Significance
Current therapy for advanced colorectal cancer (CRC) is unsatisfactory and CRC remains a major cause of cancer-related deaths. Thus, novel and ubiquitously acting oncogenic mediators that are amenable to pharmacological targeting need to be identified. We found that loss of adenomatous polyposis coli (APC), which is mutated in the majority of human CRC, results in up-regulation of the signaling protein IL-6ST/gp130. This results in activation of Src family kinases (SFKs), YAP, Notch, and STAT3, which are simultaneously activated in 64% of human CRC. In addition to better explaining how APC loss initiates colorectal tumorigenesis, we show that combined treatment with SFK and JAK inhibitors results in regression of established colorectal tumors in mice.
Keywords: colorectal cancer, adenomatous polyposis coli, IL-6ST/gp130, YAP, STAT3
Abstract
Loss of tumor suppressor adenomatous polyposis coli (APC) activates β-catenin to initiate colorectal tumorigenesis. However, β-catenin (CTNNB1) activating mutations rarely occur in human colorectal cancer (CRC). We found that APC loss also results in up-regulation of IL-6 signal transducer (IL-6ST/gp130), thereby activating Src family kinases (SFKs), YAP, and STAT3, which are simultaneously up-regulated in the majority of human CRC. Although, initial YAP activation, which stimulates IL6ST gene transcription, may be caused by reduced serine phosphorylation, sustained YAP activation depends on tyrosine phosphorylation by SFKs, whose inhibition, along with STAT3-activating JAK kinases, causes regression of established colorectal tumors. These results explain why APC loss is a more potent initiating event than the mere activation of CTNNB1.
Colorectal cancer (CRC) is the fourth leading cause of cancer-related deaths in males and third in females (1). Although early CRC (stages I and II) can be controlled by surgical resection accompanied by chemotherapy, advanced CRC (stages III and IV) is associated with high mortality rates (2). In such patients, targeted therapies, including EGF receptor and angiogenesis inhibitors, prolong survival only by several months (3). Furthermore, only a small fraction of CRC patients, whose tumors are mismatch repair-deficient, respond positively to immunotherapy (4). Undoubtedly, the future of CRC therapy depends on identification of novel and ubiquitously acting oncogenic mediators whose targeting will cause tumor regression in most patients.
CRC pathogenesis often follows a well-defined multistep genetic pathway that leads to sequential activation of several key signal transducers and transcription factors (5, 6). The most frequent tumor-initiating event is inactivation of the adenomatous polyposis coli (APC) tumor suppressor, resulting in stabilization and irreversible activation of β-catenin (CTNNB1) (7). The much higher frequency of APC loss-of-function mutations relative to CTNNB1 gain-of-function mutations (8) suggests that APC loss leads to activation of at least one more oncogenic pathway that remains to be identified. Subsequent mutations that disrupt the tumor suppressive p53 and TGF-β pathway and activate Ras-MAP kinase (MAPK) and phosphoinositide 3-kinase (PI3K) signaling mediate malignant progression (9). So far, however, MAPK and PI3K inhibition had only a marginal impact on survival in advanced CRC patients (10) and restoration of APC, p53, and TGF-β tumor suppressor activity remains an elusive goal.
CRC pathogenesis is enhanced by inflammation (11). In the case of inflammatory bowel diseases, which greatly increase CRC risk, inflammation is caused by autoimmunity (12). However, even sporadic CRC, initiated by APC loss, depends on “tumor-elicited inflammation,” which originates from localized loss of the intestinal epithelial barrier (13). Barrier disruption results in invasion of early benign tumors (adenomas) by components of the colonic microbiota, which activate IL-23–synthesizing myeloid cells and expand tumor-resident IL-17–producing T lymphocytes (13). Subsequent activation of IL-17 receptor A (IL-17RA) stimulates proliferation of early tumor progenitors and causes adenoma growth (14). Consistent with these experimental findings, epidemiological studies revealed that elevated IL-23 and IL-17 expression in low-grade human CRC predicts rapid progression to fatal metastatic disease (15).
The normal function of the Wnt–β-catenin pathway is to control the proliferation and differentiation of crypt-localized gastrointestinal epithelial stem cells (16). By activating ERK and NF-κB, engagement of IL-17RA augments epithelial proliferation and regeneration after injury (14). Other signaling pathways responsible for epithelial survival and injury repair rely on the key transcriptional regulators STAT3 and YAP (16, 17). Although the role of STAT3 in regeneration and colorectal tumorigenesis is unequivocal (18, 19), it is still debated whether YAP is a tumor suppressor (20) or an oncogenic driver (21). Here we show that Src, YAP, STAT3, and Notch are coordinately activated in mouse APC-deficient intestinal organoids and colonic tumors and in 64% of human CRC specimens. These pathways respond to the dramatic up-regulation of the IL-6 signal transducer (IL-6ST or gp130), a protein that serves as a co-receptor for IL-6, IL-11, and related cytokines. Constitutive gp130 activation in mouse intestinal epithelial cells (IEC) accelerates colorectal tumorigenesis initiated by APC loss. Conversely, inhibition of Src family kinases (SFKs) and JAK tyrosine kinases that maintain YAP and STAT3 activation results in death of CRC progenitors and regression of established tumors.
Results
Concomitant Src, YAP, Notch, and STAT3 Activation in Human CRC.
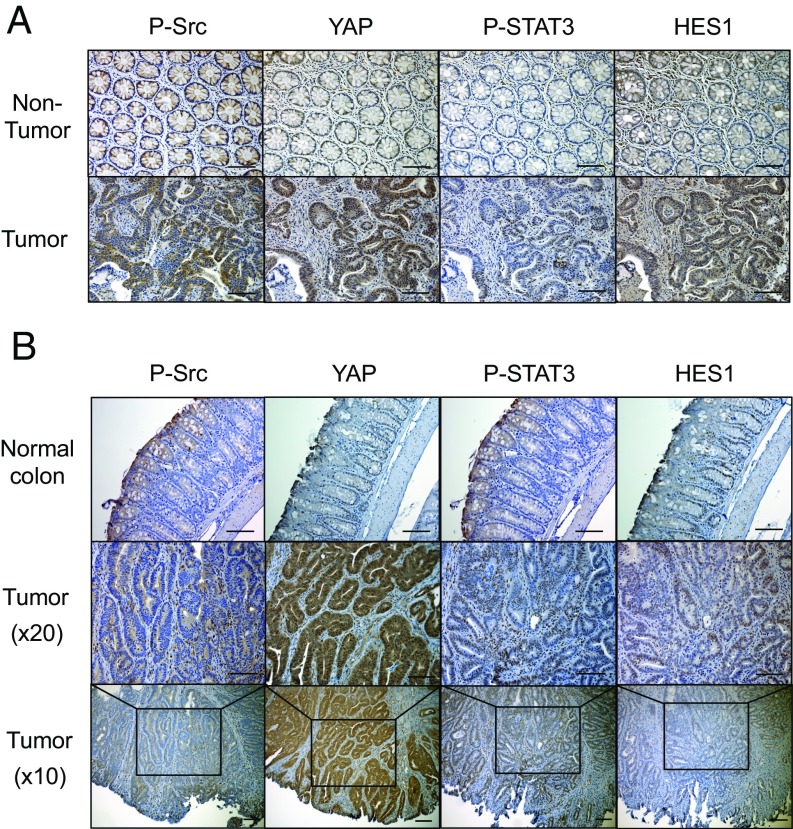
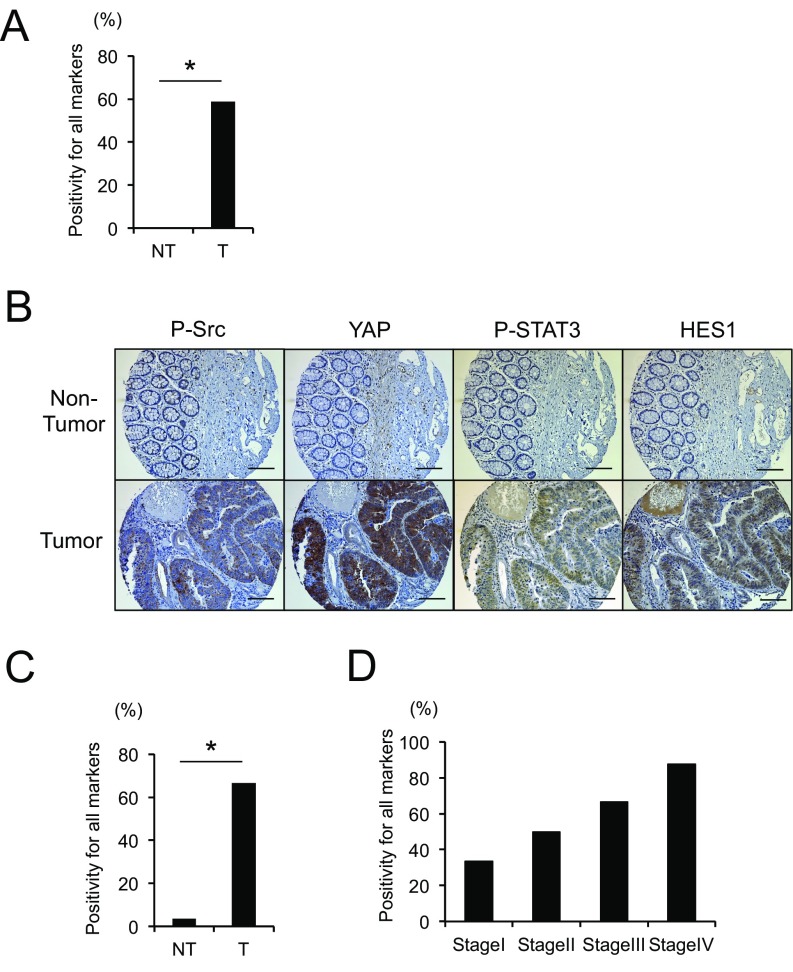
SFKs, YAP, Notch, and STAT3 are critical mediators of inflammation-driven mucosal regeneration and are activated in inflammatory bowel diseases (16, 17), which increase CRC risk (12). To query their involvement in colorectal tumorigenesis, we stained a collection of human CRC surgical specimens (n = 17) with antibodies to phosphorylated Src and STAT3, YAP, and HES1, a Notch target. Strikingly, 59% of the tumors exhibited concomitant activation and up-regulation of all four signaling molecules relative to nontumor tissue (Fig. 1A, Fig. S1A, and Table S1). Analysis of mouse colon tumors induced by APC loss revealed an identical scenario: concomitant Src, STAT3, YAP, and Notch activation in tumor specimens (Fig. 1B). We further confirmed and extended the human data using another cohort of human colon tissue microarrays (TMA). Approximately 67% of these CRC specimens (n = 27) were positive for all four markers, none of which were strongly expressed in normal tissue (Fig. S1 B and C and Table S2). Positivity of all four markers tended to be higher at advanced disease stages (Fig. S1D).
Fig. 1.
Multiple gp130-responsive signaling pathways are activated in human and mouse colorectal cancer. (A) Paraffin-embedded sections of surgically removed human CRC (n = 17) and matched normal colon tissues (n = 7) were stained with P-Src, YAP, P-STAT3, or HES1 antibodies. (B) Paraffin-embedded colon sections from tumor-bearing CPC-APC mice (n = 3) and WT controls (n = 3) were stained as above. (Scale bars, 100 μm.)
Fig. S1.
(A) Human nontumor (NT) and tumor (T) colon tissue specimens were stained with P-Src, YAP, P-STAT3, or HES1 antibodies. The percentage of tumors positive for all four markers is shown. *P < 0.05. (B and C) TMA of human CRC (n = 27) and matched normal colon sections (n = 27) were stained with P-Src, YAP, P-STAT3, or HES1 antibodies. Representative examples of one nontumor and one tumor sample are shown (B). (Scale bars, 100 μm.) The percentage of tumors positive for all four markers was determined (C). *P < 0.05. (D) The percentage of tumors positive for all four markers in each tumor-node-metastasis (TNM) stage is shown.
Table S1.
Age, gender, tumor position, TNM stage, and staining results of the CRC patients used in Fig. 1A and Fig. S1A
| Patient no. | T/NT | Age (y) | Gender | Tumor position | TNM stage | P-STAT3 | P-Src | YAP | HES1 |
| 1 | T | 52 | F | Rectum | T4N2M0 | — | + | + | + |
| 2 | T | 77 | M | Rectum | T4N2M1 | + | + | + | + |
| 2 | NT | — | — | — | — | ||||
| 3 | T | 56 | F | Rectum | T4N2M0 | — | + | + | + |
| 3 | NT | — | — | — | + | ||||
| 4 | T | 34 | M | Rectum | NA | + | + | + | + |
| 5 | T | 47 | M | Rectum | T4N2M0 | — | + | — | + |
| 6 | T | 40 | F | Rectum | T3N1M0 | — | — | + | + |
| 6 | NT | — | — | — | — | ||||
| 7 | T | 38 | F | Rectum | T4N2M0 | + | + | + | + |
| 8 | T | 45 | F | Rectum | T4N2M0 | — | + | — | + |
| 9 | T | 61 | M | Rectum | T2N2M0 | — | — | + | + |
| 9 | NT | — | — | — | — | ||||
| 10 | T | 40 | M | Rectum | NA | + | + | + | + |
| 11 | T | 57 | F | Rectum | T3N1M0 | + | + | + | + |
| 12 | T | 73 | M | Colon | T3N0M0 | + | + | + | + |
| 13 | T | 56 | F | Colon | T3N1aM0 | + | + | + | + |
| 14 | T | 66 | M | Colon | T2N0M0 | — | + | + | + |
| 14 | NT | — | — | — | — | ||||
| 15 | T | 63 | M | Colon | T3N0M1 | + | + | + | + |
| 16 | T | 63 | M | Colon | T3N0M0 | + | + | + | + |
| 16 | NT | — | — | — | — | ||||
| 17 | T | 52 | M | Colon | T3N0M0 | + | + | + | + |
| 17 | NT | — | — | — | — |
Table S2.
Age, gender, tumor position, TNM stage, and staining results of the CRC patients used in Fig. S1 B–D
| Patient no. | T/NT | Age (y) | Gender | Tumor position | Stage | TNM | P-STAT3 | P-Src | YAP | HES1 |
| 1 | T | 80 | F | Colon | I | pT2 N0 cM0 | — | + | + | + |
| 1 | NT | — | — | — | — | |||||
| 2 | T | 60 | M | Rectum | I | pT2 N0 R0 cM0 | — | + | + | + |
| 2 | NT | — | — | — | — | |||||
| 3 | T | 73 | F | Rectum | I | rpT2 V1 cM0 | + | + | + | + |
| 3 | NT | — | — | — | — | |||||
| 4 | T | 78 | M | Colon | II | pT4b pN2a pL1 pV1 Pn1 cM0 | + | + | + | + |
| 4 | NT | — | — | + | — | |||||
| 5 | T | 73 | M | Colon | II | pT3 N0 cM0 | — | + | + | — |
| 5 | NT | — | — | — | — | |||||
| 6 | T | 45 | M | Colon | II | pT4a N0 cM0 | + | + | + | + |
| 6 | NT | — | — | — | — | |||||
| 7 | T | 64 | M | Rectum | II | pT3 N0 cM0 | + | — | — | — |
| 7 | NT | + | + | + | + | |||||
| 8 | T | 55 | M | Colon | III | pT3 N2b cM0 | + | + | — | — |
| 8 | NT | — | — | — | — | |||||
| 9 | T | 68 | M | Colon | III | pT3 N1b R0 L1 V1 MX | + | + | + | + |
| 9 | NT | — | — | + | — | |||||
| 10 | T | 69 | M | Colon | III | L1 pT3 pN2a M0 | + | + | + | + |
| 10 | NT | — | — | — | — | |||||
| 11 | T | 76 | F | Colon | III | pT3 N1 R0 cM0 | — | — | + | — |
| 11 | NT | — | — | — | — | |||||
| 12 | T | 50 | F | Colon | III | pT3 N2b V1 M1 | + | + | + | + |
| 12 | NT | — | — | — | — | |||||
| 13 | T | 68 | M | Colon | III | pT3 N2a cM0 | + | — | — | — |
| 13 | NT | — | — | — | — | |||||
| 14 | T | 66 | M | Rectum | III | pT3 pN1b pL1 pV1 cM0 | + | + | + | + |
| 14 | NT | — | + | — | — | |||||
| 15 | T | 82 | M | Colon | III | pT3 N2b cM0 | + | + | + | + |
| 15 | NT | — | — | — | — | |||||
| 16 | T | 67 | M | Rectum | III | ypT3b N1a cM0 | — | — | — | — |
| 16 | NT | — | — | — | — | |||||
| 17 | T | 82 | M | Rectum | III | pT3 N1b cM0 | + | + | + | + |
| 17 | NT | — | — | — | — | |||||
| 18 | T | 72 | F | Rectum | III | pT3 N 1b | + | + | + | + |
| 18 | NT | — | — | + | — | |||||
| 19 | T | 41 | F | Colon | III | pT3 N1a R0 cM0 | + | + | + | + |
| 19 | NT | — | — | — | — | |||||
| 20 | T | 56 | M | Rectum | IV | ypT3a V1 Pn1 pN1b cM0 | — | — | — | — |
| 20 | NT | — | — | — | — | |||||
| 21 | T | 75 | M | Colon | IV | pT3 N2 cM1 | + | + | + | + |
| 21 | NT | — | — | — | — | |||||
| 22 | T | 67 | F | Colon | IV | pT3 N1b M1b | + | + | + | + |
| 22 | NT | + | — | + | — | |||||
| 23 | T | 70 | M | Rectum | IV | pT4b N1c M1a V1 | + | + | + | + |
| 23 | NT | + | + | — | — | |||||
| 24 | T | 86 | F | Colon | IV | pT3 N1b M1 | + | + | + | + |
| 24 | NT | — | — | + | + | |||||
| 25 | T | 58 | F | Colon | IV | pT3 N1b L1 V1 M1 | + | + | + | + |
| 25 | NT | — | — | — | — | |||||
| 26 | T | 72 | F | Colon | IV | pT3 N1(1/17) R0 L1 Pn1 | + | + | + | + |
| 26 | NT | — | + | — | — | |||||
| 27 | T | 74 | M | Rectum | IV | ypT3 ypN0 cM1 | + | + | + | + |
| 27 | NT | — | — | — | — |
APC Loss Results in Src, YAP, Notch, and STAT3 Activation.
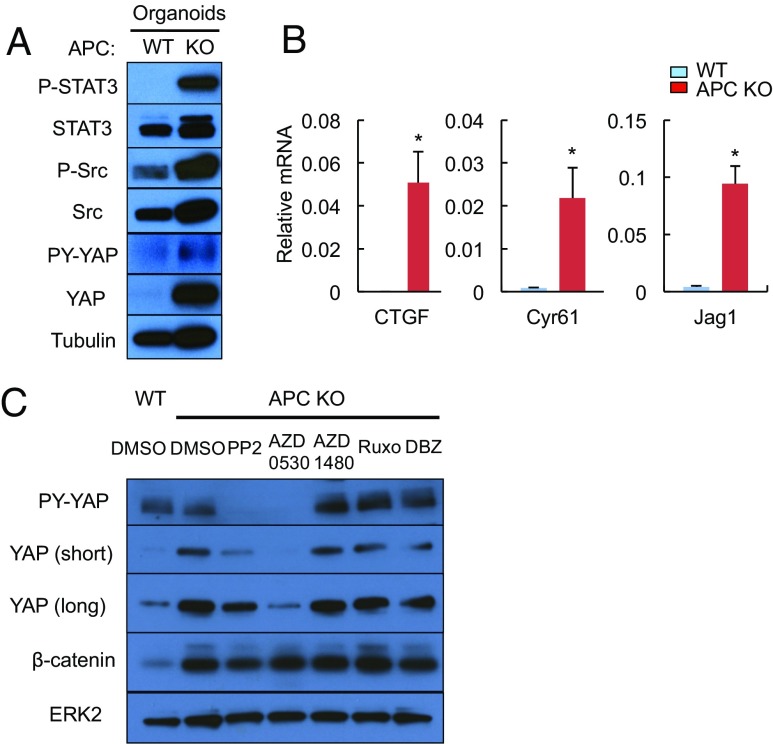
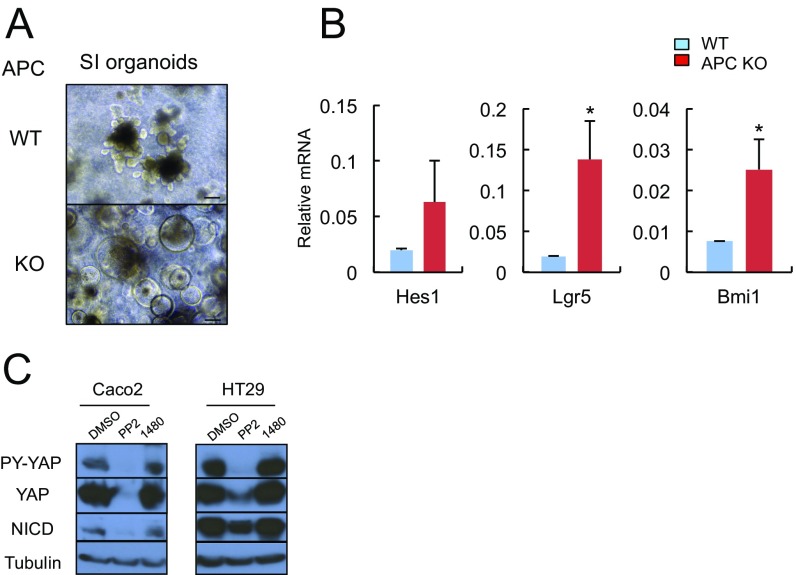
APC inactivation is the most common initiating event in human CRC development (22), which in addition to β-catenin stabilization was found to activate YAP through an ill-defined mechanism (23, 24). To examine whether APC loss is responsible for STAT3, SFK, YAP, and Notch activation, we established WT and Apc−/− mouse small intestinal (SI) organoids (enteroids) by transducing ApcF/F organoids with Adeno-Cre virus. APC-null enteroids exhibited increased STAT3 and Src tyrosine (Y) phosphorylation (Fig. 2A). YAP expression and Y phosphorylation were also up-regulated, along with mRNAs encoding YAP targets, connective tissue growth factor (Ctgf) and Cyr61, and the Notch ligand Jag1 (Fig. 2 A and B). Apc−/− organoids exhibited balloon-like morphology and up-regulation of mRNAs encoding the stem cell markers Lgr5 and Bmi1 (Fig. S2 A and B). Treatment with SFK inhibitors, PP2 and AZD0530, suppressed YAP expression and Y phosphorylation in Apc−/−enteroids, but had a minimal effect on β-catenin activation (Fig. 2C). The Src inhibitor PP2 also inhibited YAP and Notch activation in human CRC cell lines (Fig. S2C). The JAK1/2 inhibitors, AZD1480 and Ruxolitinib, or the Notch/γ-secretase inhibitor dibenzazepine (DBZ), did not affect YAP or β-catenin activation (Fig. 2C).
Fig. 2.
APC ablation results in SFK-dependent YAP activation in intestinal organoids. (A) WT and APC-null SI organoids were lysed and analyzed for expression and phosphorylation of the indicated proteins by immunoblotting (IB). (B) RNAs from WT and APC-null enteroids and indicated transcripts were analyzed by quantitative RT-PCR (qRT-PCR). Results are means ± SEM (n = 3). *P < 0.05. (C) WT and APC-null enteroids were treated with the indicated inhibitors (PP2, AZD0530, and DBZ at 10 μM, and AZD1480 and Ruxolitinib at 3 μM) or vehicle (DMSO) for 24 h. Total lysates were IB-analyzed with the indicated antibodies.
Fig. S2.
(A) Representative images of WT and Apc−/− SI organoids. (Scale bars, 100 μm.) (B) RNA extracted from WT and Apc−/− SI organoids was analyzed by qRT-PCR for expression of the indicated mRNAs. Results are means ± SEM (n = 3). *P < 0.05. (C) Caco2 and HT29 human CRC cells were treated with the indicated inhibitors (PP2, a SFK inhibitor, at 10 μM; and AZD1480, a JAK inhibitor, at 1 μM) for 24 h. Total cell lysates were IB-analyzed for expression and phosphorylation of the indicated proteins.
APC Loss Induces gp130 and IL-11R Up-Regulation.
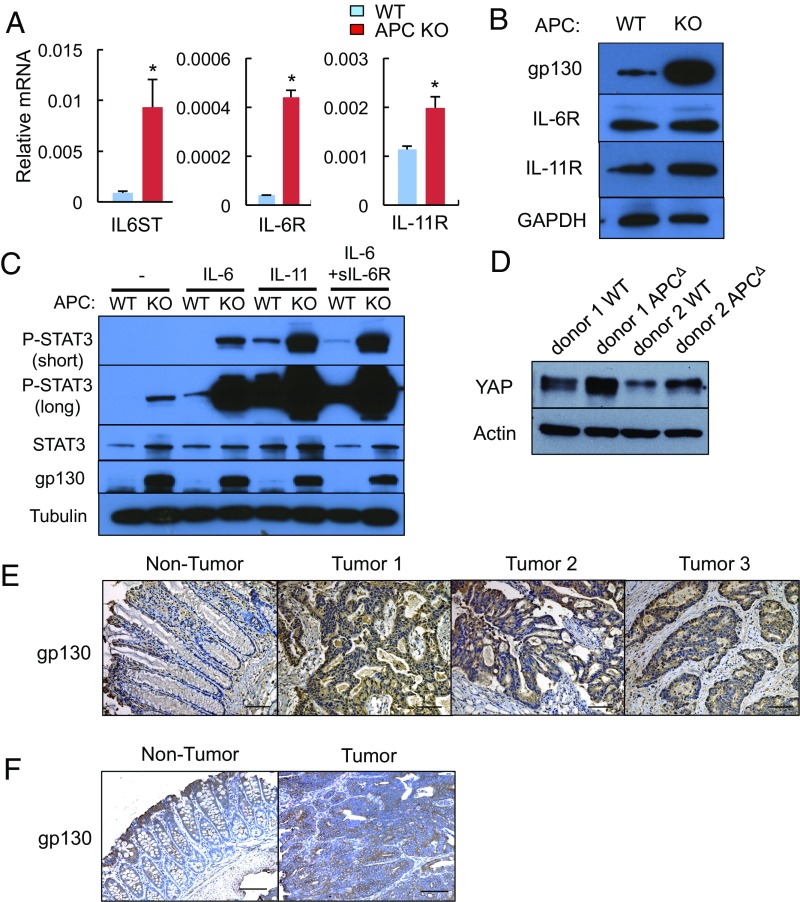
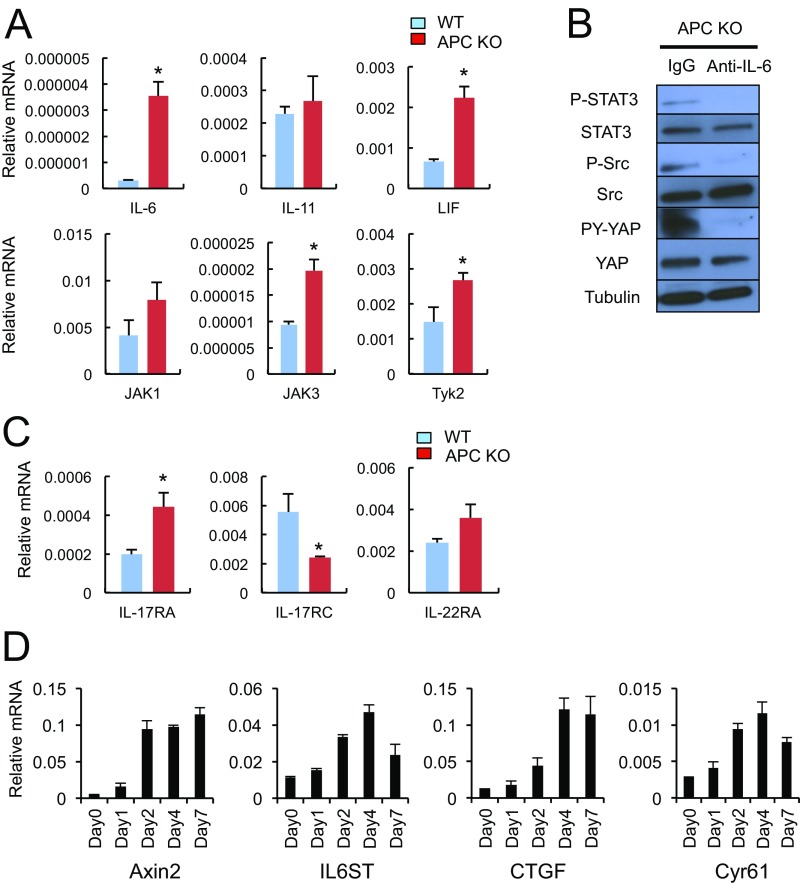
To determine how APC loss activates STAT3, SFKs, YAP, and Notch, we examined the expression status of gp130-related molecules known to activate all of the aforementioned signaling pathways (17). APC loss led to dramatic up-regulation of Il6st (gp130) and Il6r mRNAs and a more modest increase in Il11r mRNA (Fig. 3A). Substantial gp130 and IL-11R up-regulation was also observed at the protein level, whereas IL-6R expression was only modestly increased (Fig. 3B). Addition of exogenous IL-6, IL-11, or soluble IL-6R (sIL-6R) plus IL-6 to Apc−/− enteroids led to a further increase in STAT3 phosphorylation without affecting gp130 expression (Fig. 3C). APC-deficient enteroids also exhibited elevated Il6 and Lif mRNAs (Fig. S3A) and IL-6 neutralization attenuated STAT3, Src, and YAP activation (Fig. S3B). APC-deficient enteroids exhibited moderately elevated expression of Il17ra and Il22ra mRNAs, but lower expression of Il17rc mRNA (Fig. S3C). Time-course analysis using ApcF/F organoids that express 4-hydroxytamoxifen (4-OHT)–regulated Cre recombinase revealed strong induction of Axin2, a direct β-catenin target (7), peaking 2 d after 4-OHT addition (Fig. S3D). Il6st (gp130) mRNA was also up-regulated, but its induction peaked on day 4 and paralleled induction of the classic YAP targets Ctgf and Cyr61 (Fig. S3D).
Fig. 3.
APC ablation up-regulates gp130, IL-6R, and IL-11R expression. (A) WT and APC-null SI enteroids were analyzed for expression of the indicated mRNAs by qRT-PCR. Results are means ± SEM (n = 3). *P < 0.05. (B) WT and APC-null SI enteroids were lysed and IB-analyzed for the indicated proteins. (C) WT and Apc−/− SI enteroids were stimulated with IL-6 (100 ng/mL), IL-11 (100 ng/mL), or IL-6 (100 ng/mL) + sIL-6R (100 ng/mL) for 30 min, lysed, and IB-analyzed for protein expression and phosphorylation. (D) WT and APC-deficient human colon organoids were lysed and IB-analyzed for the indicated proteins. (E) Paraffin-embedded sections of surgically removed human CRC (n = 17) and matched normal colon sections (n = 7) were stained with a gp130 antibody; 65% of the cancer samples (11 of 17) showed similar gp130 positivity to the three examples depicted here. (F) Paraffin-embedded colon sections from tumor-bearing CPC-APC mice were stained with a gp130 antibody. (Scale bars, 100 μm.)
Fig. S3.
(A) RNA extracted from WT and Apc−/− SI organoids was analyzed by qRT-PCR for expression of the indicated mRNA species. Results are means ± SEM (n = 3). *P < 0.05. (B) Apc−/− SI organoids were incubated with neutralizing IL-6 antibody or control IgG for 3 d, after which expression and phosphorylation of the indicated proteins was examined by IB analysis. (C) RNA extracted from WT and Apc−/− enteroids was analyzed by qRT-PCR for expression of the indicated mRNA species. Results are means ± SEM (n = 3). *P < 0.05. (D) RNAs extracted from Villin-CreERT2 x ApcF/F enteroids treated with 4-OHT at 1 μM for the indicated times were analyzed by qRT-PCR for expression of the indicated mRNA species. Results are means ± SEM (n = 3).
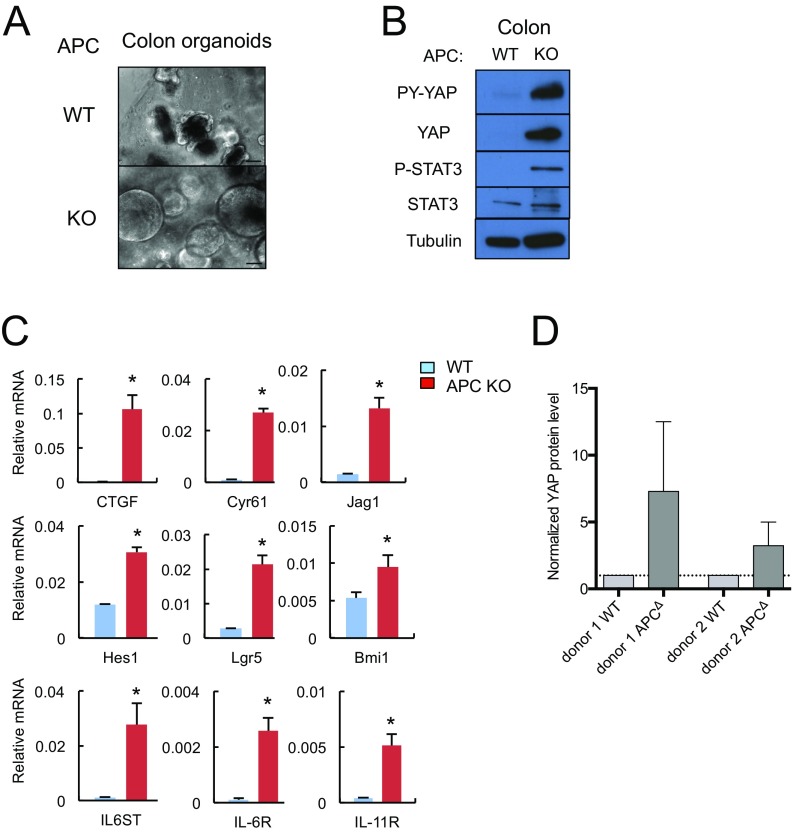
We also prepared WT and Apc−/− colon enteroids whose morphological features paralleled those of small intestinal enteroids (Fig. S4A). Apc−/− colon enteroids showed YAP and STAT3 activation and up-regulation of mRNAs encoding CTGF, Cyr61, Jag1, Hes1, Lgr5, Bmi1, gp130, IL-6R, and IL-11R (Fig. S4 B and C). Human colon organoids rendered APC deficient also showed YAP up-regulation (Fig. 3D and Fig. S4D). Of note, epithelial gp130 expression was markedly increased in both human CRC specimens and mouse colon tumors (Fig. 3 E and F). Stromal expression of gp130 remained low.
Fig. S4.
(A) Representative images of WT and Apc−/− mouse colon organoids. (Scale bars, 100 μm.) (B) WT and Apc−/− colon organoids were lysed and analyzed for expression and phosphorylation of the indicated proteins. (C) RNA extracted from WT and Apc−/− colon organoids was analyzed by qRT-PCR for expression of the indicated mRNA species. Results are means ± SEM (n = 3). *P < 0.05. (D) Densitometric measurement of YAP protein levels in human colon organoids shown in Fig. 3D was performed using ImageJ and normalized to actin. Results are means of four IB experiments ± SD (n = 4).
A Positive Autoregulatory Loop Controls gp130 and YAP Expression.
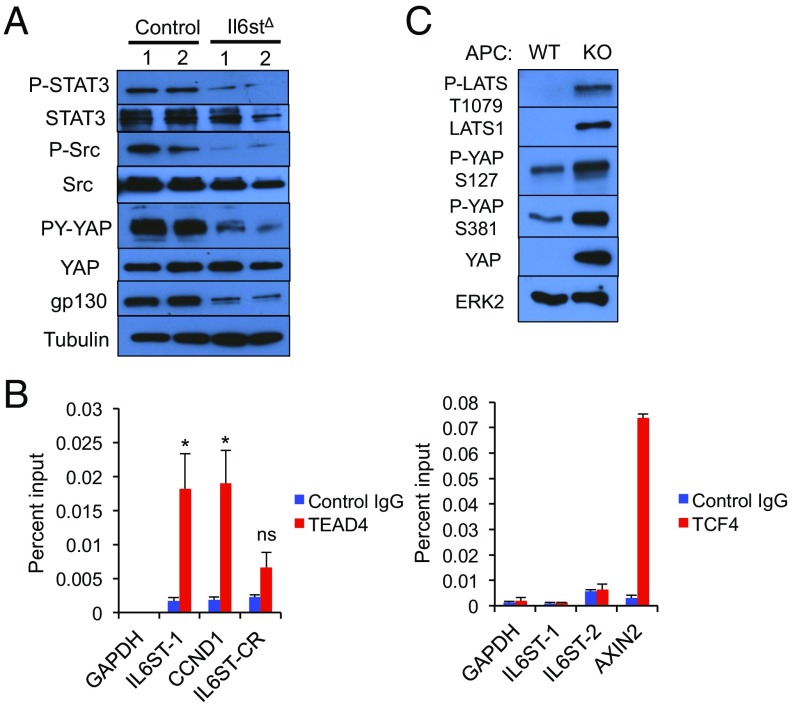
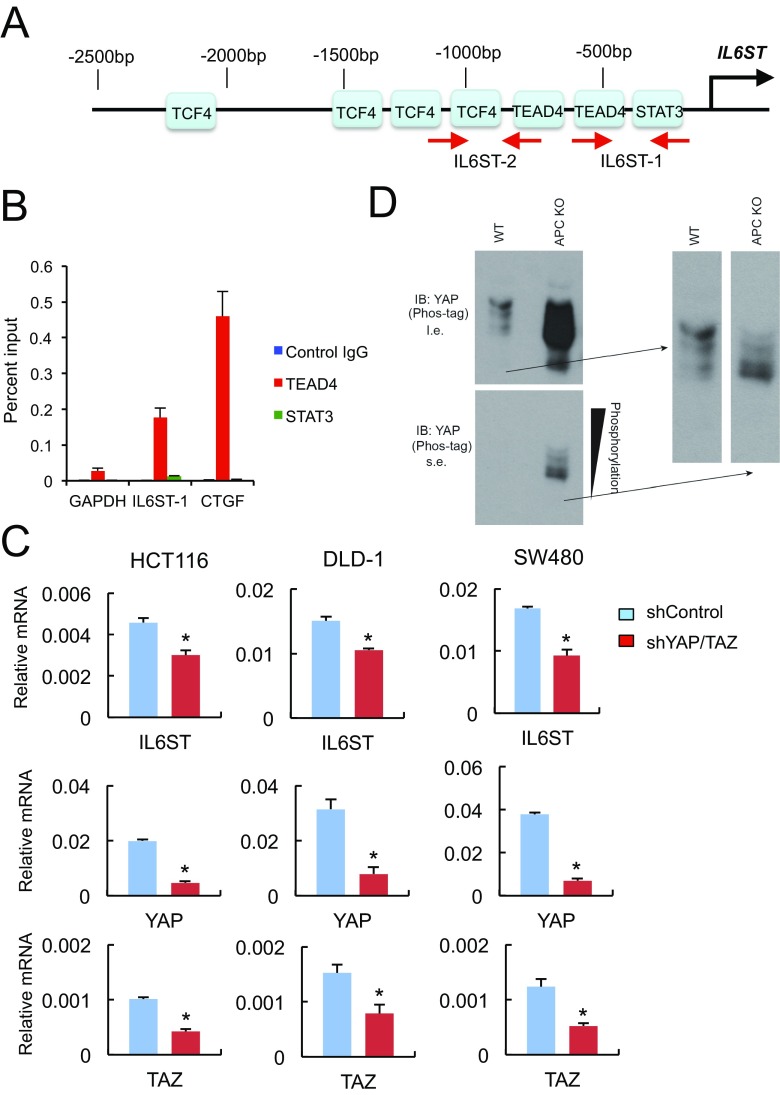
To determine the role of gp130 in activation of the signaling pathways described above, we silenced its expression in Apc−/− organoids with CRISPR/Cas9 technology. The gp130 deficiency inhibited STAT3, SFKs and the activating Y phosphorylation of YAP (Fig. 4A), suggesting that gp130 acts upstream to STAT3, SFKs, and YAP. Next, we examined the mechanisms responsible for gp130 up-regulation in APC ablated organoids. Scanning of the IL6ST gene regulatory region revealed putative binding sites for transcription factor 4 (TCF4) and TEA domain transcription factor 4 (TEAD4), which are binding partners for β-catenin and YAP, respectively, and STAT3 (Fig. S5A). To determine which of these binding sites are occupied in human CRC, we performed ChIP experiments on DLD-1 CRC cells. The only transcription factor found to occupy the IL6ST regulatory region was TEAD4 (Fig. 4B). No significant binding of TCF4 or STAT3 was observed, even after incubation of DLD-1 cells with IL-6 (Fig. 4B and Fig. S5B). To validate the importance of these results, we silenced expression of YAP and its paralog TAZ, whose expression is up-regulated in the absence of YAP (25), in different human CRC cell lines. Down-regulation of YAP and TAZ resulted in decreased IL6ST mRNA expression in these cells (Fig. S5C).
Fig. 4.
A YAP-IL-6ST autoregulatory loop. (A) Apc−/− and Il6st-ablated Apc−/− enteroids were lysed and analyzed for expression and phosphorylation of the indicated proteins. (B) DLD-1 cells were analyzed by ChIP using TEAD4 (Left) and TCF4 (Right) antibodies and control IgG for occupancy of the IL6ST regulatory region. The GAPDH, CCND1, and AXIN2 promoters served as negative and positive controls. DLD-1 cells were stimulated with 10% (vol/vol) FBS before ChIP analysis. The precipitated DNA was quantitated by real-time PCR with primers specific for the promoter regions or a control region (CR) of the indicated genes. (Left) Data are means ± SEM of three independent experiments. P values were determined using one-way ANOVA test followed by Tukey's multiple comparison test, *P < 0.05; ns, not significant (P > 0.05). (Right) Data are means ± SEM of duplicates from a representative experiment. (C) WT and APC-null enteroids were lysed and analyzed for expression and phosphorylation of the indicated proteins.
Fig. S5.
(A) Putative binding sites for TCF4, TEAD4, and STAT3 in the IL6ST regulatory region. The location of primers used in ChIP analysis is indicated with red arrows. (B) DLD-1 cells incubated with fresh 10% (vol/vol) FBS for 2 h and 50 ng/mL IL-6 for 30 min were subjected to ChIP analysis with antibodies to TEAD4, STAT3, or control IgG, and the precipitated DNA was quantitated by real-time PCR with primers specific for the promoter regions or an intergenic sequence of the indicated genes. Data are means ± SEM of duplicates from a representative experiment. The GAPDH and CTGF promoters served as negative and positive controls. (C) RNA extracted from human CRC cell lines (HCT116, DLD-1, and SW480) infected with lentiviruses expressing shRNAs against YAP and TAZ (shYAP/TAZ) or control (shControl) was analyzed by qRT-PCR for expression of the indicated mRNAs. Results are means ± SEM (n = 3). *P < 0.05. (D) Cell lysates were subjected to immunoblotting with a YAP antibody. Gels containing phos-tag were used for separation of phospho-isoforms and assessment of YAP phosphorylation status. l.e., long exposure; s.e., short exposure.
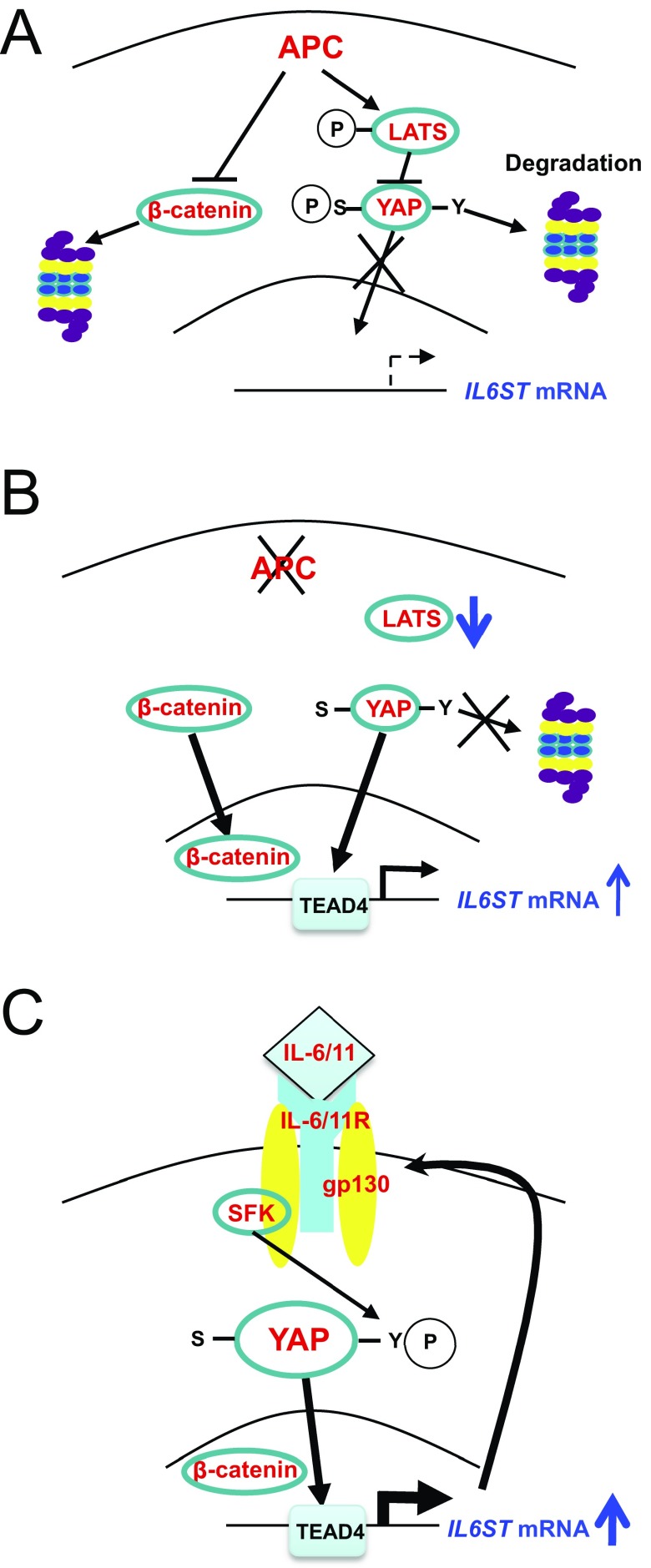
Given these results, which implicate YAP in gp130 induction and gp130 in YAP activation, we examined in closer detail whether Hippo pathway activity, which negatively regulates YAP, is perturbed by APC ablation. Immunoblot analysis indicated that APC ablation actually led to up-regulation of large tumor suppressor 1 (LATS1), a component of the mammalian Hippo pathway, which was phosphorylated at T1079 (Fig. 4C). Paralleling the increase in LATS1 expression, APC ablation increased YAP phosphorylation at the LATS phospho-acceptor sites S127 and S381, whose phosphorylation inhibits YAP nuclear translocation and promotes its proteasomal degradation (Fig. 4C). These results further support the importance of SFK-mediated Y phosphorylation, which results in YAP activation even in cells with intact Hippo signaling (17), and show that Hippo pathway inactivation cannot fully account for YAP up-regulation. However, we found that YAP was already phosphorylated on the inhibitory serines in WT enteroids and that the fold-increase in its S phosphorylation seemed lower than the overall increase in its expression (Fig. 4C). To further examine this point, we separated YAP phospho-isoforms on Phos-tag SDS/PAGE gels, which revealed that the relative ratio of underphosphorylated YAP to total YAP in APC-deficient enteroids was higher than in WT enteroids (Fig. S5D). These results are consistent with the previous report that APC ablation results in decreased LATS1 activity (23), and suggest that this may only give rise to the initial surge in YAP activity that induces Il6st transcription.
gp130 Activation Accelerates CRC Development, Which Depends on SFK and JAK Activities.
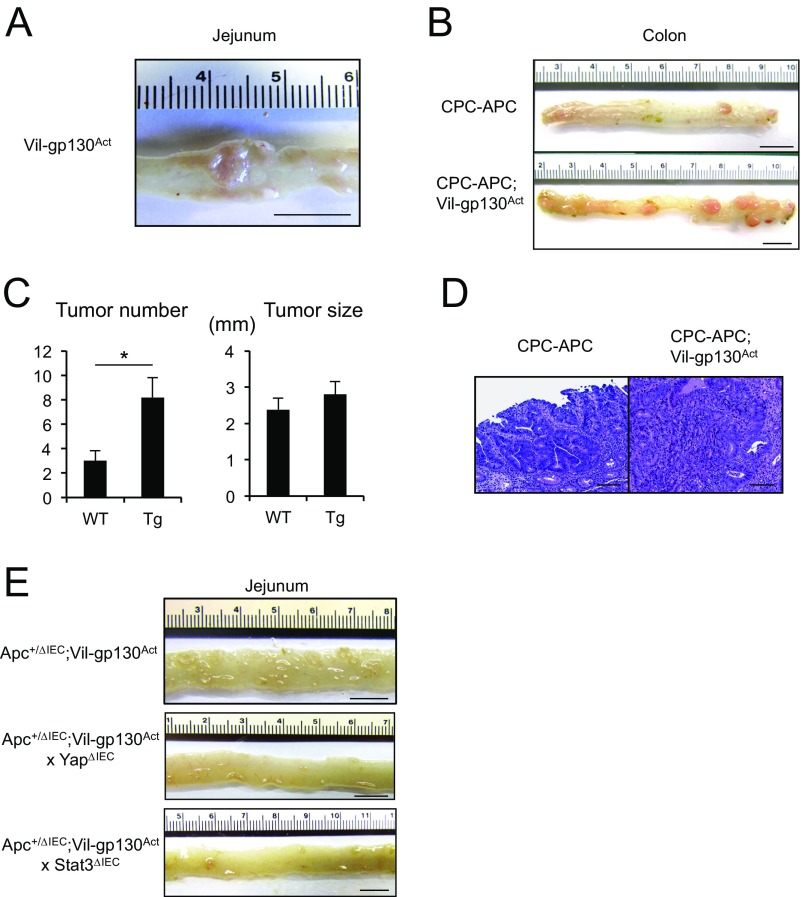
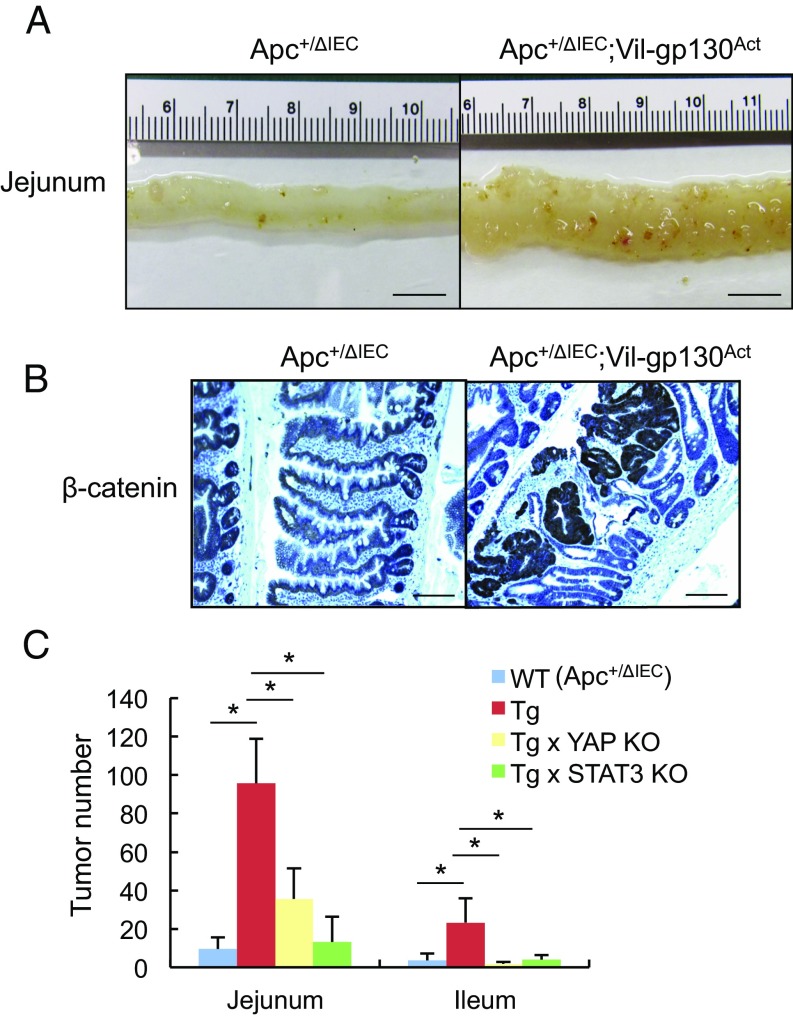
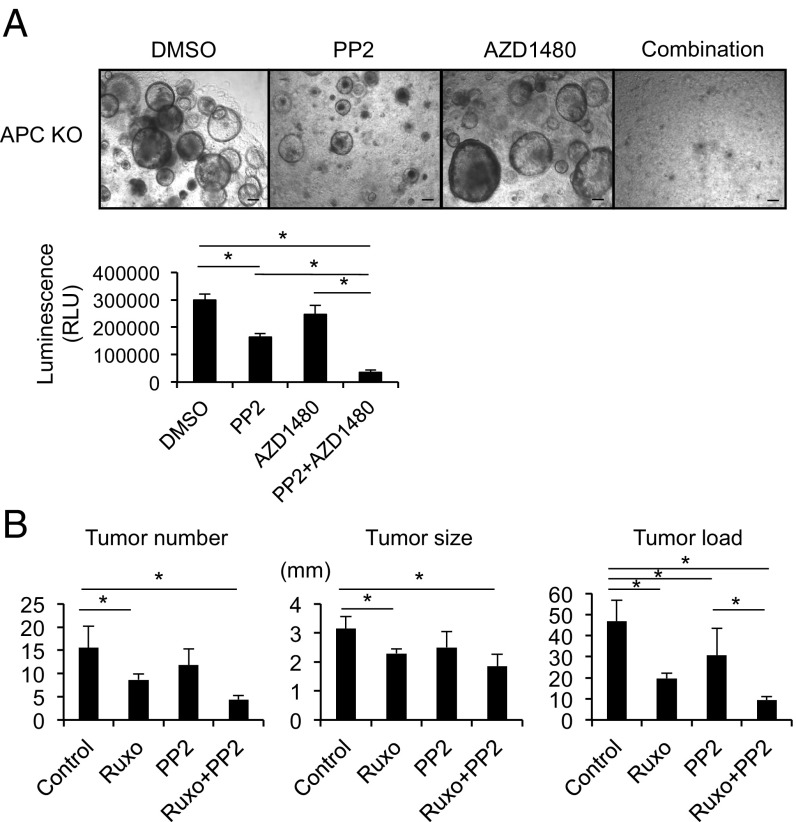
Expression of constitutively active gp130 (gp130Act) in IEC using the Villin promoter activates STAT3, SFK, YAP, and Notch signaling (17). When mice expressing gp130Act in IEC reached 12–15 mo of age, 30% of them developed SI tumors (Fig. S6A). However, introduction of the Villin-gp130Act transgene into Apc+/ΔIEC mice, in which loss of WT Apc initiates intestinal tumorigenesis (26), augmented and accelerated formation of small intestinal and colonic adenomas harboring activated β-catenin (Fig. 5 and Fig. S6 B–D). Enhanced tumor development was dependent on YAP and STAT3, as ablation of either molecule inhibited tumorigenesis in Apc+/ΔIEC;Vil-gp130Act mice (Fig. 5C and Fig. S6E). To determine the effect of simultaneous SFK and JAK inhibition on CRC development, we treated Apc−/− enteroids and organoids established from a spontaneous SI tumor in Vil-gp130Act mice with SFK and/or JAK inhibitors. Whereas SFK inhibition reduced organoid size and survival, JAK1/2 inhibition had a rather marginal effect (Fig. 6A and Fig. S7). However, together the SFK and JAK inhibitors led to extensive organoid death. To extend these results to our in vivo colon tumor system, we treated 3.5-mo-old tumor-bearing CPC-APC (Cdx2-Cre × ApcF/+) mice with PP2 and/or Ruxolitinib for 1.5 mo. Whereas each inhibitor alone led to modest tumor regression, the two inhibitors together resulted in a reduction in tumor load, although the difference between the Ruxolitinib and Ruxolitinib + PP2 groups was not statistically significant (Fig. 6B). No obvious damage to nontumor tissue was observed.
Fig. S6.
(A) A representative image showing small intestinal tumors in 12- to 15-mo-old Vil-gp130Act mice. (B) Representative images of tumor bearing CPC-APC and CPC-APC;Vil-gp130Act colons. (C) Tumor number and size in CPC-APC and CPC-APC;Vil-gp130Act colons. Results are means ± SEM (n = 5 per group). *P < 0.05. (D) H&E-stained paraffin-embedded colon sections from CPC-APC and CPC-APC;Vil-gp130Actmice. (E) Representative images of Apc+/ΔIEC;Vil-gp130Act, Apc+/ΔIEC;Vil-gp130Act;YapΔIECand Apc+/ΔIEC;Vil-gp130Act;Stat3ΔIEC SI. (Scale bars: 10 mm in A, B, and E; 100 μm in D.)
Fig. 5.
YAP or STAT3 ablation inhibits intestinal tumorigenesis. (A) Representative images of Apc+/ΔIEC and Apc+/ΔIEC;Vil-gp130Act SIs. (Scale bars, 10 mm.) (B) β-Catenin staining of paraffin-embedded SI sections from Apc+/ΔIEC and Apc+/ΔIEC;Vil-gp130Act mice. (Scale bars, 100 μm.) (C) Tumor numbers in the jejunum and ileum of the indicated mouse strains (n = 5–8 per group). Results are means ± SEM; *P < 0.05.
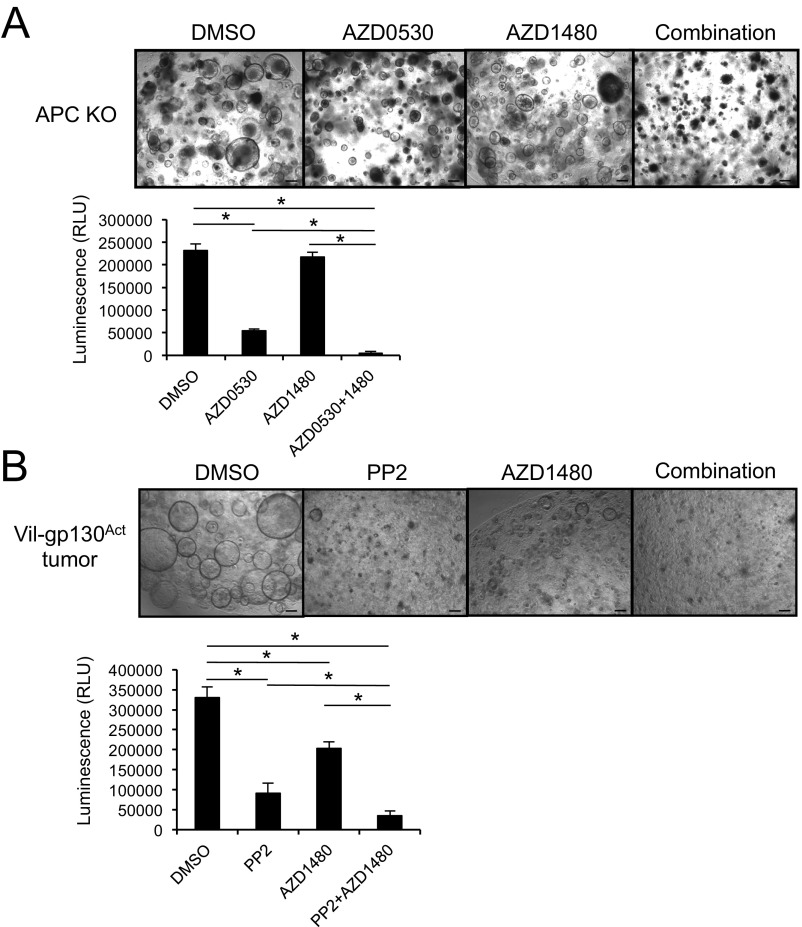
Fig. 6.
Combined treatment with SFK and JAK inhibitors causes regression of colorectal tumors. (A) Representative images of Apc−/− enteroids treated with the indicated inhibitors (PP2, a SFK inhibitor at 10 μM; and AZD1480, a JAK inhibitor at 3 μM) for 3 d. Cell viability was measured using a CellTiter-Glo assay. Results are means ± SEM; *P < 0.05. (Scale bars, 100 μm.) (B) Tumor number, size, and load in CPC-APC mice treated with the indicated kinase inhibitors for 1.5 mo starting at 3.5 mo of age (n = 5–6 per group). Results are means ± SEM; *P < 0.05.
Fig. S7.
(A) Representative images of Apc−/− enteroids treated with the indicated kinase inhibitors (AZD0530, a SFK inhibitor at 10 μM; and AZD1480 at 3 μM) for 3 d. Cell viability was measured using the CellTiter-Glo assay. Results are means ± SEM. *P < 0.05. (B) Representative images of enteroids derived from a Vil-gp130Act-induced small intestinal tumor that was treated with the indicated inhibitors (PP2 at 10 μM, and AZD1480 at 3 μM) for 3 d. Cell viability was measured using the CellTiter-Glo assay. Results are means ± SEM. *P < 0.05. (Scale bars, 100 μm.)
Discussion
The results described above shed new light on the mechanism of CRC initiation and identify novel and broadly important targets for its treatment. CRC is rather unique in its almost exclusive dependence on initiating APC loss-of-function mutations (7, 9). Although APC inactivation results in β-catenin activation, CTNNB1 gain-of-function mutations, which occur in 40% of hepatocellular carcinomas (27), are rare in CRC (8). Our results demonstrate that APC loss, as opposed to CTNNB1 activation, provides additional selective advantages to CRC-initiating cells because it makes them highly responsive to local inflammatory signals provided by members of the IL-6 cytokine family. By up-regulating gp130 expression, APC loss results in heightened sensitivity to IL-6, IL-11, and sIL-6R, which act as tumor promoters in mouse models (28, 29) and are up-regulated in human CRC (18). Whereas IL-6 is induced upon activation of IL-17RA signaling (14), IL-11 is provided by cancer-associated fibroblasts (30). The discrepancy between high IL-6R mRNA and hardly elevated cell surface protein expression might be a result of increased IL-6R shedding. Indeed, the finding that IL-6 + sIL-6R elicits a stronger response than IL-6 alone suggests more gpl30 on the cell surface than IL-6R.
In addition to activation of STAT3, a transcription factor that is of great importance in CRC establishment (18, 29), IL-6, sIL-6R, and IL-11 activate SFKs that support Hippo-independent YAP activation and thereby induce Notch receptors and ligands (17). Curiously, however, the initial signal that leads to YAP activation in APC-deficient cells and consequent induction of IL-6ST/gp130 expression seems to be loss of LATS1 activity (23), a serine/threonine kinase that promotes YAP nuclear exclusion and degradation (31). Given the small decrease in LATS activity (23), the majority of YAP activation in mouse colorectal tumors and human CRC cell lines is propagated by SFK-mediated tyrosine phosphorylation, which induces YAP nuclear translocation, even in cells that harbor active LATS1. Treatment of APC-deficient organoids or human CRC cells with SFK inhibitors results in a marked decrease in YAP expression as well as inhibition of Notch activation. Of note, an independent and unbiased screen for compounds that inhibit YAP activation in breast cancer has netted the clinically approved SFK and c-Abl inhibitor, Dasatinib (32). Thus, the initial spike in YAP activity, which was proposed to take place upon disruption of a complex between APC and LATS1 (23), gives rise to much greater Hippo-independent and SFK-dependent YAP activation through the induction of IL6ST gene transcription and up-regulation of gp130 expression (Fig. S8). Up-regulation of gp130 connects YAP and its downstream targets to localized inflammatory signals that are provided by IL-6 family members. Our findings re-enforce the important oncogenic function of YAP (21) and explain the basis for its frequent activation in CRC and possibly other epithelial tumors that are devoid of mutations that disrupt Hippo signaling. Indeed, the SFK–YAP module also functions in skin cancer (33). YAP up-regulation in epithelial cancers is also known to account for acquired drug resistance (21), but the underlying mechanisms were not reported. Our results indicate that at least in CRC, the major mechanism responsible for sustained YAP activation is the YAP–IL-6ST–SFK autoregulatory loop described above. Given the strong correlation between YAP and SFK activation in 60–70% of human CRC, this amplification loop, which links APC loss to YAP and STAT3 activation, is of great clinical relevance. Indeed, a combined treatment with SFK and JAK1/2 inhibitors that block YAP and STAT3 activation, respectively, results in substantial regression of colorectal tumors that were initiated by Apc ablation. These findings strongly support the merit of testing such compounds, several of which have been clinically approved, in advanced human CRC, which currently accounts for 700,000 yearly deaths worldwide (1).
Fig. S8.
(A–C) Schematic representation of how YAP is initially activated upon APC loss and IL6ST mRNA is induced to promote gp130 signaling, which leads to activation of SFKs, that promotes sustained activation and up-regulation of YAP.
Of further interest is the observation that two different cohorts of CRC patients, one collected in China and the other in Austria, who differ in their ethnicity and dietary habits, exhibit nearly identical rates of SFK, STAT3, YAP, and Notch activation. These findings suggest that none of these parameters are strongly influenced by the gut microbiota. Indeed, APC ablation in vivo (i.e., in the presence of microbiota) had the same effect on these signaling pathways as its ablation in germ-free organoids.
Methods
Human Colon Samples.
Human colorectal cancer tissues were collected through a study that was approved by the Institutional Review Board of Elitehealth Institute, Guangzhou, China. Written informed consent was obtained from all study participants. Tissue samples were collected and processed at the time of surgery, and were used for staining. Additional tissue specimens were obtained with informed consent from CRC patients at the Medical University of Graz, with approval from the local ethics committee and used to construct TMAs, as described in SI Methods.
Mice.
Villin-Cre mice were obtained from the Jackson Laboratory. Villin- gp130Act transgenic (Tg), Stat3F/F, YapF/F, and Villin-CreERT2 mice have been described previously (17, 34–36). ApcF/F, Cdx2-Cre (CPC), and Cdx2-CreERT mice were described previously (26, 37). All mice were on the C57BL/6 background and were maintained in filter-topped cages on autoclaved food and water at the University of California, San Diego (UCSD) according to NIH guidelines. All experimental procedures were reviewed and approved by the UCSD Institutional Animal Care and Use Committee and all experiments were performed in accordance with UCSD and NIH guidelines and regulations.
Detailed information about experimental procedures, human samples, mice, reagents, and statistical analyses can be found in SI Methods.
SI Methods
Human Colon Samples.
Human colorectal cancer tissues were collected through a study that was approved by the Institutional Review Board of Elitehealth Institute, Guangzhou, China. Written informed consent was obtained from all study participants. We used 17 fresh cancer tissues and matched normal tissue from patients with CRC who underwent surgical resection between June 1, 2009 and June 30, 2012. Tissue samples were collected and processed at the time of surgery.
Tissue Microarrays.
Tumor material was obtained with informed consent from CRC patients at the Medical University of Graz with approval from the local ethics committee and the ethics committee of the St. John of God Hospital Graz (23-015 ex 10/11). Tumor staging was reviewed by an experienced, board-certified pathologist (J.H.) using H&E-stained sections and relevant tumor areas were marked on the slide. Tissue cones of 1.2 mm in diameter were punched out from the chosen tumor area and embedded as an array in a fresh paraffin block. Tissue sections were cut at 4 µm, and mounted on adhesive-coated glass slides compatible for immunohistochemical staining and analysis. The CRC TMA was composed of 346 tissue spots including carcinoma and healthy tissue of 17 patients suffering from colon cancer and 10 patients with rectal cancer.
Mice.
Villin-Cre mice were obtained from the Jackson Laboratory. Villin- gp130Act transgenic (Tg), Stat3F/F, YapF/F, and Villin-CreERT2 mice have been described previously (17, 34–36). ApcF/F, Cdx2-Cre (CPC), and Cdx2-CreERT mice were described previously (26, 37). All mice were on the C57BL/6 background and were maintained in filter-topped cages on autoclaved food and water at the UCSD according to NIH guidelines. Beddings were interchanged between the different strains to minimize microbiome alterations. All experimental procedures were reviewed and approved by the UCSD Institutional Animal Care and Use Committee and all experiments were performed in accordance with UCSD and NIH guidelines and regulations. Analysis of spontaneous CRC tumorigenesis was done as previously described (13, 26).
In Vivo Treatment with Inhibitors.
CPC-APC (Cdx2-Cre x ApcF/+) mice (3.5-mo-old) were treated with either Ruxolitinib, PP2 alone, or in combination for 1.5 mo. PP2 (Sigma) was injected intraperitoneally at 5 mg/kg body weight every day, as previously described (38). Ruxolitinib was obtained from Incyte and was given in mouse chow at 2,000 mg/kg for the entire course of treatment. The mice were killed after the treatment and the colons were collected for further analysis.
Reagents and Plasmids.
Recombinant Noggin, IL-6, IL-11, and soluble IL-6R (sIL-6R) were purchased from Peprotech, recombinant EGF from Life Technologies, DBZ (Notch/γ-secretase inhibitor) from Axon Medchem, PP2 (Src inhibitor) from Sigma, AZD0530 (Src inhibitor) and AZD1480 (JAK1/2 inhibitor) from Selleck Chemicals, and Ruxolitinib (JAK1/2 inhibitor) from Incyte. 4-OHT was purchased from Sigma. 293T-HA-RspoI-Fc cells were kindly provided by Calvin Kuo, Stanford University, Stanford, CA (39). shYAP and shTAZ lentiviral plasmids were obtained from K.-L.G. (40).
Cell Culture.
HEK293T, DLD-1, HCT116, SW480, HT29, and Caco2 cells were cultured in high-glucose DMEM supplemented with 10% (vol/vol) FBS, penicillin, and streptomycin.
Mouse Intestinal Organoid (Enteroid) Culture.
Intestinal crypts were isolated and cultured as described previously (41, 42). In brief, crypts were isolated from small intestines of WT, ApcF/F, or Villin-CreERT2ApcF/F mice or colons of Cdx2-CreERTApcF/F mice. Organoids were plated in growth factor-reduced (GFR) Matrigel (BD Bioscience) and maintained in advanced DMEM/F12 (Life Technologies) containing B27 and N2 supplements (Life Technologies), 1.25 mM N-acetyl l-cysteine (Sigma), 100 ng/mL Noggin, 50 ng/mL EGF, and 10% (vol/vol) Rspo1-Fc–conditioned medium. APC deletion in organoids was induced with 1 μM 4-OHT (Sigma) (43) or by Adeno-Cre infection (44). After Adeno-Cre infection, the organoids were kept in the medium without R-spondin1 and Noggin for several weeks to remove APC-undeleted organoids and were used for the experiments. Intestinal organoids were treated with 100 ng/mL recombinant IL-6, IL-11, or IL-6 + sIL-6R for 30 min and lysed for immunoblot analysis. Cell viability of organoids was measured using the CellTiter-Glo assay (Promega).
Human Colon Organoid Culture.
Human colon samples were obtained from two healthy donors and organoid cultures were prepared as described previously (42). Samples were obtained with written informed consent, according to the guidelines of the University Cancer Center biobank, Frankfurt. Primary organoid lines were stably transduced with lentiCas9-Blast, a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (Addgene plasmid #52962) (45). APC loss-of-function mutations were introduced by transient transfection of the plasmid gRNA_GFP-T2, a gift from George Church, Harvard Medical School, Boston, MA (Addgene plasmid #41820) that contained the single-guide RNA sequence 5′-TTTGAGCTGTTTGAGGAGG-3′. Transfection and functional selection for growth independent of Wnt/R-spondin was performed as described previously (46). Clonal lines were expanded and confirmed by Sanger sequencing.
ChIP Assay.
DLD-1 cells were cross-linked with 1% formaldehyde for 10 min, and quenched with 0.125 M glycine for 5 min at room temperature. Harvested cells were washed twice with PBS and then lysed in ChIP cell lysis buffer (20 mM Tris⋅HCl pH 8.0, 85 mM KCl, 0.5% Nonidet P-40). The lysates were centrifuged at 2,300 × g for 5 min at 4 °C to pellet the nuclei, and the chromatinized genomic DNAs were then sheared to 300–800 base pairs in average size through sonication. The samples were then centrifuged at 16,000 × g for 10 min at 4 °C to remove debris, and the resulting supernatants were diluted 1:5 in ChIP dilution buffer (20 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). This material was used for immunoprecipitation. On day 1, antibodies to TCF4 (Cell Signaling #2569; 10 μL), TEAD4 (Abcam ab58310; 1 μg), STAT3 (Cell Signaling #12640; 10 μL), or normal control IgG were prebound to 10 μL of protein A/G magnetic beads (Thermo Fisher Scientific #88802) overnight at 4 °C. On day 2, beads were washed three times with 5 mg/mL BSA/PBS, and immunoprecipitation reactions were carried out with chromatin extracts overnight at 4 °C. Three percent of the chromatin extract was set aside for input. On day 3, beads were washed two times with low-salt wash buffer (20 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100), followed by washing twice with high-salt wash buffer (20 mM Tris⋅HCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100) and once with LiCl wash buffer (250 mM LiCl, 20 mM Tris⋅HCl pH 8.0, 1 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate). Beads were then resuspended in elution buffer (50 mM Tris⋅HCl pH 8.0, 10 mM EDTA, 1% SDS) and incubated for 30 min at 65 °C with frequent mixing. The resulting eluate and input samples from day 1 were transferred into new tubes, and reverse cross-linking reactions were carried out overnight at 65 °C. On day 4, samples were treated with RNase A (0.2 mg/mL) for 2 h at 37 °C, followed by proteinase K (0.2 mg/mL) digestion for overnight at 50 °C. On day 5, DNA was ethanol-precipitated and purified using the PCR purification kit (Qiagen). Precipitated DNA was quantitated by real-time PCR analysis. The primer sequences for ChIP analysis (forward and reverse, respectively) are 5′-TTGACTCACCCTGCCCTCAATATC-3′ and 5′-TTTCATTCCATCCAGCCTGGGG-3′ for GAPDH; 5′-CCCCGTTATTCCACAGAAAAATGG-3′ and 5′-TCCCAGACTAGATCGAAGGGTTC-3′ for IL6ST (gp130)-1; 5′-ACGCACGCACATGCCTTATC-3′ and 5′-CCTCAGAAAATGGACCTCAAGG-3′ for IL6ST (gp130)-2; 5′-AAGGCCCTGCTGTAAAAGGT-3′ and 5′-GGGGGCTTTCTTTGAAGC-3′ for AXIN2; 5′-GAGGGGACTAATATTTCCAGCAA-3′ and 5′-TAAAGGGATTTCAGCTTAGCA-3′ for CCND1 (Cyclin D1); 5′-AGGCTAAAGTCATCACGACAGTGTG-3′ and 5′-AGGCTAAAGTCATCACGACAGTGTG-3′ for IL6ST (gp130)-control region (CR); 5′-CTTTGGAGAGTTTCAAGAGCC-3′ and 5′-TCTGTCCACTGACATACATCC-3′ for CTGF. All ChIP signals were normalized to the input (labeled as percent of input on the vertical axis).
Lentiviral Infection.
For lentivirus production, HEK 293T cells were cotransfected with lentiviral vectors and the lentiviral packaging plasmids psPAX2 and pMD2.G. Forty-eight hours after transfection, lentiviral supernatants were filtered through a 0.45-μm filter, supplemented with 8 μg/mL polybrene, and used to infect organoids or human CRC cells. Seventy-two hours after infection, organoids or human CRC cells were selected with 2–4 μg/mL puromycin in culture medium.
CRISPR/Cas9-Mediated Ablation.
The Il6st (gp130) gene was knocked-out with the CRISPR/Cas9 system using two different guide sequences #1 5′-CAGTGAAGTTCGAGCCGCGC-3′ and #2 5′- GGTCTTCCACACGATGTAGC-3′. The CRISPR/Cas9 sequences were separately inserted into the lentiCRISPR v2 vector, a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (Addgene, #52961) (45), and the control vectors contained two different guide RNA sequences against GFP. Lentiviruses were prepared as described above and transduced into intestinal organoids, as previously described (47).
Immunoblot, Real-Time PCR, and Histological Analysis.
Immunochemical, histochemical and molecular biological analysis were performed as described previously (17). Histological analysis was performed with antibodies to phospho-Src (#2101), phospho-STAT3 (#9145), YAP (#4912) and HES1 (#11988) (Cell Signaling), gp130 (C-20) (Santa Cruz), and β-catenin (GTX61089) (GeneTex). Immunoblotting was performed with antibodies to phospho-YAP Y357 (ab62751) (Abcam), β-catenin (610153) (BD), phospho-Src (#2101), Src (#2110), phospho-STAT3 (#9131), phospho-YAP S127 (#4911), YAP (#4912), NICD (#3608), phospho-LATS (#8654), LATS1 (#3477) (Cell Signaling), gp130 (C-20), IL-6R (C-20), IL-11R (H-300), STAT3 (C-20), ERK2 (C-14), YAP (H-125), GAPDH (0411) (Santa Cruz), tubulin (T9026), and actin (A2066) (Sigma). The phos-tag reagents were purchased from Wako Chemicals. Gels containing phos-tag were prepared according to manufacturer’s instructions. YAP proteins can be separated into multiple bands in the presence of phos-tag because of their differential phosphorylation levels, with the more heavily phosphorylated proteins migrating more slowly.
Statistical Analysis.
Data are expressed as means ± SD or means ± SEM. Statistical analysis was conducted using Student's t test for comparison of two datasets with normal distribution, or one-way ANOVA followed by the Tukey–Kramer test for multiple comparisons. Fisher’s exact test was used for comparison of categorical variables. Statistical significance was defined as P < 0.05.
Acknowledgments
We thank Drs. D. Pan (Johns Hopkins University) and S. Akira (Osaka University) for YapF/Fand Stat3F/F mice, respectively; Drs. D. L. Gumucio (University of Michigan) for the 12.4-kb Villin promoter; C. A. O’Brien (University of Arkansas for Medical Sciences) for the gp130-luciferase plasmid; T. Sato (Keio University), H. Clevers (Hubrecht Institute), and Y. Hippo (National Cancer Center Research Institute) for protocols describing intestinal organoid culture; C. Kuo (Stanford University) for R-spondin1-producing cells; J. Zhao [University of California, San Diego (UCSD) Transgenic Mouse and Gene Targeting Core], L. Gapuz, R. Ly, and N. Varki (UCSD Histology Core), N. Hiramatsu, S. Yamachika, S. I. Grivennikov, A. Chang, and T. Lee for technical advice and assistance; and Cell Signaling, Santa Cruz Biotechnology, GeneTex, and Incyte for antibodies and reagents. This work was supported by Postdoctoral Fellowship for Research Abroad and Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science, a Uehara Memorial Foundation Fellowship, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Kanae Foundation for the Promotion of Medical Science, KAKENHI (15K21775), and the “Kibou” Projects (all to K.T.); FIRC for Abroad and iCare fellowship funded from the Italian Association for Cancer Research and Marie Curie Actions European Union–People–COFUND (G.D.C.); and Crohn’s and Colitis Foundation of America SRA#330251 (to E.R.). L.K. is supported by the European Commission Marie Curie 2020 program as a Co-Coordinator within the “ALKATRAS’ project as well as by the Austrian Science Funds FWF; and Grants P26011 and P29251. TMA generation was supported by the Innovative Medicines Initiative Joint Undertaking under Grant 115234 (OncoTrack), resources of which are composed of financial contributions from the European Union’s Seventh Framework Programme (FP7/2007–2013) and European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution (www.imi.europa.eu) (to J.H.). C.D. was supported by SPAR Austria. Research at UCSD was supported by the NIH (AI043477) and Incyte Inc. M. Karin is an American Cancer Society Research Professor and holder of the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases.
Footnotes
K.-L.G. is a co-founder and has an equity interest in Vivace Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. The other authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620290114/-/DCSupplemental.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Pohl M, Schmiegel W. Therapeutic strategies in diseases of the digestive tract—2015 and beyond targeted therapies in colon cancer today and tomorrow. Dig Dis. 2016;34(5):574–579. doi: 10.1159/000445267. [DOI] [PubMed] [Google Scholar]
- 4.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30(3):363–371. doi: 10.1002/stem.1031. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas N; Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407):330–337. [DOI] [PMC free article] [PubMed]
- 9.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 10.Shimizu T, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18(8):2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 11.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Terzic J, Grivennikov S, Karin E, & Karin M (2010) Inflammation and colon cancer. Gastroenterology 138(6):2101–2114 e2105. [DOI] [PubMed]
- 13.Grivennikov SI, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity. 2014;41(6):1052–1063. doi: 10.1016/j.immuni.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 16.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529(7586):307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi K, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519(7541):57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26(1):54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 23.Cai J, Maitra A, Anders RA, Taketo MM, Pan D. β-Catenin destruction complex-independent regulation of Hippo-YAP signaling by APC in intestinal tumorigenesis. Genes Dev. 2015;29(14):1493–1506. doi: 10.1101/gad.264515.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azzolin L, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158(1):157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Moroishi T, et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 2015;29(12):1271–1284. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinoi T, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67(20):9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 27.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46(12):1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 28.Putoczki TL, et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. 2013;24(2):257–271. doi: 10.1016/j.ccr.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Grivennikov SI. IL-11: a prominent pro-tumorigenic member of the IL-6 family. Cancer Cell. 2013;24(2):145–147. doi: 10.1016/j.ccr.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calon A, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oku Y, et al. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio. 2015;5:542–549. doi: 10.1016/j.fob.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, et al. αE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes Dev. 2016;30(7):798–811. doi: 10.1101/gad.274951.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10(1):39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19(1):27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y, et al. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. Am J Pathol. 2013;183(2):493–503. doi: 10.1016/j.ajpath.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam JS, Ino Y, Sakamoto M, Hirohashi S. Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. Clin Cancer Res. 2002;8(7):2430–2436. [PubMed] [Google Scholar]
- 39.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15(6):701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 42.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 43.Koo BK, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9(1):81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 44.Wang N, et al. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One. 2014;9(4):e93608. doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Koo BK, Sasselli V, Clevers H. Retroviral gene expression control in primary organoid cultures. Curr Protoc Stem Cell Bio. 2013;27:Unit 5A.6. doi: 10.1002/9780470151808.sc05a06s27. [DOI] [PubMed] [Google Scholar]