Significance
Reactive oxygen species (ROS) production is induced by multiple environmental stresses in various organisms. In plants, ROS transduce local and systemic signaling for adaptation and tolerance to these stresses. Here we show that red clover necrotic mosaic virus (RCNMV), a plant positive-strand RNA [(+)RNA] virus, hijacks the host’s ROS-generating machinery during infection. An RCNMV replication protein associates with host ROS-generating machinery and triggers intracellular ROS bursts. These bursts are required for robust viral RNA replication. We further show that another (+)RNA virus, brome mosaic virus, also depends on ROS for replication. This study represents an example of diversion of a plant stress-resilience system for robust virus replication.
Keywords: positive-strand RNA virus, viral RNA replication, reactive oxygen species, respiratory burst oxidase homolog, calcium-dependent protein kinase
Abstract
As sessile organisms, plants have to accommodate to rapid changes in their surrounding environment. Reactive oxygen species (ROS) act as signaling molecules to transduce biotic and abiotic stimuli into plant stress adaptations. It is established that a respiratory burst oxidase homolog B of Nicotiana benthamiana (NbRBOHB) produces ROS in response to microbe-associated molecular patterns to inhibit pathogen infection. Plant viruses are also known as causative agents of ROS induction in infected plants; however, the function of ROS in plant–virus interactions remains obscure. Here, we show that the replication of red clover necrotic mosaic virus (RCNMV), a plant positive-strand RNA [(+)RNA] virus, requires NbRBOHB-mediated ROS production. The RCNMV replication protein p27 plays a pivotal role in this process, redirecting the subcellular localization of NbRBOHB and a subgroup II calcium-dependent protein kinase of N. benthamiana (NbCDPKiso2) from the plasma membrane to the p27-containing intracellular aggregate structures. p27 also induces an intracellular ROS burst in an RBOH-dependent manner. NbCDPKiso2 was shown to be an activator of the p27-triggered ROS accumulations and to be required for RCNMV replication. Importantly, this RBOH-derived ROS is essential for robust viral RNA replication. The need for RBOH-derived ROS was demonstrated for the replication of another (+)RNA virus, brome mosaic virus, suggesting that this characteristic is true for plant (+)RNA viruses. Collectively, our findings revealed a hitherto unknown viral strategy whereby the host ROS-generating machinery is diverted for robust viral RNA replication.
Plants have evolved complicated and sophisticated strategies to survive environmental changes. The rapid generation of reactive oxygen species (ROS) is one of the hallmarks of plant responses to various biotic and abiotic stresses (1). Plant NADPH oxidase, termed RBOHs (respiratory burst oxidase homologs), localize at the plasma membrane (PM) and intracellular compartments including the Golgi apparatus (2, 3) and play a key role in ROS production upon the perception of environmental stresses (4). In Nicotiana benthamiana, an RBOHB (NbRBOHB) plays important roles in ROS production triggered by microbe-associated molecular patterns (MAMPs), such as bacterial flagellin and fungal chitin, and facilitates plant immunity against biotrophic pathogens including an oomycete pathogen Phytophthola infestance and tobacco mosaic virus (5–8). RBOH activity is tightly and coordinately regulated posttranslationally [e.g., activation by Ca2+, phosphatidic acid (PA), G proteins, or protein kinases] (9–12). It has recently been shown that receptor-like cytoplasmic kinases and calcium-dependent protein kinases (CDPKs) regulate RBOHs activity via direct phosphorylation or through modulating regulators of RBOHs during recognition of MAMPs via corresponding surface-localized pattern recognition receptors (13–16). RBOH-derived ROS induce stomatal closure to keep out invading bacteria, cross-linking the plant cell wall to block pathogen entry, and trigger local and systemic transcriptional reprogramming to activate plant immunity (1, 4, 9, 10). However, the function of ROS in plant–virus interactions remains largely unknown.
Plant viruses are obligate intracellular pathogens, and their replication cycle completely depends on host cells, thus requiring many host factors for completion (17–20). Positive-strand RNA [(+)RNA] viruses are the most abundant plant viruses and include many pathogens economically important in agriculture. They encode only a limited number of genes owing to their relatively small-sized genomes. However, they have evolved ways to use a number of host factors including proteins, lipids, and metabolites to meet their demands and reprogram host cell metabolism for their robust replication. Successful infection also involves efficient counterdefense imposed by the host (21). To achieve this, plant viruses use a variety of strategies to suppress or circumvent host defense systems and promote their infection. These include the formation of virus-induced miniorganelles like membranous structures to increase the local concentration of proviral factors and viral RNA template and to sequester newly synthesized viral genomes from host nucleases (22, 23), the exploitation of viral suppressors of RNA silencing to block the degradation or translation repression of viral RNAs, and interference with the phytohormone signaling involved in antiviral defense (21). Despite a number of recent advances that unveil mechanisms underlying plant–virus molecular arms race (reviewed in refs. 17–23), our current understanding of how (+)RNA viruses coordinate the host cell is far from complete.
Red clover necrotic mosaic virus (RCNMV) is a plant (+)RNA virus and a member of the genus Dianthovirus in the family Tombusviridae (24). The genome of RCNMV consists of RNA1 and RNA2. RNA1 encodes auxiliary replication protein p27, RNA-dependent RNA polymerase (RdRp) p88pol, and a coat protein. RNA2 encodes a movement protein that is required for viral cell-to-cell movement. p27 and p88pol target the endoplasmic reticulum (ER) membranes and form a viral replicase complex with co-opted multiple host factors (24–26). Among them are host heat shock proteins, small GTPase ADP ribosylation factor 1 (Arf1), and a PA-generating enzyme phospholipase D (PLD) hijacked by RCNMV for robust viral replication (27–29). Interestingly, affinity-purified RCNMV replicase complex from a model plant N. benthamiana (30) contains several key players in plant stress responses such as RBOH proteins and CDPK (29), suggesting that these host proteins have direct regulatory functions during a plant virus infection. Here, we present evidence that the host ROS-generating machinery is diverted by RCNMV and possibly by a well-established model plant (+)RNA virus, brome mosaic virus (BMV) (31, 32), for robust virus replication.
Results
NbRBOHB Is Required for RCNMV Replication in N. benthamiana.
Our previous proteomics analysis has identified RBOH as being associated with RCNMV replication proteins in N. benthamiana leaves (29). Most of the identified peptides by liquid chromatography (LC)-MS/MS analysis were matched with previously reported NbRBOHB (GenBank accession no. AB079499.1) (Fig. S1) except for one peptide (LGAAVPFDDNLNFHK) that was LGVAVPFDDNLNFHK in AB079499.1. We cloned NbRBOHB coding sequences from our N. benthamiana cDNA library and identified a homolog of AB079499.1 (GenBank accession no. LC156098, containing LGAAVPFDDNLNFHK sequences and showing 98.5% amino acid sequence identity to AB079499.1). Blastp search using LC156098 sequences as a query against N. benthamiana Genome v1.0.1 predicted proteins (https://solgenomics.net/organism/Nicotiana_benthamiana/genome) showed 99.47% identity between them. These slight changes may be due to differences in the N. benthamiana used for genome sequencing and this study.
Fig. S1.
Sequence alignment of NbRBOHBs. The full-length amino acid sequences of NbRBOHBs [AB079499 (6), LC156098 (this study), and Niben101Scf0258g04013.1 (https://solgenomics.net/organism/Nicotiana_benthamiana/genome)] were aligned using the Clustal omega algorithm. Equivalent residues are displayed in yellow. The peptide sequences identified by LC-MS/MS analysis (29) are highlighted as letters in red.
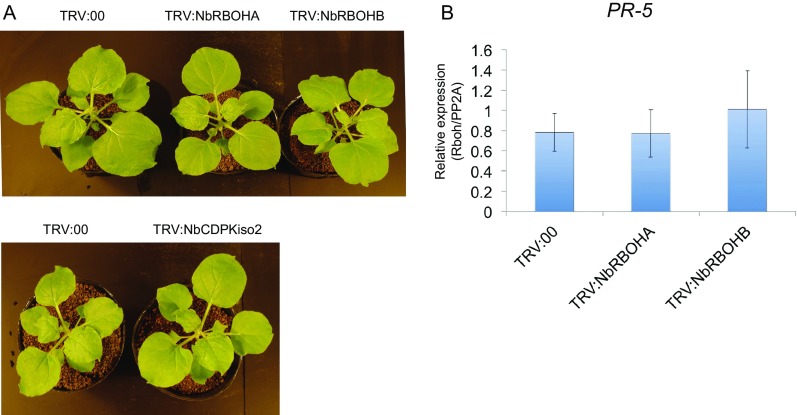
NbRBOHB-derived ROS play a positive role in plant immunity against several plant pathogens including fungi, bacteria, and viruses (5–8). Therefore, we first expected that RCNMV replication proteins would be recognized by RBOHs to trigger ROS production to inhibit viral infection, or that RCNMV replication proteins act as a suppressor of RBOHs to counteract RBOH-mediated antiviral immunity. To examine the function of NbRBOHB in RCNMV infection, we down-regulated NbRBOHB expression in N. benthamiana by tobacco rattle virus (TRV)-mediated virus-induced gene silencing (VIGS) and tested plants for susceptibility to RCNMV infection by analyzing viral RNA accumulation by Northern blotting. We also investigated the effects of VIGS of an NbRBOHB homolog, NbRBOHA, on RCNMV infection, because NbRBOHA is required for ROS production in response to P. infestance infection (6). Significant morphological defects such as chlorotic and stunted phenotypes were not observed in either NbRBOHA- or NbRBOHB-silenced plants (Fig. S2A). To our surprise, RCNMV RNA accumulations were significantly inhibited in NbRBOHA- and NbRBOHB-silenced plants, an unexpected phenomenon (Fig. 1A). In Arabidopsis thaliana, rbohD knockout mutant plants accumulate high levels of the defense hormone salicylic acid (SA) and show increased expression of the SA marker gene PR-1 upon pathogen challenge (33, 34). However, it has been reported that no enhancement of PR-1 expression is observed in NbRBOHA- or NbRBOHB-silenced N. benthamiana upon MAMPs treatment (35). Consistent with the latter finding, the expression of another SA-marker gene (PR-5) was not increased in NbRBOHA- and NbRBOHB-silenced N. benthamiana compared with control plants at 2 d after RCNMV inoculation (Fig. S2B). The results indicated that both NbRBOHA and NbRBOHB were required for RCNMV leaf-level infection.
Fig. S2.
NbRBOHA-, NbRBOHB-, and NbCDPKiso2-knockdown plants do not exhibit any symptoms. (A) Pictures of silenced N. benthamiana plants. The TRV vector harboring a partial fragment of N. benthamiana RBOHA (TRV:NbRBOHA), RBOHB (TRV:NbRBOHB), or CDPKiso2 (TRV:NbCDPKiso2) was expressed in N. benthamiana by Agrobacterium infiltration. The empty TRV vector (TRV:00) was used as a control. Pictures were taken at 20 d after infiltration. Note that the infiltrated plants showed no morphological defects at this stage. (B) Expression of SA-marker gene PR-5 in NbRBOHA- or NbRBOHB-silenced N. benthamiana plants. Total RNA was extracted from NbRBOHA- or NbRBOHB-silenced leaves, and the expression of SA-signaling marker gene PR-5 was analyzed by RT-qPCR. Graphed data are means ± SE from three independent experiments. PP2A was used as an internal control.
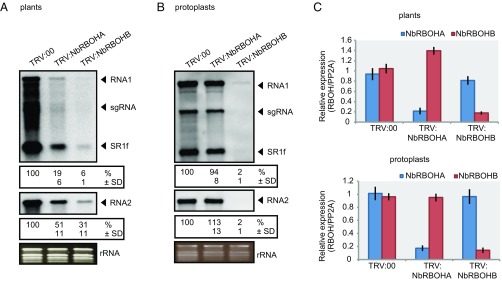
Fig. 1.
NbRBOHB is essential for robust RCNMV RNA replication. (A) Accumulations of RCNMV RNA in N. benthamiana leaves silenced for NbRBOHA and NbRBOHB. Total RNA was extracted from the mixture of three independent inoculated leaves 2 d after RCNMV inoculation and used for Northern blot analysis. (B) Accumulations of RCNMV RNAs in protoplasts isolated from N. benthamiana leaves silenced for NbRBOHA and NbRBOHB. Total RNA was extracted at 16 h postinoculation and used for Northern blot analysis. Ethidium bromide-stained rRNAs are shown below the Northern blots, as loading controls. The numbers below the blots represent the relative accumulation levels (means ± SD) of viral RNAs (RNA1 and RNA2, respectively) using ImageJ software (NIH), which were calculated based on three independent experiments. sgRNA, subgenomic RNA; SR1f, a small RNA fragment that derived from noncoding region of RNA1. (C) Efficiencies of gene silencing as assessed by RT-qPCR analysis. Graphed data are means ± SE from three independent experiments. PP2A was used as an internal control.
We next examined whether protoplasts isolated from NbRBOHA- or NbRBOHB-silenced N. benthamiana can support RCNMV replication. RCNMV RNA accumulation was significantly lower in NbRBOHB-silenced protoplasts compared with that in control protoplasts (Fig. 1B). In contrast, RCNMV RNA accumulation in NbRBOHA-silenced protoplasts was comparable to that in control protoplasts, suggesting that whereas NbRBOHB is essential for RCNMV cellular-level replication NbRBOHA is involved in postreplication step(s) in RCNMV infection. Reverse transcription quantitative PCR (RT-qPCR) analyses confirmed the specific reduction of NbRBOHA or NbRBOHB mRNAs in NbRBOHA- or NbRBOHB-silenced plants, respectively (Fig. 1C).
RCNMV p27 Replication Protein Associates with NbRBOHB.
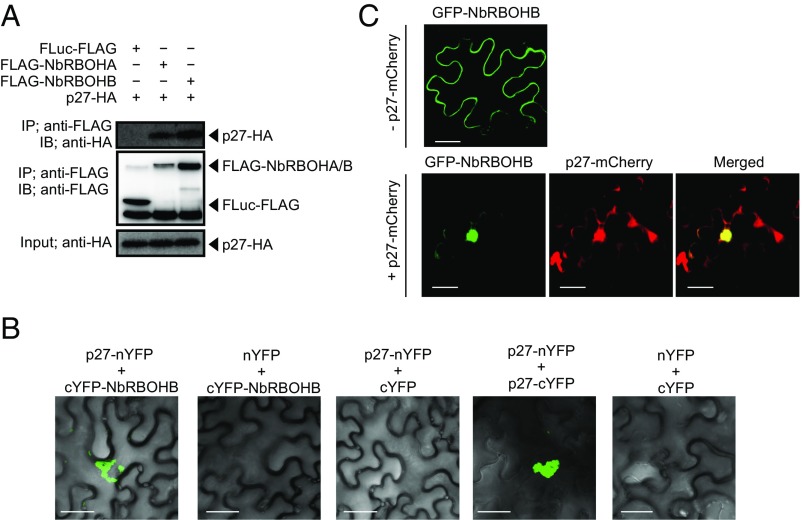
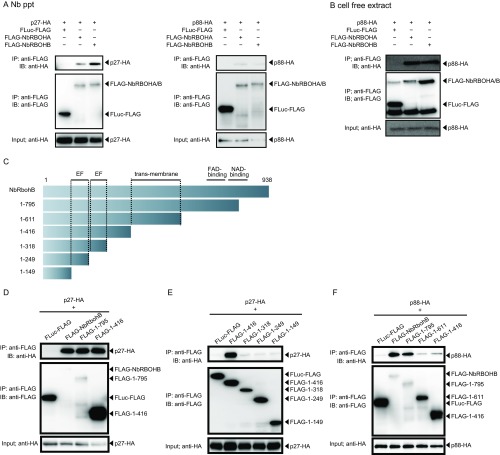
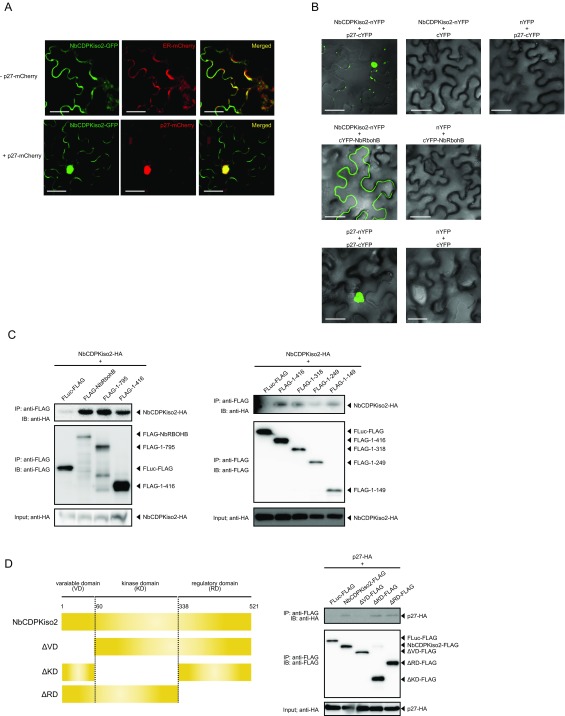
To investigate whether RCNMV replication protein p27 interacts with NbRBOHA and NbRBOHB we performed a coimmunoprecipitation (co-IP) assay in cell lysates prepared from evacuolated tobacco BY-2 protoplasts (BYL) (29, 36). C-terminally HA-tagged p27 was coexpressed with N-terminally FLAG-tagged NbRBOHs from in vitro synthesized transcripts in BYL, and the extracts were subjected to IP using FLAG-affinity resin. Consistent with our previous LC-MS/MS analysis (29), p27-HA was copurified with FLAG-NbRBOHB or FLAG-NbRBOHA but not with C-terminally FLAG-tagged firefly luciferase (FLuc-FLAG), which was used as a negative control (Fig. 2A). The p27–NbRBOH interaction was confirmed in transfected protoplasts using a co-IP assay (Fig. S3A). Furthermore, p88pol interacted with NbRBOHB in vitro and in protoplasts (Fig. S3 A and B). Deletion analysis indicated that p27 and p88pol interacted with different regions of NbRBOHB (Fig. S3 C–F). We also tested the p27–NbRBOHB interaction in plants with a bimolecular fluorescent complementation (BiFC) assay. A BiFC signal was detected as an aggregated structure in an intracellular space (Fig. 2B), which was reminiscent of the sites of RCNMV replication (26). The fluorescence of N-terminally GFP-fused NbRBOHB showed its localization at the cell periphery in N. benthamiana epidermal cells (Fig. 2C). However, coexpression of C-terminally mCherry-fused p27 (p27-mCherry) caused redistribution of the fluorescence of GFP-NbRBOHB from the cell periphery to the intracellular aggregated structure, in which they colocalized (Fig. 2C).
Fig. 2.
RCNMV p27 replication protein associates with NbRBOHB. (A) A co-IP of p27 and NbRBOHs in BYL (evacuolated tobacco BY-2 cell extracts). After in vitro translation, the extract was solubilized and subjected to immunoprecipitation by anti-FLAG antibody, and immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis. (B) A BiFC of p27 and NbRBOHB in N. benthamiana epidermal cells. The coexpression of p27-nYFP and p27-cYFP was used as a positive control. (Scale bars, 30 μm.) (C) Confocal laser scanning microscopy of N. benthamiana epidermal cells transiently expressing GFP-NbRBOHB alone or coexpressing GFP-NbRBOHB with p27-mCherry. The superimposition of the green (GFP) and red (mCherry) fluorescence is shown as a yellow color. (Scale bars, 30 μm.)
Fig. S3.
Both p27 and p88pol interact with NbRBOHs in vitro and in protoplasts. (A) Co-IPs of RCNMV replication proteins (p27 and p88pol) and NbRBOHs. The indicated constructs were transiently expressed in N. benthamiana protoplasts. Total protein was extracted and subjected to the immunoprecipitation of FLAG-fused proteins by anti-FLAG antibody. Immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis. (B) A co-IP of p88pol and NbRBOHs in BYL. The indicated capped mRNAs were incubated in BYL for in vitro translation. The extract was solubilized and subjected to immunoprecipitation by anti-FLAG antibody. Immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis. (C) The schematic representation of NbRBOHB or its deletion derivatives used for co-IP assays. (D–F) Co-IPs of p27 and NbRBOHB or its deletion derivatives in N. benthamiana protoplasts. The indicated constructs were transiently expressed in N. benthamiana protoplasts. Total protein was extracted and subjected to the immunoprecipitation of FLAG-fused proteins by anti-FLAG antibody. Immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis.
RCNMV Triggers an Intracellular ROS Burst via the p27 Replication Protein in an RBOH-Dependent Manner.
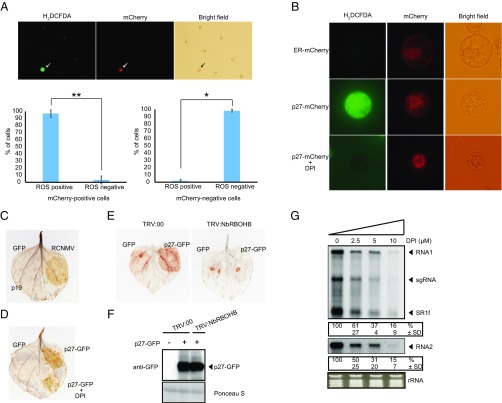
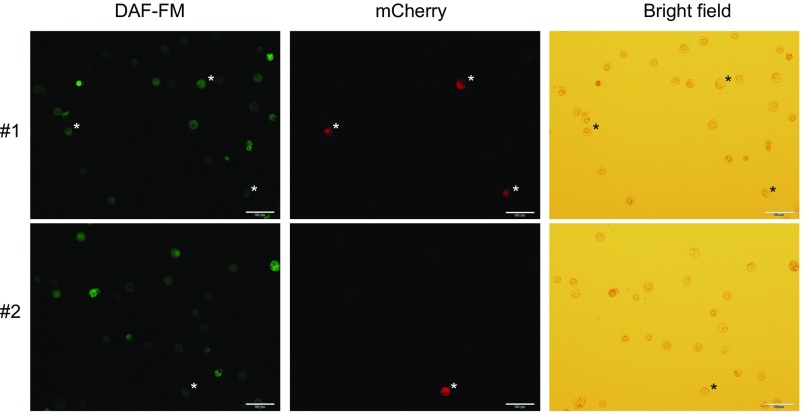
We next asked whether RCNMV induces ROS production in an infected cell using an intracellular ROS staining dye CM-H2DCFDA and a recombinant RCNMV that expresses mCherry serving as an infection marker (37). RCNMV-infected cells showed extremely high ROS signal compared with uninfected cells (Fig. 3A). In contrast, RCNMV infection did not promote DAF-FM-DA (4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate) signaling, which indicates the presence of intracellular reactive nitrogen species (RNS) (Fig. S4). These results indicate that RCNMV infection promotes intracellular ROS production. Furthermore, the expression of p27-mCherry but not ER-retention signal-containing mCherry (ER-mCherry) induced high intracellular ROS production (Fig. 3B). Importantly, this p27-induced high ROS accumulation was canceled by diphenyliodide (DPI), a well-known inhibitor of RBOH activity (Fig. 3B). These results were confirmed in N. benthamiana leaves infected by RCNMV or transiently expressing p27-GFP, using the DAB (3,3′-diaminobenzidine) assay for ROS staining. Again, DPI inhibited, albeit incompletely, the ROS accumulation induced by p27-GFP (Fig. 3 C and D). We also found that p27-induced ROS generation was inhibited in NbRBOHB-silenced leaves (Fig. 3E). The accumulations of p27-GFP were comparable between control and NbRBOHB-silenced leaves, thus excluding the possibility that lower p27-induced ROS production in silenced leaves was due to absence of the viral protein (Fig. 3F). These results strongly suggest that p27 induces high intracellular ROS accumulations in an RBOH-dependent manner. Importantly, treatment of N. benthamiana protoplasts with the RBOH inhibitor DPI led to the dose-dependent inhibition of RCNMV replication (Fig. 3G), suggesting that RCNMV replication depends on RBOH-derived ROS, which are highly induced by RCNMV p27.
Fig. 3.
RCNMV promotes ROS production in an infected cell via the p27 replication protein in an RBOH-dependent manner. (A) The H2DCFDA-stained tobacco BY-2 protoplasts inoculated with recombinant RCNMV that expresses mCherry. RCNMV-infected cells are marked by arrows. H2DCFDA-positive and H2DCFDA-negative cells were counted for mCherry-positive (about 40 cells per experiment) and mCherry-negative (about 300 cells per experiment) cells, respectively. Graphed data are means ± SD from three independent experiments; *P < 0.005, **P < 0.001, Student’s t test. (B) The H2DCFDA-stained tobacco BY-2 protoplasts inoculated with p27-mCherry-expressing plasmids. Protoplasts were treated with DMSO or 10 μM RBOH inhibitor DPI for 16 h postinoculation. A plasmid expressing mCherry containing ER-targeting signal (ER-mCherry) was used as a negative control. (C and D) DAB-stained N. benthamiana leaves transiently expressing RCNMV or p27-GFP by Agrobacterium-mediated infiltration. Leaves were treated with DPI (50 μM) at 1 d after infiltration and photographed at day 4. GFP or p19 was used as a negative control. (E) DAB-stained leaves of NbRBOHB-silenced and control N. benthamiana plants (TRV:00) expressing p27-GFP. (F) Accumulation of p27-GFP in N. benthamiana leaves silenced for NbRBOHB. Total protein was extracted and analyzed by immunoblot using an anti-GFP antibody. (G) Accumulation of RCNMV RNA in N. benthamiana protoplasts. The protoplasts were inoculated with in vitro synthesized RNA1 and RNA2 and incubated at 17 °C for 16 h in the presence of various concentrations of DPI. Ethidium bromide-stained rRNA is shown below the Northern blots, as loading controls. The numbers below the images represent the relative accumulation levels (means ± SD) of viral RNAs (RNA1 and RNA2, respectively) from three independent experiments.
Fig. S4.
RCNMV does not promote the production of RNS in infected cells. Tobacco BY-2 protoplasts were inoculated with recombinant RCNMV that expresses mCherry, and RNS accumulations were visualized by DAF-FM staining. RCNMV-infected cells are marked by an asterisk. (Scale bars, 100 μm.)
NbCDPKiso2 Is Required for p27-Induced ROS Burst and RCNMV RNA Replication.
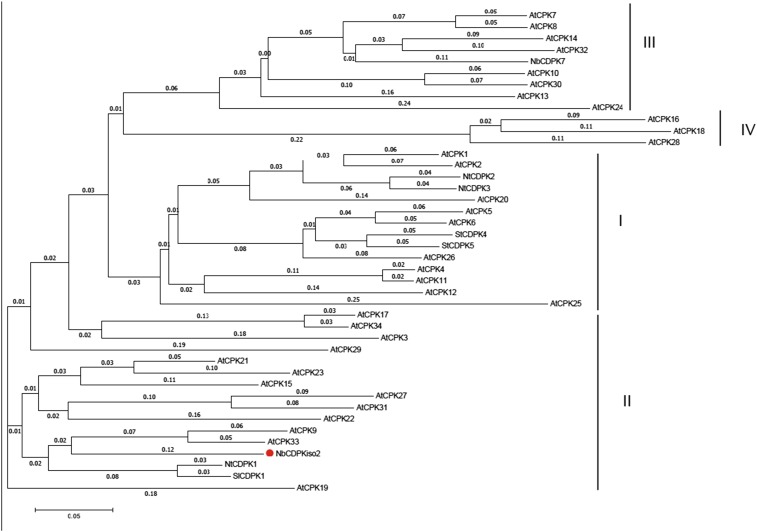
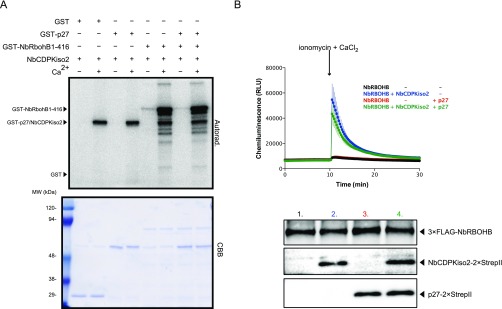
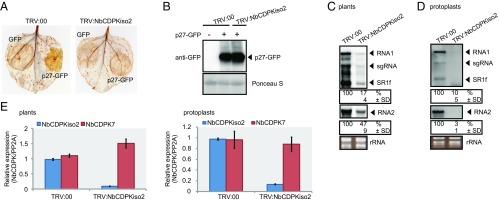
Our previous proteomics analysis showed that NbCDPKiso2 was present in the affinity-purified fraction of RCNMV replication complex (29). CDPK is a calcium sensor protein and comprises a multigene family (e.g., 34 members in A. thaliana and 29 members in tomato), which is divided into four subgroups (38–40). Phylogenetic analysis revealed that NbCDPKiso2 belongs to subgroup II CDPK (Fig. S5). Tobacco CDPK1 (NtCDPK1), a close relative of NbCDPKiso2, regulates the function of a transcription factor in response to a phytohormone, gibberellin (41). Interestingly, it has been proposed that A. thaliana subgroup II CDPK (AtCPK3) may be involved in MAMP responses (38). Furthermore, it is known that some isoforms of CDPK phosphorylate and activate RBOH (13–15). Therefore, we investigated whether NbCDPKiso2 is involved in RBOH-dependent ROS production triggered by p27. We found that NbCDPKiso2 phosphorylated an N-terminal fragment of NbRBOHB in a Ca2+-dependent manner in an in vitro kinase assay (Fig. S6A) and enhanced NbRBOHB activity in mammalian cells in response to the calcium ionophore ionomycin (Fig. S6B). These results suggest that NbCDPKiso2 can phosphorylate and activate NbRBOHB. Therefore, we predicted that NbCDPKiso2 is co-opted by RCNMV to activate NbRBOHB during viral infection. Unexpectedly, in these in vitro and mammalian systems p27 failed to promote NbCDPKiso2-mediated NbRBOHB phosphorylation and activation (Fig. S6 A and B). Nevertheless, importantly, we found that silencing NbCDPKiso2 expression inhibited p27-induced ROS generation without affecting the accumulation of the viral replication protein (Fig. 4 A and B). Furthermore, RCNMV accumulation was inhibited in NbCDPKiso2-silenced leaves and protoplasts (Fig. 4 C and D). RT-qPCR analyses confirmed the specific reduction of NbCDPKiso2 in the NbCDPKiso2-silenced plants (Fig. 4E). No morphological defects were observed in these plants (Fig. S2A). These results suggest that NbCDPKiso2 mediates p27-induced ROS generation and that it plays a positive role in the RNA replication of RCNMV in a plant cell.
Fig. S5.
NbCDPKiso2 is subgroup II CDPK. The full-length amino acid sequences of CDPKs from Arabidopsis (At), tobacco (Nt), potato (St), tomato (Sl), and N. benthamiana (Nb) were aligned and analyzed with ClustalW and MEGA7 algorithms. The CDPK family is divided into four subgroups (I–IV). The branched lengths are proportional to divergence and the scale of 0.05 indicates 5% change.
Fig. S6.
NbRBOHB is directly phosphorylated and enhanced by subgroup II CDPK, NbCDPKiso2. (A) The autoradiograph of the in vitro kinase assay performed by incubating purified NbCDPKiso2 with GST-p27, GST-NbRBOHB 1-416 or both for 30 min. The CBB-stained loading control is shown below the autoradiograph. (B) ROS productions in the transfected HEK293T cells in 96-well plates determined by luminol-amplified chemiluminescence. The data are presented at an average of relative luminescence units (RLU) in three wells ± SD (n = 3). One micromolar ionomycin and 1 mM CaCl2 was added at the time indicated by arrow. Accumulations of the proteins in HEK293T cells were confirmed by immunoblot analysis.
Fig. 4.
NbCDPKiso2 is required for p27-induced ROS bursts and RCNMV RNA replication. (A) DAB-stained leaves of NbCDPKiso2-silenced and control N. benthamiana plants (TRV:00) expressing p27-GFP. (B) Accumulation of p27-GFP in N. benthamiana leaves silenced for NbCDPKiso2. Total protein was extracted and analyzed by immunoblot using an anti-GFP antibody. (C) Accumulations of RCNMV RNAs in N. benthamiana leaves silenced for NbCDPKiso2. (D) The accumulation of RCNMV RNAs in protoplasts isolated from N. benthamiana leaves silenced for NbCDPKiso2. The relative accumulation levels of viral RNAs were determined as described in the legend to Fig. 1. (E) Efficiencies of gene silencing as assessed by RT-qPCR analysis. NbCDPK7 expression was also analyzed to evaluate silencing specificity. Graphed data are means ± SEs from three independent experiments. PP2A was used as an internal control.
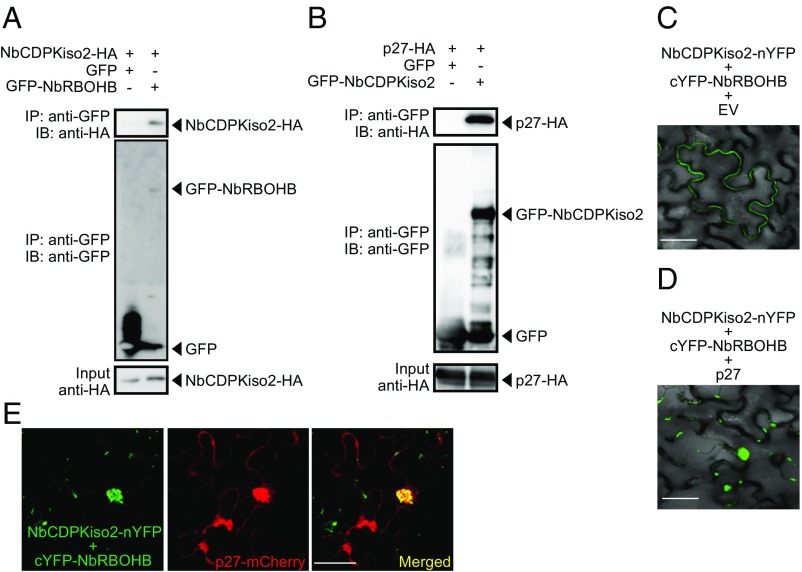
Next, we asked whether NbCDPKiso2 is incorporated into the p27–NbRBOHB complex. We found that NbCDPKiso2 formed complexes with NbRBOHB in a co-IP assay (Fig. 5A). In addition to NbRBOHB, NbCDPKiso2 also interacted with and colocalized with p27 (Fig. 5B and Fig. S7 A and B). Using BiFC, we showed that the NbRBOHB–NbCDPKiso2 interaction, which typically occurred at the cell periphery (Fig. 5C), was redistributed to the intracellular aggregated structure by coexpression with p27 (Fig. 5D). Furthermore, the intracellular NbRBOHB–NbCDPKiso2 signal was colocalized with p27-mCherry (Fig. 5E). We also found that NbCDPKiso2 was copurified with the N-terminal short fragments of NbRBOHB, which were not associated with p27 in co-IP assays in N. benthamiana protoplasts (Figs. S3E and S7C), suggesting that NbCDPKiso2 and p27 bind to different regions of NbRBOHB. These results suggest that p27 can form a ternary complex with NbRBOHB and NbCDPKiso2, which is required to activate NbRBOHB to generate ROS in the cytoplasm.
Fig. 5.
RCNMV p27 colocalizes with the NbRBOHB—NbCDPKiso2 complex at the intracellular aggregated structure. (A and B) Co-IPs of NbCDPKiso2 and NbRBOHB (A) and NbCDPKiso2 and p27 (B). The indicated constructs were transiently expressed in N. benthamiana. Total protein was extracted and subjected to the immunoprecipitation of GFP-fused proteins by anti-GFP antibody. Proteins were analyzed by immunoblot using antibodies, as indicated on the left. (C) BiFC of NbRBOHB and NbCDPKiso2 in N. benthamiana epidermal cells. EV, empty vector. (Scale bar, 30 μm.) (D) BiFC of NbRBOHB and NbCDPKiso2 in the presence of p27 N. benthamiana epidermal cells. (Scale bar, 30 μm.) (E) Confocal laser scanning microscopic images of N. benthamiana epidermal cells transiently expressing NbCDPKiso2-nYFP, cYFP-NbRBOHB and p27-mCherry. The superimposition of the green (YFP) and red (mCherry) fluorescence is shown as a yellow color. (Scale bar, 30 μm.)
Fig. S7.
NbCDPKiso2 is recruited by p27. (A) Confocal laser scanning microscopic images of N. benthamiana epidermal cells transiently expressing NbCDPKiso2-GFP. NbCDPKiso2-GFP was coexpressed with mCherry fused with ER-targeting signal (ER-mCherry) and RNA silencing suppressor of tomato bushy stunt virus (p19) or C-terminally mCherry-fused p27 (p27-mCherry) and p19 in N. benthamiana leaves by Agrobacterium infiltration. Fluorescence was visualized by confocal laser scanning microscopy 4 d after infiltration. The superimposition of the green (GFP) and red (mCherry) fluorescence is shown as a yellow color. (Scale bars, 30 μm.) (B) BiFC assays in N. benthamiana epidermal cells. BiFC signal was reconstituted by coexpression of NbCDPKiso2-nYFP with p27-cYFP or cYFP-NbRBOHB. Coexpression of p27-nYFP with p27-cYFP was used as a positive control. (Scale bars, 30 μm.) (C) Co-IPs of NbCDPKiso2 and NbRBOHB or its deletion derivatives. The indicated constructs were transiently expressed in N. benthamiana protoplasts. Total protein was extracted and subjected to the immunoprecipitation of FLAG-fused proteins using anti-FLAG antibody. Immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis. (D) The schematic representation of NbCDPKiso2 or its deletion derivatives used for a co-IP assay (Left). The indicated constructs were transiently expressed in N. benthamiana protoplasts. Total protein was extracted and subjected to the immunoprecipitation of FLAG-fused proteins using anti-FLAG antibody. Immunoprecipitates were probed with anti-FLAG and anti-HA immunoblots after gel electrophoresis (Right).
Each CDPK consists of a kinase domain, an autoinhibitory pseudosubstrate domain, multiple Ca2+-binding EF-hand motifs, and an N-terminal variable domain (V domain) (38). It is proposed that the V domain determines the substrate specificity of CDPKs (42, 43). Deletion analysis indicated that the N-terminal variable domain of NbCDPKiso2 is required for the p27–NbCDPKiso2 interaction (Fig. S7D).
RNA Replication of BMV Depends on RBOH-Derived ROS.
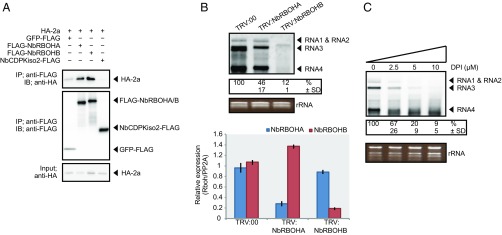
The results described above suggest that RCNMV hijacks the host’s ROS-generating machinery for viral RNA replication. To investigate whether this phenomenon is also observed in other plant (+)RNA viruses, we tested BMV, another (+)RNA plant virus, for its dependency on host ROS-generating enzyme. We found that BMV replication protein 2a was efficiently copurified with NbRBOHA and NbRBOHB but not with NbCDPKiso2 in a co-IP assay in N. benthamiana protoplasts (Fig. 6A). BMV failed to accumulate in NbRBOHA- or NbRBOHB-silenced N. benthamiana leaves (Fig. 6B), and DPI inhibited BMV replication in N. benthamiana protoplasts in a dose-dependent manner (Fig. 6C). These results, similar to those observed for RCNMV (Figs. 2A and 3E), indicate that BMV replication depends on RBOH-derived ROS.
Fig. 6.
RBOH-derived ROS is required for RNA replication of BMV in N. benthamiana. (A) Co-IPs of the BMV replication protein 2a and NbRBOHs. The indicated constructs were transfected into N. benthamiana protoplasts. Total protein was extracted and subjected to the immunoprecipitation of FLAG-fused proteins using anti-FLAG antibody. Proteins were analyzed by immunoblotting using the antibodies indicated on the left. (B) Accumulations of BMV RNAs in N. benthamiana leaves silenced for NbRBOHA and NbRBOHB. Total RNA was extracted from the mixture of three independent inoculated leaves 2 d after BMV inoculation and used for Northern blot analysis. Efficiencies of gene silencing were assessed by RT-qPCR analysis. Graphed data are means ± SEs from three independent experiments. PP2A was used as an internal control. (C) Accumulation of BMV RNAs in N. benthamiana protoplasts. The protoplasts were inoculated with in vitro synthesized BMV RNAs and incubated at 17 °C for 24 h in the presence of various concentrations of DPI. Ethidium bromide-stained rRNAs are shown below the Northern blots, as loading controls. The numbers below the images represent the relative accumulation levels (means ± SD) of RNA3 from three independent experiments.
Discussion
It is known that ROS play a critical role in stress acclimation in animals and plants. During pathogen infection plants rapidly produce ROS to induce local or systemic signaling through the activation of cell surface-localized RBOH proteins. ROS signaling mediates systemic resistance against plant viruses (7, 8) and is therefore considered to be a positive regulator of plant antiviral defense. However, the precise function of ROS in plant–virus interaction has not been extensively studied. In this study, we showed that RBOH-derived ROS production is required for robust RNA replication of RCNMV. Because ROS have generally short half-lives and high reactivity within cells, spatial control of ROS generation sites is important for physiological ROS signaling (44). Here, we showed that the p27 replication protein of RCNMV interacts with and redistributes NbRBOHB from the PM to intracellular aggregate structures to induce ROS production in an RBOH activity-dependent manner (Figs. 2 and 3). Because most, if not all, p27-containing perinuclear ER aggregate structures are believed to be the sites of de novo viral RNA synthesis (28), the presented data suggest that a large proportion of NbRBOHB molecules are retargeted to the viral replication site from the PM through interaction with p27. The ER membrane is known to associate with the PM (45), and therefore it is possible that p27 recognizes NbRBOHB at or near the ER–PM contact sites and then recruits it to sites of viral RNA replication. Alternatively, p27 may recruit RBOH proteins from the Golgi apparatus, where a population of RBOH molecules resides (3). In this context, it should be noted that p27 binds to Arf1, which is a key regulator of vesicle trafficking from the Golgi apparatus, and interferes with host secretory pathways (28, 37).
The data presented here suggest that BMV, another plant (+)RNA virus, also uses the host ROS-generating machinery for viral RNA replication (Fig. 6). Recently, it has been shown that RBOH proteins are also copurified with bamboo mosaic virus RdRp (46). Generation of ROS is a common response during pathogen infection, and RBOH is highly conserved in plants. Therefore, plant (+)RNA viruses may have evolved a sophisticated strategy to co-opt this highly conserved plant immune system. Recent findings have suggested that ROS play a positive role in mammal and algal viral infections. The production of influenza A virus particles in lung epithelial cells depends on NOX4-derived ROS (47). Emiliania huxleyi virus, a large dsDNA virus, which infects coccolithophores, induces ROS production and uses it for successful viral lytic infection (48). Collectively, these studies suggest that the host ROS-generating machinery may be a common target shared by diverse viruses, regardless of their hosts.
ROS can mediate a diverse array of reversible or irreversible modifications on biomolecules, including proteins and lipids. They govern the functions of various proteins, including redox-sensitive chaperone Hsp33 and calcium/calmodulin-dependent protein kinase CaMKII, through regulation of disulfide bond formation or oxidation of methionine residues (49, 50). Oxidizing agents enhance the guanyltransferase activity of flavivirus NS5 or alphavirus nsP1 in vitro (51). Therefore, it is possible that ROS-mediated oxidation of viral and/or co-opted host factors would facilitate viral replication. However, the mode of action of ROS in viral replication in vivo remains unexplored. Further studies are needed to understand how ROS boost plant viral RNA replication.
RBOH activity is regulated posttranslationally by a number of factors, such as Ca2+, PA, G proteins, and a number of kinases (9–13). Several CDPKs are involved in biotic and abiotic stresses as positive or negative regulators of RBOH proteins. For example, several A. thaliana subgroup I CDPKs, in particular AtCPK1 and AtCPK2, phosphorylate AtRBOHD and AtRBOHF and are required for the ROS accumulation induced by pathogen effectors such as avrRpm1 and avrRpt2 (14). The A. thaliana subgroup IV CDPK, AtCPK28, phosphorylates and regulates the turnover of receptor-like cytoplasmic kinase BIK1 (16), which acts upstream of AtRBOHD during MAMPs-triggered immunity and is required for ROS bursts (34, 52). However, their involvement in plant–virus interactions has not been reported previously. In this study, we identified the N. benthamiana subgroup II CDPK, NbCDPKiso2, which phosphorylates and enhances NbRBOHB activity (Fig. S6). Interestingly, NbCDPKiso2 was shown to be co-opted by RCNMV and essential for robust viral RNA replication (Fig. 4). Importantly, NbCDPKiso2 is required for p27-induced ROS accumulation. Therefore, our findings suggest that RCNMV indirectly activates NbRBOHB with the help of NbCDPKiso2. In contrast, the BMV replication protein 2a was not efficiently copurified with NbCDPKiso2 (Fig. 6A), suggesting that BMV and RCNMV use different strategies to harness RBOH enzymes.
In in vitro and mammalian cell assays we failed to detect further enhancement of NbCDPKiso2-mediated NbRBOHB activation by p27 (Fig. S6). Although we do not exclude the possibility that p27 is not functional in vitro or in the heterologous in vivo system, this may imply that an additional host factor could be involved in virus-mediated NbRBOHB activation in plant cells. Recently, it has been reported that NtCDPK1 can act as a kinase and also as a scaffold protein to bridge a transcription factor, repression of shoot growth (RGS), and 14-3-3 protein (53). Phosphorylation and 14-3-3 binding cooperatively regulate the function of RGS in response to gibberellins (41). Our proteomics analysis identified several known RBOH activators, such as PA-generating enzyme PLD and heterotrimeric G protein subunit β (29). Recently, it was reported that A. thaliana heterotrimeric G protein forms complexes with AtPLDα1 (54) and AtRBOHD (12) and is required for AtRBOHD phosphorylation and ROS bursts triggered by the flagellin epitope. In addition to NbCDPKiso2, RCNMV may also co-opt these RBOH activators for optimal activation of RBOH proteins.
Plant RNA viruses must interact and use host factors at every step of the replication cycle (e.g., RNA synthesis, virus assembly, movement, counterdefense, and transmission). Examples include eukaryotic translation initiation factors, RNA-binding proteins, membrane trafficking machinery, and lipid-generating enzymes (17, 19, 20, 55, 56). These host factors simultaneously serve as virus restriction determinants if they have mutations that are unfavorable for virus replication (56). This study represents an example of the hijack of a host defense-associated pathway by a plant virus and provides an interesting insight into the coevolution of viruses/hosts (i.e., the endless arms race between them).
Experimental Procedures
Plant Growth Conditions.
N. benthamiana plants were grown on soil at 22 ± 2 °C and 12 h light/12 h dark per day.
Gene Cloning.
RNA extraction from N. benthamiana leaves was performed using an RNeasy Plant Mini-Kit (Qiagen). Reverse-transcription PCR (RT-PCR) was carried out as described earlier (28). Superscript III reverse transcriptase (Invitrogen) and oligo (dT) were used for RT. Primers to amplify coding sequences (CDSs) of NbRBOHA, NbRBOHB, and NbCDPKiso2 were designed based on the N. benthamiana RNA sequencing data (Transcriptome version 5: benthgenome.qut.edu.au/) (57). The full-length CDSs of NbRBOHA, NbRBOHB, and NbCDPKiso2 were amplified from N. benthamiana cDNA and cloned into pBYL2 (25) (Table S1). All plasmids constructed in this study were verified by sequencing.
Table S1.
Primers used for gene cloning and construction in this study
| Plasmid | Primer | Sequence |
| pBYL FLAG-NbRBOHA | AscI FLAG-NbRBOHA Fwd | CTGGCGCGCCATGGATTACAAGGACGACGATGACAAGAGAGGGTTACCGGGGCATGAACGCCGGTGG |
| AscI NbRBOHA Rev | CTGGCGCGCCCTAAAAATGTTCTTTGTGAAACTCGAACTTTG | |
| pBYL FLAG-NbRBOHB | AscI FLAG-NbRBOHB Fwd | TGGCGCGCCATGGATTACAAGGACGACGATGACAAGCAAAATTCTGAAAATCATCATCCGCACCACC |
| AscI NbRBOHB Rev | CTGGCGCGCCCTAAAAATTTTCTTTATGGAAATCAAACTTGG | |
| pBYL NbCDPKiso2-myc | AscI NbCDPKiso2 Fwd | CTGGCGCGCCATGTGTAGGGCTATTGTCAATGTTGTGCACG |
| AscI-myc-NbCDPKiso2 Rev | CTGGCGCGCCCTACAGGTCCTCCTCGCTGATCAGCTTCTGCTCGAAGAGTTTGCCTGGCTGTTTGACTCCACTTCTC | |
| pTV:NbRBOHA | HindIII NbRBOHA Fwd | ATCAAGCTTGCATTGTGCGAAATCGGAACGGTAAAGATAACCG |
| ClaI NbRBOHA Rev | GGTATCGATTGTGATTAACATTGACGCTCCTGATCTTCCCG | |
| pTV:NbRBOHB | HindIII NbRBOHB Fwd | ATCAAGCTTTGCATTCTCCAAATTTGACACGAGGAAGCAAGCC |
| ClaI NbRBOHB Rev | GGTATCGATGTTGAATAAACGAGGCGGCAAAAAGAGTGCG | |
| pTV:NbCDPKiso2 | XmaI NbCDPKiso2 Fwd | ATCCCCCGGGGGCTGGATCGCCTATTCCATGCTCTTCC |
| XmaI NbCDPKiso2 Rev | GCAGCCCGGGGAAGAAATTCACGGACTCAAAGCAATGTTCC |
The restriction enzyme sites are underlined.
Transient Expression in N. benthamiana.
Agrobacterium tumefaciens strain GV3101 carrying various binary constructs was used for transient expression. The CDSs of NbRBOHA, NbRBOHB, and NbCDPKiso2 were amplified from corresponding pBYL vectors and inserted into binary vector pBICP35S (with the 35S promoter and HA, nYFP, cYFP, or GFP fusion tag) using an in-fusion cloning kit (TaKaRa Bio, Inc.). Standard electroporation was used for transformation of A. tumefaciens GV3101. Transformed bacterial cells were grown in liquid Luria broth medium supplemented with appropriate antibiotics for 24 h. Cells were harvested and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone, pH 5.7). Suspensions were mixed at final OD600 of 0.05 for each construct for the BiFC experiments and an OD600 of 0.2 each for subcellular localization or co-IP experiments with the RNA silencing suppressor of tomato bushy stunt virus, p19. Four- to five-week-old N. benthamiana plants were infiltrated with 1-mL needleless syringes. The infiltrated N. benthamiana leaves were taken off 4 d postinfiltration (dpi) and used for subsequent analysis.
VIGS in N. benthamiana.
The partial sequences of CDSs from NbRBOHA (a 499-bp fragment), NbRBOHB (a 500-bp fragment), or NbCDPKiso2 (a 333-bp fragment) were amplified from corresponding pBYL plasmids and inserted into pTV00 (58) (Table S1). Appropriate combinations of silencing vectors were inoculated via Agrobacterium infiltration into N. benthamiana plants at 2–3 wk postgermination. At 18 dpi, the leaves located above the infiltrated leaves were challenged with in vitro synthesized RCNMV RNA1 and RNA2 (0.5 μg each). At 2 d after inoculation, inoculated leaves from three different plants infected with the same inoculum were pooled, and total RNA was extracted using plant RNA purification reagents (Invitrogen), treated with RQ1 RNase-free DNase (Promega), purified by phenol–chloroform extraction, and precipitated with ethanol. Viral RNAs were detected by Northern blotting, as described previously (27, 28).
Protoplast Experiments.
Young expanded leaves from 5- to 6-wk-old N. benthamiana were used for protoplast preparation. N. benthamiana protoplasts were inoculated with RCNMV RNA1 and RNA2 (0.5 μg each) and incubated with 0, 2.5, 5, or 10 μM DPI (diphenyleneiodonium; Sigma-Aldrich) at 17 °C for 16 h. For the BMV replication assay, N. benthamiana protoplasts inoculated with BMV RNA1, RNA2, and RNA3 (1.5 μg each) were incubated with 0, 2.5, 5, or 10 μM DPI at 17 °C for 24 h. DPI was dissolved in DMSO (Sigma-Aldrich). Total RNA was extracted and subjected to Northern blotting, as described previously (28). Each experiment was repeated at least three times using different batches of protoplasts.
For VIGS experiments, protoplasts isolated from TRV-infected N. benthamiana plants at 21 dpi were used for viral RNA inoculation. Total RNA was extracted at 16 h postinoculation and subjected to Northern blotting. Each experiment was repeated at least three times using different batches of protoplasts.
Quantitative RT-PCR.
Total RNA was isolated from leaves or protoplasts after treatment with plant RNA purification reagents (Invitrogen). cDNA was synthesized from 0.5 μg of total RNA with a ReverTra Ace qPCR RT kit (Toyobo). RT-qPCR analysis was carried out using the StepOnePlus Real-Time PCR system (Applied Biosystems) with THUNDERBIRD SYBR qPCR Mix (Toyobo) using specific gene primers (Table S2). The relative expression values of genes were determined using the comparative Ct method (2ΔCt) with PP2A as a reference gene.
Table S2.
Primers used for RT-qPCR analysis
| Gene | Forward primer | Reverse primer |
| NbRBOHA | GAGCAAGCAGATTTAACCTCAGA | CTTCTGATCAAGTTCAGCCACTT |
| NbRBOHB | TTTTCTCTGAGGTTTGCCAGCCACCA | GCCTTCATGTTGTTGACAATGTCTTT |
| NbCDPKiso2 | ACCTGAACAGTCAAATTCCAACG | GAATTGTACCGTGTGCAGAAACT |
| NbCDPK7 | TGGAAATGGTGGGTACAAATCC | CCTCTCTCCTCACATCCTCAAT |
| NbPR-5 | GCAGGTGGTGAATGTTCCTT | CCACCACCAAAAGGACTGAT |
| NbPP2A | GACCCTGATGTTGATGTTCGCT | GAGGGATTTGAAGAGAGATTTC |
Co-IP Experiments.
For a co-IP assay using evacuolated tobacco BY-2 protoplast lysate (BYL), FLAG- or HA-tagged proteins were expressed in BYL by adding capped in vitro synthesized transcripts. After incubation at 25 °C for 120 min, a 10-μL bed volume of ANTI-FLAG M2-Agarose Affinity Gel (Sigma-Aldrich) was added to the BYL and further incubated for 1 h with gentle mixing. The resin was washed three times with 200 μL of TR buffer [30 mM Hepes, 100 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, 150 mM NaCl, and 0.5% Triton X-100, pH 7.4].
For a co-IP assay in protoplasts, fragments or full-length CDSs of NbRBOHA, NbRBOHB, and NbCDPKiso2 were amplified from corresponding pBYL vectors and inserted into pUBP35S (with the 35S promoter and HA or FLAG fusion tag) using an in-fusion cloning kit (TaKaRa Bio, Inc.). N. benthamiana protoplasts were transfected with 10 μg of the indicated plasmids then incubated for 16 h at 17 °C. Total protein was extracted with buffer A [50 mM Hepes, 150 mM NaCl, 0.1% 2-mercaptoethanol, 0.5% Triton X-100, and 5% (vol/vol) glycerol, pH 7.5], incubated with 20 μL ANTI-FLAG M2-Agarose Affinity Gel (Sigma-Aldrich) at 4 °C for 2 h with gentle mixing, and washed with 1 mL of buffer A three times.
Details of a co-IP experiment following Agrobacterium-mediated transient expression in N. benthamiana were described previously (29). Total protein was extracted from 0.33 g of leaves in 1 mL of buffer A containing a mixture of protease inhibitors (Roche), incubated with GFP-Trap agarose beads (10 μL) (ChromoTek) at 4 °C for 1 h with gentle mixing. The beads were washed three times with 1 mL of buffer A. The bound proteins were eluted by addition of 1× SDS gel loading buffer, followed by incubation at 95 °C for 3 min. Protein samples were subjected to SDS/PAGE, followed by immunoblotting with anti-FLAG M2 (Sigma-Aldrich), anti-HA 3F10 (Roche), or anti-GFP JL8 (Clontech).
Subcellular Localization Assays.
Appropriate combinations of fluorescent protein-fused proteins were expressed in N. benthamiana leaves by Agrobacterium infiltration. The fluorescence of GFP and mCherry was visualized with confocal laser scanning microscopy at 4 dpi as described previously (59).
Detection of ROS Production.
For ROS staining assays with CM-H2DCFDA [5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate; Invitrogen], tobacco BY-2 protoplasts were inoculated with in vitro synthesized RCNMV-mCherry transcripts or plasmids expressing ER-mCherry or p27-mCherry, as described previously (60). After 16 h, the protoplasts were incubated for 10 min at room temperature in 500 nM CM-H2DCFDA. Fluorescence was observed by fluorescent microscopy (Olympus).
For histological H2O2 production in N. benthamiana leaves upon infiltration with Agrobacterium, the leaves were excised and immersed in DAB solution [1 mg/mL DAB (Wako), 10 mM NaHPO4, and 0.01% Nonidet P-40] with vacuum infiltration, followed by an overnight incubation at room temperature in the dark. The stained leaves were cleared in boiled 70% (vol/vol) ethanol.
The measurement of ROS production in HEK293T cells was performed essentially as described previously (11, 61). SI Experimental Procedures for details.
SI Experimental Procedures
RNA Preparation.
pUCR1 (62) and pRC2_G (63) are full-length cDNA clones of RNA1 and RNA2, respectively, of the RCNMV Australian strain. pB1TP3, pB2TP5, and pB3TP8 are full-length cDNA clones of RNA1, RNA2, and RNA3 of the BMV M1 strain, respectively (64). RCNMV RNA1 and RNA2 were transcribed from SmaI-linearized pUCR1 and pRC2_G, respectively, using T7 RNA polymerase (TaKaRa Bio, Inc.). BMV RNAs were transcribed from EcoRI-linearized pB plasmids using T7 RNA polymerase and capped with a ScriptCapm7G capping system (Epicentre Biotechnology). Capped mRNAs were transcribed from NotI-linearized pBYL plasmids using T7 RNA polymerase (TaKaRa Bio, Inc.) and capped with a ScriptCapm7G capping system (Epicentre Biotechnology). All transcripts were purified with a Sephadex G-50 fine column (Amersham Pharmacia Biotech.). RNA concentration was determined spectrophotometrically, and its integrity was verified by agarose gel electrophoresis.
Detection of RNS Production.
Tobacco BY-2 protoplasts were inoculated with in vitro synthesized RCNMV-mCherry transcripts or plasmids expressing ER-mCherry or p27-mCherry as described previously (60). After 16 h, the protoplasts were incubated for 10 min at room temperature in 10 μM DAF-FM-DA (4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate; Goryo Chemical, Inc.). Fluorescence was observed by fluorescent microscopy (Olympus).
Measurement of ROS Production in HEK293T Cells.
The CDS of NbRBOHB was amplified and subcloned into pcDNA3.1 vector (11) to generate pcDNA 3xFLAG-NbRBOHB. The CDSs of NbCDPKiso2 and p27 were amplified and subcloned into pEF1 2xStrepII-C (61) to construct pEF1 NbCDPKiso2–2xStrepII and pEF1 p27–2xStrepII, respectively. HEK293T cells were maintained and transiently transfected as described previously (11). Cells in each single well of a 96-well plate were transfected with 160 ng of total plasmid DNA consisting of the pcDNA 3xFLAG-NbRBOHB (100 ng), pEF1 NbCDPKiso2–2xStrepII (40 ng), and pEF1 p27–2xStrepII (20 ng) or filled up to 160 ng with the empty vector, pEF1/myc–His. Each plasmid combination was transfected into three replicate wells of a single 96-well plate. ROS production in response to adding 1 μM of ionomycin and 1 mM CaCl2 was measured by the peroxidase-dependent luminol-amplified chemiluminescence technique as described previously (11). Experiments were repeated six times with similar results.
An in Vitro Kinase Assay.
Full-length NbCDPKiso2 or an N-terminal region of NbRBOHB (1–416 aa) was amplified from corresponding pBYL plasmids and subcloned into pCOLDI (TaKaRa Bio, Inc.) or pCOLDI GST (65), respectively. Proteins were expressed in Escherichia coli BL21 (DE3) and purified with Ni-NTA agarose. Purified NbCDPKiso2 (0.1 μg) was incubated with GST (0.5 μg), GST-p27 (1 μg) (65), or GST-NbRBOHB 1–416 (1.5 μg) proteins in kinase reaction buffer (20 mM Hepes, pH 7.5, 1 mM DTT, 5 mM MgCl2, 50 μM ATP, and 10 μCi/mL [γ-32P]ATP) in the presence or absence of 0.5 mM CaCl2 for 30 min at room temperature. The reactions were stopped by the addition of SDS gel loading buffer and subjected to SDS/PAGE followed by the visualization of 32Pi-phosphorylated polypeptides using BAS 2000 (Fuji Film). As a loading control, proteins were separated in independent gels and stained with Coomassie brilliant blue R-250 (CBB). Experiments were repeated two times using different batches of purified proteins.
Acknowledgments
We thank Steven A. Lommel (North Carolina State University) and Paul Ahlquist (University of Wisconsin) for providing plasmids. This work was supported by an Okayama University Startup fund and Japan Society for the Promotion of Science (JSPS) KAKENHI Grant 15H06420 (to K. Hyodo), a grant for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Grant 16H06436 (to N.S.), and JSPS KAKENHI Grants 22248002 and 15H04456 (to T.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NbRBOHB and NbCDPKiso2 have been deposited in the DNA Data Bank of Japan (accession nos. LC156098 and LC156099, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610212114/-/DCSupplemental.
References
- 1.Sewelam N, Kazan K, Schenk PM. Global plant stress signaling: Reactive oxygen species at the cross-road. Front Plant Sci. 2016;7:187. doi: 10.3389/fpls.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao H, et al. Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell. 2014;26(4):1729–1745. doi: 10.1105/tpc.113.122358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noirot E, et al. Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J Exp Bot. 2014;65(17):5011–5022. doi: 10.1093/jxb/eru265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki N, et al. Respiratory burst oxidases: The engines of ROS signaling. Curr Opin Plant Biol. 2011;14(6):691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Segonzac C, et al. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 2011;156(2):687–699. doi: 10.1104/pp.110.171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshioka H, et al. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell. 2003;15(3):706–718. doi: 10.1105/tpc.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng XG, et al. Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci Rep. 2016a;6:20579. doi: 10.1038/srep20579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng XG, et al. Orchestration of hydrogen peroxide and nitric oxide in brassinosteroid-mediated systemic virus resistance in Nicotiana benthamiana. Plant J. 2016b;85(4):478–493. doi: 10.1111/tpj.13120. [DOI] [PubMed] [Google Scholar]
- 9.Adachi H, Yoshioka H. Kinase-mediated orchestration of NADPH oxidase in plant immunity. Brief Funct Genomics. 2015;14(4):253–259. doi: 10.1093/bfgp/elv004. [DOI] [PubMed] [Google Scholar]
- 10.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56(8):1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- 11.Ogasawara Y, et al. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem. 2008;283(14):8885–8892. doi: 10.1074/jbc.M708106200. [DOI] [PubMed] [Google Scholar]
- 12.Liang X, et al. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife. 2016;5:e13568. doi: 10.7554/eLife.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubiella U, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA. 2013;110(21):8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013;9(1):e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi M, et al. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 2007;19(3):1065–1080. doi: 10.1105/tpc.106.048884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monaghan J, et al. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe. 2014;16(5):605–615. doi: 10.1016/j.chom.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Nagy PD, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2011;10(2):137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyodo K, Okuno T. Host factors used by positive-strand RNA plant viruses for genome replication. J Gen Plant Pathol. 2014;80(2):123–135. [Google Scholar]
- 19.Hyodo K, Okuno T. Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr Opin Virol. 2016;17:11–18. doi: 10.1016/j.coviro.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Wang A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu Rev Phytopathol. 2015;53:45–66. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- 21.Mandadi KK, Scholthof KB. Plant immune responses against viruses: how does a virus cause disease? Plant Cell. 2013;25(5):1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 23.Laliberté JF, Zheng H. Viral manipulation of plant host membranes. Annu Rev Virol. 2014;1(1):237–259. doi: 10.1146/annurev-virology-031413-085532. [DOI] [PubMed] [Google Scholar]
- 24.Okuno T, Hiruki C. Molecular biology and epidemiology of dianthoviruses. Adv Virus Res. 2013;87:37–74. doi: 10.1016/B978-0-12-407698-3.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mine A, et al. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus. J Virol. 2010;84(12):6070–6081. doi: 10.1128/JVI.00054-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusumanegara K, et al. Identification of domains in p27 auxiliary replicase protein essential for its association with the endoplasmic reticulum membranes in Red clover necrotic mosaic virus. Virology. 2012;433(1):131–141. doi: 10.1016/j.virol.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Mine A, et al. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J Virol. 2012;86(22):12091–12104. doi: 10.1128/JVI.01659-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyodo K, et al. ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J Virol. 2013;87(1):163–176. doi: 10.1128/JVI.02383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyodo K, et al. Phosphatidic acid produced by phospholipase D promotes RNA replication of a plant RNA virus. PLoS Pathog. 2015;11(5):e1004909. doi: 10.1371/journal.ppat.1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact. 2008;21(8):1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 31.Diaz A, Wang X. Bromovirus-induced remodeling of host membranes during viral RNA replication. Curr Opin Virol. 2014;9:104–110. doi: 10.1016/j.coviro.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 33.Pogány M, et al. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009;151(3):1459–1475. doi: 10.1104/pp.109.141994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadota Y, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54(1):43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Fang Q, Zhang Z, Wang Y, Zheng X. The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J Exp Bot. 2009;60(11):3109–3122. doi: 10.1093/jxb/erp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komoda K, Naito S, Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc Natl Acad Sci USA. 2004;101(7):1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyodo K, Kaido M, Okuno T. Traffic jam on the cellular secretory pathway generated by a replication protein from a plant RNA virus. Plant Signal Behav. 2014;9(3):e28644. doi: 10.4161/psb.28644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18(1):30–40. doi: 10.1016/j.tplants.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JP, Xu YP, Munyampundu JP, Liu TY, Cai XZ. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol Genet Genomics. 2016;291(2):661–676. doi: 10.1007/s00438-015-1137-0. [DOI] [PubMed] [Google Scholar]
- 40.Schulz P, Herde M, Romeis T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013;163(2):523–530. doi: 10.1104/pp.113.222539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishida S, Yuasa T, Nakata M, Takahashi Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell. 2008;20(12):3273–3288. doi: 10.1105/tpc.107.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asai S, et al. The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J Biol Chem. 2013;288(20):14332–14340. doi: 10.1074/jbc.M112.448910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Alteration of substrate specificity: The variable N-terminal domain of tobacco Ca(2+)-dependent protein kinase is important for substrate recognition. Plant Cell. 2010;22(5):1592–1604. doi: 10.1105/tpc.109.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7(8):504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefano G, Hawes C, Brandizzi F. ER - The key to the highway. Curr Opin Plant Biol. 2014;22:30–38. doi: 10.1016/j.pbi.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee CC, et al. Promotion of Bamboo Mosaic Virus accumulation in Nicotiana benthamiana by 5′→3′ exonuclease NbXRN4. Front Microbiol. 2016;6:1508. doi: 10.3389/fmicb.2015.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amatore D, et al. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell Microbiol. 2015;17(1):131–145. doi: 10.1111/cmi.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 2016;10(7):1742–1754. doi: 10.1038/ismej.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135(4):691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erickson JR, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gullberg RC, Jordan Steel J, Moon SL, Soltani E, Geiss BJ. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology. 2015;475:219–229. doi: 10.1016/j.virol.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15(3):329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Scaffold function of Ca2+-dependent protein kinase: Tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol. 2014;165(4):1737–1750. doi: 10.1104/pp.114.236448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy Choudhury S, Pandey S. The role of PLDα1 in providing specificity to signal-response coupling by heterotrimeric G-protein components in Arabidopsis. Plant J. 2016;86(1):50–61. doi: 10.1111/tpj.13151. [DOI] [PubMed] [Google Scholar]
- 55.Hyodo K, Kaido M, Okuno T. Host and viral RNA-binding proteins involved in membrane targeting, replication and intercellular movement of plant RNA virus genomes. Front Plant Sci. 2014;5:321. doi: 10.3389/fpls.2014.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanfaçon H. Plant translation factors and virus resistance. Viruses. 2015;7(7):3392–3419. doi: 10.3390/v7072778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakasugi K, et al. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One. 2013;8(3):e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001;25(2):237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- 59.Kaido M, et al. GAPDH--a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PLoS Pathog. 2014;10(11):e1004505. doi: 10.1371/journal.ppat.1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyodo K, et al. Identification of amino acids in auxiliary replicase protein p27 critical for its RNA-binding activity and the assembly of the replicase complex in Red clover necrotic mosaic virus. Virology. 2011;413(2):300–309. doi: 10.1016/j.virol.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 61.Drerup MM, et al. The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol Plant. 2013;6(2):559–569. doi: 10.1093/mp/sst009. [DOI] [PubMed] [Google Scholar]
- 62.Takeda A, et al. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24(17):3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong ZG, Lommel SA. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology. 1991;182(1):388–392. doi: 10.1016/0042-6822(91)90687-7. [DOI] [PubMed] [Google Scholar]
- 64.Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cdna and effects of 5′ extensions on transcript infectivity. Virology. 1987;158(1):259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 65.Mine A, et al. Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology. 2010;407(2):213–224. doi: 10.1016/j.virol.2010.07.038. [DOI] [PubMed] [Google Scholar]