 | Co-localization | Co-Mentioned in PubMed Abstracts |
| CD36 -> Lpl | Co-expression | Co-Expression and |
| CD36 -> Uqcrc2 | Co-expression | Co-Mentioned in PubMed Abstracts |
| CD36 -> Scab1 | Predicted shared protein domain | - |
| CD36 -> Scap2 | Predicted, shared protein domain co-expression | - |
| CD36 -> Mertk | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species ; Co-Mentioned in PubMed Abstracts |
| CD36 -> CD9 | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species Co-Mentioned in PubMed Abstracts Association in Curated Databases |
| CD36 -> Hand1 | Predicted | - |
| CD36 -> Rag1 | Predicted | - |
| CD36 -> Map3K2 | Predicted | - |
| CD36 -> Il2 | Predicted | - |
| CD36 -> Ifg | Predicted | - |
| CD36 -> Thbs1 | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species Co-Mentioned in PubMed Abstracts Association in Curated Databases |
| CD36 -> Sdc4 | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species Co-Mentioned in PubMed Abstracts Association in Curated Databases |
| CD36 -> Irs1 | Predicted | - |
| CD36 -> Pdagfra | Predicted and Co-expression | - |
| CD36 -> TnnT3 | Co-expression | |
| CD36 -> TnnT1 | Co-expression | |
| CD36 -> Sdc1 | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species Co-Mentioned in PubMed Abstracts Association in Curated Databases |
| CD36 -> Sdc3 | Predicted | Experimental/Biochemical Data : putative homologs were found interacting in other species Co-Mentioned in PubMed Abstracts Association in Curated Databases |
| TNNT2 ->MYL4 | Co-expression | Co-Expression Association in Curated Databases; Co-Mentioned in PubMed Abstracts |
| UQCRC1 ->UQCRH | Co-expression | - |
| CD9->UQCRH | Co-expression | - |
| PDHB->UQCRH | Co-expression | - |
| UQCRC1->PDHB | Co-expression | - |
| HAND1->TNNT2 | Co-expression | - |
| PDGFRA->THBS1 | Co-expression | - |
| LPL->THBS1 | Co-expression | - |
| IRS1->PDGFRA | Co-expression | - |
| LPL->MYL4 | Co-expression | - |
| SDC4->MERTK | Co-expression | - |
| SDC4->PDGFRA | Co-expression | - |
| SDC4->IFNG | Co-expression | - |
| HAND1->MYL4 | Co-expression | - |
| HAND1->PDGFRA | Co-expression | - |
| HAND1->THBS1 | Co-expression | - |
| HAND1->IRS1 | Co-expression | - |
| HAND1->LPL | Genetic interactions | - |
| UQCRC1 ->DGAT1 | Genetic interactions | - |
| HAND1->ADH4 | Genetic interactions | - |
| HAND1 ->IL2 | Genetic interactions | - |
| LPL ->FAT4 | Genetic interactions | - |
| PDGFRA->FAT4 | Genetic interactions | - |
| LPL ->TNNT2 | Genetic interactions | - |
| MAP3K2 ->IDE | Genetic interactions | - |
| LPL->MYL4 | Genetic interactions | - |
| TNNT2->MYL4 | Genetic interactions | - |
| UQCRC1 ->RAG1 | Genetic interactions | - |
| IFNG->THBS1 | Genetic interactions | - |
| SDC4 ->API5 | Genetic interactions | - |
| UQCRC1->API5 | Genetic interactions | - |
| LPL->API5 | Genetic interactions | - |
| MERTK ->RAG1 | Genetic interactions | - |
| IFNG->UQCRC1 | Genetic interactions | - |
| MYL4->FAT4 | Genetic interactions | - |
| UQCRH->API5 | Genetic interactions | - |
| IFNG->UQCRH | Genetic interactions | - |
| IRS1->API5 | Genetic interactions | - |
| PDGFRA->MAP3K2 | Genetic interactions | - |
| MAP3K2->FAT4 | Genetic interactions | - |
| MERTK->PDLIM3 | Genetic interactions | - |
| CD9->MERTK | Genetic interactions | - |
| PDGFRA ->IDE | Genetic interactions | - |
| FAT4 ->API5 | Genetic interactions | - |
| IRS1->FAT4 | Genetic interactions | - |
| PDGFRA->PDHB | Genetic interactions | - |
| PDHB ->FAT4 | Genetic interactions | - |
| SDC4->THBS1 | Pathway | - |
| TNNT2->MYL4 | Pathway | - |
| IFNG->PDGFRA | Pathway | - |
| IFNG ->IL2 | Pathway | - |
| IRS1->IL2 | Pathway | - |
| UQCRC1->IDE | Shared protein domains | - |
| PDGFRA->MERTK | Shared protein domains | - |
| IFNG->IL2 | Shared protein domains | - |
| THBS1->FAT4 | Shared protein domains | - |
Discussion
In this report, we were able to show that our data is consistent with the role CD36 protein in the FA metabolism, angiogenesis/apoptosis and structure. In cardiac muscle cells, CD36 is known for its lead role as a receptor / transporter of long-chain fatty acids in heart cells and involvement in metabolism in general. Indeed, the ability of cardiomyocytes to adapt its energy demand is a determinant for myocardial function and the Fatty acids are the main energy source of the heart with oxygen. Thus, the CD36 absence results in a loss of absorption of long chain fatty acids and cause deleterious effects on the heart muscle, while a decrease in the availability of acyl- CoA causes inhibition of B-oxidation. This leads to a decrease in free carnitine and accumulation of intra-mitochondrial acylcarnitine long chain and an important membrane alteration. These disturbances result in a significant decrease in energy production as ATP, which causes the use of an alternative energy source such as glucose. In 1963, Randle and his colleagues [24], have brought to light, using in vitro experiments on rat isolated and perfused hearts, the existence of competition between glucose and fatty acids with long chain fatty acids being preferentially metabolized. Moreover, according to Dyck et al. [25], CD36 plays an important role in the choice of substrate in the heart [25]. WT-CD36 which normally draws more of its energy from fatty acids is forced to uses more glucose in the absence of CD36 (The heart of the CD36-KO). Our work confirms these results as we see the over-expression of genes that stimulate glucose metabolism such as IRS1, IRS2, IRS3, IDE etc.
On the other hand, in endothelial cells where angiogenesis/apoptosis are critical mechanisms for cell survival, CD36 has been described as a thrombospondin membrane receptor (TSP-1) [26]. TSP-1 plays a role in the inhibition of endothelial cell migration and apoptosis induction. Observations indicate that CD36 does not contain the motif CxxC shown to be necessary for the LCK-CD4 association, which allows to think that other proteins can be associated to CD36 to facilitate signal transduction induced by the CD36_TSP1 binding [26].
Our results confirmed that showing that CD9 that encodes the CD9 protein and CD36 share the same expression profiles. Studies on platelets (for solubilization of the platelet membrane) identify CD9; a11b3 and a6b11 integrins are CD36 partners [27]. Immunofluorescence studies show that CD9 and CD36 alpha6belt1 are co-located in endothelial cells. Thus it suggests that CD9, CD36, and alpha 6 beta 1 might form a complex in endothelial cells upon the binding of TSP1. Our data also a showed a differential expression of MAP3K2 (also known as MEKK2), a member of the serine /threonine kinase family, and an activator of P38 which in turns activates a cascade that results in increased cell size [28]. In monocytic cells, the same results were observed since it was determined the existence of a physical association between the intra-cytoplasmic carboxyl terminus domain of CD36 and a signaling complex containing Lyn and MEKK2, upstream of the P38 MAP kinase cascade was de [29].
Data analysis showed that many Structural genes were altered in CD36-KO. Indeed, failing heart differs from the normal heart in function as well as in structure as failing heart is most often remodeled with hypertrophy. Cardiac imaging with the increases of the ventricular wall thickness and smaller ventricular chambers can clinically recognize hypertrophy. Our work confirms cardiac such remodeling as we see the expression of genes that stimulate cardiac cell structure such as: MYL4, TNNT2, HAND1, PDLIM3 AND PDGFRA.
Conclusion:
In summary, we utilized the strengths of existing network/pathway tools and databases to gain insight into processes related to cardio-myopathy have distinct molecular interaction patterns visible from various systems levels, including gene (microarray analysis), gene set, molecular pathway, and gene networks have shown that cardio-myopathy could involve three pathways; FA metabolism, angiogenesis/apoptosis and structure. Discriminating between the three pathways can help to improve the understanding of a drug’s mechanism and further improve targeting in therapeutics drug research.
Table 3. Reference list to Table 1.
| Reference [R] | URL |
| [a] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=948 |
| [b] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=8694 |
| [c] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=127 |
| [d] | http://www.uniprot.org/uniprot/O5 |
| [e] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=3667 |
| [f] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=3558 |
| [g] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=79633 |
| [h] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=5162 |
| [i] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=4023 |
| [j] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=4023 |
| [k] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=440567 |
| [l] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=7384 |
| [m] | http://string-db.org/newstring_cgi/show_network_section.pl |
| [n] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=7057 |
| [o] | http://www.uniprot.org/uniprot/B4E1S6 |
| [p] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=928 |
| [q] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=5156 |
| [r] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=9421 |
| [s] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=9915 |
| [t] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=5896 |
| [u] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=8539 |
| [v] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=8539 |
| [w] | http://www.uniprot.org/uniprot/B2RE75 |
| [x] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=4635 |
| [y] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=7139 |
| [z] | http://www.uniprot.org/uniprot/D6RAF1 |
| [aa] | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=3458 |
| [bb] | http://www.uniprot.org/uniprot/D6RAF1 |
Edited by P Kangueane
Citation: Sabaouni et al. Bioinformation 12(6): 332-339 (2016)
References
- 1.Richardson P,, et al. Circulation. 1996;93(5):841. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, JAMA. 2002;287(10):1308. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 3.Sherrid MV, et al. Ann ThoracSurg. 2003;75(2):620. doi: 10.1016/s0003-4975(02)04546-0. [DOI] [PubMed] [Google Scholar]
- 4.Wigle ED, et al. 1985;28(1):1. [Google Scholar]
- 5.Wigle ED, et al. 1995;92(7):1680. doi: 10.1161/01.cir.92.7.1680. [DOI] [PubMed] [Google Scholar]
- 6.Maron BJ, et al. J Am Coll Cardiol. 2003;42(9):1687. doi: 10.1016/s0735-1097(03)00941-0. [DOI] [PubMed] [Google Scholar]
- 7.Maron BJ, et al. Circulation. 1996;94(4):850. doi: 10.1161/01.cir.94.4.850. [DOI] [PubMed] [Google Scholar]
- 8.Halabchi Farzin, Seif-Barghi Tohid, Mazaheri Reza. Sudden Cardiac Death in Young Athletes; a Literature Review and Special Considerations in Asia-Asian J Sports Med. 2011;2(1):1. doi: 10.5812/asjsm.34818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abumrad N, et al. J Lipid Res. 1998;39(12):2309. [PubMed] [Google Scholar]
- 10.Ibrahimi A, Abumrad NA, Proc Nat Acad Sci USA. 2002;93:2646. doi: 10.1073/pnas.93.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahimi A.,, Sfeir Z.,, maghaais H. Proc Nat Acad Sci USA. 1993;93:2646. [Google Scholar]
- 12.Asch AS, et al. J.Clin.Invest. 1984;79:1054. doi: 10.1172/JCI112918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajri TR, et al. J Biol Chem. 2001;276:23661. doi: 10.1074/jbc.M100942200. [DOI] [PubMed] [Google Scholar]
- 14.Fukuchi K, et al. J Nucl Med. 1999;40:239. [PubMed] [Google Scholar]
- 15.Sabaouni I, et al. Bioinformation. 2013;9(17):849. doi: 10.6026/97320630009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, et al. Proc Nat Acad Sci USA. 2005;102(43):15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, et al. Bioinformatics. 2007;23(23):3251. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 18.Merico D, et al. PLoS ONE. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuberi K, et al. Nucleic Acids Research. 2013;41(1):W115. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warde-Farley D, et al. Nucleic Acids Research 38(supplement 2) 2010:W214. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, et al. Nucleic Acids Research 39(supplement 1) 2011:D561. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franceschini A, et al. Nucleic Acids Research. 2013;41(1):D808. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma'ayan A. Sci Signal. 2011;4:190. doi: 10.1126/scisignal.2001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randle PJ, et al. 1993;1:785. [Google Scholar]
- 25.Kuang M, et al. Circulation. 2004;109:1550. doi: 10.1161/01.CIR.0000121730.41801.12. [DOI] [PubMed] [Google Scholar]
- 26.Asch AS, et al. J.Biol.Chem. 1991;266:1740. [PubMed] [Google Scholar]
- 27.Miao WM, et al. Blood. 2001;97(6):1689. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- 28.Khatri P, Draghici S, Bioinformatics. 2005;21:3587. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge H, et al. Nat. Genet. 2001;29:482. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]


