Abstract
Eight types of short-chain lysine (Lys) acylations have recently been identified on histones: propionylation, butyrylation, 2-hydroxyisobutyrylation, succinylation, malonylation, glutarylation, crotonylation and β-hydroxybutyrylation. Emerging evidence suggest that these histone modifications affect gene expression and are structurally and functionally different from the widely studied histone Lys acetylation. In this review, we discuss the regulation of non-acetyl histone acylation by enzymatic and metabolic mechanisms, acylation “reader” proteins that mediate the effects of different acylations, and their physiological functions, including in signal-dependent gene activation, spermatogenesis, tissue injury and metabolic-induced stress. We propose a model to explain our present understanding of how differential histone acylation is regulated by metabolism of the different acyl-CoA forms, which in turn modulate the regulation of gene expression.
Introduction
Histone proteins are subject to a wide variety of post-translational modifications (PTMs), which, alone or in combination, characterize and shape functional chromatin states. Misregulation of histone PTM patterning has been intimately linked to a number of diseases, including developmental and neurological disorders as well as various etiologies of cancer1-5.
With the application of high-sensitivity mass spectrometry to the identification of histone PTMs, the current catalogue of observed histone modifications is immense and growing6. The majority of novel histone PTMs is classified as short-chain Lys acylations. These modifications are similar to the well-studied Lys acetylation (Kac) in their ε-amine linkage, yet distinct in their hydrocarbon chain length, hydrophobicity or charge. So far the newly discovered short-chain Lys acylations include histone Lys propionylation (Kpr)7, butyrylation (Kbu)7, 2-hydroxyisobutyrylation (Khib)8, succinylation (Ksucc)9, malonylation (Kma)9, glutarylation (Kglu)10, crotonylation (Kcr)11, and β-hydroxybutyrylation (Kbhb)12. Although non-histone proteins, including cytoplasmic and mitochondrial proteins, also harbor Lys acylations13, this review will focus specifically on histone Lys acylations.
Histone Kac was first discovered in 1963 (REF. 14) and functionally characterized as a positive regulator of transcription by Vincent Allfrey and colleagues in 1964 (REF. 15). Over the decades the regulation and function of Kac has been further established by the identification of three major classes of proteins, colloquially termed: (1) “writers,” the enzymes that catalyze the covalent modification of specific residues, (2) “erasers,” the enzymes that remove the modification and (3) “readers,” the effector proteins that bind histones in a modification-specific manner. More recently it has become established that the metabolic state of the cell regulates histone Kac by influencing the activities of writers and erasers16-18. The identification and characterization of histone Kac writers, readers and erasers and their interplay with cellular metabolism has made Kac one of the best characterized histone PTMs. Here, we describe recent studies that have broadened this Kac-established paradigm to histone Lys acylations and discuss its implications.
Since the discovery of novel histone Lys acylations, the major question has been whether they represent an expanding repertoire of functional modifications or are functionally redundant with histone acetylation. In this review we discuss recent studies, which provide evidence that non-acetyl histone acylations are functionally distinct from histone Kac and have a unique role in modulating transcription and chromatin structure. We first describe the discovery and characterization of non-acetyl histone Lys acylations and the recent literature on the writers, erasers, and readers of these acylations. We discuss the evidence that histone acylations are regulated by the relative concentrations of different acyl-CoA forms present in the nucleus. And finally, we discuss recent evidence to suggest that non-acetyl acylations are functionally distinct from acetylation and can illicit unique consequences on the regulation of transcription and chromatin structure. Thus, histone Lys acylations connect acyl-CoA metabolism and gene regulation.
The discovery of histone acylations
Over the past decade, eight types of short-chain Lys acylations that are distinct from Kac have been discovered on histone proteins, using mass spectrometry-based approaches, which have become the method of choice for identifying novel PTMs. Mass spectrometry depends on the identification of modification-induced shifts in the mass of residues19,20. Two general approaches have been used to discover these new Lys acylations (FIG.1). The first is a candidate approach based on high structural similarity among acyl-CoAs. The predicted structures of Kpr and Kbu are so similar to Kac that a Kac antibody could theoretically be used to enrich Kpr- and Kbu-modified peptides alongside the intended Kac-modified peptides (FIG.1a). Indeed, analysis of tandem mass spectrometry datasets of Kac immuno-affinity enriched tryptic peptides21 revealed the existence of Kpr and Kbu on histone H4 (REF. 7)7. Likewise, after the discovery of Ksucc, the structurally similar Kmal22 and Kglu10 were predicted to exist and subsequently detected. The second approach relies on unbiased, systematic screening for all the possible mass shifts caused by previously uncharacterized PTMs (FIG.1b). The modification-induced mass shifts can be identified from the changes in the masses of the peptides, and these mass differences can be localized to certain residues through aligning spectra of tandem mass spectrometry against the existing protein sequence database. Algorithms were developed to carry out such an analysis for tandem mass spectrometry data derived from low-resolution mass spectrometers23-25. This approach was used to identify four novel Lys acylations: Ksucc26, Kcr11, Kbhb12 and Khib8. In addition, high-resolution tandem mass spectrometry data can also be used in combination with a mass-tolerant database search to detect unknown mass shifts caused by a PTM26. Once such a shift is detected, its chemical structure can be deduced and further validated by chemical and biochemical methods, including peptide synthesis, high performance liquid chromatography (HPLC) co-elution, tandem mass spectrometry analysis, isotopic labeling, and immunochemistry using an appropriate antibody.
Figure 1. Discovering Lys acylations.
(a) An antibody against a certain Lys acylation (for example, acetylation) can be used to enrich for peptides with structurally-related acylations, for example, that differ in only one (propionylation) or two (butyrylation) hydrocarbon groups. (b) Alternatively, spectra obtained by tandem mass spectrometry can be unbiased analyzed using a nonrestrictive protein-sequence alignment algorithm to detect a mass shift in a substrate peptide, which is in turn used to locate a new modification in the peptide. In the left panel, the top diagram represents the peptide backbone cleavage; blue-colored bars represent peaks from a theoretical peptide, and red-colored bars represent peaks from an experimentally-detected peptide with a mass shift; the y-axis represents the relative abundance of peptide ions; and the m/z ratio at x-axis represents mass-to-charge ratio of peptide ions. The accurate molecular weight of a modification is used to deduce its theoretical chemical structures (middle panel). The candidate peptides with such modifications are synthesized and used to validate the in vivo-derived peptides by high performance liquid chromatography (HPLC) -coelution and tandem mass spectrometry (right panel; HPLC peaks from an in vivo peptide (blue), synthetic peptide (red) and their mixture (green) are illustrated).
The chemical structure of acylations
The functional basis for any PTM is the chemical change made to the modified amino acid. For example, Kac neutralizes the positive amine group of the Lys residue, enhances hydrophobicity and increases the size of the Lys side chain. These changes not only compromise the capability of the Lys side chain to form hydrogen bonds and electrostatic interactions with negatively charged residues, but also increase van der Waals interactions with other proteins. The chemical properties of the eight recently described short-chain Lys acylations are variations on the theme established by Kac and can be classified into three groups based on their chemical structures in comparison to Kac (FIG. 2a): 1) hydrophobic acyl groups, which include propionyl, butyryl, and crotonyl, have extended hydrocarbon chains, which increase the hydrophobicity and bulk of the Lys residue beyond that of Kac. Additionally, the crotonyl group contains a C-C π-bond that results in a rigid and planar configuration, which is unique among histone acylations. 2) Polar acyl groups, which include β-hydroxybutyryl and 2-hydroxyisobutyryl, have a hydroxyl group that enables the modified Lys to form hydrogen bonds with other molecules. 3) Negatively charged acidic acyl groups include malonyl, succinyl, and glutaryl, which change the charge at the Lys residue from +1 to −1 at a physiological pH level; this is comparable to the change caused by protein phosphorylation (from 0 to - 2). This group of Lys acylations is the most different from other short-chain acylations owing to its negative charge. So far 246 histone sites bearing new Lys acylations have been detected (FIG. 2b). The discovery of such functionally diverse PTMs has dramatically increased the potential complexity of combinatorial chemical modification of histones.
Figure 2. Chemical structures of nine Lys acylations and their distributions on histones.
a) Illustration of Lys acetylation and eight new types of short-chain Lys acylations, clustered into three groups according to their chemical properties. b) A map of histone sequences showing the distributions of Lys acylations on the different histones from mouse. Sequences in boxes refer to the global domains of the five histones. Modified with permission from REF. 6. Ac, acetylation; Bhb, β-hydroxybutyrylation; Bu, butyrylation; Cr, crotonylation; Glu, glutarylation; Hib, 2-hydroxyisobutyrylation; Ma, malonylation; Pr, propionylation; Succ, succinylation.
Writers and erasers of Lys acylation
Histone Kac is enzymatically regulated by the dynamic balance between writers and erasers. Here we review several studies that describe writer and eraser activities for the histone Lys acylations (FIG. 3). Additionally, Lys acylations have been proposed to occur non-enzymatically, particularly within the mitochondria where relatively high pH and high concentrations of acyl-CoAs would favor it27-29, but conditions in the nucleus are less favorable to this process30.
Figure 3. Writers and erasers for Lys acylations.
a) Illustration of the structures of acyl-Coenzyme A (acyl-CoA) and of the enzymatic reactions of adding or removing acyl-lysine by two groups of enzymes. b) A table showing the enzymatic activities of Lys acyltransferases (writers) and deacylases (erasers). CBP, CREB-binding protein; GNATs, Gcn5-related N-acetyltransferases; HAT, histone acetyltransferase; HDAC, histone deacetylase; Kbhb, Lys β-hydroxybutyrylation; Kbu, Lys butyrylation; Kcr, Lys crotonylation; Kglu, Lys glutarylation; Khib, Lys 2-hydroxyisobutyrylation; Kma, Lys malonylation; Kpr, Lys propionylation; Ksucc, Lys succinylation; MYSTs, MOZ, Ybf2, Sas2, Tip60; NA, not available; NAD+, nicotinamide adenine dinucleotide; p300, E1A binding protein p300; sirtuins, Sir2 (silent information regulator 2)-related enzymes.
Writers
Writers that are specific to non-acetyl acyls have not yet been identified; rather, previously characterized histone acetyltransferases (HATs) were shown to have an expanded repertoire of acyl-transferase activities. There are three major families of HATs, which are characterized by their sequence and structural features: GNAT (Gcn5-related N-acetyltransferases), MYST (named for founding members: MOZ, Ybf2 (Sas3), Sas2 and Tip60), and the p300–CBP (CREB-binding protein) family31,32. HATs from all three families have been shown to utilize several different acyl-CoAs as substrates for histone Lys acylation (FIG. 3a)
The HAT p300, a well-studied transcription co-activator, has emerged as the most promiscuous acyl-transferase to date. Beyond its originally described acetyltransferase activity33,34, p300 can catalyze histone Kpr7,35, Kbu7, Kcr36, Kbhb37 as well as Ksucc38 and Kglu10 (FIG. 3b). Furthermore, p300 can accommodate artificial acyl-CoA analogs, such as 4-pentynoyl-CoA39. Kinetic analysis of the Kac, Kpr, Kbu and Kcr activities of p300 confirm that the enzyme catalyzes these acylations, but reveal progressively slower rates commensurate with acyl-chain lengthening37. Members of the GNAT and MYST families have a more limited range of non-acetyl acylation activities. The GNAT family HATs, GCN5 (also known as KAT2A) and PCAF (also known as KAT2B), and the MYST family HAT, Esa1 (also known as MEAF6), can catalyze Kpr in vitro, albeit with reduced kinetics compared to the catalysis of Kac40-42. Both GCN5 and PCAF can also catalyze Kbu, yet this activity is even more impaired than Kpr catalysis41,42.
Recent structural studies on GCN542 and p30037 bound to a variety of acyl-CoAs revealed molecular mechanisms behind the differing abilities of these HATs to catalyze acylations. Since GCN5 binds to the invariable coenzyme A section of acyl-CoAs (FIG. 3a), it has similar affinities for acetyl-CoA, propionyl-CoA and butyryl-CoA. However, whereas acetyl is positioned perfectly for catalysis, a catalytic water molecule blocks the addition of any extra methyl groups. Thus, acyl chains longer than acetyl must rotate their orientation to fit in the active site, thereby reducing the efficiency of catalysis. Although the acyl-chains of propionyl-CoA and butyryl-CoA are flexible enough to fit into the active site, the rigid acyl structure of crotonyl-CoA would not be accommodated at all42, consistent with the inability of GCN5 to catalyze crotonylation36.
Although p300 also exhibits reduced enzymatic kinetics with increased acyl-chain length, it is able to catalyze diverse acylations, as discussed above. Structural studies of p300 reveal that it contains a deep aliphatic pocket within the active site, a feature absent in GCN5 and other HATs such as TIP60 and MOF37. Structure-based mutating of a key Ile residue to Met, which was predicted to open the pocket further, led to enhanced catalysis of Kbu and Kcr. Furthermore, mutation of the same Ile to the bulkier Phe residue reduced the catalysis of Kbu and Kcr, indicating that the conformation of this aliphatic pocket is crucial for catalysis of short-chain acylations with extended chains37. A valuable tool for future functional studies will be active-site engineering to either broaden or sharpen acyl-CoA discrimination by p300 and other HATs. A summary of acyltransferase activities of the tested HATs is shown in FIG. 3b.
Erasers
There are two major families of Lys deacetylases, the Zn2+-dependent histone deacetylases (HDACs) and the nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins43,44. Soon after the first discovery of Kpr and Kbu, several sirtuin enzymes were described as having depropionylase and debutyrylase activities, albeit weaker than their deacetylase activities35,45. Since then several observations have been made regarding the members of the mammalian sirtuin family in regards to varied deacylase activities. Although all seven sirtuins (SIRT1-7) were originally annotated as histone deacetylases based on sequence homology to the yeast sir2 histone deacetylase46, growing evidence suggests that the sirtuin enzymes have an expanded repertoire of deacylation activities. For example, SIRT5 has robust desuccinylase, demalonylase and deglutarylase activities (FIG. 3b), but little deacetylase activity10,22,47,48. A systematic survey of the seven mammalian sirtuins against peptides synthesized with twelve different short-chain and long-chain acylations at position H3K9 highlighted the diversity of their deacylase activities49. This survey corroborated the desuccinylase activity of SIRT5, but also demonstrated that SIRT1, SIRT2 and SIRT3 have broad-range deacylase activities (FIG. 3b). Specifically, SIRT1 and SIRT2 have depropionylase, debutyrylase and decrotonylase activities. In these assay conditions, SIRT7 did not exhibit any measurable activity, likely due to its preference for nucleosomal histones over peptide substrates50. Using cell-based proteomics methods, SIRT7 was characterized as a histone desuccinylase with function in the DNA damage response51. Another study demonstrated that SIRT3 has decrotonylase activity in vitro and that knockdown of SIRT3 increased histone Kcr levels, which correlated with increases in gene expression52. Early proteomic studies questioned whether histone Kac is a major sirtuin substrate53,54; indeed, the more recent observations that knockdowns of SIRT3 and SIRT7 have specific effects on non-acetyl histone acylations raise the interesting possibility that histone acylations might be the relevant sirtuin substrates.
Compared to the sirtuins, relatively little is known about the capacity of Zn2+-dependent HDACs to deacylate the full repertoire of histone acylations. One study profiled HDAC substrates and included Kcr and Ksucc 55. None of the Zn2+-dependent HDACs showed activity for Ksucc and only HDAC3 demonstrated measurable activity for Kcr, albeit significantly lower than its deacetylase activity55. The activities of Zn2+-dependent HDACs on the other acylations remain unknown.
Readers of histone Lys acylation
There are three major classes of histone Kac readers: bromodomain, YEATS (Yaf9, ENL, AF9, Taf14, and Sas5) and double plant homeodomain (PHD) finger proteins. Each class utilizes a distinct structural mechanism to bind acetylated Lys with greater affinity compared to the unmodified form. In this section we review studies that investigate the ability of these domains to read the expanded class of short-chain Lys acylations.
Bromodomain
The discovery of histone Lys acylations prompted interest in whether specific protein domains existed that could recognize their unique and varied structures. Given that bromodomains are the archetypal histone acetylation readers, an early study focused on the bromodomains of BRD4 and found that although they could bind Kpr and Kbu, they did so with significantly reduced binding affinities compared to Kac at the same residues56. In a more comprehensive survey of the capacity of bromodomains to recognize non-acetyl acylations, 49 bromodomains were assayed for binding to peptides bearing Kac, Kpr, Kbu, Kcr, and Ksucc57. Kpr was generally bound by the bromodomains but only three of the bromodomains could bind Kbu, and only one exhibited measurable binding to the rigid Kcr, albeit with reduced affinity compared to Kac. None of the tested bromodomains showed affinity for Ksucc. A recent report focusing on the role of histone Kbu in mouse spermatogenesis revealed that the situation could be more complex. Indeed, the first bromodomain of bromodomain testis-specific protein (BRDT), which is known to bind simultaneously acetylated histone h4 Lys 5 (H4K5ac) and H4K8ac58, could also accommodate H4K5ac and H4K8bu but not H4K5bu and H4K8ac or H4K5bu and H4K8bu59. The general conclusion from these studies is that bromodomains can bind Kac and Kpr, but, aside from notable exceptions, do not tolerate binding of Kbu, Kcr or acidic acyls. However, since bromodomains are known to engage combinations of Kac, their capacity to bind combinations of Kac and non-acetyl acylations remains to be established.
YEATS
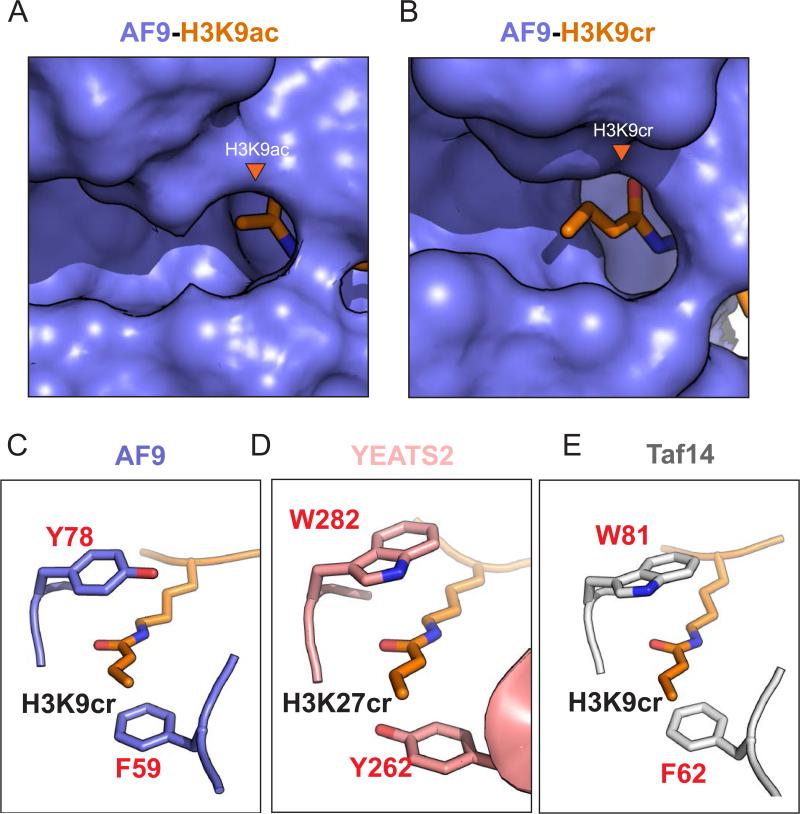
The YEATS domain was recently identified as a novel Kac reader by studies of human AF960,61 and later corroborated by studies of yeast Taf1462. The YEATS domain is a highly conserved protein domain present in proteins implicated in the regulation of transcription62. The co-crystal structures of YEATS bound to H3K9ac revealed a binding pocket with an open end, where the acetyl group is positioned, suggesting that an extended acyl chain could be spatially accommodated63 (FIG. 4a). Three recent studies have shown that the YEATS domain, in contrast to the bromodomain, has enhanced binding affinities for Kpr, Kbu, and Kcr over Kac, with the highest affinity for Kcr63-65. YEATS domains tested by isothermal titration calorimetry showed a 2- to 4- fold increase in affinity for Kcr over Kac63. The co-crystal structure of AF9 YEATS in complex with H3K9cr revealed that Kcr was indeed accommodated into the tunnel-like YEATS binding pocket (FIG. 4b) and bound in orientation similar to Kac, with only minor conformational changes. Preferential binding to Kcr is a result of π-aromatic interactions of the planar crotonylamide group with two sandwiching aromatic AF9 pocket residues (FIG. 4c), as well as by hydrophobic interactions introduced by hydrocarbon extension63. This type of π-stacking interaction is also observed in the co-crystal structures of human YEATS264 (FIG. 4d) and yeast Taf1465 (FIG. 4e) bound to Kcr, as well as in an independently-derived AF9-Kcr structure66. These structural studies demonstrate how multiple members of the YEATS domain family, from yeast to human, have protein structures that specifically take advantage of the unique double bond of Kcr.
Figure 4. Co-Crystal structures of the YEATS domain bound to crotonyl-Lys reveal a common mechanism for crotonyl-Lys preference.
The YEATS (Yaf9, ENL, AF9, Taf14, and Sas5) domain binds with higher affinity to histone Lys crotonylation compared to histone Lys acetylation. Crystal structures of three different YEATS domains reveal a conserved mechanism for Lys crotonylation recognition and preference. a) Close-up of the AF9-YEATS (purple) binding pocket bound to an acetylated histone h3 Lys 9 (H3K9ac) peptide (PDB: 4TMP). b) Close-up of the AF9-YEATS binding pocket bound to a crotonylated H3K9 (H3K9cr) peptide. The extended hydrocarbon chain of the crotonyl group is accommodated within the YEATS binding pocket (PDB: 5HJB). c-e) Co-crystal structures of the YEATS domain of (c) AF9 (purple), (d) YEATS2 (pink) and (e) Taf14 (grey) bound to the indicated crotonylated histone peptides (orange) reveal a common aromatic-sandwich that clamps the crotonylamide by π-aromatic interactions. Each structure reveals the use of a different combination of two aromatic residues to sandwich Kcr: F59 and Y78 for AF9 (c), Y262 and W282 for YEATS2 (d), and F62 and W81 for Taf14 (e). This π -stacking exploits the unique double bond of crotonyl and explains the preference of the YEATS domain for Lys crotonylation over Lys acetylation. (c) PDB: 5HJB, (d) PDB: 5IQl, (e) PDB: 5IOK.
Double PHD finger
Although the PHD finger was originally identified as a methyl-lysine reader67-70, it was later discovered that tandem PHD fingers, or double PHD finger (DPF), bind to Kac71. To date, DPF domains from five human proteins have been characterized as Kac readers, including MOZ and MORF (paralogous MYST family members), and DPF1, DPF2 and DPF3 (all subunits of the BAF chromatin remodeling complex)72-75. Focusing on the DPFs of MOZ and DPF2, a recent study demonstrated that, like the YEATS domain, DPF domains have greater binding preference for acyl-chains that are longer than acetyl, with the strongest affinity measured for Kcr76. DPF proteins tested by isothermal titration calorimetry showed a 4- to 8-fold greater affinity for Kcr over Kac. X-ray crystal structures of the DPF of MOZ bound to an H3K14cr peptide revealed a largely hydrophobic binding pocket, lacking aromatic sandwiching residues. Whereas the Kcr-selectivity of the YEATS domain is a result of π-π sandwiching interactions, the Kcr-selectivity of the DPF domain is brought about by a distinct mechanism of hydrophobic encapsulation and coordinated hydrogen bonding76. Thus, at least two reader domains exhibit preference for non-acetyl Lys acylations, by different molecular mechanisms.
Metabolic regulation of Lys acylations
Under the assumption that previously characterized HATs can also catalyze various acylations, the differential acylation state of proteins should be established by the competition of the different acyl-CoAs for these promiscuous enzymes. Therefore, the relative concentrations of acyl-CoAs will influence the levels of the various histone acylations. According to this model there are two means to enhance non-acetyl histone acylation levels: 1) decrease the concentration of acetyl-CoA or other dominant CoAs, thereby reducing the competition with less dominant acyl-CoAs 2) increase the concentration of a specific acyl-CoA.
Competition with acetyl-CoA
In metazoans, the major source of acetyl-CoA that is used for histone Kac is derived from citrate by ATP Citrate Lyase (ACL) 77 (FIG. 5). Depletion of ACL is known to reduce histone Kac77. In agreement with the model that the different acyl-CoAs compete for acyl-transferase activity, depletion of the cytoplasmic and nuclear pool of acetyl-CoA by knockdown of ACL also leads to an increase in p300-catalyzed Kcr36. Furthermore, this increase in Kcr can be reduced by replenishing acetyl-CoA pools through acetate treatment36, demonstrating the responsiveness of p300 to the shifting levels of acyl-CoA pools. Because the citrate used by ACL is derived from glucose (FIG. 5), in the high glucose conditions of most cell cultures, cells will be replete with acetyl-CoA, which may be artificially dampening the levels of non-acetyl histone acylations. These data suggest that in conditions where acetyl-CoA concentrations are depleted, either by low glucose levels or by glucose carbons being directed away from ACL, other acyl-CoA forms will have less competition for acyl-transferase activity and will be used more often to modify histones. This suggests that non-acetyl acylations may be more abundant in tissues in vivo than in cultures cells. In fact, histone Kcr can be readily detected by tandem mass spectrometry in cardiac cells78 and neuronal cells79, whereas immuno-affinity enrichment is required for detecting Kcr in cells grown in high glucose cell culture media. Additional measures of acyl-CoA concentrations and histone acylation levels in tissues and in other physiological contexts are needed to consolidate these initial observations (see future perspectives).
Figure 5. Metabolic regulation of histone acylation.
The levels of any particular histone acylation depend on the relative concentration of its respective acyl-CoA (R-CoA) and of the other acyl-CoAs. For simplicity, this is represented here as the ratio between R-CoA and the most abundant acyl-CoA, acetyl-CoA (AcCoA). This ratio will determine how much of a particular acyl-CoA will be used by promiscuous histone acyl-transferases (writers) to acylate histone Lys residues. Acyl-CoAs can be generated through various intermediate metabolic pathways. Here we highlight pathways discussed in this review, which contribute either to R-CoA (non-acetyl CoAs) or AcCoA. For a more comprehensive discussion of these pathways see REF. 108108. In metazoans, Acetyl-CoA used for histone acetylation is synthesized from citrate (derived from glucose through the tricarboxylic acid (TCA) cycle) by ATP-citrate lyase (ACL). Many acyl-CoAs can be derived from their cognate short-chain fatty acid (SCFA) by Acyl-CoA Synthetase 2 (ACSS2), including acetate and its CoA form. The pathway generating the SCFA β-hydroxybutyrate (bhb) involves the breakdown of long-chain fatty acids (LCFA) through β-oxidation and subsequent ketogenesis. Malonyl-CoA (Mal-CoA) is generated as the first step in LCFA synthesis. It is still unclear what other cytoplasmic or nuclear pathways exist that generate the acyl-CoAs needed for the diversity of the observed histone acylations (represented as a question mark).
Short chain fatty acids are a source of short chain acyl-CoAs
The cellular concentration of specific non-acetyl acyl-CoAs and therefore the levels of histone acylation are also influenced by short-chain fatty acids (SCFAs). Treating cells with either isotopic or radio-labeled SCFAs leads to the labeling of histone proteins by the heavy or radioactive acyl, suggesting that the SCFA is converted into its cognate acyl-CoA, which is then used directly as a co-factor in a histone acylation reaction11,12,22 (FIG. 5). For example, SCFAs such as crotonate or β-hydroxybutyrate dramatically increase cellular concentrations of their respective acyl-CoAs and the steady-state levels of their respective histone acylation, in a dose-dependent manner12,36.
The modulation of the concentration of particular acyl-CoA by a SCFA is not just a useful experimental tool, but is likely a physiologically-relevant source of acyl-CoA within the cell. Acyl-CoA synthetase short-chain family member 2 (ACSS2; also known as AceCS1, acetyl-coenzyme A synthetase, cytoplasmic), for example, has been implicated as the enzyme generating crotonyl-CoA from crotonate (FIG. 5). Depletion of ACSS2 leads to reduced levels of histone Kcr, suggesting that SCFAs like crotonate might be an endogenous source of non-acetyl acyl-CoAs36. Intriguingly, ACSS2 is overexpressed in a large proportion of human tumors and its expression levels correlate with tumor grade80,81. However, the impact of ACSS2 expression on non-acetyl histone acylations was not explored. Furthermore, acetylated ACSS2 is itself activated by SIRT1-mediated deacetylation82. As sirtuins are NAD+-dependent enzymes, their activity is regulated by the NAD+/NADH ratio. Therefore low nutrient conditions, where the NAD+/NADH ratio is high, will promote ACSS2-deacetylation by SIRT183 and potentially increase the synthesis of acyl-CoAs from endogenous SCFAs. Taken together, these studies suggest that low nutrient conditions will favor non-acetyl acylations by both depleting the levels of ACL-derived acetyl-CoA and by activating ACSS2 by SIRT1-mediated deacetylation.
Ketogenesis and Lys β-hydroxybutyrylation
A dramatic example of an endogenous SCFA used for histone acylation is the role of β-hydroxybutyrate in driving Kbhb during ketogenesis12. Ketogenesis is a physiological response to low levels of blood glucose and liver glycogen. Ketone bodies (acetoacetate, acetone, and β-hydroxybutyrate) are generated from acetyl-CoA that is derived from the breakdown of fatty acids (β-oxidation) (FIG. 5). β-hydroxybutyrate and other ketone bodies are generated in the mitochondria of the liver and secreted into the blood stream. In ketogenic conditions, such as low carbohydrate diet, starvation, or prolonged exercise, serum levels of β-hydroxybutyrate can increase from uM to mM concentrations84,85. The canonical role for β-hydroxybutyrate is as an energy source, but a recent study has discovered that it can also be used as a source for the novel histone acylation, Kbhb. In this study, mice made to undergo ketogenesis by 48hr starvation showed the expected elevation of β-hydroxybutyrate in liver and kidney tissue, but the investigators also observed a previously unidentified increase in histone Kbhb12. Further experiments using culture cells confirmed that the levels of histone Kbhb were elevated by the addition of β-hydroxybutyrate12. The functional consequences of elevated β-hydroxybutyrate and histone Kbhb are discussed below.
Studies on the metabolic regulation of histone acylation are converging on a common theme: that non-acetyl histone acylations are more common in conditions of low glucose and are likely under-represented in the artificially high glucose concentrations of most cell cultures. The following evidence supports this claim: 1) Low glucose reduces acetyl-CoA concentrations86, 2) extended periods of low glucose will lead to ketogenesis, which produces β-hydroxybutyrate that was shown to increase histone Kbhb12, and 3) high NAD+/NADH ratio, which is associated with nutrient deprivation, activates ACSS2, a known source of acyl-CoAs36,82,83.
Differential acylation is demonstrably regulated by acyl-CoA metabolism and physiological conditions have been identified where acyl-CoA metabolism is in flux, but what is the consequence of shifting the proportion of Lys acylation towards any of the novel acylations? We discuss this question next.
The function of differential acylation
Histone Kac marks active regulatory elements across the genome and promotes gene expression through the neutralization of the positive charge of Lys, leading to 1) electrostatic and structural changes in the chromatin fiber (effects in cis) and 2) generation of a platform for the recruitment of reader proteins (effects in trans). ChIP-seq experiments for Kcr11,36,87, Kbu59, Khib8 and Kbhb12 illustrated that active regulatory elements are also marked by diverse histone acylations. The evidence from these studies points to a model where genomic loci are distinguished not by the presence or absence of one particular acylation, but rather by the proportional mixture of distinct acylations, established by the relative concentrations of acyl-CoAs. Several recent studies have demonstrated that alterations in the proportional mixture of histone acylations have functional consequences and correlate with distinct physiological states11,12,36,59,63,87,88. These studies have focused on three area of biology: signal-dependent gene activation, the developmental process of spermatogenesis, tissue injury and metabolic-induced stress (FIG. 6).
Figure 6. The functions of differential histone acylation.
Examples of how different histone acylations are involved in various biological processes. a) Signal-dependent gene activation. Lys crotonylation (Kcr) and Lys acetylation by the histone acyl transferase p300 are induced by the inflammatory signal lipopolysaccharide (not shown) at target genes. Increasing the concentration of crotonyl-CoA (CrCoA) leads to an increase in the levels of p300-catalyzed Kcr, leading to increased recruitment of AF9 and enhanced gene expression. b) Spermatogenesis. Kcr marks X-linked genes that escape meiotic sex chromosome inactivation (MSCI). Histone butyrylation (Kbu) disrupts BRDT binding to chromatin, thereby preventing the histone-to-protamine transition in specific loci in elongating spermatids. c) Metabolic-induced stress. During prolonged starvation, when glucose molecules are scarce, fatty acids replace glucose as the major energy source in the liver. Through the process of ketogenesis, acetyl-CoAs (AcCoA) are converted to ketone bodies, such as β-hydroxybutyrate. β-hydroxybutyrate can then be charged to free coenzyme A to form β-hydroxybutyrate-CoA (bhb-CoA), the high energy donor for the histone modification β-hydroxybutyrylation (Kbhb) is induced in response to ketogenesis and marks a class of activated starvation-response genes.
Signal-dependent gene activation
The HAT activity of p300 is necessary for its role in gene activation in vitro89,90 and in vivo91. More recently, utilizing cell-free transcription assays, p300-catalyzed histone Kbu and Kcr were shown to promote transcription, as effectively as or better than Kac 36,59. In a cell-based model of the inflammatory response, lipopolysaccharide (LPS) is used to activate toll-like receptor 4 signaling, a process known to involve p300-mediated gene activation92. Upon LPS stimulation, p300-targeted genes exhibit an increase in both Kac and Kcr, commensurate with the level of p300 recruitment and their activation36. Furthermore, increasing crotonyl-CoA concentrations prior to stimulation leads to more Kcr at p300-target genes and greater expression of those genes36. The increase in histone Kcr also leads to an increase in the levels of the YEATS domain protein AF963, a positive regulator of transcription93. Knockout of AF9 significantly reduced the effect of increased Kcr, but did not abolish it completely, suggesting that other reader proteins could be involved, or that Kcr has reader-independent cis-effects63. These studies demonstrate that gene activation can be modulated by acyl-CoA metabolism through the interaction of the YEATS domain of AF9 with histone Kcr (FIG. 6a). The focus of these studies was histone Kcr, but given the preference of YEATS-domain proteins for Kpr and Kbu over Kac, the mechanism described above is likely relevant to those acylations as well.
Taken together, these studies provide a proof-of-principle that dynamic changes in acyl-CoA metabolism can modulate signal-dependent gene activation. Moreover, studies focused on spermatogenesis and the stress conditions of acute kidney injury and starvation-induced ketogenesis, discussed below, provide early evidence that histone acylations are involved in diverse physiological processes.
Spermatogenesis
Several intriguing histone acylation patterns have been observed in murine spermatocytes undergoing transcriptional meiotic sex chromosome inactivation (MSCI). MSCI is associated with depletion of histone Kac, Khib and Kbu8,59, but not Kcr11. Interestingly, in post-meiotic round spermatids, Kcr and Khib become specifically associated with the transcription start site of the fraction of sex chromosome-linked genes that escape MSCI8,11,94, suggesting that these marks are able to sustain transcription in a generally repressive environment95,96 (FIG. 6b).
Unique histone acylation patterns have also been observed in late stages of spermatogenesis. Compared to Kac, Kbu is enriched in the later steps of spermatogenesis and persists on chromatin further into differentiation than Kac. As discussed above, BRDT, a testis-specific bromodomain protein that is crucial for transcription and the histone to protamine transition88, is unable to bind H4K5bu-contaning peptides, suggesting that a potential function of site-specific Kbu would be to reduce BRDT binding and slow histone-to-protamine exchange59 (FIG. 6b). Thus, Kbu could have an important role in the genomic reorganization observed during late spermatogenesis and might mark loci that maintain histones on chromatin through this developmental process. These studies raise the interesting question of whether a unique metabolic state or acyl-specific writers exist during germ-cell development.
Kidney injury
A recent study found that Kcr levels in murine kidney tissue increase as a result of acute kidney injury (AKI) induced by folic acid or cisplatin87. Following the induction of AKI, Kcr levels increased at the promoters of AKI stress induced genes, in correlation with their elevated expression. Intraperitoneal injection of crotonate increased Kcr in the kidney tissue, expression of the AKI stress-induced genes and protection against the induced AKI. Thus, stress-induced shifts in histone acylation patterns can modulate the response to AKI.
Ketogenesis
As discussed in the section metabolic regulation of Lys acylations, elevated histone Kbhb levels were observed in liver tissue of mice kept in ketogenic conditions (48hr starvation)12. Comparative ChIP-seq of H3K9bhb, H3K9ac, and H3K4me3 in combination with RNA-seq analysis of liver tissue from starved vs. fed mice showed striking correlation between enhanced H3K9bhb levels and starvation-induced gene expression12. Interestingly, many H3K9bhb-induced genes exhibited little to no change in H3K9ac levels. Furthermore, genes exhibiting changes in H3K9bhb and those exhibiting changes in H3K9ac were distinct with regard to gene-ontology classification, which suggests that the two histone acylations have non-redundant functions in this context. This study suggests a feedback exists in which accumulated β-hydroxybutyrate induces the formation of histone Kbhb modifications that, in turn, activate expression of physiologically relevant genes (FIG. 6c). Whether a Kbhb-selective reader or writer is responsible for process is unknown.
Although β-hydroxybutyrate is a SCFA that has been usually regarded as an energy source, a number of surprising and important physiological functions have been ascribed to it, for which mechanisms have remained elusive. For example, the ketogenic diet and β-hydroxybutyrate are used to treat epilepsy97, are protective against neurodegenerative diseases98,99, and are currently being tested in about 30 clinical trials (https://clinicaltrials.gov). In addition to promoting Kbhb, β-hydroxybutyrate has been reported to repress oxidative stress in mouse kidney by inhibiting HDAC activity and thus enhancing histone Kac100. However, quantitative experiments, in both cell culture and animals, showed that β-hydroxybutyrate has a much more profound impact on the levels of histone Kbhb than on histone Kac12. Thus, the discovery of histone Kbhb and its role in starvation-induced gene expression provides a potential mechanism for the various physiological properties ascribed to β-hydroxybutyrate.
Conclusion and future perspectives
The relatively recent discovery of short-chain acylations on histones significantly expands the complexity of histone PTMs as well as their interconnections with cellular metabolism. Several principles regarding the regulation and function of histone acylations have emerged: 1) Although there is evidence that some erasers have acyl-specificity, the writers are generally shared; 2) acylations are regulated by acyl-CoA metabolism, and low-nutrient conditions will give preference to non-acetyl acylations; 3) readers with preferential binding to non-acetyl acylations have been identified, demonstrating that protein domains have evolved to exploit the diversity of acylation forms. The studies discussed in this review provide a glimpse into the complex regulation and function of differential acylation. Nevertheless, many questions remain.
Differential regulation of histone acylations
We present a model in which the nuclear and cytoplasmic pools of acyl-CoAs determine the activity of promiscuous acyl-transferases such as p300. In this model, genomic loci will be modified by a mixture of histone acylations in proportion to the relative nuclear concentrations of the acyl-CoAs. But some genomic regions are enriched for a specific histone acylation, for example the “escapee” spermatogenesis genes and the starvation-response genes. How are the various acylations differentially regulated in these contexts? Several metabolic enzymes are known to “moonlight” in the nucleus, often as part of transcription-associated complexes101, raising the possibility that they could be acting locally as “feeders” for compartmentalized acyl-transferases. Local synthesis of a particular acyl-CoA could lead to locus-specific acylation, which might explain ChIP-seq observations of regions of enriched histone acylation. It is also possible that novel acyl-selective writers have yet to be discovered or that the acyl-promiscuity of already characterized acyl-transferases is regulated. Many HAT enzymes belong to multi-subunit complexes31,102 and are themselves modified with regulatory PTMs103. Do components of these complexes or PTMs influence the relative affinities of HATs for acyl-CoAs?
Acyl-CoA dynamics
Another major question is whether the concentrations of the non-acetyl acyl-CoAs are dynamic and high enough to impact physiological processes. Early examples, such as the recently reported Kbhb and bhb-CoA link to the ketogenic state, provide evidence that the concentrations of acyl-CoAs are highly dynamic in response to physiological changes, but finding additional examples will be crucial for moving forward.
Methods for the purification, stabilization and measurement of acyl-CoA species have been developed104 and successfully utilized in cell cultures to study the metabolic regulation of histone modifications12,36,105,106. Applying these techniques to a broader range of physiological states should provide valuable information about the dynamic range of the lesser-studied acyl-CoAs. Candidates for comparative acyl-CoA profiling would be cases where differential histone acylation has already been observed or would be predicted, for example at different stages of spermatogenesis, in fed versus starved mice, or across tumors with varying ACSS2 expression levels.
Additional candidates for acyl-CoA profiling could be any number of congenital diseases that are caused by mutations in acyl-CoA pathways, including succinyl-CoA synthetase deficiency (OMIM 612073), malonyl-CoA decarboxylase deficiency (OMIM 248360), glutaric aciduria I (OMIM 231670) and short-chain acyl-CoA dehydrogenase deficiency (OMIM 201470). Whether these defects in acyl-CoA metabolism affect the regulation of gene expression through differential histone acylation, remains to be explored.
Another potential source of dynamic acyl-CoA metabolism is the microbiota. SCFAs can be generated by microbial fermentation and have been regarded as metabolites with broad function in host metabolism and immunity 107. That SCFAs are precursors of Lys acylations suggests that SCFAs may function through regulating histone acylation. Acyl-CoA and histone acylation profiling of colon tissue of mice with different microbiota could unravel the role of microbial fermentation in acyl-CoA metabolism and histone acylation of the host.
We have presented the current evidence to support a functional role for non-acetyl histone acylations in gene regulation and presented the current challenges to the field. The proportional mixture of histone acylations is controlled by the integration of environmental and metabolic cues. Several lines of evidence demonstrate that modulating this proportional mixture of histone acylations has functional consequences on gene expression and chromatin structure. Future functional characterization of the writers, erasers and readers as well as the influence of acyl-CoA metabolism on these proteins will pave the way for a deeper understanding of the regulation and function of differential histone acylation.
Online summary.
Eight new types of histone short-chain Lys acylations have been discovered in histones during past few years, including propionylation (Kpr), butyrylation (Kbu), 2-hydroxyisobutyrylation (Khib), succinylation (Ksucc), malonylation (Kma), glutarylation (Kglu), crotonylation (Kcr) and β-hydroxybutyrylation (Kbhb).
Histone Lys acylations are regulated by acyltransferases and deacylases.
Histone Lys acylations are modulated by cellular metabolism of cognate short-chain acylCoA species.
Histone Lys acylations are recognized by specific protein domains, and can be differentiated from acetylation.
Histone Lys acylations mark transcriptionally active genes and function in different physiological processes, such as signal-dependent gene activation, spermatogenesis, tissue injury and metabolic-induced stress.
Ontology terms.
Biological sciences / Molecular biology / Chromatin / Histone post-translational modifications [URI /631/337/100/2285
Biological sciences / Genetics / Gene regulation [URI /631/208/200]
Biological sciences / Molecular biology / Post-translational modifications / Acetylation [URI /631/337/458/1275]
Biological sciences / Chemical biology / Metabolic pathways [URI /631/92/1643]
ACKNOWLEDGMENTS
We would like to thank He Huang, Saadi Khochbin and members of the Allis lab for comments and scientific input. This work was funded by support from The Rockefeller University and from the National Cancer Institute to C.D.A. (CA204639), and by NIH grants GM105933, DK107868 and GM115961 to Y.Z. B.R.S was supported by the National Science Foundation Graduate Research Fellowship Program (DGE-1325261).
Glossary
- Isothermal titration calorimetry
A technique to analyze intermolecular interactions by directly measuring the heat generated or absorbed when molecules interact.
- Lys acetylation
The addition of an acetyl group onto ε-amine of a Lys amino acid residue.
- Short-chain Lys acylations
The addition of a short-chain acyl group (less than six carbon atoms) other than formyl or acetyl group onto ε-amine of a Lys residue.
- ε-amine
The amino group at the side chain of Lys, where most post-translational modifications occur.
- Mass-tolerant database search
An algorithm that allows an unbiased way to search unexplained mass spectrometry data and detect a mass shift at a specific amino acid.
- van der Waals interactions
Refers to the attraction between molecules that is not generated from covalent bonds or ionic interactions.
- C-C π-bond
Covalent chemical bonds that are formed when two atomic orbitals overlap side-to-side along a plane perpendicular to a line connecting the nuclei of the atoms.
- Long-chain Lys acylations
Here refers to Lys acylations with longer hydrocarbon chains, such as Lys myristoylation and palmitoylation.
- Zn2+-dependent histone deacetylases
Classes I, II, and IV of histone deacetylases, which require Zinc ions to remove acetyl groups from ε-amine of Lys on histones.
- Bromodomains
protein modules of ~ 110 amino acids that mediate interaction with acetylated Lys; often found in acetyltransferases and ATP-dependent chromatin remodeling factors.
- Cell-free transcription assays
A system comprised of a reconstituted chromatin template and recombinant transcription factors and cofactors, which when incubated with nuclear extracts, allows RNA synthesis to occur in vitro.
- Histone to protamine transition
A process during spermatogenesis in which histones are replaced gradually by arginine-rich protamine, which packages the sperm tightly.
Biography
Benjamin R. Sabari is a graduate student in the Laboratory of Chromatin Biology and Epigenetics at The Rockefeller University. His graduate studies focused on the regulation and function of histone lysine crotonylation and its role in gene activation.
Di Zhang obtained his Ph.D. degree from Peking University, China, where he studied molecular mechanism in breast cancer development under supervision of Dr. Yongfeng Shang. He is currently working as a postdoctoral scholar in Yingming Zhao's lab at the University of Chicago. His studies focused on metabolic regulation of histone acylations using mass spectrometry and molecular biology methods.
C. David Allis is the Joy and Jack Fishman Professor and Head of the Laboratory of Chromatin Biology and Epigenetics at The Rockefeller University. He is a member of the US National Academy of Sciences and the American Academy of Arts and Sciences. His laboratory currently studies the role of histone proteins in normal cellular physiology and human disease.
Yingming Zhao received his Ph.D. degree from the Rockefeller University under Professor Brian Chait. He is currently a Professor in the Ben May Department for Cancer Research at the University of Chicago. His laboratory is interested in applying mass spectrometry-based proteomics technologies to identify new types of protein posttranslational modifications and characterizing their functions in different biological systems.
Footnotes
FURTHER INFORMATION
Online Mendelian inheritance in man (OMIM) website: http://www.omim.org/
References
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458. e451. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [Describes the first identification and validation of histone propionylation and butyrylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai LZ, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol. 2014;10:365–U373. doi: 10.1038/nchembio.1497. [The first study describing the identification and characterizing the epigenetic function of histone 2-hydroxyisobutyrylation.] [DOI] [PubMed] [Google Scholar]
- 9.Xie Z, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan MJ, et al. Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan MJ, et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell. 2011;146:1015–1027. doi: 10.1016/j.cell.2011.08.008. [Identifies and characterizes the epigenetic function of histone Lys crotonylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z, et al. Metabolic Regulation of Gene Expression by Histone Lysine beta-Hydroxybutyrylation. Mol Cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [Reports Lys β-hydroxybutyrylation as a new type of histone modification that is closely associated with ketone body metabolism.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 14.Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in Regulation of Rna Synthesis. P Natl Acad Sci USA. 1964;51:786–+794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. Acs Chem Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etchegaray JP, Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Molecular cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., 3rd Protein analysis by shotgun/bottom-up proteomics. Chemical reviews. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Molecular & cellular proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & cellular proteomics. 2011;10:M111 012658. doi: 10.1074/mcp.M111.012658. [The first study demonstrating Lys malonylaton as a new type of protein modification.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications by blind search of mass spectra. Nature biotechnology. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 24.Hansen BT, Davey SW, Ham AJ, Liebler DC. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. Journal of proteome research. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Chen W, Cobb MH, Zhao Y. PTMap--a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc Natl Acad Sci U S A. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chick JM, et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nature biotechnology. 2015;33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner GR, Payne RM. Widespread and Enzyme-independent N-is an element of- Acetylation and N-is an element of-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. Embo J. 2015;34:2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinert BT, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Bio. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 32.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 33.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 34.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Z, et al. Molecular characterization of propionyllysines in non-histone proteins. Molecular & cellular proteomics. 2009;8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabari BR, et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300- Catalyzed Histone Crotonylation. Molecular cell. 2015;58:203–215. doi: 10.1016/j.molcel.2015.02.029. [Demonstrates that p300-catalyzed histone crotonylation activates gene transcription, which is regulated by the cellular concentration of crotonylcoA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaczmarska Z, et al. Structure of p300 in complex with Acyl-CoA variants. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2217. http://dx.doi.org/10.1038/nchembio.2217. [DOI] [PMC free article] [PubMed]
- 38.Hu A, Britton LM, Garcia BA. Investigating the specificity of histone acetyltransferase activity for producing rare modifications on histones using mass spectrometry.. The 62nd Annual American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics; Baltimore, MD. 2014. [Google Scholar]
- 39.Yu-Ying Y, Markus G, Howard HC. Identification of lysine acetyltransferase p300 substrates using 4-pentynoyl-coenzyme A and bioorthogonal proteomics. Bioorg Med Chem Lett. 2011;21:4976–4979. doi: 10.1016/j.bmcl.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry-Us. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leemhuis H, Packman LC, Nightingale KP, Hollfelder F. The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. Chembiochem. 2008;9:499–503. doi: 10.1002/cbic.200700556. [DOI] [PubMed] [Google Scholar]
- 42.Ringel AE, Wolberger C. Structural basis for acyl group discrimination by human Gcn5L2. Acta Crystallogr D Struct Biol. 2016;72:841–848. doi: 10.1107/S2059798316007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing H, Lin H. Sirtuins in epigenetic regulation. Chem Rev. 2015;115:2350–2375. doi: 10.1021/cr500457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith BC, Denu JM. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J Biol Chem. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 46.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 47.Du JT, et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [Together with REF. 22, shows that SIRT5 has robust desuccinylase and demalonylase activities.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molecular cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [A systematic study of the enzymatic activities of mammalian sirtuins agaisnt different lysine acylations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barber MF, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L, et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao XC, et al. Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholz C, et al. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, et al. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics. 2012;11:1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angewandte Chemie. 2012;51:9083–9087. doi: 10.1002/anie.201203754. [DOI] [PubMed] [Google Scholar]
- 56.Vollmuth F, Geyer M. Interaction of Propionylated and Butyrylated Histone H3 Lysine Marks with Brd4 Bromodomains. Angew Chem Int Edit. 2010;49:6768–6772. doi: 10.1002/anie.201002724. [DOI] [PubMed] [Google Scholar]
- 57.Flynn EM, et al. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure. 2015;23:1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Moriniere J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 59.Goudarzi A, et al. Dynamic Competing Histone H4 K5K8 Acetylation and Butyrylation Are Hallmarks of Highly Active Gene Promoters. Molecular cell. 2016;62:169–180. doi: 10.1016/j.molcel.2016.03.014. [Demonstrates in mice that dynamic competition between histone acetylation and butyrylatoin modulates the binding of BRDT, which in turn controls gene expression and chromatin reorganization during spermatogenesis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li YY, et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shanle EK, et al. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 2015;29:1795–1800. doi: 10.1101/gad.269977.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Molecular cell. 2016;62:181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao D, et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016;26:629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrews FH, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12:396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q, et al. Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain. Structure. 2016 doi: 10.1016/j.str.2016.05.023. [REFS 63–67 identified the YEATS domain as a crotonylation-specific reader.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 68.Li HT, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi XB, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lange M, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Gene Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu Y, et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Gene Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreveny I, et al. The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Research. 2014;42:822–835. doi: 10.1093/nar/gkt931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng L, et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–U138. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ali M, et al. Tandem PHD Fingers of MORF/MOZ Acetyltransferases Display Selectivity for Acetylated Histone H3 and Are Required for the Association with Chromatin. J Mol Biol. 2012;424:328–338. doi: 10.1016/j.jmb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiong X, et al. Selective Recognition of Histone Crotonylation by Double PHD Fingers of MOZ and DPF2. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2218. http://dx.doi.org/10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed]
- 77.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhanu NA-S, L., Garcia BA. Quantification of lysine crotonylation during in vitro human myogenic differentiation. The 61st Annual American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics. 2013 [Google Scholar]
- 79.Tweedie-Cullen RY, et al. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PloS one. 2012;7:e36980. doi: 10.1371/journal.pone.0036980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Comerford SA, et al. Acetate Dependence of Tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mashimo T, et al. Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 84.Cahill GF., Jr. Fuel metabolism in starvation. Annual review of nutrition. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 85.Robinson AM, Williamson DH. Physiological Roles of Ketone-Bodies as Substrates and Signals in Mammalian-Tissues. Physiol Rev. 1980;60:143–187. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 86.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz-Andres O, et al. Histone lysine-crotonylation in acute kidney injury. Dis Model Mech. 2016;9:633–645. doi: 10.1242/dmm.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaucher J, et al. Bromodomain-dependent stage-specific male genome programming by Brdt. Embo J. 2012;31:3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Molecular cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 90.Kundu TK, et al. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Molecular cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 91.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–U225. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 93.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 94.Sin HS, et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012;26:2737–2748. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montellier E, Rousseaux S, Zhao Y, Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays. 2012;34:187–193. doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 96.Rousseaux S, Khochbin S. Histone Acylation beyond Acetylation: Terra Incognita in Chromatin Biology. Cell J. 2015;17:1–6. doi: 10.22074/cellj.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McNally MA, Hartman AL. Ketone bodies in epilepsy. Journal of Neurochemistry. 2012;121:28–35. doi: 10.1111/j.1471-4159.2012.07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kashiwaya Y, et al. D-beta-hydroxybutyrate protects neurons in models of Alzheimer's and Parkinson's disease. Proc Natl Acad Sci USA. 2000;97:5440–5444. doi: 10.1073/pnas.97.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim S, et al. D-beta-Hydroxybutyrate Is Protective in Mouse Models of Huntington's Disease. Plos One. 2011;6 doi: 10.1371/journal.pone.0024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shimazu T, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends Biochem Sci. 2016;41:712–730. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 102.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsuchiya Y, Pham U, Gout I. Methods for measuring CoA and CoA derivatives in biological samples. Biochem Soc T. 2014;42:1107–1111. doi: 10.1042/BST20140123. [DOI] [PubMed] [Google Scholar]
- 105.Lee JV, et al. Akt-Dependent Metabolic Reprogramming Regulates Tumor Cell Histone Acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu X, et al. High-Resolution Metabolomics with Acyl-CoA Profiling Reveals Widespread Remodeling in Response to Diet. Molecular & cellular proteomics. 2015;14:1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 108.Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. Acs Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]