Significance
Newly replicated sister chromatids are held together by cohesin to facilitate chromosome segregation and DNA repair. The ring-shaped cohesin complex encircles DNA at specific chromosomal sites, including sites that encode genes. Transcription by RNA polymerase pushes cohesin complexes off genes, but the effect of mobilization on cohesin function is not known. Here a system to study the fate of cohesin during transcription in yeast is described. The data indicate that transcription moves cohesin off genes by sliding the complexes along DNA. The mobilized complexes continue to hold sister chromatids together while being moved. This study demonstrates that sister chromatid cohesion and transcription are mutually compatible because cohesin complexes retain their function when mobilized by RNA polymerase.
Keywords: cohesin, sister chromatid cohesion, transcription, URA3, poly(dA:dT)
Abstract
The ring-shaped cohesin complex orchestrates long-range DNA interactions to mediate sister chromatid cohesion and other aspects of chromosome structure and function. In the yeast Saccharomyces cerevisiae, the complex binds discrete sites along chromosomes, including positions within and around genes. Transcriptional activity redistributes the complex to the 3′ ends of convergently oriented gene pairs. Despite the wealth of information about where cohesin binds, little is known about cohesion at individual chromosomal binding sites and how transcription affects cohesion when cohesin complexes redistribute. In this study, we generated extrachromosomal DNA circles to study cohesion in response to transcriptional induction of a model gene, URA3. Functional cohesin complexes loaded onto the locus via a poly(dA:dT) tract in the gene promoter and mediated cohesion before induction. Upon transcription, the fate of these complexes depended on whether the DNA was circular or not. When gene activation occurred before DNA circularization, cohesion was lost. When activation occurred after DNA circularization, cohesion persisted. The presence of a convergently oriented gene also prevented transcription-driven loss of functional cohesin complexes, at least in M phase-arrested cells. The results are consistent with cohesin binding chromatin in a topological embrace and with transcription mobilizing functional complexes by sliding them along DNA.
The protein complex known as cohesin organizes eukaryotic genomes into structures that segregate faithfully between dividing cells. The complex was first discovered for its role in mediating sister chromatid cohesion, but it is now known to participate in numerous aspects of chromosome biology (1, 2). Cohesin is clinically relevant because mutations in subunits of the complex or in factors that load and activate the complex lead to developmental disorders such as Cornelia de Lange syndrome, Roberts syndrome, and Warsaw breakage syndrome (3, 4).
Cohesin is a ring-shaped complex composed of four subunits named Smc1, Smc3, Scc3, and Mcd1/Scc1 in budding yeast. The complex is thought to embrace DNA topologically with chromatin fibers passing through a central open channel (5–7). A substantial body of work supports a model in which cohesion arises from single cohesin complexes that embrace both sister chromatids [the double embrace model (6)]. Whether the central channel is large enough to accommodate both chromatids has recently been brought into question (8). In addition, the behavior of cohesin has not always been consistent with a double embrace by single complexes (for examples, see refs. 9–11). These situations usually involve a perturbation that causes loss of cohesion but not a loss of cohesin binding from chromatin. Such results force consideration of alternative models in which cohesin complexes embrace only one chromatid topologically and interact with other cohesin complexes or chromatin features to mediate cohesion (9, 11, 12).
Cohesin binds densely in centromeric regions of chromosomes where it helps mount sister chromatids onto spindle microtubules from opposing poles (biorientation). Cohesin also participates in the repair of damaged DNA, binding large regions surrounding double-stranded breaks. In the absence of damage, cohesin still binds chromosome arms at somewhat regular intervals, averaging about once per 10–15 kb in yeast (13–16). In both Saccharomyces cerevisiae and Schizosaccharomyces pombe, many of these sites correspond to locations where transcription units converge. Because cohesion occurs along the entire length of sister chromatids, it might be assumed that all cohesin on chromosome arms participates in cohesion. Rarely, however, has the contribution of complexes at individual sites been tested (for exceptions, see refs. 9, 17, 18). Binding at only a subset of chromosomal sites would likely be sufficient to hold chromosome arms together (9). To complicate matters, at any given site, cohesion might occur in some cells but not in others. That the cellular level of cohesin can be lowered by 85% in yeast without an overt increase in sister chromatid separation indicates that there are more complexes than necessary to achieve genome-wide cohesion (19).
The tight topological embrace of chromatin by cohesin raises the question of whether transcriptional elongation and sister chromatid cohesion are compatible. Work with representative RNA polymerase II genes showed that transcription causes cohesin to move from initial positions on ORFs, often leading to enrichment at downstream regions (15, 16, 20–22). Mobilization by an advancing RNA polymerase has been invoked to account for the redistribution. One model holds that RNA polymerase slides cohesin to new locations. An alternative model holds that RNA polymerase passage evicts cohesin complexes from the DNA, followed by subsequent rebinding downstream. Partial or complete disassembly of cohesin during transcription would lead to cohesion loss in an M phase cell because cohesion is only established during S phase progression (22–25).
In this study we sought to characterize the function and fate of cohesin complexes on a model eukaryotic gene in the yeast S. cerevisiae. We evaluated cohesion by converting a genomic region of interest into an extrachromosomal DNA circle in M phase-arrested cells. We used the approach to identify sequences responsible for loading functional cohesin complexes onto DNA and then investigated the fate of those complexes upon transcriptional induction. The closed circular nature of the DNA templates after DNA circularization allowed us to distinguish between models for cohesin mobilization. Our data are consistent with the transcription-driven repositioning of functional cohesin complexes by sliding.
Results
Cohesion of the URA3 Gene.
This study focuses on the well-studied reporter gene URA3 that encodes orotidine-5′-phosphate decarboxylase of the pyrimidine biosynthetic pathway. The gene is expressed at low basal levels in rich media and induced severalfold by a transcriptional activator upon uracil depletion (26). The domain of chromosome V that bears URA3 has frequently been used as a reporter of genome-wide cohesion (1). Cohesin binds in and around the gene, but whether these specific complexes contribute to cohesion of the chromosomal domain is not known (15, 16, 27).
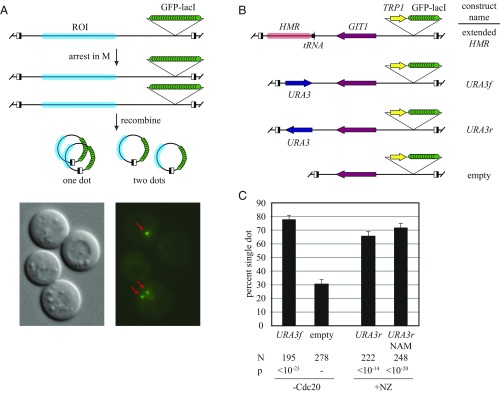
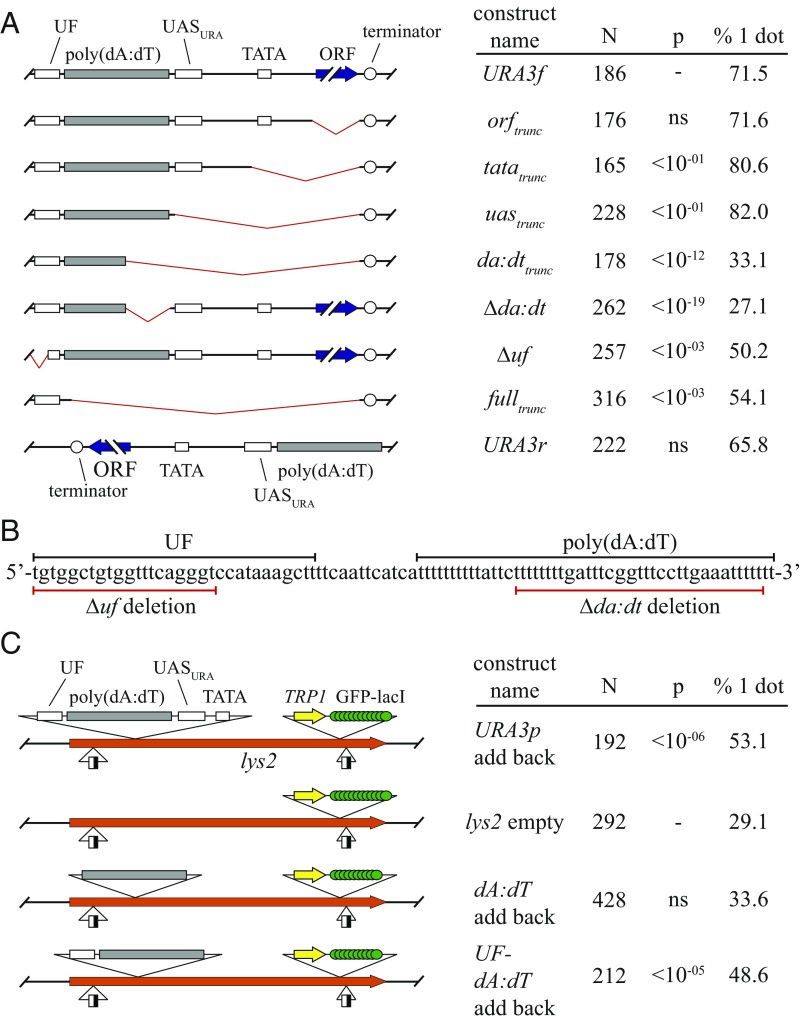
An inducible recombination assay was used to study cohesion mediated by URA3. In the assay, a chromosomal region of interest (ROI) is tagged with GFP-lacI and flanked by RS target sites for the R site-specific recombinase (Fig. 1A). Such constructs are termed excision cassettes. Induction of the recombinase in M phase-arrested cells creates a pair of DNA circles that yield one dot of GFP fluorescence if they cohere or two foci if they do not. In this way, the cohesive behavior of a specific chromosomal domain can be evaluated independently of other domains of chromosomal cohesion. Previous work in our laboratory used the assay to study the heterochromatic HMR locus (9, 28). The work described herein started with an HMR excision cassette (Fig. 1B). All sequences corresponding to HMR and the adjacent tRNA gene were replaced with a 1.1 kb DNA segment containing the URA3 ORF and all known URA3 regulatory elements (construct URA3f, Fig. 1B). A similar construct with a slightly smaller DNA segment was generated with the gene in the opposite orientation (construct URA3r). URA3 was induced normally in these ectopic positions (Fig. S1A). Unless otherwise specified, uracil was included in the growth media to suppress transcriptional activation. A third construct, named empty, was generated by omitting all URA3 sequences.
Fig. 1.
URA3 supports cohesion. (A) Schematic of experimental protocol. ROI, region of interest. Half-filled boxes represent RS sites for the R site-specific recombinase. Fluorescence micrographs of strain JSC97 are provided. Two large-budded, M phase-arrested cells are shown, one with a single focus of GFP-lacI fluorescence and the other with two foci of GFP-lacI fluorescence (red arrows). (B) Schematic of the constructs used. (C) Cohesion measurements. Strains MSB7 (URA3f) and MSB13 (empty) were arrested in M phase by Cdc20 depletion, whereas JSC97 (URA3r) was arrested with nocodazole (+NZ). Nicotinamide (NAM; final concentration, 2 mM) was added to half of the JSC97 culture 2 h after DNA circularization, and cells were harvested 2 h later. N denotes the number of cells examined. P values for pairwise χ2 tests are presented relative to a benchmark strain designated with a dash.
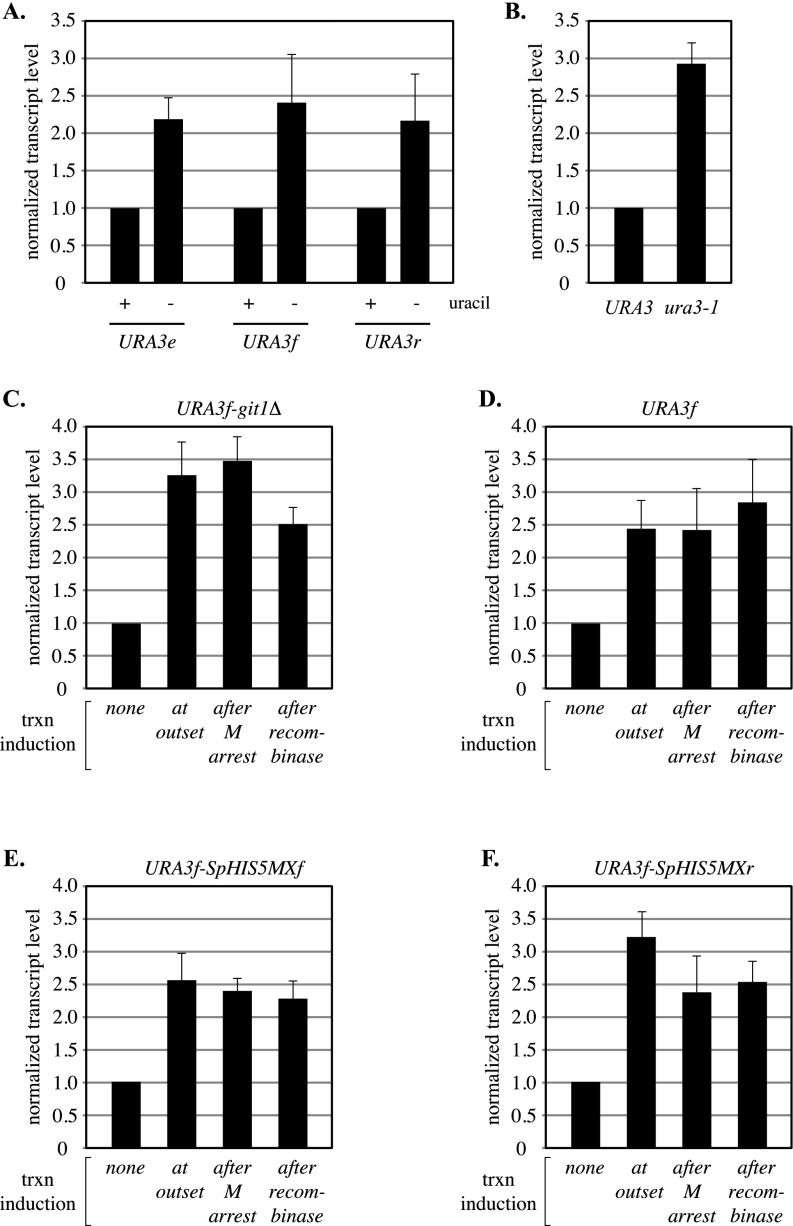
Fig. S1.
Transcript analysis of URA3. (A) Relative induction levels of ring-borne URA3 genes and the endogenous locus. Control strain PJ1 with URA3 at its normal location (labeled URA3e) was grown in SC media either containing or lacking uracil. Strains MSB14 (URA3f) and MSB20 (URA3r) were grown similarly, but methionine was withheld until midlog phase, when methionine was added to induce M phase arrest by Cdc20 depletion. DNA circles were formed by the addition of galactose and RNA extracts were prepared two hours later. URA3 transcript levels were normalized first to an internal control (ACT1 mRNA) and then to the uninduced condition for each strain (+uracil). The data show that ring-borne URA3 genes were induced at endogenous levels. (B) Effect of Ura3 activity on transcription. The ura3-1 mutation of strain W303-1A (glycine to glutamate at amino acid 234) causes uracil auxotrophy by disrupting the Ura3 active site (51). Isogenic strains PJ1 (URA3) and W303-1A (ura3-1) were grown in YPDA media. After normalization to ACT1 mRNA, samples were normalized to strain PJ1. The data show that ura3 transcription elevated in the absence of Ura3 activity. (C–F) Effect of induction timing on URA3 transcriptional output. Induction (uracil depletion) was initiated at each of the following stages: (i) at the outset of the experiment, (ii) following M phase arrest but before DNA circle formation, or (iii) following DNA circle formation (Fig. 6A). Strains MSB105 (URA3f-Δgit1), MSB14 (URA3f), MSB110 (URA3f-SpHISMXf), and MSB112 (URA3f-SpHISMXr) were used. The data show that transcriptional output of URA3 was not affected by the time of induction during the cohesion assay. Note that URA3r DNA circles did not induce fully after circularization and were therefore not used for this set of experiments.
DNA circles were generated efficiently from the excision cassettes in M phase-arrested cells (Fig. S2). Fig. 1C shows that DNA circles bearing the uninduced URA3 gene, irrespective of orientation, were cohered in the majority of cells examined. On the other hand, the empty DNA circles lacking URA3 yielded a low level of cohesion, a level that was comparable to earlier negative control DNA circles (28). Sir2 inhibitors, like nicotinamide (NAM), abolish heterochromatic cohesion (9). That NAM did not diminish cohesion here indicated that the mechanism holding these DNA sequences together did not stem from unanticipated residual heterochromatin (Fig. 1C). These data indicate that the URA3 gene alone is sufficient to impart cohesion to a chromosomal domain.
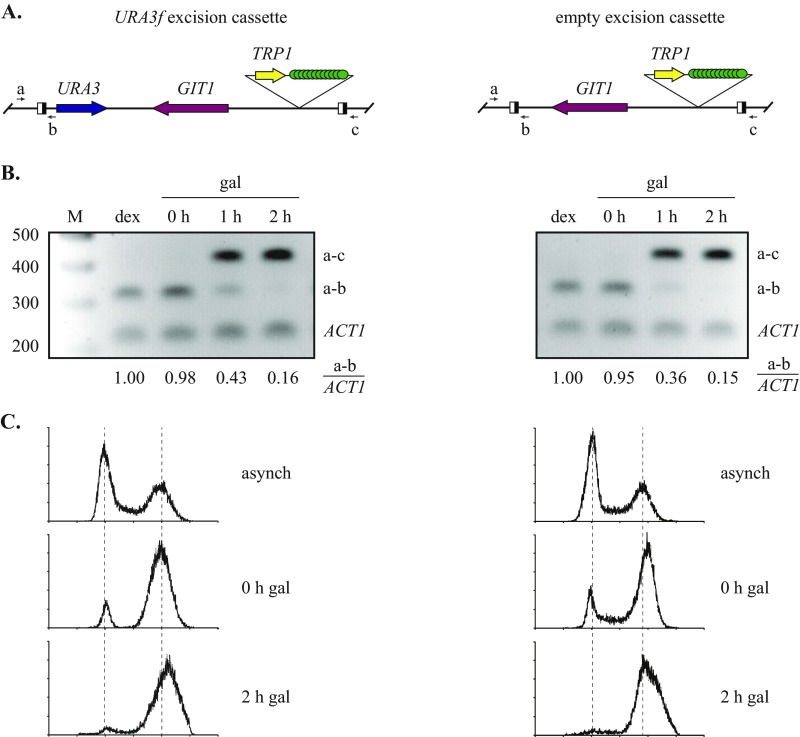
Fig. S2.
Validation of DNA excision and cell cycle arrest. (A) Diagram of the binding sites for the primers used to monitor excision (Table S2). (B) Time course of excision. Strains MSB7 (URA3f) and MSB13 (empty) were first grown asynchronously in dextrose, subcultured in raffinose, and then arrested in M phase by Cdc20 depletion. At 0, 1, and 2 h after the addition of galactose, aliquots of cell culture were harvested. One part was used to make DNA extracts, and the other was fixed with ethanol and stained with SYTOX Green (Molecular Probes). qPCR of the extracts was used to determine the extent of excision (primers a and b) using ACT1 as an internal control. The values reported under each gel lane are the average of two independent trials. The data indicate that excision reached 85% completion in the 2-h time frame of a typical cohesion assay. Ethidium bromide-stained agarose gels show the progression of excision. Each lane contains three pooled PCR reactions (a–c, a–b, and ACT1) after 24 amplification cycles. Lane M, 100 bp ladder. (C) DNA histograms during the cell cycle arrest. Flow cytometry was performed at the Rutgers Environmental and Occupational Health Sciences Institute core facility. The results show that a significant M phase arrest was maintained over the 2-h time frame of a typical cohesion assay.
URA3 Cohesion Requires the Cohesin Pathway.
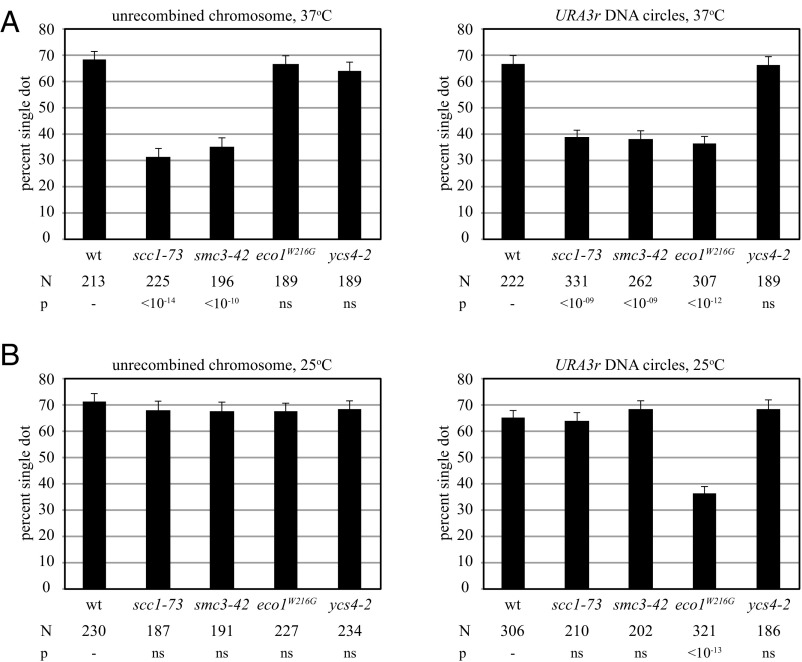
Cohesion of the left arm of chromosome V near the endogenous URA3 locus relies on cohesin, as well as condensin (27). To determine whether these protein complexes participated in cohesion of URA3 specifically, URA3r DNA circles were monitored in strains with conditional mutations in the cohesion and condensation pathways. Cultures were shifted from permissive to nonpermissive temperature (from 25 to 37 °C) after initiating recombination during the M phase arrest. The unrecombined chromosomal arm was also evaluated by omitting the recombination step. Fig. 2A shows that inactivation of Mcd1 and Smc3 cohesin subunits caused loss of cohesion for both the DNA circles and chromosome arm. Inactivation of condensin subunit Ycs4, on the other hand, did not disrupt cohesion of either. These results indicate that cohesion of URA3 requires cohesin but not condensin at this ectopic location.
Fig. 2.
URA3 cohesion requires activated cohesin but not condensin. Strains JSC97 (wt), JSC96 (scc1-73), MSB147 (smc3-42), MSB157 (eco1W216G), and JSC164 (ycs4-2) bearing the URA3r excision cassette were grown at 25 °C and arrested in M phase with nocodazole. To examine the unrecombined chromosome, dextrose was added, and cells were fixed 2 h later. To examine DNA circles, galactose was added. After 2 h, the cultures were split, and half was shifted to 37 °C for 2 additional hours (A). The other half was left at 25 °C for 2 h (B). ns, nonsignificant.
ECO1 acetylates residues in the cohesin subunit Smc3 to establish cohesion (29, 30). The conditional eco1W216G allele contains a missense mutation that corresponds to a pathogenic change in the orthologous human ESCO2 gene that causes Roberts syndrome (31, 32). Inactivation of Eco1 did not lead to an obvious defect in cohesion of the chromosomal arm (Fig. 2A) (also see ref. 33), nor did it affect cohesion of a circular, CEN/ARS minichromosome (Fig. S3A). These results are expected because Eco1 normally establishes cohesion during S phase. Inactivation of the establishment factor during M phase should not alter preexisting cohesion.
Fig. S3.
Effect of the eco1W216G mutation on cohesion. (A) Analysis of a CEN/ARS minichromosome. A GFP-tagged, CEN/ARS minichromosome (pJSC2) was constructed in yeast by joining plasmids pRS416 and pSR10 via in vivo recombination, according to the method of ref. 52. pJSC2 is comparable in size to DNA circles bearing either HMR or URA3. Stains JSC209 (wt), JSC81 (eco1W216G), and JSC208 (smc3-42) containing pJSC2 were grown overnight in SC-trp plus dextrose at 25 °C. An equal volume of YPAD was added when the cultures reached midlog, and 20 min later, nocodazole was added. After 3 h, half of the culture was shifted to 37 °C. Cohesion was evaluated after 2 additional hours. The data show that cohesion of a circular, CEN/ARS minichromosome was not diminished by eco1W216G. (B) Analysis of HMR. DNA circles bearing HMR were examined in nocodazole-arrested strains RDY215 (wt) and JSC52 (eco1W216G) at 25 °C per the cohesion assay. To examine the unrecombined chromosome, cells were grown to midlog in dextrose at 25 °C and arrested with nocodazole. The data show that cohesion of HMR DNA circles was abolished in the eco1W216G mutant, whereas cohesion of the unrecombined chromosomal arm was not.
Surprisingly, cohesion of URA3r DNA circles was diminished in the eco1W216G mutant, even at the permissive temperature for the strain (Fig. 2). Similar results were obtained with DNA circles bearing heterochromatic HMR, indicating that the effect was not specific to URA3 (Fig. S3B). The results demonstrate that the eco1W219G allele is not fully functional at 25 °C. We hypothesize that the DNA circles of the cohesion assay are sensitive reporters of eco1 impairment because they permit evaluation of cohesion at individual loci. The redundancy of sites where cohesin acts in an unrecombined chromosome arm, as well as the robust binding of cohesin near a centromere, may mask partial loss of Eco1 activity on chromosomes.
Sequence Determinants for Cohesion of URA3.
Deletions were made within the URA3 locus to identify elements required for cohesion. Note that ura3 mutants accumulate upstream pathway metabolites that induce the activity of Ppr1, the transcriptional activator that operates on URA3 (Fig. S1B) (34). As discussed in depth below, URA3 induction alters cohesion of the locus. Therefore, all experiments using truncated ura3 constructs were performed in strains that contained a second functional URA3 allele either from S. cerevisiae or Kluyveromyces lactis.
Deletion of the URA3 ORF alone did not disrupt cohesion of the locus (Fig. 3A, construct orftrunc). Additional truncations through the URA3 promoter showed that the TATA element and the binding site for Ppr1 (UASURA) were also not required (constructs tatatrunc and uastrunc, respectively). By contrast, a more extreme truncation that deleted part of a poly(dA:dT) tract caused complete loss of cohesion (construct da:dttrunc). Moreover, deletion of part of the 62 bp poly(dA:dT) tract from an otherwise intact URA3 locus also caused complete loss of cohesion (construct Δda:dt, Fig. 3 A and B). Homopolymeric dA:dT tracts commonly reside in yeast promoters where their unique structural and mechanical properties, along with the RSC chromatin remodeling complex, help form nucleosome free regions (35, 36). The poly(dA:dT) tract of URA3 plays precisely this role in organizing chromatin (37). The cohesin loading complex (Scc2-Scc4) has been found at A-T rich DNA both in vitro and in vivo (8, 38). Together, these findings suggest that the poly(dA:dT) tract helps load functional cohesin complexes that generate cohesion at URA3.
Fig. 3.
Sequence elements in the URA3 promoter are necessary and sufficient for cohesion. (A) The poly(dA:dT) tract is necessary for cohesion. Strains MSB7 (URA3f), HJ5 (orftrunc), HJ6 (tatatrunc), HJ7 (uastrunc), HJ19 (da:dttrunc), HJ4 (fulltrunc), and HJ24 (Δuf) were arrested in M phase by Cdc20 depletion. Strains HJ6388 (Δda:dt) and JSC97 (URA3r) were arrested with nocodazole. (B) DNA sequence surrounding the poly(dA:dT) element in the URA3 promoter. Underlines highlight the portions of the element that were deleted in the Δda:dt and Δuf constructs. (C) The poly(dA:dT) and UF elements generate cohesion in the lys2 context. Strains CSW91 (lys2 cassette alone), HJ22 (URA3 promoter add back), HJ23 (UF-dA:dT add back), and MSB155 (dA:dT alone add back) were arrested in M phase with nocodazole.
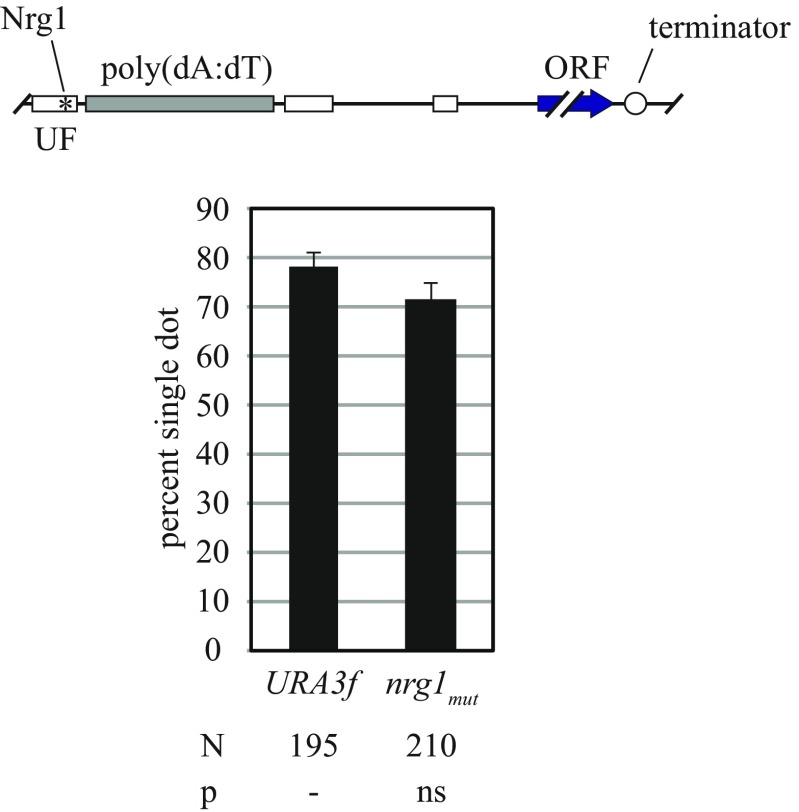
Evidence that the poly(dA:dT) did not act alone was obtained from experiments in which URA3 promoter fragments were transferred to a different excision cassette assembled at the lys2 locus (Fig. 3C). Transfer of the entire promoter was sufficient for cohesion of lys2 DNA circles. By contrast, transfer of the poly(dA:dT) tract yielded only basal cohesion levels. When the poly(dA:dT) tract was transferred along with 31 bps of DNA that flank the upstream end of the tract (designated the UF, hereafter), cohesion was restored. These data suggest that the UF augments the function of the poly(dA:dT) tract in cohesion. In support, disruption of the UF in an otherwise intact URA3f excision cassette (construct Δuf) reduced cohesion to an intermediate level (Fig. 3 A and B). URA3r lacks the UF entirely, yet the construct supports cohesion, suggesting the UF might not always be required (Fig. 3A). It is not clear what features of the UF are relevant. The element contains a putative binding site for Nrg1, a transcriptional repressor that regulates carbon response (39). However, cohesion was not altered significantly when the Nrg1 binding site was mutated (Fig. S4). Conceivably, the UF recruits a factor that facilitates activity of poly(dA:dT) or instead promotes a favorable chromatin context for the nucleosome-free poly(dA:dT) tract.
Fig. S4.
Effect of the Nrg1 site in the URA3 promoter on cohesion. DNA circles in strains MSB7 (URA3f) and HJ25 (nrg1mut) were compared in the cohesion assay after M phase arrest by Cdc20 depletion. In strain HJ25, the Nrg1 binding site of the URA3f excision cassette was mutated from 5′-AGGGTCC-3′ to 5′-AaaaTCC-3′. The data show that the Nrg1 site did not contribute to URA3 cohesion.
One construct in this series of experiments stood out as an outlier (construct fulltrunc). In this case, everything between the UF and the URA3 terminator was deleted, including the poly(dA:dT) tract. Nevertheless, an intermediate level of cohesion was observed (Fig. 3A). We speculate that the large deletion placed the UF in sufficient proximity to (dA:dT) features in the URA3 terminator to yield cohesion.
Cohesin Binding to URA3.
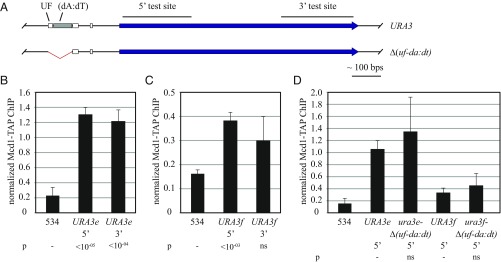
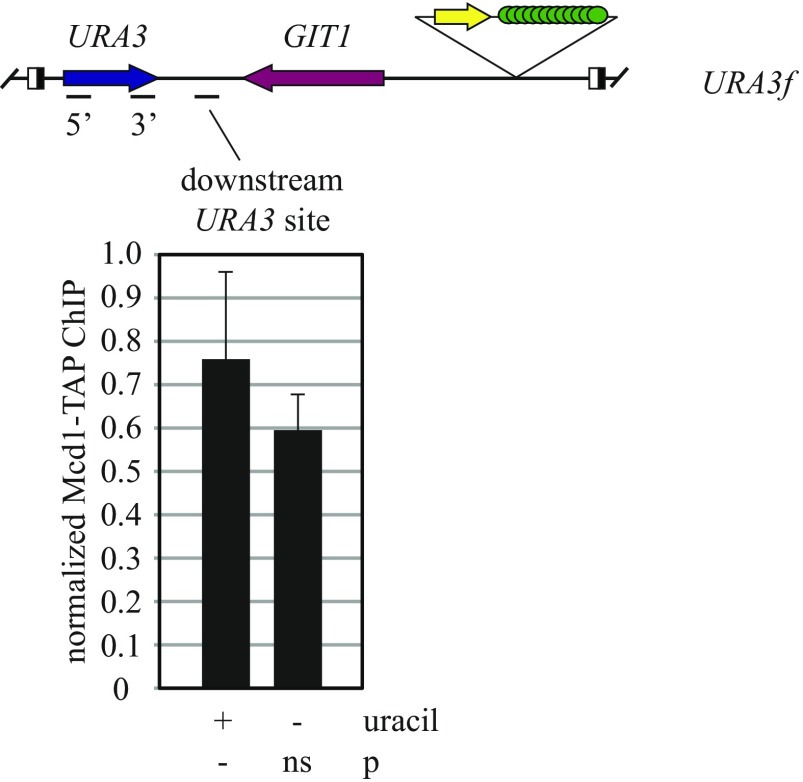
Chromatin immunoprecipitation (ChIP) was used to measure binding of cohesin to URA3 using a TAP-tagged Mcd1 cohesin subunit. Binding was evaluated in M phase-arrested cells under growth conditions in which the gene was not induced. Binding to the URA3 ORF at both the 5′ and 3′ ends was normalized to a well-documented binding site and compared with a negative control site (549.7 kb and 534 kb on chromosome V, respectively). At the endogenous location of URA3 (labeled URA3e for clarity), cohesin was detected at both 5′ and 3′ ends of the gene (Fig. 4 A and B). At the URA3f excision cassette, lower but measurable levels were detected, at least at the 5′ end of the gene (Fig. 4C). Despite the drop, the inclusion of URA3 in the cassette provides sufficient functional cohesin for cohesion (Fig. 1C).
Fig. 4.
Binding of cohesin to URA3. (A) Maps of the constructs used with regions tested by ChIP highlighted. (B and C) ChIP of TAP-tagged Mcd1 cohesin subunit. Binding was measured in strains MSB37 (URA3e, where e denotes the endogenous locus) and MSB19 (URA3f). Primers used to evaluate the 5′ and 3′ test sites are listed in Table S2. P values were obtained by Student’s t tests and are presented relative to a negative control site designated with a dash. ns, not significant. (D) Effect of the Δ(uf-da:dt) deletion on cohesin binding. Strains MSB37, MSB19, MSB153 [URA3e Δ(uf-da:dt)], and MSB156 [URA3f Δ(uf-da:dt)] were used as in B and C.
To measure the effect of the URA3 poly(dA:dT) tract on cohesin binding, the region spanning the poly(dA:dT) and UF elements was deleted, and the ChIP analysis was repeated. Surprisingly, the deletions had no measurable effect on the level of cohesin binding, either at the endogenous locus or the URA3f excision cassette (Fig. 4D). These findings indicate that the bulk of the cohesin in and about URA3 arises from complexes that load independently of the poly(dA:dT) tract. Moreover, based on the cohesion assays in Fig. 3A, the data suggest these complexes do not participate in cohesion of DNA circles. These results indicate that the DNA ring assay for cohesion provides a more reliable measure of functional cohesin complexes than ChIP, which yields a positive signal whether the complexes are functional or not.
Transcription and the Fate of URA3 Cohesion.
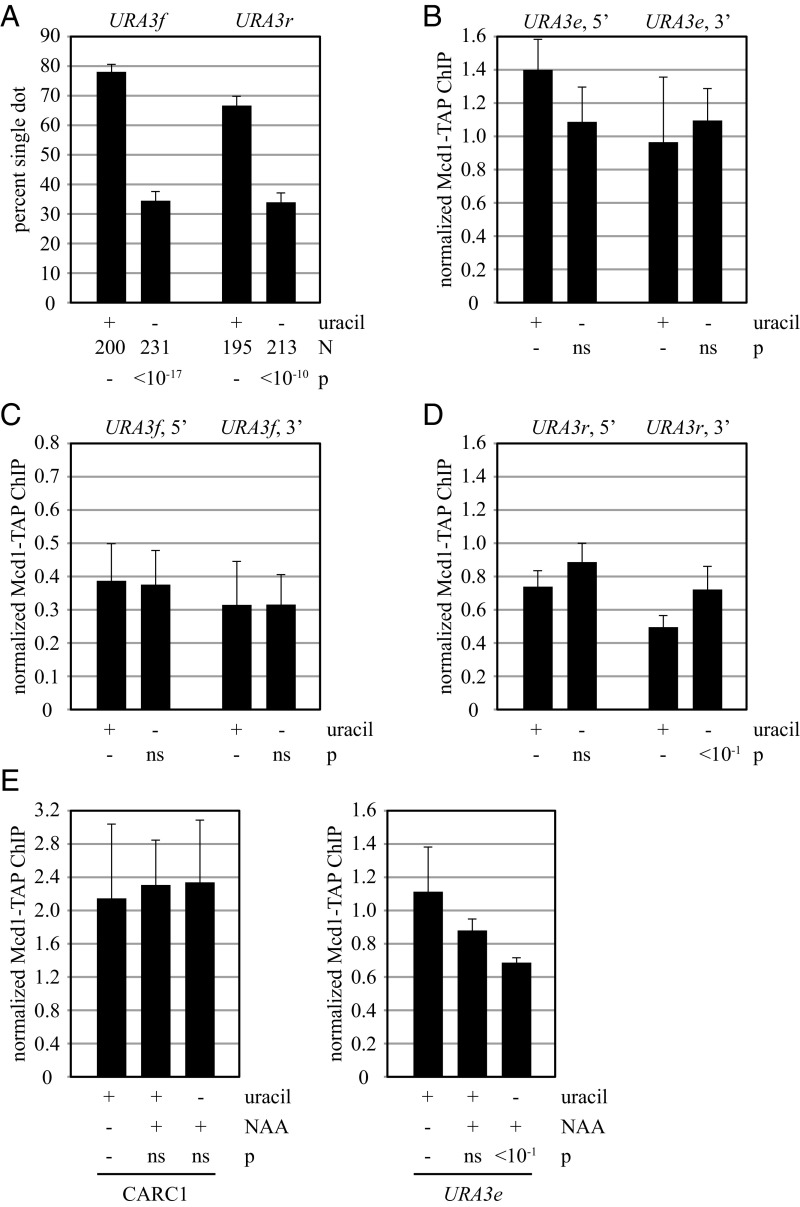
Transcription of URA3 increases roughly 2.5-fold when uracil is omitted from the growth media (Fig. S1A). To determine whether transcriptional induction alters cohesion of the gene, DNA circles were formed in strains that had been grown continuously in the absence of uracil. Strikingly, the modest increase in URA3 transcription abolished cohesion of both the URA3f and URA3r DNA circles (Fig. 5A). The data suggest that transcription prevents the accumulation of functional cohesin complexes on the URA3 excision cassettes. The results are consistent with previous reports of transcription altering the distribution of cohesin on chromatin (15, 16, 20–22, 40).
Fig. 5.
Transcriptional induction of URA3 abolishes cohesion but not cohesin binding. (A) Cohesion of DNA circles bearing the URA3 gene. Strains MSB7 (URA3f) and MSB20 (URA3r) were grown in media containing or lacking uracil from the outset of the experiment. (B) ChIP of Mcd1-TAP at chromosomal URA3 genes. Strain MSB37 (URA3e) was grown in media containing or lacking uracil from the outset and cross-linked with formaldehyde following M phase arrest by Cdc20 depletion. (C and D) Strains MSB19 (URA3f) and JSC230 (URA3r) were evaluated as in B. (E) Transcription-dependent loss of cohesin from URA3 in the absence of Scc2. Strain MSB128 (SCC2-AID-9myc ADH1p-OsTIR1) was arrested in M phase by Cdc20 depletion. Three hours later the culture was split into thirds. DMSO was added to one third, and 500 μM NAA was added to another. Uracil was removed from the final third when 500 μM NAA was added. Cohesin levels were assessed 2 h later at the 5′ end of the endogenous URA3 gene, as well as at a site on chromosome III (CARC1) that binds cohesin stably (42).
ChIP of Mcd1-TAP was used to measure how transcriptional induction affected cohesin binding. As in the case of deleting the poly(dA:dT) and UF elements, cohesin levels at the 5′ and 3′ ends of the endogenous URA3 gene were largely unchanged by elevated transcription (Fig. 5B). Similar results were obtained from analysis of the same sites in the URA3f excision cassette (Fig. 5C), as well as a site downstream of URA3 in the cassette (Fig. S5). When the experiment was repeated with the URA3r excision cassette, the cohesin level at the 5′ end of the gene similarly did not change, and the level at the 3′ end actually increased slightly (Fig. 5D). These results indicate that nonfunctional cohesin complexes remain associated with URA3 when transcription increases and cohesion falters.
Fig. S5.
Cohesin binding between URA3 and the convergent GIT1 gene. ChIP was performed with strain MSB19 (URA3f) as in Fig. 5C. Primers used are listed in Table S2. The data show that transcriptional induction (uracil depletion) does not affect cohesin binding levels downstream of URA3.
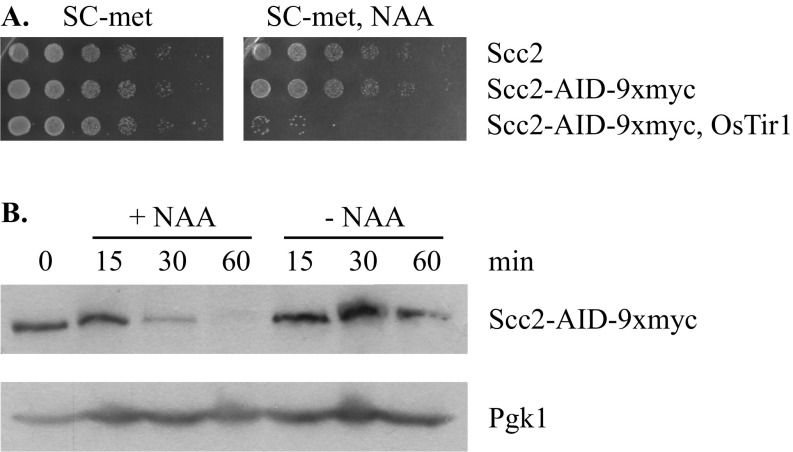
Typically, cohesin loads onto chromosomes from late G1 phase until anaphase onset, but only those complexes that load before S phase passage acquire cohesive function (ref. 25 and references therein). If transcription was mobilizing cohesin, then new complexes arriving after S phase passage would yield ChIP signals but not cohesion. To test whether this process contributed to the cohesin landscape on URA3, cohesin loading was blocked conditionally by depleting Scc2, a subunit of the cohesin loading complex. The protein was fused to an auxin-inducible degron (AID) in strains expressing the SCFOsTir1, an alternative F box protein required for SCF-mediated degradation (41). Addition of the synthetic auxin analog 1-naphthaleneacetic acid (NAA) blocked colony formation of a strain expressing both Scc2-AID and OsTir1 and caused clearance of the Scc2-AID protein in 60 min (Fig. S6). In this experiment, NAA was added simultaneously with the induction of URA3 transcription after cells had been arrested in M phase. Cohesin levels were assessed 2 h later at the endogenous URA3 gene, as well as at a site on chromosome III (CARC1) that binds cohesin stably (42). At CARC1, loss of Scc2 did not affect cohesin binding irrespective of whether the media contained uracil or not (Fig. 5E). At URA3e, on the other hand, simultaneous induction of transcription and depletion of Scc2 lowered the level of bound cohesin more than 1.5-fold (Fig. 5E). Even without transcriptional induction, depletion of Scc2 caused a slight reduction of cohesin, perhaps due to basal URA3 expression. Taken together, these results suggest that elevated transcription causes displacement of at least some cohesin from URA3. In M phase-arrested cells, Scc2 normally replaces the displaced cohesin with new nonfunctional complexes.
Fig. S6.
Conditional depletion of Scc2-AID. (A) Effect of Scc2 depletion on viability. Fivefold serial dilutions of strains MSB37 (SCC2), MSB123 (SCC2-AID-9xmyc), and MSB128 (SCC2-AID-9xmyc OsTIR1) were plated on SC-met media containing or lacking 500 μM NAA. The data show that cell growth ceased when Scc2 was depleted. (B) Time course of Scc2-AID-9xmyc depletion. Strain MSB128 was arrested in M phase by Cdc20 depletion for 3 h. Thereafter, Scc2-AID-9xmyc levels were measured at intervals following the addition of 500 μM NAA. Immunoblotting was performed as described in ref. 53 using anti-myc (9E10; Roche) and anti-Pgk1 (ab-113687; Abcam) antibodies. The data show that Scc2 levels were greatly reduced by 60 min.
Transcription Displaces Functional Cohesin Complexes by Sliding.
The experiments of the last section indicated that elevated transcription was incompatible with cohesion. They did not shed light on whether functional cohesin complexes were displaced by sliding or by eviction, as discussed in the Introduction. A cohesion assay based on DNA circles allowed us to distinguish between these two models. In a covalently closed DNA circle, transcription-driven sliding of functional cohesin complexes should not alter cohesion. If, on the other hand, transcription causes eviction of functional complexes from one or both sister chromatid circles, then cohesion should be abolished. Even if cohesin were to rebind following release, cohesion would be permanently lost in these M phase-arrested cells.
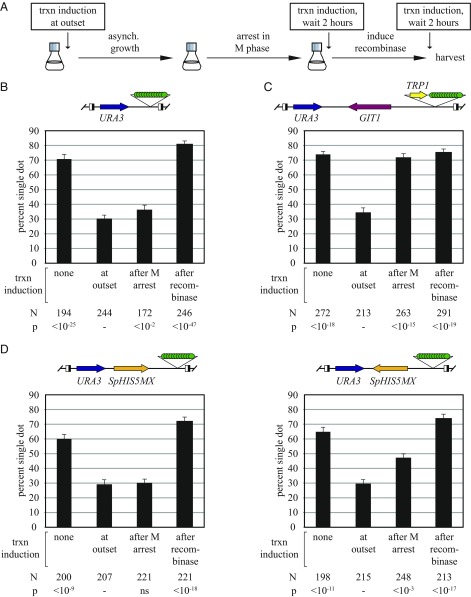
Transcription of URA3 was induced at three different stages during the cohesion assay, either (i) at the outset of the experiment, (ii) following M phase arrest but before DNA circle formation, or (iii) following DNA circle formation (Fig. 6A). The final level of transcriptional induction in each of these scenarios was comparable for the constructs examined (Fig. S1 C–F). To simplify the study, we first generated a URA3 excision cassette that lacked all other transcribed genes (construct URA3f-git1Δ). Fig. 6B shows that the resulting DNA circles were cohered in the absence of transcriptional induction and that transcriptional induction at any time before DNA circularization abolished cohesion. Cohesion was not abolished if the DNA was circularized before transcriptional induction. These results show that topological closure of DNA prevents transcription-driven loss of functional cohesin complexes.
Fig. 6.
DNA circularization or an opposing transcribed gene prevents transcription-dependent loss of URA3 cohesion. (A) Experimental flowchart. Media containing uracil was replaced with media lacking uracil to induce URA3 transcription at the various times indicated during cell growth for the cohesion assay. (B) DNA circularization before URA3 induction prevents cohesion loss. Strain MSB35 (URA3f-git1Δ) was used. (C) A convergently oriented GIT1 gene prevents cohesion loss if URA3 transcription is induced after M phase. Strain MSB7 (URA3f) was used. (D) Orientation dependence of the SpHIS5MX gene in preventing transcription-dependent loss of URA3 cohesion. Strains MSB30 (URA3f-SpHIS5MXf) and MSB31 (URA3f-SpHIS5MXr) were used.
The experiments were repeated with the URA3f excision cassette that contains the GIT1 gene oriented convergently with URA3 (Fig. 6C). Transcriptional induction from the outset of the experiment abolished cohesion, as shown first in Fig. 5A. Like the URA3f-git1Δ construct, prior DNA circularization of URA3f blocked subsequent transcription-driven loss of cohesion. The presence of the GIT1 gene imparted one important difference. Transcriptional induction after M phase arrest yet before DNA circularization also failed to abolish cohesion. The results suggest that the opposing GIT1 gene presents a barrier to transcription-driven loss of functional cohesin complexes in M phase. That escape is blocked in M phase is consistent with the maturation of DNA-bound cohesin during cell cycle progression, as discussed in more depth below.
The striking accumulation of cohesin between convergently transcribed genes in genome-wide S. cerevisiae studies suggested that gene orientation might contribute to the barrier created by GIT1. To test this notion, GIT1 was replaced with the SpHIS5MX module, a synthetic gene fusion built entirely from non-S. cerevisiae sequences (43). SpHIS5MX is transcribed in S. cerevisiae, conferring prototrophy to his3 mutants. Irrespective of orientation of the integrated SpHIS5MX module, induction of URA3 transcription from the outset of the experiment abolished cohesion, whereas induction following circularization did not (Fig. 6D). When transcription was induced following M phase arrest yet before DNA circularization, the orientation of the SpHIS5MX module affected the fate of cohesion. If SpHIS5MX was oriented in tandem with URA3, cohesion was abolished by URA3 induction. If SpHIS5MX was oriented convergently with URA3, measurable cohesion persisted despite URA3 induction. That GIT1 was more effective than SpHIS5MX at preventing cohesion loss may be related to differences in the strength of the two genes. These results show that a convergently oriented gene presents a barrier to transcription-driven loss of functional cohesin complexes in M phase-arrested cells.
Discussion
Transcription Causes Sliding of Functioning Cohesin Complexes.
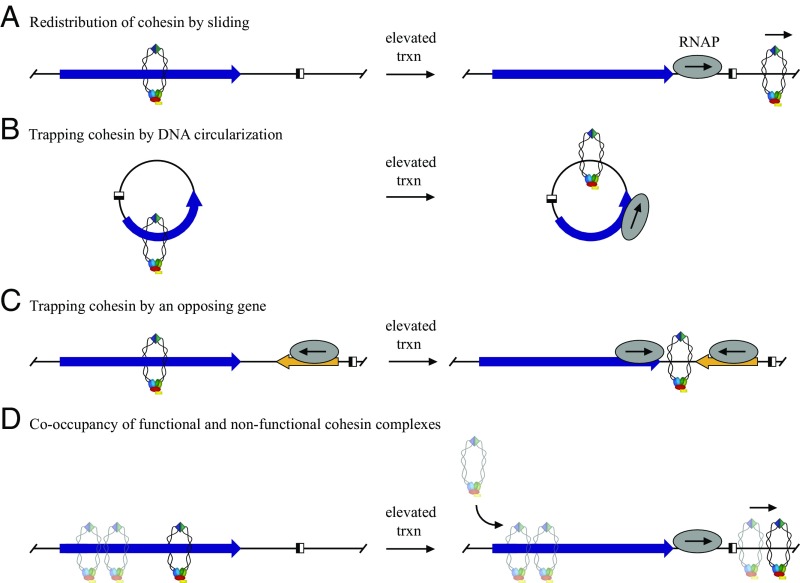
Previous studies showed that transcription mobilizes cohesin, causing a redistribution of complexes on genomic DNA. Previous studies also showed that cohesin binds DNA through a stable, topological embrace, a binding conformation that would support mobilization by sliding along DNA. Although some studies provided strong evidence for sliding of complexes in vivo (21), other studies found evidence for both sliding and eviction (22). With the DNA circle assay, we were able to use the persistence of cohesion as a definitive measure of whether functional cohesin complexes remain on DNA during transcription. We showed that URA3, a model euchromatic gene, was cohered by functional cohesin before transcriptional induction (Figs. 1 and 2) and that steady-state transcription abolished cohesion of the gene (Fig. 5). Critically, we found two conditions that prevented transcription-driven cohesion loss in M phase cells: DNA circularization before transcription and the presence of a convergently transcribed gene (Fig. 6). The two conditions are related in that both would prevent escape of cohesin complexes via sliding: topologically linked cohesin complexes would be trapped by either topological closure of DNA or by a large roadblock to cohesin passage, such as an opposing RNA polymerase (see models in Fig. 7 A–C). We conclude that transcription during M phase causes mobilization of functional cohesin complexes by sliding without dissociation from DNA. In a typical chromosomal setting, transcription would cause a localized redistribution of functional complexes without loss of sister chromatid cohesion. Although these data are consistent with a continuous topological embrace of DNA during transcription, the experiments do not address how many chromatids are held by cohesin; cohesion could arise from single complexes that embrace both chromatids or from multiple, interacting complexes that each embrace a single chromatid.
Fig. 7.
Model for binding and sliding of cohesin at URA3. Only one sister chromatid is shown for clarity. (A) Before transcriptional activation, functional cohesin binds the gene, but the complex is displaced by sliding upon collision with RNA polymerase. (B) Circularization of DNA prevents escape of functional cohesin complexes that are mobilized by transcription. (C) An opposing gene prevents escape of functional cohesin complexes. (D) Cooccupancy of functional and nonfunctional cohesin. Nonfunctional complexes are depicted as semitranslucent because their true nature of binding is not known. Upon URA3 induction, cohesin slides downstream, but additional cohesin is loaded onto the gene without acquiring cohesive function in M phase-arrested cells.
Convergently oriented genes blocked cohesion loss only when transcription was induced after M phase arrest (Fig. 6). Why did the same genes not create a barrier before M phase arrest? A likely explanation is related to the increase in cohesin binding stability achieved during S phase (44, 45). Coincident with DNA replication, acetylation of cohesin by Eco1 blocks opening of the DNA exit gate between the Smc3 and Mcd1 interface (29, 30, 46, 47). In yeast, acetylated complexes that are locked in a chromatid embrace are released only at anaphase onset by proteolytic cleavage. Before Eco1 acts in S phase, it is conceivable that unstable complexes slide without sensing roadblocks to passage. Indeed, a recent study showed that transcription redistributes cohesin on DNA even before S phase (21). After M phase arrest, however, it is clear that stabilized functional complexes cannot slide efficiently past a convergently transcribed barrier (Fig. 6). In agreement, the Gerton and Uhlmann laboratories found that cohesin mobilized by transcription in M phase-arrested cells accumulated at a convergently oriented gene (21, 22).
Comparison with the Dynamics of Cohesin in Vitro and in Vivo.
The dynamics of purified cohesin on DNA were reportedly recently (8). Using microscopy of single complexes, Stigler et al. (8) showed that after DNA binding, cohesin diffused rapidly along the contour of the DNA molecule without dissociating until reaching a DNA end. Dissociation was blocked when both DNA ends were immobilized, a constraint similar to DNA circularization. Moreover, cohesin complexes could be pushed by a simple motor protein along DNA without dissociation. In a related fashion, the heterologous T7 RNA polymerase was shown to mobilize cohesin in S. cerevisiae (21). When mobilized by yeast RNA polymerase, DNA-bound complexes did not exchange with their soluble counterparts, a result also consistent with sliding (21). Our work extends these findings by showing that cohesin can also be pushed by yeast RNA polymerase in vivo without losing the ability to hold sister chromatids together.
In the purified system, cohesin complexes migrated with a rate of about 7 kb/h on naked DNA and much slower on chromatinized template (8). In even the simplest excision cassette used here, the URA3 ORF constitutes only a small fraction of the total DNA (800 bp of 13,500 in the URA3f-git1Δ cassette; Fig. 6B). How transcription of the short gene can cause mobilization over such a great distance to escape the cassette is unclear. Inefficient termination of transcription at coding genes could result in the sliding of complexes beyond the limits of typical 3′-untranslated regions, and transcription of additional noncoding elements could move complexes still farther. Other DNA tracking enzymes might also contribute to cohesin mobility once complexes have been set in motion by transcription.
Poly(dA:dT) as a Point of Entry for Functional Cohesin Complexes.
Our analysis of URA3 truncations and deletions identified a promoter proximal poly(dA:dT) tract as a site critical for cohesion of the locus (Fig. 3). A small DNA segment immediately upstream of the tract (the UF) was also found to be important and might provide a proper context in which the poly(dA:dT) tract operates. In principle, the A-T rich sequences could act in one of two ways: as a site where functional cohesin complexes are loaded or a site where preloaded, nonfunctional complexes are made competent for cohesion. Recent work showed that the Scc2-Scc4 cohesin loading complex of S. cerevisiae associated with and helped maintain nucleosome free regions like those created by poly(dA:dT) tracts (38). Indeed, the data of Lopez-Serra et al. (38) showed a strong correlation between Scc2-Scc4 binding and the presence of poly(dA:dT) tracts. Work with proteins purified from S. pombe also showed that both cohesin and the cohesin loading complex possess an affinity toward A-T rich sequences (8). The loader increased the lifetime of complexes loaded at these sites (8, 48). Presumably, the long-lived complexes described in vitro are those that go on to create cohesion in vivo. We propose that the cohesin complexes that create cohesion at URA3 load at the poly(dA:dT) tract in the URA3 promoter.
A Distinction Between Functional and Nonfunctional Cohesin Complexes.
Despite the effect of the poly(dA:dT) tract on cohesion, deletion of the element did not reduce the level of measurable cohesin at URA3 (Fig. 4). This indicates that the complexes detected by ChIP were not those that yielded cohesion. If functional complexes were present at URA3 then why was ChIP not more informative? The simplest explanation is that functional complexes were outnumbered by nonfunctional complexes. The origin of nonfunctional complexes is not yet known, but they must accumulate via a path independent of the poly(dA:dT) tract.
Additional insight into the nonfunctional complexes came from the transcription experiments of Fig. 5. In most cases, induction of URA3 abolished cohesion without loss of cohesin (Fig. 5 A–D and Fig. S5). Only when Scc2 was depleted did cohesin levels diminish (Fig. 5E). This result suggests that transcription mobilized some of the nonfunctional complexes. Continuous loading of new nonfunctional complexes during M phase would thus mask changes in cohesin binding caused by URA3 transcription. Our findings at URA3 may seem at odds with previous studies of cohesin mobility, where transcriptional induction of model MET and GAL genes (25- and 1000-fold induction, respectively) generated cohesin-free ORFs (21, 22). In these cases, the rate of cohesin clearance by highly elevated transcription likely exceeded the rate of cohesin reloading. In the model in Fig. 7D, functional complexes are depicted alongside nonfunctional complexes (drawn as semitranslucent). Although transcription slides functional complexes (and presumably nonfunctional complexes) downstream, additional complexes that do not support cohesion are loaded after S phase.
It should be noted that nonfunctional complexes have been documented before. Upon Scc2 inactivation, which abolishes cohesion (49), cohesin still accumulated near Scc2 binding sites, perhaps owing to abortive loading reactions (15, 25). Nonfunctional cohesin was also mapped to a site downstream of HMR (denoted as tpx in ref. 28). Like URA3, cohesin levels at this site were not diminished by deleting the likely cohesin loading site (a neighboring Scc2-bound tRNA gene). The existence of nonfunctional complexes demands caution when interpreting the genomic distributions of cohesin on chromosome arms. Simply mapping binding sites does not inform about whether the complexes confer cohesion at any given position.
Materials and Methods
Yeast Strains.
Details of strain construction are provided in SI Materials and Methods. Strains are listed in Table S1, and oligonucleotides are listed in Table S2.
Table S1.
Yeast strains
| Name | Genotype | Progenitor | Ref. |
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 54 | |
| PJ1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 | W303-1A | 55 |
| RDY213 | MATa hmlΔ::(loxP-kanMX-loxP)2 hmrΔ::RS-HMR-tT(AGU)C-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 scc1-73 can1-100 his3-11,15 trp1-1 ura3-1 bar1Δ::hisG | 28 | |
| RDY215 | MATa hmrΔ::RS-HMR-tT(AGU)C-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | 28 | |
| CSW91 | MATα lys2Δ::RS-lys2-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 can1-100 his3-11,15 trp1-1 ura3-1 | 56 | |
| K5824 | MATa smc3-42 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 psi+ | 1 | |
| SG179.4 | MATα eco1W216G::hphMX ade2-1 bar1Δ can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 33 | |
| JSC52 | MATa hmrΔ::RS-HMR-tT(AGU)C-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 eco1W216G::hphMX bar1Δ can1-100 his3-11,15 trp1-1 ura3-1 | cross of RDY215 and SG179.4 | This study |
| JSC81 | MATa hmr-git1Δ::RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 eco1W216G::hphMX bar1Δ can1-100 his3-11,15 trp1-1 ura3-1 [pJSC2 (CEN/ARS/TRP1/256xlacop)] | cross of RDY215 and SG179.4 | This study |
| JSC96 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS Δhml::(loxP-kanMX-loxP)2 ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 scc1-73 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | RDY213 and PCR template strain RRY5 (H. Madhani) | This study |
| JSC97 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | RDY215 and PCR template strain RRY5 (H. Madhani) | This study |
| JSC157 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | JSC97 | This study |
| JSC164 | MATα hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 ycs4-2::natMX bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | ycs4-2 from PCR template pSO6 (B. Lavoie) | This study |
| JSC183 | MATa hmrΔ::RS-URA3f-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | RDY215 and PCR template pER7 (A. Bresson) | This study |
| JSC196 | MATa hmrΔ::RS-6lexopssEB-a2a1–256lacop-TRP1-Δhmri-RS ade2-1::HIS3p-lacGFP::ADE2 (LEU2::GAL1p-R)2::leu2-3,112 kanMX::MET3p-CDC20 can1-100 his3-11,15 trp1-1 ura3-1 | CSW19 (56) | This study |
| JSC208 | MATa hmrΔ::RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 smc3-42 can1-100 his3-11,15 trp1-1 ura3-1 [pJSC2 (CEN/ARS/TRP1/256xlacop)] | CSW48 (56) | This study |
| JSC209 | MATa hmr-git1Δ::RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 [pJSC2 (CEN/ARS/TRP1/256xlacop)] | RDY215 | This study |
| JSC230 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 MCD1-TAP::natMX kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | JSC157 | This study |
| MSB7 | MATa hmrΔ::RS-URA3f-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | cross of JSC183 and JSC196 | This study |
| MSB13 | MATa hmrΔ::RS-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5 | JSC97 | This study |
| MSB14 | MATa hmrΔ::RS-URA3f-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | MSB7 | This study |
| MSB19 | MATa hmrΔ::RS-URA3f-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 MCD1-TAP::natMX kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | MSB14 | This study |
| MSB20 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | JSC157 | This study |
| MSB30 | MATa hmrΔ::RS-URA3f-loxP-SpHIS5MXf-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | MSB7 | This study |
| MSB31 | MATa hmrΔ::RS-URA3f-loxP-SpHIS5MXr-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | MSB7 | This study |
| MSB35 | MATa hmrΔ::RS-URA3f-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | MSB31 | This study |
| MSB37 | MATa MCD1-TAP::natMX kanMX::MET3p-CDC20 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 | PJ1 | This study |
| MSB105 | MATa hmrΔ::RS-URA3f-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::SpHIS5MX | MSB35 | This study |
| MSB110 | MATa hmrΔ::RS-URA3f-loxP-SpHIS5MXf-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::hphMX | MSB30 | This study |
| MSB112 | MATa hmrΔ::RS-URA3f-loxP-SpHIS5MXr-loxP-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::hphMX | MSB31 | This study |
| MSB123 | MATα SCC2-AID-9myc::kanMX MCD1-TAP::natMX kanMX::MET3p-CDC20 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 | cross of MSB37 and YFC89 | This study |
| MSB128 | MATα SCC2-AID-9myc::kanMX trp1-1::ADH1p-OsTIR1::TRP1 MCD1-TAP::natMX kanMX::MET3p-CDC20 ade2-1 can1-100 his3-11,15 leu2-3,112 | MSB123 | This study |
| MSB147 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 smc3-42 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | smc3-42 crossed in from strain K5824 | This study |
| MSB153 | MATa MCD1-TAP::natMX kanMX::MET3p-CDC20 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3e-Δ(uf-da:dt)-loxP [pXR-klURA3-HIS3] | MSB37 | This study |
| MSB155 | MATα lys2Δ::RS-lys2-dA:dT-loxP-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 can1-100 his3-11,15 trp1-1 | CSW91 | This study |
| MSB156 | MATa hmrΔ::RS-URA3f-Δ(uf-da:dt)-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 MCD1-TAP::natMX kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3Δ::hphMX [pXR-klURA3-HIS3] | MSB19 | This study |
| MSB157 | MATa hmrΔ::RS-loxP-URA3r-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 eco1W216G::hphMX bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | cross of JSC97 and SG179.4 | This study |
| HJ4 | MATa hmrΔ::RS-ura3f-fulltrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 | MSB7 | This study |
| HJ5 | MATa hmrΔ::RS-ura3f-orftrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 | MSB7 | This study |
| HJ6 | MATa hmrΔ::RS-ura3f-tatatrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 | MSB7 | This study |
| HJ7 | MATa hmrΔ::RS-ura3f-uastrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 | MSB7 | This study |
| HJ12 | MATa hmrΔ::RS-ura3f-fulltrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 | MSB7 | This study |
| HJ19 | MATa hmrΔ::RS-ura3f-da:dttrunc-loxP-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 | MSB7 | This study |
| HJ22 | MATα lys2Δ::RS-lys2-URA3p-loxP-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 can1-100 his3-11,15 trp1-1 | CSW91 | This study |
| HJ23 | MATα lys2Δ::RS-lys2-UF-dA:dT-loxP-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 can1-100 his3-11,15 trp1-1 | CSW91 | This study |
| HJ24 | MATa hmrΔ::RS-ura3f-Δuf-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 [pXR-URA3-HIS3] | HJ12 | This study |
| HJ25 | MATa hmrΔ::RS-ura3f-nrg1mut-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 kanMX::MET3p-CDC20 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 [pXR-URA3-HIS3] | HJ12 | This study |
| HJ6388 | MATa hmrΔ::RS-ura3f-Δda:dt-GIT1-TRP1-256xlacop-RS ade2-1::HIS3p-GFPlacI::ADE2 leu2-3,112::(GAL1p-RecR::LEU2)2 bar1Δ::hisG can1-100 his3-11,15 trp1-1 ura3-1 [pXR-URA3-HIS3] | RDY215 and PCR template pER7-Δpoly(dA-dT)1 (A. Bresson) | This study |
| YFC89 | MATα SCC2-AID-9myc:kanMX ura3-1::ADH1p-OsTIR1::URA3 MCD1-TAP::natMX ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 | W303-1A | This study |
All strains were derived from W303-1A.
Table S2.
Oligonucleotides
| Name | Sequence (5′-3′) | Use | Comment |
| 5′-URA3 F | TAAGGAACGTGCTGCTACTC | ChIP-qPCR | Figs. 4 B and C and 5 B–E |
| 5′-URA3 R | TTGGCGGATAATGCCTTTAGCG | ChIP-qPCR | Figs. 4 B and C and 5 B–E |
| 5′-ScURA3 F | CATGTCGAAAGCTACATATAAGGAAC | ChIP-qPCR | Fig. 4D |
| 5′-ScURA3 R | CTTAACTGTGCCCTCCATGGAA | ChIP-qPCR | Fig. 4D |
| 3′-URA3 F | GCTCAAAGAGACATGGGTGaA (lowercase “a” mismatches in S288c) | ChIP-qPCR | Figs. 4 B and C and 5 B–D |
| 3′-URA3 R | GCCGCATCTTCTCAAATATGC | ChIP-qPCR | Figs. 4 B and C and 5 B–D |
| 549.7kb S | TGAAGATGACATTGCTCCTTT | ChIP-qPCR | All ChIP |
| 549.7kb AS | TGGATAATGGATCTGAAACCG | ChIP-qPCR | All ChIP |
| 534kb S | ACAAGCATCATTCATAGCCT | ChIP-qPCR | All ChIP |
| 534kb AS | ATCGTGGCTAGGACATTTTG | ChIP-qPCR | All ChIP |
| SL-LEU-R31F | CCTGGAATGGAGACCGTTTTCT | ChIP-qPCR | Fig. 5E |
| SL-LEU-R31B | GGGTTGTGCTATTGTCTTTATGCC | ChIP-qPCR | Fig. 5E |
| atest | TAAAATCCTCGCACTATCGCTG | qPCR | Fig. S2 |
| btest | CTATATGCAATAAAAAGGGTACAAGA | qPCR | Fig. S2 |
| ctest | GAATACAAGGTTATCAGCCGTC | qPCR | Fig. S2 |
| 5′ ACT1 (F) | CGTTACCCAATTGAACACGGT | qPCR, RT-qPCR | Figs. S1 and S2 |
| 3′ ACT1 (R) | AAGGACAAAACGGCTTGGAT | qPCR, RT-qPCR | Figs. S1 and S2 |
| URA3 RT-qPCR F | AGAATTGTCATGCAAGGGCTC | RT-qPCR | Fig. S1 |
| URA3 RT-qPCR R | TCAAATATGCTTCCCAGCCT | RT-qPCR | Fig. S1 |
| URA3f downstream F | CAAAGTTGTGCTTTGGAT | ChIP-qPCR | Fig. S5 |
| URA3f downstream R | ACTTTCTAAGACGCTAGGA | ChIP-qPCR | Fig. S5 |
| URA3p-LoxP-SpHis5MX-LoxP R | ATTTGTGAGTTTAGTATACATGCATTTACTTATAATACAGATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCT | Strain construction | Strains HJ4, HJ5, HJ6, HJ7, and HJ19 |
| URA3p-LoxP-SpHis5MX-LoxP F | ACTGCACAGAACAAAAACCTGCAGGAAACGAAGATAAATCATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain HJ5 |
| URA3pΔTATA1-LoxP-SpHis5MX-LoxP F | CAGAAGGAAGAACGAAGGAAGGAGCACAGACTTAGATTGGATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain HJ6 |
| URA3pΔUAS-LoxP-SpHis5MX-LoxP F | GTTCTTTTTTTTGATTCCGGTTTCTTTGAAATTTTTTTGA ATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain HJ7 |
| URA3pΔdA-dT1-LoxP-SpHis5MX-LoxP F | TTCAGGGTCCATAAAGCTTTTCAATTCATCTTTTTTTTTTTTGTTCTTTTATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain HJ19 |
| ΔURA3p::LoxP-SpHis5MX-LoxP F | GCATTTCTTTGTGGCTGTGGTTTCAGGGTCCATAAAGCTTATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain HJ4 |
| dAdT-LoxP-HIS5-LoxP in LYS2 F | CAAGATTTCAAATTAGACGAGTTCAAGCATCATTTAGTGGACTTTGCTTTCAATTCATCTTTTTTTTTTTTGTTCTTTTTTTTGATTCCG | Strain construction | Strain MSB155 |
| pURA3-LoxP-HIS5-LoxP in LYS2 F | CAAGATTTCAAATTAGACGAGTTCAAGCATCATTTAGTGGACTTTGCTTTTGTGGCTGTGGTTTCAGGGTC | Strain construction | Strains HJ22 and HJ23 |
| Lys2-dAdT-LoxP R | GGTGATCAGAGAGATTTTAGTTATGCAATTGGATGGATATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCTA | Strain construction | Strains HJ22, HJ23, and MSB155 |
| git1-trp1Δ::LoxP-SpHIS5MXf-LoxP F | AGTGCACAAACAATACTTAAATAAATACTACTCAGTAATAACATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain MSB30 |
| git1-trp1Δ::LoxP-SpHIS5MXf-LoxP R | AAAAGAAATTTTGAGGTTAAAGGATGGAGGAAGAAATCGATACCAATTATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCT | Strain construction | Strain MSB30 |
| git1-trp1Δ::LoxP-SpHIS5MXr-LoxP F | AATTTTGAGGTTAAAGGATGGAGGAAGAAATCGATACCAATTATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain MSB31 |
| git1-trp1Δ::LoxP-SpHIS5MXr-LoxP R | CAGGCAAGTGCACAAACAATACTTAAATAAATACTACTCAGTAATAACATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCT | Strain construction | Strain MSB31 |
| LoxP-MX-LoxP URA3 R | AGTCTGTGCTCCTTCCTTCGTTCTTCCTTCTGCTCGGAGATTACCGAAATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCT | Strain construction | Strains MSB153 and MSB156 |
| LoxP-amdMX-LoxP URA3fΔrdAdT F | TAATCCTTCACATCATGAAATATAAGCTAAATCGCATTTCTTATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain MSB156 |
| LoxP-HIS3MX-LoxP URA3ΔrdAdT F | ATTTATGGTGAAGGATAAGTTTTGACCATCAAAGAAGGTTAAATAACTTCGTATAGCATACATTATACGAAGTTATATTAAGGGTTCTC | Strain construction | Strain MSB153 |
| URA3pDupstreamdAdT F | GGTCAAATAATCCTTCACATCATGAAATATAAGCTAAATCGCATTTCTTTCCATAAAGCTTTTCAATTCATC | Strain construction | Strain HJ24 |
| upstream URA3 NRG1 to AAA | ACATCATGAAATATAAGCTAAATCGCATTTCTTTGTGGCTGTGGTTTCAAAATCCATAAAGCTTTTCAATTCATC | Strain construction | Strain HJ25 |
| frURA3t R | GACTTATGGAACATGGAACC | Strain construction | Strains HJ24 and HJ25 |
Cell Growth and Arrest.
Strains were grown for the cohesion assay in the following manner unless specified otherwise. Cell growth was initiated in YPA or SC-met drop-out media containing 2% (wt/vol) dextrose for 6 to 8 h before inoculating similar media containing 2% (wt/vol) raffinose. Additional drop-outs were imposed as necessary to select for plasmids. After back-dilution the following morning, M phase arrest was initiated when the cultures reached midlog. Cultures grown in YPA were arrested with 10 μg/mL nocodazole, a microtubule depolymerization agent. Cultures grown in SC-met were arrested by the addition of 2 mM methionine, which shut off MET3p-driven expression of Cdc20, a subunit of the anaphase promoting complex. After 3 h when roughly 80% of the cells attained a dumbbell shape, DNA circularization was triggered by the addition of 2% (wt/vol) galactose, which induced the expression of a GAL1p-R recombinase gene fusion. Cells were harvested 2 h thereafter and fixed with fresh 4% (wt/vol) paraformaldehyde. Transcriptional induction of URA3 was achieved by either growing cells from the outset in SC-ura media or by washing cells grown with uracil twice with water before resuspension in SC-ura media. After switching to SC-ura media, cultures were incubated for 2 h before proceeding to the next step of the protocol.
Fluorescence Microscopy.
Paraformaldehyde fixation, slide preparation, and fluorescence microscopy were performed as described in ref. 9. All data sets were based on at least three independent trials for a total of 100–300 cells per condition. Roughly 10% of cells were excluded from analysis because they either (i) were unbudded, (ii) contained a new bud, or (iii) contained single dots on either side of the bud neck or other features indicative of escape from M phase arrest. In each case, data from at least three independent trials were pooled because they satisfied χ2 tests of the homogeneity of proportions. Error bars represent the SEs of proportions. Each reported value was compared with that for an appropriate control by a χ2 test.
Chromatin Immunoprecipitation.
ChIP was performed as described in ref. 50 using primers listed in Table S2. Primers that distinguish between the S. cerevisiae and K. lactis URA3 genes were used with strains that carry both URA3 alleles. The reported normalized values correspond to the signal at a given site relative to an internal positive control (549.7) divided by the same ratio of sites within the input. The mean and SD of three or more biological replicates are presented. Statistical significance was determined by pairwise Student’s t tests.
qRT-PCR.
RNA extraction and reverse transcription-PCR (RT-PCR) were performed as described in ref. 50 using the oligonucleotides listed in Table S2. The mean and SD of three or more biological replicates are presented.
SI Materials and Methods
URA3f and URA3r excision cassettes contain URA3 segments of chromosome V (SGD coordinates 115919–117111 and 116011–117110, respectively). Modifications of URA3f were made by integrating rebuilt loci into the empty cassette (strains HJ24, HJ25, and HJ6388). Additional modifications were made by PCR-mediated gene replacement (PMGR) using a loxP-SpHIS5MX-loxP module, which was either left in place (strains MSB30, MSB31, MSB110, and MSB112) or reduced to a single loxP site by Cre (strains MSB35, HJ5, HJ6, HJ7, HJ19, and HJ4). Oligonucleotides for strain construction are listed in Table S2. URA3 promoter elements were moved to the lys2 cassette as loxP-SpHIS5MX-loxP hybrids by PMGR, after which SpHIS5MX-loxP was removed by Cre (strains HJ22, HJ23 and MSB155). In strains bearing partial URA3 loci, a second copy of a functional URA3 gene was integrated at the endogenous location or provided with plasmids bearing S. cerevisiae or K. lactis URA3 alleles (pXRH3-URA3 or pXRH3-KlURA3, respectively). All excision cassette constructions were confirmed by DNA sequencing. Other strain modifications were made by standard PCR-mediated methods, knocking in plasmids or by genetic crosses.
Acknowledgments
We thank Anne Bresson, Jennifer Gerton, and Brigitte Lavoie for materials. This work was funded by United States Public Health Service Grants NIGMS 51402 (to M.R.G.) and NIGMS 2T32A100740319 (to M.S.B.), March of Dimes Grant 1-FY08-481 (to M.R.G.), and an Aresty Undergraduate Research Fellowship (to H.J.). The Flow Cytometry Core Facility of Robert Wood Johnson Medical School and The Rutgers Cancer Institute of New Jersey was supported by grants from the National Cancer Institute (Grant P30CA072720) and NIH (Grant 1 S10 RR025468-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617309114/-/DCSupplemental.
References
- 1.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 2.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91(1):47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J Cell Biol. 2010;189(2):201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cucco F, Musio A. Genome stability: What we have learned from cohesinopathies. Am J Med Genet C Semin Med Genet. 2016;172(2):171–178. doi: 10.1002/ajmg.c.31492. [DOI] [PubMed] [Google Scholar]
- 5.Onn I, Heidinger-Pauli JM, Guacci V, Ünal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 7.Uhlmann F. SMC complexes: From DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17(7):399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 8.Stigler J, Çamdere GO, Koshland DE, Greene EC. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Reports. 2016;15(5):988–998. doi: 10.1016/j.celrep.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CR, Wu CS, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19(24):3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma U, Stefanova D, Holmes SG. Histone variant H2A.Z functions in sister chromatid cohesion in Saccharomyces cerevisiae. Mol Cell Biol. 2013;33(17):3473–3481. doi: 10.1128/MCB.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng T, Guacci V, Koshland D. Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol Biol Cell. 2015;26(23):4224–4235. doi: 10.1091/mbc.E15-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, et al. A handcuff model for the cohesin complex. J Cell Biol. 2008;183(6):1019–1031. doi: 10.1083/jcb.200801157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98(2):249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 14.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151(5):1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430(6999):573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glynn EF, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2(9):E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Megee PC, Koshland D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285(5425):254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj S, Schlackow M, Rabajdova M, Gullerova M. Transcription facilitates sister chromatid cohesion on chromosomal arms. Nucleic Acids Res. 2016;44(14):6676–6692. doi: 10.1093/nar/gkw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20(10):957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132(6):983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 21.Ocampo-Hafalla M, Muñoz S, Samora CP, Uhlmann F. Evidence for cohesin sliding along budding yeast chromosomes. Open Biol. 2016;6(6):150178. doi: 10.1098/rsob.150178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bausch C, et al. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(24):8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haering CH, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15(6):951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16(6):1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 25.Lengronne A, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23(6):787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics. 2012;190(3):885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam WW, Peterson EA, Yeung M, Lavoie BD. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 2006;20(21):2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubey RN, Gartenberg MR. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 2007;21(17):2150–2160. doi: 10.1101/gad.1583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolef Ben-Shahar T, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321(5888):563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 30.Ünal E, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321(5888):566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 31.Vega H, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37(5):468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 32.Gordillo M, et al. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet. 2008;17(14):2172–2180. doi: 10.1093/hmg/ddn116. [DOI] [PubMed] [Google Scholar]
- 33.Gard S, et al. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187(4):455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynn PJ, Reece RJ. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol. 1999;19(1):882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal E, Widom J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorch Y, Maier-Davis B, Kornberg RD. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 2014;28(22):2492–2497. doi: 10.1101/gad.250704.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen A, van der Zande E, Meert W, Fink GR, Verstrepen KJ. Distal chromatin structure influences local nucleosome positions and gene expression. Nucleic Acids Res. 2012;40(9):3870–3885. doi: 10.1093/nar/gkr1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Serra L, Kelly G, Patel H, Stewart A, Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat Genet. 2014;46(10):1147–1151. doi: 10.1038/ng.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309(5740):1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 42.Eng T, Guacci V, Koshland D. ROCC, a conserved region in cohesin’s Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Mol Biol Cell. 2014;25(16):2351–2364. doi: 10.1091/mbc.E14-04-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13(11):1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16(15):1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 45.McNairn AJ, Gerton JL. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res. 2009;17(2):155–163. doi: 10.1007/s10577-008-9007-9. [DOI] [PubMed] [Google Scholar]
- 46.Chan KL, et al. Cohesin’s DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150(5):961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huis in ’t Veld PJ, et al. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346(6212):968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- 48.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505(7483):367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5(2):243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 50.Chen M, Gartenberg MR. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 2014;28(9):959–970. doi: 10.1101/gad.236729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a proficient enzyme: The structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc Natl Acad Sci USA. 2000;97(5):2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohner S, Gasser SM, Meister P. Modules for cloning-free chromatin tagging in Saccharomyces cerevisae. Yeast. 2008;25(3):235–239. doi: 10.1002/yea.1580. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y-F, Chou C-C, Gartenberg MR. Determinants of Sir2-mediated, silent chromatin cohesion. Mol Cell Biol. 2016;36(15):2039–2050. doi: 10.1128/MCB.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 55.Cheng T-H, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 2000;14(4):452–463. [PMC free article] [PubMed] [Google Scholar]
- 56.Wu CS, Chen YF, Gartenberg MR. Targeted sister chromatid cohesion by Sir2. PLoS Genet. 2011;7(2):e1002000. doi: 10.1371/journal.pgen.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]