Significance
The rate of genomic adaptation is determined by the rate of environmental change, the availability of beneficial mutations, and the efficiency of positive selection. The relative importance of these factors has been actively discussed. We address the questions using whole genome sequences of great apes, which have very different population sizes whereas their genomic architectures are highly similar. We infer that the impact of selection on the genomic diversity of a species increases with the effective population size, most likely due to the differential influx rate of beneficial mutations. This explanation is, among other possibilities, expected if adaptive evolution is limited by the waiting time for new favorable mutations in great apes.
Keywords: selective sweep, population size, great ape, adaptive evolutionary rate, mutation limitation
Abstract
Quantifying the number of selective sweeps and their combined effects on genomic diversity in humans and other great apes is notoriously difficult. Here we address the question using a comparative approach to contrast diversity patterns according to the distance from genes in all great ape taxa. The extent of diversity reduction near genes compared with the rest of intergenic sequences is greater in a species with larger effective population size. Also, the maximum distance from genes at which the diversity reduction is observed is larger in species with large effective population size. In Sumatran orangutans, the overall genomic diversity is ∼30% smaller than diversity levels far from genes, whereas this reduction is only 9% in humans. We show by simulation that selection against deleterious mutations in the form of background selection is not expected to cause these differences in diversity among species. Instead, selective sweeps caused by positive selection can reduce diversity level more severely in a large population if there is a higher number of selective sweeps per unit time. We discuss what can cause such a correlation, including the possibility that more frequent sweeps in larger populations are due to a shorter waiting time for the right mutations to arise.
The number of strong selective sweeps in human evolution and their collected impact on nucleotide diversity have proven difficult to establish (1, 2). From human diversity data we can robustly infer selective sweeps only within the last ∼250,000 y (3), yet recent evidence suggests that hundreds of sweeps have each affected the diversity in regions of several hundreds of kilobases (kb), mostly in or around genes (4, 5). It is conceivable that humans have adapted very quickly to the emergence of new biotic and abiotic challenges and that the number of strong sweeps has mainly been determined by the rate of changes to the environment. Alternatively, the number of strong sweeps could have been determined by the rate at which new beneficial mutations have entered the gene pool (6). The waiting time for a new beneficial mutation to arise depends on the mutation rate and the size of the mutational target, i.e., the number of nucleotides, where different mutations have equivalent effects. The selection coefficient s of new mutations determines their fate, because approximately a proportion 2s of new beneficial mutations with additive effects is expected to escape initial drift and eventually go to fixation. Because the rate at which new mutations enter a population is expected to be proportional to the census population size Nc, one way to investigate whether mutational input limits the rate of strong sweeps is to search for a correlation between the number of strong sweeps and Nc. Evidence has been presented that the rate of adaptive evolution, which involves strong, weak, and soft sweeps added together (7), or the total impact of cumulative positive and purifying selection (8), scales with effective population size Ne across eukaryotes. These studies aim to control for the action of other evolutionary factors such as demographic differences and the reduction in diversity at neutral sites due to purifying selection at linked sites, i.e., background selection (9, 10). Due to the broad taxonomic scale of these previous studies, however, it is difficult to ascribe different polymorphism patterns to the rate of adaptive evolution because these species also differ in genomic features such as gene density, recombination rate, and mutation rate, all of which affect diversity patterns and hence inferences of selection.
We present a comparative analysis of the genomic impact of selection using genome sequences from all great ape species. The great apes have almost the same set of genes, which are generally in synteny, and the overall amount of recombination per generation per megabase is very similar among humans (11), chimpanzees (4), bonobos, and gorillas (12), as well as in the common ancestor of human and chimpanzee (13). Because these species differ in their demographic histories and in overall genomic diversity by up to a factor of about three (14), great apes constitute a perfect study system for relating signatures of selection to demographic differences (15). Previous analyses based on pairwise sequentially Markovian coalescent have shown that orangutans have the largest long-term Ne, followed by gorillas, the chimpanzee subspecies, and finally humans and bonobos (14). Here we assume that the targets for most strong sweeps are within or very close to genes (5), enabling us to study how linked diversity is affected by sweeps by contrasting how intergenic diversity increases as a function of distance to the closest genes.
We find that the extent of diversity reduction near genes is positively correlated with the diversity far away from genes in great apes. This result suggests that selection reduces genomic diversity more severely in species with larger Ne. Using simulations, we show that this correlation is unlikely to be caused by background selection but can be created by a higher rate of selective sweeps in larger populations.
Results
Differences Among Species in Reductions of Diversity Around Genes.
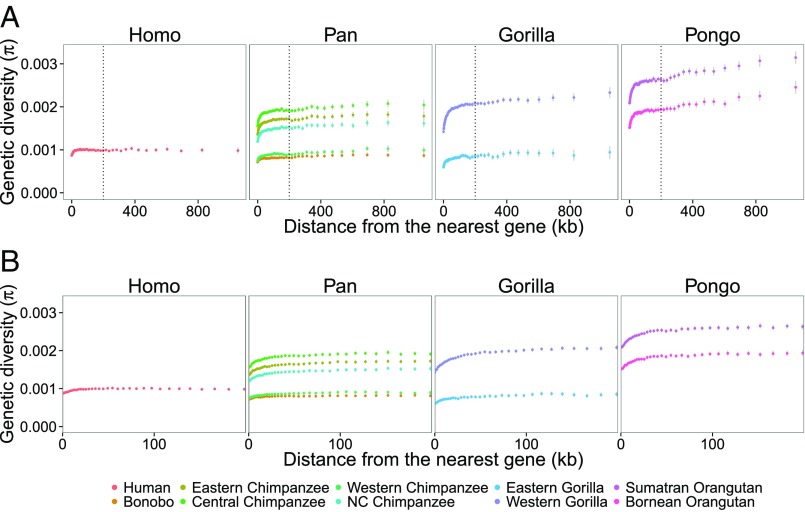
The overall genomic nucleotide diversity (π) varies by a factor of about three among the great ape species (14, 16). In all species, diversity is lowest in exons, intermediate in introns, and highest in intergenic sequences (SI Appendix, Fig. S1). Fig. 1A shows how π increases with physical distance from genes up to 1 Mb away, with Fig. 1B zooming in on the closest 200 kb. The 50 bins were chosen such that each bin contains approximatively the same number of nucleotides. We considered only sites that were called in all species and removed all sites with a PhastCons (a posterior probability that each nucleotide belongs to a conserved element) score larger than 0.25 to avoid sites under purifying selection (92% of sites remaining).
Fig. 1.
Reduction of diversity around genes. Plots show the relationship between the nucleotide diversity, π, and the physical distance to the nearest genes for each genus of the great apes. (A) For distances up to 1 Mb. (B) For distances up to 200 kb. Error bars show 95% confidence intervals calculated from bootstrapping with 1,000 replicates from 1-Mb windows. The name of each (sub) species is shown at Bottom.
The patterns of π at increasing distance from genes are strikingly different among the species both in terms of the relative reduction in π near genes and the distance from genes where a reduction is still visible. Gorillas show the largest reduction in diversity in regions of ∼1 kb around genes (26.0–26.8%) compared with the rest of the intergenic regions, followed by orangutans (18.6–19.5%), chimpanzees (15.6–18.3%), bonobos (11.7%), and humans (11.4%). The reduction in diversity reaches furthest away in orangutans (∼1 Mb), followed by gorillas (∼300 kb), chimpanzees (∼200 kb), bonobos (∼100 kb), and humans (∼100 kb). SI Appendix, Fig. S2 shows these relationships normalized with diversity far from genes. We used physical distance rather than genetic distance because a pedigree-based genetic map is available only for humans and because species-specific polymorphism-based maps are biased by the levels of diversity along the genome. Using physical distance is not expected to systematically bias our results because the recombination landscape in great ape species is well conserved at the megabase scale (4, 12) and because the concentration of recombination hotspots is similar, although most are positioned differently on the fine scale (4, 17). Mutation rates may also depend on distance from genes due to a higher repeat density around genes (18) and putative differences in chromatin structure away from genes. Therefore, we normalized π with the divergence between humans and macaques. This result is qualitatively similar, but shows a slight increase in π around 100 kb from genes where repeat density is highest (SI Appendix, Fig. S3). Because conserved intergenic sites were removed, we expect that only a small part of the reduction in diversity near genes is due to direct effects of purifying selection (19) and we, therefore, suggest that the difference among species is rather due to positive or purifying selection on linked sites.
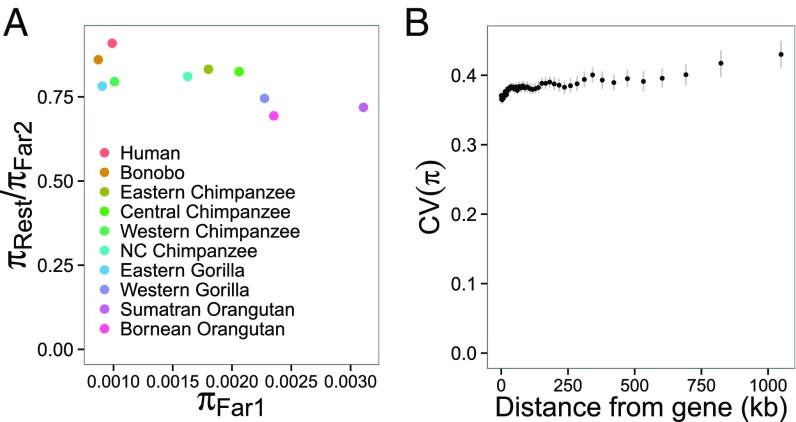
To quantify the reduction in genomic diversity caused by selection, we assumed that the genomic regions furthest from genes are affected least by selection and that their diversity levels thus best reflect diversity in the absence of selection. We denote this level of diversity πFAR and estimated πFAR from the genomic regions corresponding to the two rightmost points in each panel of Fig. 1. These regions are at least 823 kb away from genes and constitute 50.57 Mb in total (median length of regions is 390 kb) scattered throughout 86 gene deserts on 18 chromosomes (SI Appendix, Fig. S4). These gene deserts do not display an elevated mutation rate, and the single-nucleotide polymorphism quality and coverage within these regions are similar to the rest of the intergenic sequences for all great ape species (SI Appendix, Figs. S5 and S6). We contrasted the reduction in diversity, πREST/πFAR, where πREST is the average diversity levels from the rest of the genome. The πREST/πFAR is smallest in orangutans (0.69–0.72), followed by gorillas (0.75–0.78), chimpanzees (0.80–0.83), bonobos (0.86), and humans (0.91). To test a correlation between πREST/πFAR and πFAR, we randomly divided the regions used to calculate πFAR into two groups, from which we calculated two independent estimates of the diversity, πFAR1 and πFAR2, with 1,000 replications. The πREST/πFAR1 is inversely correlated with πFAR2 (Pearson’s r = −0.70, P = 0.0237) (Fig. 2A). Because diversity patterns in closely related species/subspecies are not independent, we merged estimates in closely related groups and calculated the correlation using human, bonobo, average across chimpanzee, average across gorilla, and average across orangutan, and still found a significant negative correlation (r = −0.89, P = 0.0407). We, therefore, conclude that diversity is reduced relatively more in species where the diversity far from genes is high. We also note that Western chimpanzees, Eastern gorillas, and Bornean orangutans went through recent population bottlenecks (12, 14, 20), and that πFAR might, therefore, underestimate long-term Ne during the past few hundred thousand years. In line with this notion, these three taxa have a relatively strong reduction in genomic diversity near genes compared with their present genomic diversity (Fig. 2A).
Fig. 2.
Reduction of diversity as a function of genomic diversity. (A) Relationship between diversity estimated from genomic regions far (>823 kb) from genes (πFar1) and diversity ratio of the rest of the genome to the genomic region far from genes (πRest/πFar2). All positions far from genes are randomly assigned into two groups used to calculate πFar1 and πFar2. Error bars indicate 95% confidence intervals for this randomization. (B) Relationship between the distance from genes and the coefficient of variance (CV) of π. Error bars show 95% confidence intervals calculated by 1,000 bootstrapping replicates from 1-Mb windows.
If selection has a homogenizing effect on diversity level by reducing diversity level more severely when Ne is larger, we expect diversity to be more similar across species in regions that are affected more by selection, i.e., regions near genes. We quantified the heterogeneity of diversity levels among species by the coefficient of variation of π, CV(π). We find that CV(π) is lowest in exons, intermediate in introns, and highest in intergenic sequences (SI Appendix, Fig. S7). The intergenic CV(π) shows a strong positive correlation with the distance from genes (r = 0.90, P < 2.2 × 10−16) (Fig. 2B).
All variants were called by mapping sequencing reads against the human reference genome. Thus, the varying phylogenetic distances of the different great ape species to human could have affected our inferences. To test this possibility, we considered the phylogenetically most distant genus from humans, orangutans, and mapped sequencing reads from the orangutan samples against the orangutan reference genome. We observe very similar diversity patterns as a function of distance from genes (SI Appendix, Fig. S8) and a similar ratio of intergenic diversity to πFAR (SI Appendix, Fig. S9). Thus, mapping artifacts are unlikely to cause the differences in diversity patterns among species.
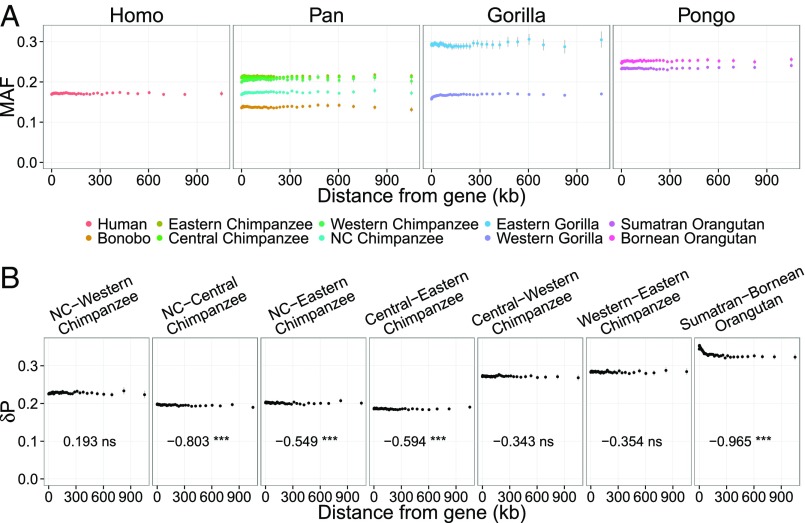
Selective sweeps are expected to distort the site frequency distribution and increase population differentiation more dramatically than background selection (21, 22). Fig. 3A shows the average minor allele frequency as a function of distance from genes. Western gorillas show a significant decrease in minor allele frequency near genes (Spearman’s rho (ρ) = 0.92, Bonferroni-adjusted P < 0.0001) and a weaker signal is seen for Nigeria–Cameroon chimpanzees (ρ = 0.71, adjusted P < 0.0001), western chimpanzees (ρ = 0.68, adjusted P < 0.0001), and Bornean orangutans (ρ = 0.63, adjusted P < 0.0001). For the other taxa, minor allele frequency is not detectably lower near genes (adjusted P > 0.05). These observations are generally consistent with a larger number of selective sweeps in western gorillas, orangutans, and chimpanzee subspecies than in humans and bonobos. The two orangutan species show elevated population differentiation in regions reaching up to 250 kb away from genes (Fig. 3B and SI Appendix, Fig. S10 in which the results are robust to species mapping reference). Only a slight decrease in population differentiation away from genes is found among pairs of chimpanzee subspecies.
Fig. 3.
Signatures of selective sweeps. (A) Relationship of minor allele frequency and the distance from the nearest gene for each genus of the great apes. The name of each (sub) species is shown at Bottom. Significant positive correlations were observed from Western gorillas (Bonferroni-adjusted P value <0.0001), Nigeria–Cameroon chimpanzees (adjusted P value <0.0001), western chimpanzees (adjusted P value <0.0001), and Bornean orangutans (adjusted P value <0.0001). (B) Relationship of population divergence levels with the distance from the nearest genes. Spearman’s correlation coefficient and the significance are shown in each panel (***, **, *, and ns indicate Bonferroni-corrected P values with <0.001, <0.01, <0.05, and ≥0.05, respectively). Error bars show 95% confidence intervals calculated by 1,000 bootstrapping replicates from 1-Mb windows.
Simulations.
We investigated to which extent background selection and selective sweeps reduce diversity under realistic population parameters and to which extent diversity reduction depends on the population size (N) through forward simulations. According to theoretical predictions (9, 10, 23), the relative decrease in diversity due to background selection is independent of N and depends only on the distribution of s. The effect of background selection on diversity is expected to increase when s decreases (23), because weakly deleterious mutations segregate longer in populations and thus affect the fitness of more individuals. This prediction, however, assumes that selection is sufficiently strong that the effect of genetic drift can be ignored (24). It is not known to which extent background selection is able to reduce diversity near genes when drift also plays a role.
Using forward simulations, we investigated (i) how the strength of purifying selection affects the reduction in diversity due to background selection and (ii) whether this diversity reduction depends on N. Specifically, we assumed genes of 100 kb length flanked by 200 kb of intergenic regions, using mutation and recombination parameters chosen to represent humans (Methods). In each simulation, all nonsynonymous mutations in the genes were assumed to have the same s. We tested a wide range of s (between 0.0001 and 0.05) for two population sizes, n = 1,000 and n = 2,000 of diploid individuals.
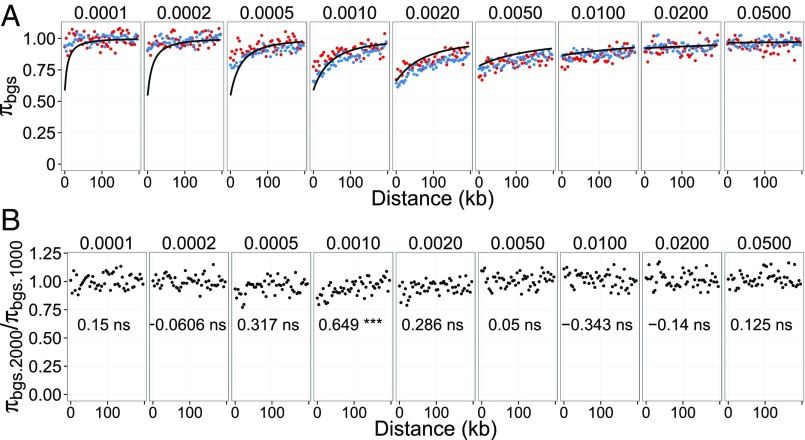
We find that the reduction in diversity levels near the gene is undetectable when s is very small (s = 0.0001, i.e., Ns < 1) or very high (s > 0.02, i.e., Ns > 20) (Fig. 4A). The theoretical prediction by Durrett (23) (solid line in Fig. 4A) is accurate for Ns > 2 but breaks down for smaller Ns where genetic drift is more important than selection. As a result, the predicted strong reduction in diversity close to genes for small s is not recovered in simulated results. To test if the strength of background selection is dependent on N, we performed a correlation test between the distance from the gene and the ratio of the diversity for n = 2,000 to that for n = 1,000. As expected, the relative reduction in diversity due to background selection is independent of N for most of the range of s (Fig. 4B) and therefore the reduction in diversity can be predicted by s alone. The notable exception is when Ns is close to one. In simulations using s = 0.001 (corresponding to Ns = 1 and Ns = 2 for the two population sizes), the diversity ratio is positively correlated with distance from genes. Hence, N effects the strength of background selection only when Ns is very close to one. It is still an open question how large a fraction of mutation have Ns ∼1 in great apes. Studies aimed at inferring the distribution of fitness effects find a large variance and heavy mass at high values of s (25, 26). In a study of three chimpanzee subspecies, Bataillon et al. (25) showed that more than 80% of deleterious point mutations have Ns ≫ 1. Thus, it is unlikely that enough deleterious mutations are found in a narrow region around Ns = 1 to create the observed differences in diversity reduction among the great apes.
Fig. 4.
Simulations of background selection. The diversity pattern in the sequences flanking the genic region under evolutionary constraint for two populations in which N = 1,000 and 2,000, respectively, is shown. Selection coefficients range from 0.0001 to 0.05. For each set of parameters, we performed 1,000 independent simulations and report the average π. (A) Relationship between the distance from genes and the relative reduction in π due to background selection, πBGS, together with theoretical expectation (based on ref. 23) (black lines). Blue and red points represent when n = 1,000 and n = 2,000, respectively. (B) Relationship between the distance from genes and the ratio of πBGS with n = 2,000 to πBGS with n = 1,000. Pearson’s correlation coefficient and the significance are shown in each panel (***, **, *, and ns indicate Bonferroni-corrected P values with <0.001, <0.01, <0.05, and ≥0.05, respectively).
Recent changes in population size may affect the proportion of segregating deleterious mutation over time and thus the strength of background selection. To test this possibility, we performed separate sets of simulations where the population experiences either an expansion (from n = 1,000 to n = 2,000) or a shrinkage (n = 2,000 to n = 1,000) 100 generations ago. For each demographic scenario we tested a wide range of s (between 0.0001 and 0.05). We find that neither a twofold expansion nor a twofold shrinkage in N affects qualitatively the relationship between N and strength of background selection (SI Appendix, Fig. S11). However, site frequency spectrum is greatly distorted (SI Appendix, Fig. S12).
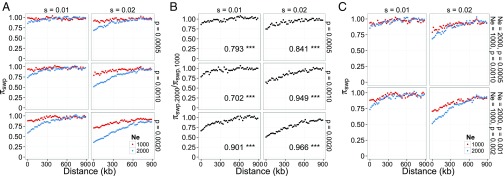
To study the qualitative effect of selective sweeps, we allowed beneficial mutations with s = 0.01 and s = 0.02 and a proportion, p, of mutations to be beneficial of 0.05%, 0.1%, and 0.2% of all nonsynonymous mutations. In this scenario, we found positive correlations between diversity and distance from genes for all combinations of s and p (Fig. 5A). In contrast to the simulations of background selection alone, the diversity ratio of n = 2,000 to n = 1,000 is positively correlated with distance from genes with all combinations of parameters (Fig. 5B). How much diversity is reduced as a function of N depends on the extent of diversity reduction from each sweep, the rate of sweeps, and the time span over which the reduction by a sweep is detectable. In our simulations, the larger population (n = 2,000) experiences twice as many beneficial mutations per unit time. Each of these sweeps is expected to reduce diversity less in a larger population because in a larger population it takes longer time for a new beneficial mutation to fix (27). The effect of population-wide influx of beneficial mutations on diversity is seen more clearly when we compare simulations with different N (and thus different Ns) and the same beneficial mutation rate: a population with n = 2,000 and P = 0.05% and a population with n = 1000 and P = 0.1%. We found that the larger populations shows a stronger reduction in diversity by 4.2–28.0% (Fig. 5C). This greater reduction is not due to a higher fixation probability of beneficial mutations in a larger population (effect of Ns), because the number of positively selected sites is not different between the large and the small populations (SI Appendix, Table S3). In a large population a single selective sweep can be detected in diversity patterns for a longer time, because it takes a longer time for diversity levels to reach an equilibrium state. As a consequence, we expect a greater reduction in diversity in the large population for the same number of sweeps per unit of time. However, the magnitude of this effect does not account for the very large differences in how much diversity is reduced across the great apes. Whereas simulations of twofold different population sizes causes 4.2–28% difference in diversity reduction, the observed difference between human and gorilla is 169%, even though the population size of gorillas (πFAR) is only 1.61 times larger than that of humans. Thus, in great apes the reduction in diversity is predominantly determined by the number of experienced sweeps unless the s is larger in larger populations. Background selection expects to reduce diversity level at physical distances 200 kb–1 mb from genes only by 0.14–3.5% with a range of s (0.0001–0.05) based on the prediction of Durrett (23). This observation contrasts with what we observe in several species (Fig. 1). We also performed simulations with different parameters for the mutation rate, the length of sequences, and p (a proportion of beneficial mutations), and observed the same patterns (SI Appendix, Fig. S13).
Fig. 5.
Simulations of selective sweeps. The selection coefficients of adaptive mutations, s, are 0.01 and 0.02; the proportion of all genic mutations that are beneficial (p) is 0.0005, 0.001, and 0.002. For each set of parameters, we performed 1,000 independent simulations and report the average π. (A) πSWP, diversity reduction due to selective sweeps, as a function of distance from genes for different combinations of N, s, and p. (B) The ratio of πSWP with n = 2,000 to π with n = 1,000 as a function of distance from genes. Pearson’s correlation coefficient and the significance are shown in each panel (***, **, *, and ns indicate Bonferroni-corrected P values with <0.001, <0.01, <0.05, and ≥0.05, respectively). (C) πSWP as a function of distance from genes when the number of beneficial mutations per generation is the same but N differs by a factor of two.
Discussion
In great apes, genetic diversity is reduced around genes. This reduction is positively correlated with the expected nucleotide diversity in the absence of selection, assuming that the average diversity in regions furthest from genes is a good proxy for the diversity in the absence of selection. The relative reduction in diversity is thus larger in species with larger Ne. As a consequence, estimates of Ne based on whole genome diversity data are expected to be more homogeneous among species than the actual differences in the numbers of individuals that effectively contribute to the next generation.
Simulations showed that the reduction of diversity by background selection depends on the underlying distribution of fitness effects but generally not on N. N will only affect the extent of reduction when a large fraction of deleterious mutations has s very close to 1/N. Because the location and function of genes are very similar across great ape species, it is reasonable to assume that the distribution of s for deleterious mutations is also very similar. Thus, it is unlikely for background selection alone to produce the large differences in the extent of diversity reduction around genes among great ape species. This may appear at odds with McVicker et al. (28) who found that background selection can explain the reduction in genomic diversity in the hominid lineage by 19–26%. However, their analysis ignored the effects of drift and assumed s being exponentially distributed down to 10−5. If Ne is smaller than 100,000, as has been estimated in the great apes (14), many of these mutations will be subject to strong drift and therefore do not contribute to a decrease in diversity due to background selection.
Our simulations showed that, in contrast to background selection, selective sweeps affect diversity several hundreds of kilobases away from the targets of selection and their impact on diversity is correlated with N. However, a single sweep is not likely to reduce diversity more in a larger population because beneficial mutations reside on average longer in a population before fixation (proportional to logN) (27). Thus, if strong sweeps are the main force homogenizing diversity levels among species (29), larger populations must experience either more or stronger sweeps (with larger s). Most models of adaptation predict that larger populations are closer to their fitness maximum and that the proportion of adaptive mutations with large s is lower than in smaller populations (30), arguing against larger s in larger populations. Even though we find in simulations that larger populations have a larger reduction in diversity for the same rate of sweeps over time, perhaps because a single sweep affects diversity for a longer time in the larger population, the quantitative effect is much smaller than the population size effect we observe in great apes. We, therefore, conclude that the number of experienced sweeps per unit of time increases with population size. Assuming the same mutation rate across populations, the number of beneficial mutations arising each generation should scale with Nc rather than Ne, but because we cannot observe Nc over an evolutionary timescale, our interpretation is based on the assumption that long-term Nc is proportional to long-term Ne. One possible explanation for a correlation between the rate of strong selective sweeps and population size is that the waiting time for beneficial mutations to occur is different between species with different Ne and that this waiting time may limit the rate of adaptation to different degrees.
In species with much larger Ne than the great apes, such as Drosophila, the rate of adaptive evolution is likely not limited by the waiting time for new mutations (31, 32) due to a higher number of segregating polymorphisms that may have advantageous effects, depending upon changes in the environment. In such cases, we expect that soft sweeps are relatively more important in large populations (33), which would cause a smaller reduction in diversity at linked sites. In humans, such alternative modes of selection may play a significant role in recent adaptation (2, 34). A simulation study shows that soft sweeps with s = 0.05 and plausible parameters for humans reduce π 100 kb away from the beneficial allele if a beneficial allele has a frequency of 1% at the onset of positive selection (35). Thus, soft sweeps are likely to affect diversity close to genes but do not easily explain the far reaching depressions in diversity that we observe. But we cannot exclude a possibility that a large fraction of soft sweeps involves s much larger than 0.05 in great apes. Due to the computational feasibility, our simulation is limited to a small number of demographic scenarios and the input parameters of Ne are much lower than the Ne of the great apes. Simulations with more realistic ranges of demographic parameters and varying s would enable one to infer quantitative effects of hard/soft selective sweeps and background selection on genetic diversity levels more precisely in future.
Mutation limitation may appear at odds with the well-known recent sweep on the lactase gene in humans. At least four different mutations within a 100-bp interval have been selected for in different populations (36). However, the lactase example might be atypical because there are potentially many possible mutations in the regulatory region of the lactase gene that might cause the altered phenotype. Furthermore, the population-specific sweeps at the lactase gene are so recent that Nc of humans was much larger than Nc of the other great apes.
We can make a rough estimation of how often a specific beneficial mutation appears and is fixed. If we consider a mutation rate of u = 4.8 × 10−10 per base pair per year (corresponding to 1.2 × 10−8 per site per generation (37), assuming a generation time of 25 y) and Nc of 200,000 (4–20 times larger than the estimated Ne of great ape species), the expected waiting time for a mutation on a single nucleotide to occur is 1/2uNc = 5,208 y in a diploid genome. Thus, the expected time until fixation of a beneficial mutation with additive s = 0.01 is 5,208/2s = 206,400 y, assuming that the fixation probability is 2s. If there are, in contrast to the lactase example, only a few solutions to a given evolutionary pressure and new adaptive opportunities arise on the scale of tens of thousands of years, it is plausible that a large fraction of adaptation by strong positive selection is limited by the availability of beneficial mutations in great apes.
Methods
We downloaded SNP data from the Great Ape Genome Project homepage (biologiaevolutiva.org/greatape/) (14). These SNPs are based on high coverage (∼25×) whole genome sequencing data of 87 great ape individuals (SI Appendix, Table S1 shows the number of individuals per species/subspecies). Our analysis is based on 1.93-Gb sites that have been called with high-quality scores in all species. This size corresponds to 67.4% of the total length of autosomes. In total 83,266,775 SNPs were analyzed (SI Appendix, Table S2 shows a break down into species/subspecies). For the intergenic sequences that are used to analyze the relationship between physical distance from genes and diversity levels, we excluded CpG islands and conserved noncoding sequences (PhastCons score >0.25) obtained from the University of California, Santa Cruz Genome Bioinformatics Site (https://genome.ucsc.edu/index.html).
To calculate the physical distance from the nearest genes, we used the gene annotation of RefSeq and Ensembl for the data mapped against the human reference genome (hg18) and the orangutan reference genome (ponAbe2), respectively. We calculated nucleotide diversity (π), minor allele frequency, population divergence (ΔP, see ref. 16), and average distance from the nearest gene in 1-kb windows. We then sorted these windows into 50 bins at increasing distance from the closest gene, each bin having similar number of nucleotides, and calculated π for each bin.
We used SLiM v1.7 software (38) to estimate the reduction in diversity due to background selection or selective sweeps and to count the number of fixations by positive selection. We simulated 2N haploid sequences with a 100-kb genic region flanked by 200-kb and 900-kb intergenic regions for the models of background selection and selective sweeps, respectively. The recombination rate was set to 1.1 × 10−8 per site per generation and the mutation rate to 2.5 × 10−8 per site per generation (39, 40). We treated 72.15% of genic mutations as nonsynonymous and the remaining 27.85% as synonymous. The nonsynonymous mutations were assumed to be deleterious with a uniform s. When sweeps were included, a fraction of nonsynonymous mutations (0.05% or 0.1%) was assumed to have either s = 0.01 or s = 0.02 (Ns thus always greater than or equal to 10). Population sizes were either 1,000 or 2,000 diploid individuals. Each population was simulated for 105 generations. We calculated π and counted the number of positively selected sites from a sample of 500 diploid individuals for each simulation. For each combination of population size, selection coefficient, and the proportion of beneficial mutations, we performed 1,000 independent simulations, followed by averaging π and the number of positively selected sites. All statistical analyses were performed with R (www.r-project.org).
Supplementary Material
Acknowledgments
This study was supported by the European Commission-funded 7th Framework Programme for Research and Technological Development NEXTGENE Project (T.M. and M.H.S.), Grant 1323-00076A from the Danish Research Council for Independent Research (to M.H.S.), as well as by European Molecular Biology Organization (Young Investigator Programme-2013), Ministerio de Economía y Competitividad BFU2014-55090-P (Fondo Europeo de Desarrollo Regional), BFU2015-7116-European Research Council (ERC), and BFU2015-6215-ERC and Fundacio Zoo Barcelona (PRIC 2016) (to T.M.-B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.P. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605660114/-/DCSupplemental.
References
- 1.Enard D, Messer PW, Petrov DA. Genome-wide signals of positive selection in human evolution. Genome Res. 2014;24(6):885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez RD, et al. 1000 Genomes Project Classic selective sweeps were rare in recent human evolution. Science. 2011;331(6019):920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160(3):1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman SR, et al. 1000 Genomes Project Identifying recent adaptations in large-scale genomic data. Cell. 2013;152(4):703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29(1):33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Gossmann TI, Keightley PD, Eyre-Walker A. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol Evol. 2012;4(5):658–667. doi: 10.1093/gbe/evs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13(4):e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth B. The effects of deleterious mutations on evolution at linked sites. Genetics. 2012;190(1):5–22. doi: 10.1534/genetics.111.134288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134(4):1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVean GAT, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304(5670):581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 12.Stevison LS, et al. Great Ape Genome Project The time scale of recombination rate evolution in great apes. Mol Biol Evol. 2016;33(4):928–945. doi: 10.1093/molbev/msv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munch K, Mailund T, Dutheil JY, Schierup MH. A fine-scale recombination map of the human-chimpanzee ancestor reveals faster change in humans than in chimpanzees and a strong impact of GC-biased gene conversion. Genome Res. 2014;24(3):467–474. doi: 10.1101/gr.158469.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499(7459):471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 2013;14(4):262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam K, et al. Great Ape Genome Diversity Project Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc Natl Acad Sci USA. 2015;112(20):6413–6418. doi: 10.1073/pnas.1419306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: Trends and variations with respect to genomic composition. Bioinformatics. 2004;20(6):813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 19.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015;47(3):276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nater A, et al. Reconstructing the demographic history of orang-utans using Approximate Bayesian Computation. Mol Ecol. 2015;24(2):310–327. doi: 10.1111/mec.13027. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20(3):393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y. Allele frequency distribution under recurrent selective sweeps. Genetics. 2006;172(3):1967–1978. doi: 10.1534/genetics.105.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durrett R. Natural Selection. Probability Models for DNA Sequence Evolution. 2nd Ed. Springer; New York: 2008. pp. 211–218. [Google Scholar]
- 24.Good BH, Walczak AM, Neher RA, Desai MM. Genetic diversity in the interference selection limit. PLoS Genet. 2014;10(3):e1004222. doi: 10.1371/journal.pgen.1004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bataillon T, et al. Inference of purifying and positive selection in three subspecies of chimpanzees (Pan troglodytes) from exome sequencing. Genome Biol Evol. 2015;7(4):1122–1132. doi: 10.1093/gbe/evv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173(2):891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23(1):23–35. [PubMed] [Google Scholar]
- 28.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5(5):e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neher RA. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu Rev Ecol Evol Syst. 2013;44(1):195–215. [Google Scholar]
- 30.Lourenço JM, Glémin S, Galtier N. The rate of molecular adaptation in a changing environment. Mol Biol Evol. 2013;30(6):1292–1301. doi: 10.1093/molbev/mst026. [DOI] [PubMed] [Google Scholar]
- 31.Karasov T, Messer PW, Petrov DA. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 2010;6(6):e1000924. doi: 10.1371/journal.pgen.1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galtier N. Adaptive protein evolution in animals and the effective population size hypothesis. PLoS Genet. 2016;12(1):e1005774. doi: 10.1371/journal.pgen.1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermisson J, Pennings PS. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169(4):2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20(4):R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59(11):2312–2323. [PubMed] [Google Scholar]
- 36.Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- 37.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messer PW. SLiM: Simulating evolution with selection and linkage. Genetics. 2013;194(4):1037–1039. doi: 10.1534/genetics.113.152181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156(1):297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.