Abstract
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is a powerful molecular mapping technology that offers unbiased visualization of the spatial arrangement of biomolecules in tissue. Although there has been a significant increase in the number of applications employing this technology, the extracellular matrix (ECM) has received little attention, likely because ECM proteins are mostly large, insoluble and heavily cross-linked. We have developed a new sample preparation approach to enable MALDI IMS analysis of ECM proteins in tissue. Prior to freezing and sectioning, intact tissues are decellularized by incubation in sodium dodecyl sulfate. Decellularization removes the highly abundant, soluble species that dominate a MALDI IMS spectrum while preserving the structural integrity of the ECM. In situ tryptic hydrolysis and imaging of tryptic peptides are then carried out to accommodate the large sizes of ECM proteins. This new approach allows the use of MALDI IMS for identification of spatially specific changes in ECM protein expression and modification in tissue.
Keywords: imaging mass spectrometry, extracellular matrix, MALDI, FTICR, MALDI imaging
Introduction
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS)[1] combines the tremendous sensitivity and selectivity of mass spectrometry with the spatial analysis provided by traditional histology, offering unbiased visualization of the spatial arrangement of biomolecules in tissue. As such, MALDI IMS is a powerful new molecular visualization technology for the biological and clinical sciences. With a significant increase in the number and variety of applications employing this technology, there is ongoing interest in MALDI IMS, and considerable effort has gone into significantly improving both methods and instrumentation in order to meet the performance needs of researchers.[2,3]
One important area that has received only limited attention is the extracellular matrix (ECM). The ECM provides structural support to surrounding cells in tissue and plays important roles in cell adhesion, migration, and differentiation.[4] Moreover, the importance of the ECM in cellular processes is evident by the range of diseases that accompany defects in ECM proteins.[5] Given its prominent role in cellular biology and pathogenesis, the ECM is an important target for MALDI IMS. However, the physical and chemical properties of the ECM pose significant challenges to current MS technology, thus far preventing effective and routine analysis. For example, ECM components are heavily cross-linked and highly insoluble. Moreover, signals from soluble cellular proteins dominate the mass spectrum, such that ECM proteins are difficult to detect. Finally, most ECM constituents are high-molecular weight glycoproteins (>100 kDa) that are currently beyond the practical mass range for protein imaging experiments.
We have developed a novel sample preparation procedure to overcome the challenges of ECM analysis. It involves the removal of cellular material using sodium dodecyl sulfate (SDS), an idea previously developed in biomedical engineering.[6] Perfusion with chemical detergents removes cellular material, leaving a native three-dimensional scaffold, which can be repopulated with human cells to build functioning organs.[6] Along these lines, we sought to use detergents to remove cellular material to enhance signal from ECM components and make the ECM accessible to MALDI IMS analysis. Initial attempts to use washes of thin tissue sections were insufficient to remove all cellular material. Instead, a long incubation of intact tissue in cold SDS solution was necessary to maintain the integrity of the tissue. Following decellularization, an in situ tryptic digest was performed to hydrolyze large ECM proteins into tryptic peptides that are more amenable to analysis. Our results show that this preparation protocol can be applied to a variety of tissue types and is crucial to successfully analyze ECM proteins by MALDI IMS.
Methods
Samples and materials
Fresh frozen mouse kidney, rat heart and rat liver were purchased from Pel-Freez Biologicals (Rogers, AR). A human biopsy of a renal cell carcinoma was obtained from the Vanderbilt University Ingram Cancer Center-Human Tissue Acquisition and Pathology Resource and the National Institutes of Health Cooperative Human Tissue Network, from a patient undergoing a nephrectomy. Ethanol, methanol, acetonitrile (ACN) and acetic acid were purchased from Fisher Scientific (Suwanee, GA), trifluoroacetic acid from Acros (Morris Plains, NJ) and chloroform and α-cyano-4-hydroxycinnamic acid from Sigma-Aldrich (Milwaukee, WI). Conductive indium tin oxide (ITO)-coated microscope glass slides were purchased from Delta Technologies (Stillwater, MN).
Decellularization and washing of the tissue
Intact, frozen tissue was thawed in 1× phosphate-buffered saline for 10 min at 4 °C. Decellularization was performed as described in the succeeding text. Decellularized and control tissues were sectioned at 12 µm thickness using a Leica CM3050 cryostat (Leica Microsy stems GmbH, Wetzlar, Germany). Frozen tissue sections were thaw mounted on cold ITO-coated microscope slides, with two sections on each slide, and stored in a desiccator until needed. Tissue was washed in a six-step rinse protocol: 70% ethanol, 100% ethanol, Carnoy’s fluid, 100% ethanol, H2O and 100% ethanol. Carnoy’s fluid was prepared from 60 ml of ethanol, 30 ml of chloroform, and 10 ml of acetic acid. All rinse steps were carried out for 30 s except for the step with Carnoy’s solution, which was carried out for 2 min.
In situ tryptic digest
A 0.1 µg/1 µl solution of trypsin was prepared in 10% ACN and 90% 20 mm ammonium bicarbonate buffer. Trypsin was applied using the SunCollect robotic sprayer (SunChrome) in eight passes with a 7.5 µl/min solution flow rate at medium speed. Samples were incubated in a hydration chamber for 6 h in an oven at 37 °C. The hydration chamber consisted of a plastic Petri dish with a small piece of tissue paper, to which 1 ml of ammonium bicarbonate was added. The ITO slide was placed on top of the wet paper with the tissue section facing up, and the chamber was carefully sealed shut with tape to keep vapor from escaping.
Tissue microextractions and LC–MS analysis
One tissue section on each slide was used for tissue microextractions, following tryptic digest, but prior to matrix application. For microextraction of tryptic peptides, solutions of 5% and 50% ACN in water were prepared. Using a pipette, 1 µl of each ACN solution was pipetted onto the surface of the tissue and then drawn back into the pipette, three times in several spots around the tissue. Extracts were combined and collected for LC–MS analysis.
The peptide solutions collected from on-tissue digestions were diluted with 0.1% formic acid and were loaded onto a reverse-phase capillary trap column using a helium-pressurized cell. The trap column (360 µm OD × 150 µm ID) was fritted with a filter end-fitting (IDEX Health & Science) and packed with 4 cm of C18 reverse-phase material (Jupiter, 5 µm beads, 300 Å, Phenomenex). Once the sample was loaded, an M-520 microfilter union (IDEX Health & Science) was used to connect the trap column to a capillary analytical column (360 µm OD × 100 µm ID), equipped with a laser-pulled emitter tip and packed with 18 cm of C18 material (Jupiter, 3 µm beads, 300 Å, Phenomenex). Using an Eksigent NanoLC Ultra High-performance liquid chromatography (HPLC), peptides were gradient-eluted at a flow rate of 500 nl/min, and the mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% ACN (solvent B). A 90-min gradient was performed, consisting of the following: 0–14 min, 2% B; 14–64 min, 2–40% B; 64–72 min, 40–90% B; 72–74 min, 90% B; 74–75 min 90–2% B; and 75–90 min, 2% B. Upon gradient elution, proteins were mass analyzed on an LTQ Orbitrap Velos mass spectrometer, equipped with a nanoelectrospray ionization source (Thermo Scientific). The instrument was operated using a data-dependent method with dynamic exclusion enabled. Full scan (m/z 300–2000) spectra were acquired with the Orbitrap (resolution 60 000), and the top 16 most abundant ions in each MS scan were selected for fragmentation via collision-induced dissociation in the LTQ ion trap. An isolation width of 2 m/z, activation time of 10 ms and 35% normalized collision energy were used to generate MS2 spectra. Dynamic exclusion settings allowed for a repeat count of 1 within a repeat duration of 10 s, and the exclusion duration time was set to 15 s. For identification of peptides, tandem mass spectra were searched with Sequest (Thermo Fisher Scientific) against a rat subset database created from the UniprotKB protein database (www.uniprot.org). Search results were assembled using Scaffold 3.0 (Proteome Software).
MALDI matrix application and sample analyses
Matrix was applied immediately following trypsin hydrolysis. A 5 mg/ml solution of α-cyano-4-hydroxycinnamic acid in 50% ACN and 0.5% formic acid in water was prepared. The solution was sonicated to ensure dissolution of the matrix in solution. The matrix solution was applied to the tissue surface using the TM sprayer (HTX technologies). The matrix was sprayed with a solvent flow rate of 0.1 ml/min, with a track spacing of 1.2 mm and nozzle speed of 700 mm/min, for a total of four passes.
Samples were analyzed on either a 9.4T Fourier transform ion cyclotron resonance (FTICR) MS (Bruker Daltonics SolariX) or an Ultraflex MALDI TOF-MS (Bruker Daltonics) in positive ion mode. For FTICR experiments, data were acquired using ftmsControl software. Approximately 100 shots per spot were acquired with a 1 kHz repetition rate Smartbeam II Nd:YAG laser. Image acquisition was carried out using FlexImaging 3.0. For Ultraflex MALDI time-of-flight (TOF) analysis, the instrument was operated in LIFT mode for MS/MS analysis. Approximately 15 000 shots were summed for the species of interest and acquired using FlexControl software. MS analysis was performed with FlexAnalysis 3.3 (Bruker).
Identification of tryptic peptides
Using the results generated from the LC–MS/MS experiment and database search as a guide, masses of tryptic peptides from proteins identified in the LC–MS/MS experiment were searched against peaks within the average spectrum generated by the MALDI FTICR IMS experiment. Peak picking of FTICR data was performed using mMass software.[7] An error of <5 ppm was required for a peptide to be positively identified. In addition to this peptide mass fingerprint-style approach, a targeted analysis was also performed to demonstrate the ability to identify a single peptide by MALDI MS/MS, directly from the tissue, as described earlier. In this case, peak picking of LIFT data generated by the MS/MS experiment was performed manually in FlexImaging, and peak lists were exported to Biomap (Bruker Daltonics) and searched using the MASCOT database.
Results and discussion
ECM enrichment
In the MALDI IMS workflow, careful sample preparation is an essential step for a successful experiment and must be optimized for the specific analyte and tissue of interest. Chemical washing of tissue sections is often performed to enhance signal for specific analyte classes, including lipids[8] and proteins and peptides.[9,10] Because of this, we first performed detergent washes of tissue sections on slides, in order to remove cellular material and enhance signal from ECM proteins. Immediately after sectioning, slides containing sections of mouse kidney were washed with SDS, Triton X-100 or sodium deoxycholate in a Petri dish. Dishes containing the slide and detergent solution were gently swirled for up to 6 min. The tissue section detached from the slide at longer incubation times. After the detergent wash, sections were washed in the following sequence: water for 30 s, 70% ethanol for 30 s, 100% ethanol for 30 s, Carnoy’s solution for 120 s, water for 30 s, 100% ethanol for 30 s and water for 30 s. Control sections were washed using the same washing sequence but excluding the detergent wash.
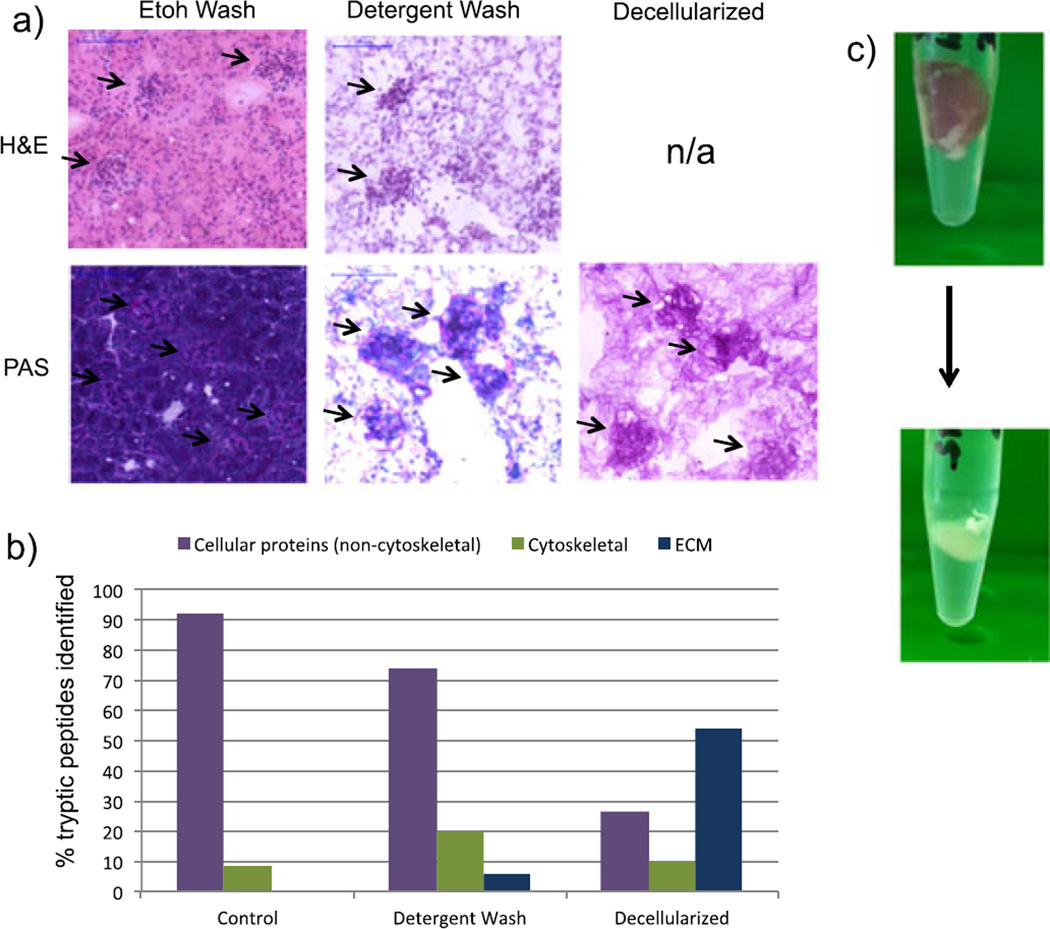
Figure 1a shows images of control and SDS-washed renal tissue sections. The hematoxylin-stained and eosin-stained control sample shows staining of cytoplasmic and eosinophilic components, while the detergent-washed section shows only nuclear components. This indicates that the detergent wash removes cytoplasmic material, although cell nuclei persist. Sections were also stained with a periodic acid–Schiff (PAS) stain. The PAS stain is commonly used in histopathological analysis of renal tissue and stains glycans (pink) and counterstains nuclei (dark purple). As expected, the control and detergent-washed samples show the presence of nuclei (Fig. 1a). This was confirmed by LC–MS/MS analysis of tissue extracts, following an in situ tryptic digest of each prepared section. Fig. 1b shows the percentage of tryptic peptides that derived from cellular (non-cytoskeletal), cytoskeletal and ECM proteins. LC–MS/MS analysis of tissue extracts did not identify any peptides derived from ECM proteins in the control tissue. Even though ECM-derived peptides were present in the detergent-washed sample, tryptic peptides of cellular proteins, especially nuclear proteins such as histones, were still dominant. In addition, signals from cytoskeletal proteins in SDS-washed sections increased compared with control sections. Similar results were obtained for the Triton X-100-washed and sodium deoxycholate-washed tissue sections (data not shown).
Figure 1.
(a) Stained sections of ethanol-washed mouse renal tissue, detergent-washed tissue, and decellularized tissue. Top: H&E stain, staining nuclei purple and cytoplasm pink. Bottom: PAS, staining nuclei dark purple and glycans/glycoproteins dark pink. (b) Percent of tryptic peptides identified in LC–MS/MS analysis of mouse kidney tissue extracts from ethanol-washed (‘control’), detergent-washed, and decellularized tissue. The graph shows peptides classified by non-cytoskeletal, cellular proteins, cytoskeletal proteins, and ECM proteins. (c) Mouse kidney before (top) and after (bottom) 4 days of incubation in 2% SDS.
Taken together, these results suggest that detergent washing of tissue sections is not an effective approach for ECM enrichment. Thus, we sought a more efficient decellularization method. As described earlier, longer tissue washes in detergent solutions resulted in the detachment of the tissue from the surface of the slide. Therefore, we developed an alternative method of decellularizing the intact tissue prior to sectioning. This was accomplished by incubating the tissue in a 2% SDS solution at 4 °C for 4 days. The tissue and solution were placed in a tube that allowed the tissue to float freely in solution, and the tube was mechanically rotated for the duration of the incubation. The SDS solution was replaced daily with fresh solution. After the tissue became completely opaque (~4 days, Fig. 1c), it was rinsed three times in a 10 mm ammonium bicarbonate solution, to remove excess SDS solution at 4 °C for 15 min, and frozen for sectioning.
When sections of decellularized tissue were stained with hematoxylin and eosin, no stained features were observed, indicating the removal of both cytoplasmic material and cell nuclei (Fig. 1a). In the PAS-stained sections, the decellularized tissue contains fibrillar pink material, indicating the presence of glycans and glycoproteins, which are major constituents of the ECM, but no nuclei. Furthermore, the sample contains bundles of glycan-rich tissue in the kidney cortex ~80 µm in diameter representing intact, decellularized glomeruli, thus indicating that the structural integrity of the tissue has been preserved. These results were confirmed by LC–MS/MS analysis of extracts from decellularized tissue (Fig. 1b). Over 50% of the tryptic peptides identified in the extracts were derived from ECM-related proteins, compared with less than 30% for non-cytoskeletal, cellular proteins. Table 1 lists ECM-related proteins that were identified in the decellularized sample, including several collagens and laminins. Based on these results, decellularization of intact tissue was used to prepare samples for MALDI IMS experiments.
Table 1.
ECM proteins identified by LC MS/MS analysis of tissue extracts following in situ tryptic digestion of decellularized mouse kidney
| Identified proteins | Approximate molecular weight (kDa) |
Number of unique peptides |
|---|---|---|
| Collagen α1(I) | 138 | 30 |
| Collagen α1(III) | 139 | 20 |
| Heparan sulfate | 469 | 52 |
| Collagen α2(I) | 130 | 25 |
| Collagen α2(IV) | 167 | 14 |
| Laminin subunit α5 | 404 | 41 |
| Laminin subunit gamma-1 | 177 | 37 |
| Collagen α3(VI) | 354 | 33 |
| Collagen α1(IV) | 161 | 7 |
| Laminin subunit beta-1 | 197 | 26 |
| Fibronectin | 273 | 29 |
| Laminin subunit beta-2 | 197 | 22 |
| Nidogen-1 | 137 | 17 |
| Protein Fga | 87 | 15 |
| Collagen α1(VI) | 108 | 12 |
| Collagen α2(VI) | 110 | 9 |
| Collagen alpha-4(IV) chain | 164 | 2 |
| Collagen α1(V) | 184 | 5 |
| Aminopeptidase N | 110 | 7 |
| Collagen α3(IV) | 162 | 2 |
| Nidogen-2 | 154 | 6 |
| Collagen α5(IV) | 162 | 2 |
| Collagen α2(V) | 145 | 2 |
| Collagen α1(XII) | 340 | 2 |
| Laminin α1 | 338 | 2 |
MALDI imaging mass spectrometry of ECM proteins
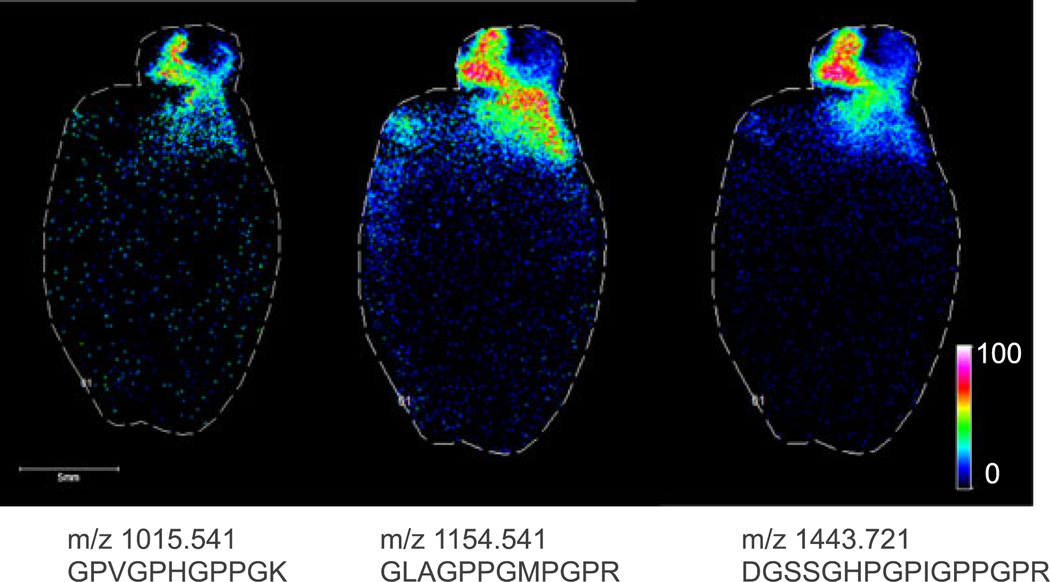
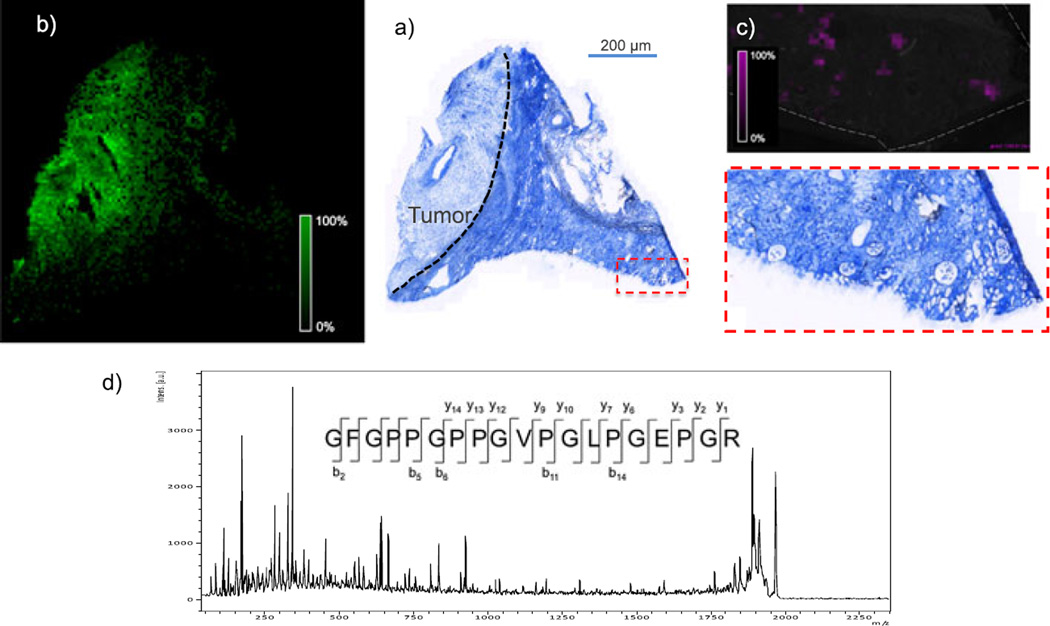
Because of the large size of many ECM-related proteins, it was necessary to analyze tryptic peptides from the decellularized tissue rather than intact proteins. Therefore, in each case, an in situ trypsin digest[11] was performed, and the tryptic peptides were imaged. Figure 2 shows MALDI FTICR IMS images of three signals at m/z 1015.541, 1154.541 and 1443.721 in rat heart. These m/z values match those of collagen III tryptic peptides, as identified in the LC–MS experiment. All three peptides co-localize to the aorta of the decellularized heart. Collagen III is known to be enriched in the aortic wall and plays crucial roles in the function and mechanical properties of the aorta. The highlighted imaging results provide confident identification of collagen III based on the detection of three separate tryptic peptides, high mass accuracy (<5 ppm) for each of the peptides, and observed spatial localization that is consistent with previous reports.[12] In a second example, Fig. 3 shows MALDI FTICR images of signals at m/z 1323.789 and m/z 1245.61 in a human renal cell carcinoma biopsy. The signal at m/z 1323.789 is located primarily in the area of the tumor and was identified as fibronectin, based on accurate mass and LC–MS/MS analysis of tissue extracts. Fibronectin levels have been shown to be elevated in renal cell carcinoma, similar to this result.[13,14] The signal at 1245.61 was identified as collagen IV α1 and is abundant in glomeruli within the healthy tissue section. Collagen IV α1 is a known component of the glomerular mesangial matrix. These images demonstrate the efficacy of this sample preparation protocol to measure histologically significant changes to ECM proteins in disease. Moreover, the decellularization does not disrupt the localization of the proteins.
Figure 2.
MALDI FTICR images (100 µm step size) of three unique tryptic peptides of collagen III, which is known to be enriched in the aortic wall.[12] The experimental m/z and sequence of each peptide are listed below each corresponding image.
Figure 3.
(a) Trichrome blue stain of a decellularized human renal cell carcinoma biopsy. Blue indicates the presence of collagen (not specific to type); dark purple and red indicate the presence of nuclei and muscle cells (not present because of decellularization). The biopsy contains tumor (labeled and indicated by dashed line) and adjacent healthy tissue (area to the right of dashed line). (b and c) MALDI MS images of an adjacent section showing signal for m/z 1323.789 (b, pink; identified as fibronectin) and m/z 1245.61 (green; identified as collagen IV α1) from MALDI TOF IMS analysis of an adjacent section (100 µm). (d) MALDI TOF MS/MS spectrum of the tryptic peptide GFGPPGPPGVPGLPGEPGR from collagen 1 α1 from TOF analysis of tissue.
Ideally, peptide identification in these experiments would be performed directly from the tissue surface by MALDI MS/MS. There are several factors, such as limited dynamic range and the generation of low-charge state ions, that make this type of analysis challenging. Nevertheless, Fig. 3c shows the MALDI TOF MS/MS spectrum of a signal at m/z 1939. The peptide was identified as a peptide of collagen I α1 chain. This protein is ubiquitous in the tissue, allowing for the collection of many MS/MS spectra from one tissue section, thus providing sufficient signal for a strong tandem MS spectrum. For less abundant ions, which may be distributed diffusely or in small areas of the tissue, identification of peptides by accurate mass would be necessary, followed by confirmation using other methods (e.g. LC–MS/MS or immunohistochemistry).
Conclusions
We have presented a simple, novel sample preparation protocol that enables MALDI IMS analysis of the ECM. The images presented here show enhanced detection of ECM-related signals and biologically relevant localization of ECM proteins. Moreover, targeted MALDI MS/MS of tryptic peptides was demonstrated, and multiple peptides from the same protein may be detected for greater confidence in identification. Overall, this protocol will allow investigators to take full advantage of the powerful chemical and spatial analysis afforded by IMS technology. Given the emerging consensus that the ECM plays a central role in many fundamental biological processes in health and disease, these analyses will be of great interest to the biological research community.
Acknowledgments
The authors gratefully acknowledge a grant from the National Institutes of Health GM8P41GM103391 to R. C. and DK65138 to B.H. and F32 DK97875 to M.G. We thank K. L. Rose and P. Angel for the helpful discussions about our approach. LC MS/MS work was performed by K. L. Rose in the Proteomics Core of the Vanderbilt University Mass Spectrometry Research Center.
References
- 1.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Gessel MM, Norris JL, Caprioli RM. MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery. J Proteomics. 2014;107C:71–82. doi: 10.1016/j.jprot.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom. Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 4.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011;3:1–24. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvelainen H, Sainio A, Koulu M, Wight TN, Penttinen R. Extracellular matrix molecules: potential targets in pharmacotherapy. Pharmacol. Rev. 2009;61:198–223. doi: 10.1124/pr.109.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faulk DM, Johnson SA, Zhang L, Badylak SF. Role of the extracellular matrix in whole organ engineering. J Cell. Physiol. 2014;229:984–989. doi: 10.1002/jcp.24532. [DOI] [PubMed] [Google Scholar]
- 7.Strohalm M, Kavan D, Novák P, Volný M, Havlíček V. mMass 3: a cross-platform software environment for precise analysis of mass spectro metric data. Anal. Chem. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 8.Mainini V, Angel PM, Magni F, Caprioli RM. Detergent enhancement of on-tissue protein analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. RCM. 2011;25:199–204. doi: 10.1002/rcm.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicklay JJ, Harris GA, Schey KL, Caprioli RM. MALDI imaging and in situ identification of integral membrane proteins from rat brain tissue sections. Anal. Chem. 2013;85:7191–7196. doi: 10.1021/ac400902h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am. Soc. Mass Spectrom. 2008;19:1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 12.Gay S, Balleisen L, Remberger K, Fietzek PP, Adelmann BC, Kühn K. Immunohistochemical evidence for the presence of collagen type III in human arterial walls, arterial thrombi, and in leukocytes, incubated with collagen in vitro. Klin. Wochenschr. 1975;53:899–902. doi: 10.1007/BF01468981. [DOI] [PubMed] [Google Scholar]
- 13.Chaves KCB, Turaça LT, Pesquero JB, Mennecier G, Dagli MLZ, Chammas R, Schor N, Bellini MH. Fibronectin expression is decreased in metastatic renal cell carcinoma following endostatin gene therapy. Biomed. Pharmacother. Bioméd. Pharmacothérapie. 2012;66:464–468. doi: 10.1016/j.biopha.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Steffens S, Schrader AJ, Vetter G, Eggers H, Blasig H, Becker J, Kuczyk MA, Serth J. Fibronectin 1 protein expression in clear cell renal cell carcinoma. Oncol. Lett. 2012;3:787–790. doi: 10.3892/ol.2012.566. [DOI] [PMC free article] [PubMed] [Google Scholar]