Significance
The pKa of Glu325 in the lactose permease is very alkaline, and protonation is essential for effective binding of galactopyranosides because a negative charge at this position is incompatible with binding. Thus, Glu325 plays a central role in “coupling.”
Keywords: transport, membrane proteins, lactose permease, protonation, surface-enhanced infrared spectroscopy
Abstract
Lactose permease (LacY), a paradigm for the largest family of membrane transport proteins, catalyzes the coupled translocation of a galactoside and a H+ across the cytoplasmic membrane of Escherichia coli (galactoside/H+ symport). One of the most important aspects of the mechanism is the relationship between protonation and binding of the cargo galactopyranoside. In this regard, it has been shown that protonation is required for binding. Furthermore when galactoside affinity is measured as a function of pH, an apparent pK (pKapp) of ∼10.5 is obtained. Strikingly, when Glu325, a residue long known to be involved in coupling between H+ and sugar translocation, is replaced with a neutral side chain, the pH effect is abolished, and high-affinity binding is observed until LacY is destabilized at alkaline pH. In this paper, infrared spectroscopy is used to identify Glu325 in situ. Moreover, it is demonstrated that this residue exhibits a pKa of 10.5 ± 0.1 that is insensitive to the presence of galactopyranoside. Thus, it is apparent that protonation of Glu325 specifically is required for effective sugar binding to LacY.
The lactose permease of Escherichia coli (LacY), the most intensively studied membrane transport protein in the major facilitator superfamily (MFS)(1), catalyzes galactopyranoside/H+ symport by a mechanism proposed recently (2). LacY is composed of N- and C-terminal domains, each with six mostly irregular transmembrane helices linked by a relatively long cytoplasmic loop with the N and C termini on the cytoplasmic face of the membrane. Structures of two conformations of LacY have been solved: (i) an inward-open conformer with a large aqueous cavity open to the cytoplasmic side and a tightly sealed periplasmic side (3–6); and (ii) an outward-open, occluded conformer with a tightly sealed cytoplasmic side and a bound lactose homolog (7, 8) or a nanobody (9). As extensively documented (10), LacY operates by an alternating access mechanism. By this means, substrate- and H+-binding sites in the middle of the molecule become alternatively accessible to either side of the membrane as the result of reciprocal opening/closing of periplasmic and cytoplasmic cavities.
Each of the 417 residues in LacY has been mutated (11), and remarkably, only 9 amino acyl side chains are irreplaceable with respect to lactose/H+ symport. Seven side chains are directly involved in galactoside binding and specificity, whereas Glu325 (helix X) and possibly Arg302 (helix IX) are involved in coupled H+ translocation (2, 11, 12). LacY mutants with neutral replacements for Glu325 (helix X) bind galactosides with normal affinity and catalyze equilibrium exchange and counterflow of galactosides, but do not catalyze any reaction involving H+ symport (13, 14).
The affinity of WT LacY for galactosides (Kd) varies with pH and exhibits an apparent pK (pKapp) of ∼10.5 (12, 15, 16). Therefore, over the physiological range of pH, LacY is protonated. Furthermore, sugar binding to purified LacY in detergent does not induce a change in ambient pH under conditions where binding or release of 1 H+/LacY can be measured (15). These observations and many others (reviewed in refs. 17, 18) provide strong evidence for a symmetrical ordered mechanism in which protonation precedes galactoside binding on one side of the membrane, and follows sugar dissociation on the other side. A similar ordered mechanism may also be common to other members of the MFS (19–23).
Dramatically, the pKapp titration is abolished in LacY mutants with neutral replacements for Glu325, and high-affinity binding is observed up to pH 11. This behavior is unique and suggests the possibility that Glu325 may be the sole residue directly involved in H+ binding and coupled transport (2). In any case, the observations indicate that Glu325 is directly involved in coupling between galactoside and H+ translocation. LacY cannot sustain a negative charge on Glu325 and bind galactoside simultaneously or, stated conversely, Glu325 must be protonated to bind sugar. Of course, LacY must also deprotonate for turnover to occur. Because certain Arg302 LacY mutants cannot catalyze active transport but catalyze equilibrium exchange, it has been postulated that positively charged Arg302 (helix IX) may be important with respect to deprotonation (24).
In this study, Glu325 is identified in situ, and the pKa of this intriguing residue is determined by monitoring pH-induced changes in purified LacY by infrared (IR) spectrometry. Reaction-induced IR is an established tool for studying protonation changes in proteins. The technique has been used successfully to identify several critical residues in the proton path of membrane proteins (25, 26), as well as water molecules (27). Among several specific examples, Zscherp et al. (28) determined the pKa of Asp96 in bacteriorhodopsin, and the protonation state of the central glutamic acid in cytochrome c oxidase was also identified by IR (29). Here we study pH- and substrate-dependent conformational changes in a monolayer of immobilized LacY on a modified gold layer in an attenuated total reflectance (ATR) cell. Surface-enhanced IR absorption spectra are presented for WT LacY and mutant E325A, as well as alkali-stable mutants G46W/G262W (LacYww) and LacYww/E325A, which allow identification of Glu325 in situ and the direct demonstration that Glu325 has a pKa of 10.5. ± 0.1.
Results
Perfusion-Induced IR Spectroscopy.
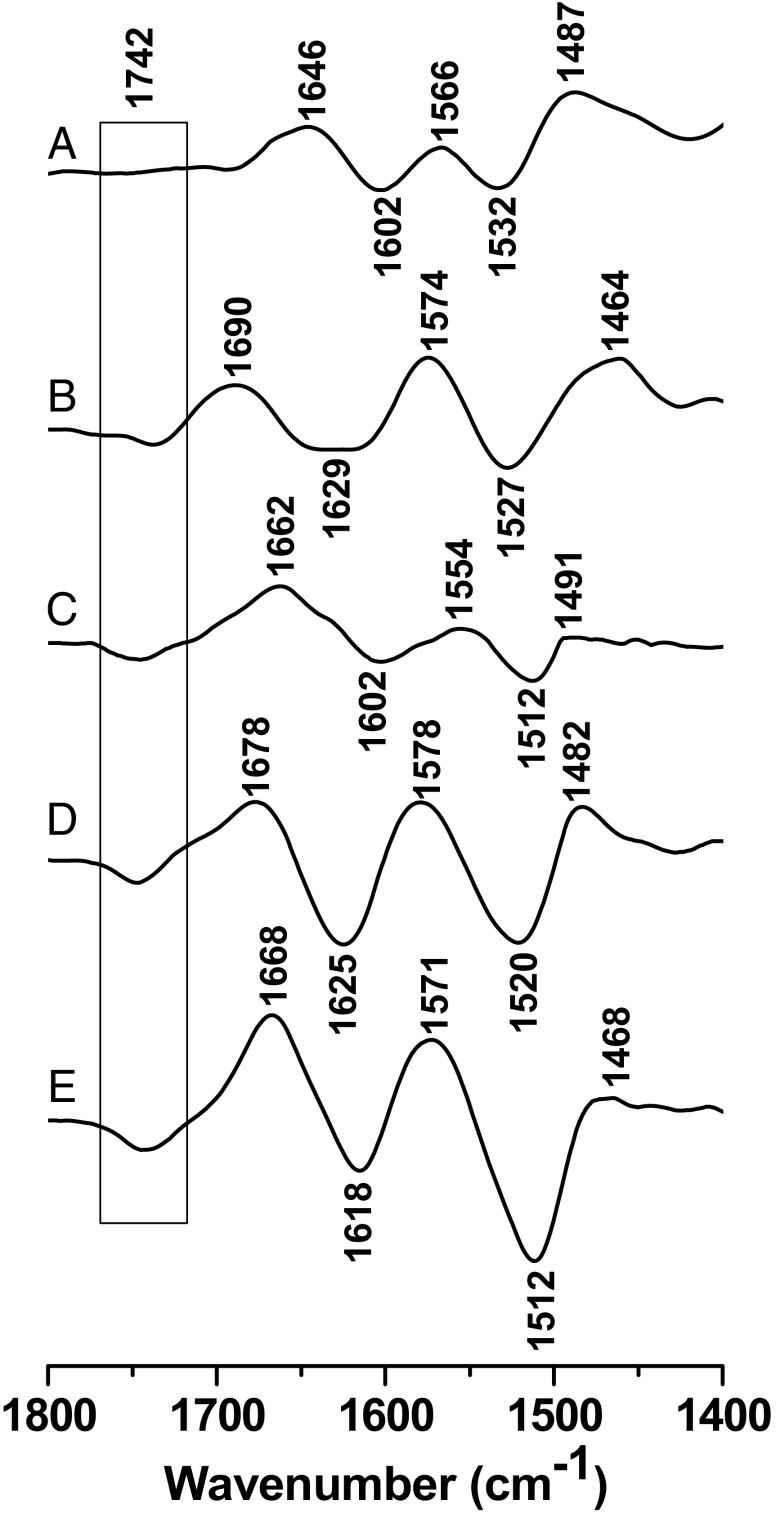
A stable monolayer with LacY immobilized via a C-terminal His-tag to a modified gold layer was obtained as described (30) (Fig. S1), and the effect of pH changes was monitored by surface enhanced IR spectroscopy in an ATR perfusion cell. Fig. 1 shows the reversible perfusion-induced IR difference spectra obtained from pH 9–10.5 and back with WT LacY in the presence of p-nitrophenyl-α-d-galactopyranoside (NPG). A broad peak centered at 1,695 cm−1 and 1,675 cm−1 is observed in the amide I region, which ranges from 1,700 cm−1 to 1,600 cm−1 and includes conformational changes in the C=O groups in the protein backbone. For a typical α-helical structure, signals at ∼1,650–1,660 cm−1 are expected (31) (see absorption spectra in Fig. S2). However, conformational changes in parts of a helix can strongly shift this vibrational mode because both dipole–dipole coupling and the hydrogen bonding environment may be perturbed (31). Therefore, changes in the amide I signature are informative with respect to conformational rearrangements. When comparing spectra obtained stepwise at pHs up to 10.5 (Fig. 1 A–C), it is apparent that the broad amide I absorbance becomes sharper with increasing pH, and at pH 10.5 it is centered at 1,668 cm−1 with a shoulder at 1,632 cm−1, indicating a conformational change related to the increase in pH.
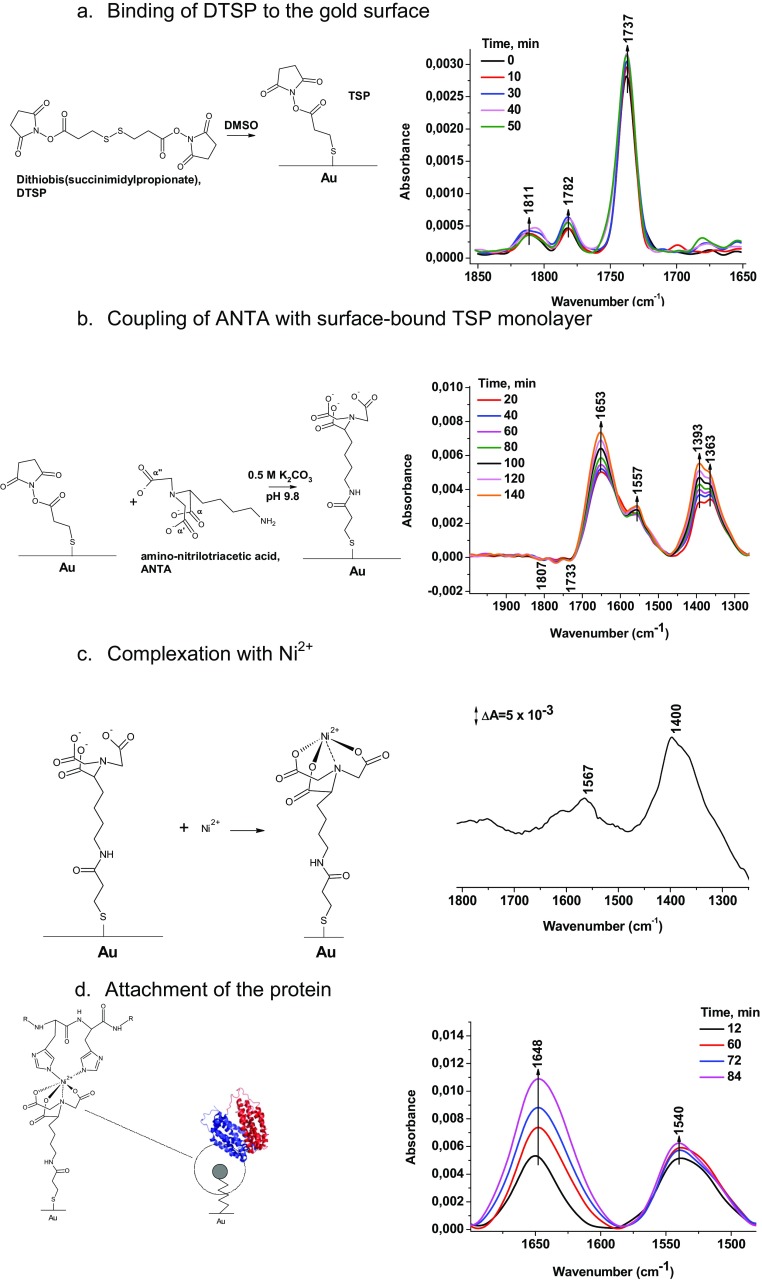
Fig. S1.
Immobilization of the protein on the silicon crystal. A–D describe the observed reactions and the surface-enhanced infrared spectra measured in order to follow the reaction.
Fig. 1.
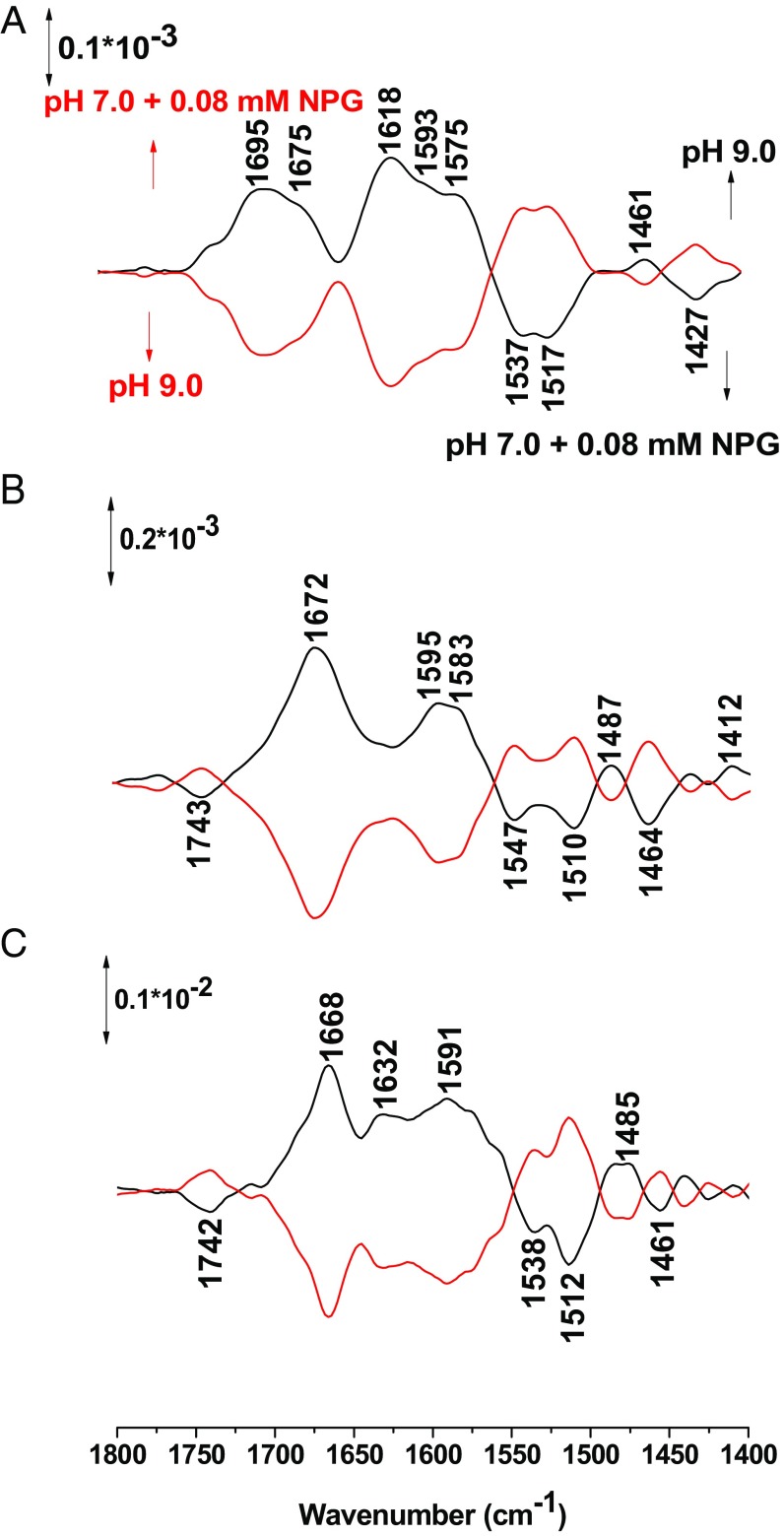
Perfusion-induced FTIR difference spectra of Lac Y obtained from the sample equilibrated at pH 7 in the presence of NPG subtracted from the sample equilibrated at pH 9 (A), 10 (B), and 10.5 (C), respectively (black line) and reverse (red line).
Fig. S2.
Absorption IR spectra of LacY and E325 mutant.
The amide II range (between 1,590 cm−1 and 1,450 cm−1) involves coupled CN/NH vibrational modes of the protein backbone (31), as well as contributions from individual amino acids reorganizing or changing protonation states with the alkaline pH shift. Significant shifts and increases in intensity are also observed with increasing pH.
The most interesting change with increasing pH is observed at 1,742 cm−1. Signals at this position are characteristic of protonated Asp or Glu residues in a hydrophobic environment as shown in IR spectra of model compounds and in IR difference spectra obtained with a number of membrane proteins (32). A negative signal is observed here, reflecting deprotonation. As shown, the signal is absent at pH 9 (Fig. 2A), begins to appear at pH 10 (Fig. 2B), and increases in intensity at pH 10.5 (Fig. 2C), clearly indicating deprotonation of an acidic side chain(s). Protonation and deprotonation are fully reversible and correlate with reorganization of the polypeptide backbone. Although signals for deprotonated acidic side chains are expected to be ∼1,575 cm−1 and 1,400 cm−1 (33, 34), these signals are obscured by the amide II band, and they are difficult to assign unambiguously.
Fig. 2.
Perfusion-induced FTIR difference spectra of LacY wild type equilibrated in the presence of 0.08 mM NPG at pH 7 subtracted from the sample equilibrated at pH 9.0 (A), 10.0 (B), and 10.5 (C), respectively.
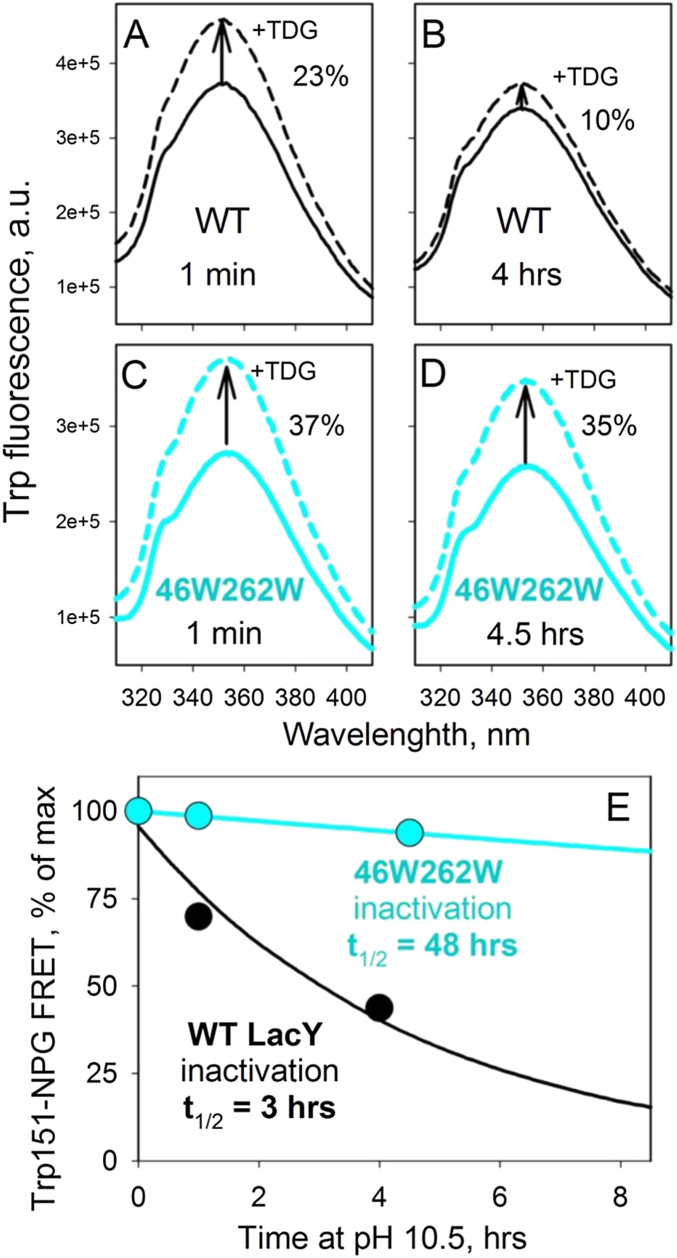
Galactoside binding by WT LacY becomes unstable at highly alkaline pH (>11) (12, 15, 16). Because the studies presented here necessitate exposure of the protein to high pH, the alkali stability of WT LacY was compared with that of the double-Trp mutant G46W/G262W (LacYww), which is more stable than WT LacY, but exhibits the identical pKapp for sugar binding (35). WT LacY exhibits a half-life (t1/2) with respect to NPG binding of 3 h at pH 10.5, whereas the t1/2 for LacYww is 48 h (Fig. S3). Difference spectra obtained from this mutant show the same signal at 1,742 cm−1 very clearly (Fig. 3).
Fig. S3.
Stability of WT LacY and 46W/262W mutant at pH 10.5. Sugar binding was measured as Trp151-NPG Förster resonance energy transfer (44) after preincubation of reconstituted PLs (with 14 mM protein at a lipid-to-protein ratio of 5 (wt/wt) in 50 mM CAPS pH 10.5 for given time at room temperature. (A–D) Increase of Trp fluorescence intensity (excitation at 295 nm) resulting from displacement of bound NPG (0.1 mM) by excess of TDG (6 mM) measured at pH 7.5 in 50 mM NaPi with 0.4 mM LacY. (E) Time course of LacY inactivation at pH 10.5.
Fig. 3.
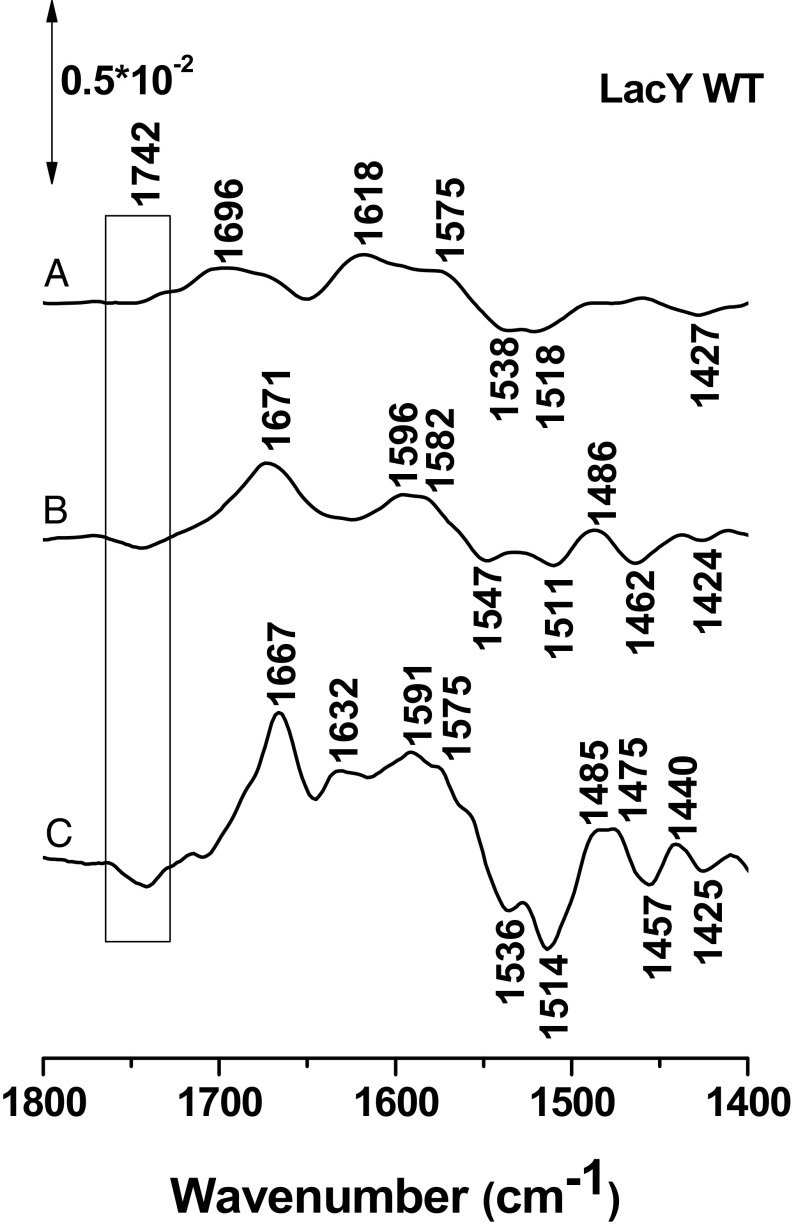
Perfusion-induced FTIR difference spectra of LacY G46W/G262W in the presence of NPG in both perfusion solutions. The sample equilibrated at pH 7.0 was subtracted from the sample equilibrated at 9.0 (A), 10.0 (B), 10.5 (C), 10.9 (D), and 11.5 (E), respectively.
Identification of Glu325.
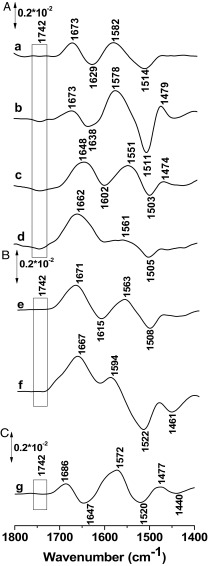
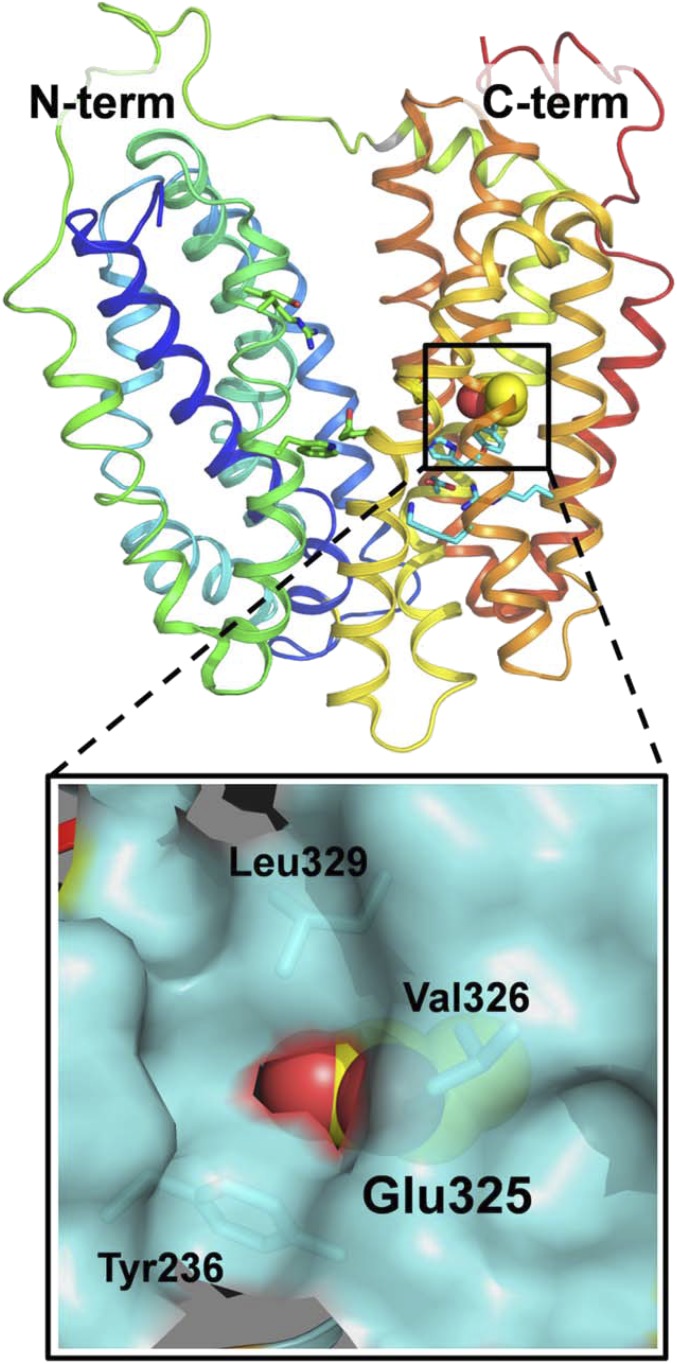
The previous experiments were repeated with LacYww and with LacYww carrying the E325A mutation. Both mutants are significantly more stable at high pH than WT LacY. The difference signal at 1,742 cm−1 becomes significantly stronger in LacYww as pH increases from 10.5 to 10.9–11.5 (Fig. 4A). However, in perfusion-induced IR difference spectra of the LacYww/E325A and the E325A mutants, the signal at 1,742 cm−1 is totally absent (Fig. 4 B and C, respectively). Glu325, which is clearly involved in coupling between H+ and galactoside translocation (2), is located within the membrane bilayer in helix X and surrounded by hydrophobic side chains (Fig. 5). Previous observations show that this side chain has a pKapp of ∼10.5 based on pH titrations of galactoside affinity (12, 15, 16). The current findings unequivocally confirm that Glu325 is responsible for the 1,742-cm−1 band.
Fig. 4.
Perfusion-induced FTIR difference spectra of LacY G46W/G262W in the absence of NPG obtained from samples equilibrated at pH 7 subtracted from the samples equilibrated at pH 8.0 (A,a), 10.5 (A, b), 10.9 (A, c), and 11.5 (A, d), respectively. Difference spectra of the LacY G46W/G262W/E325A variant for the step from to pH 7.0 to pH 11.5 (B, e) and 10.9 (B, f), and for the simple LacY E325A mutant for the step from pH 6.0 to pH 10.0 (C, g).
Fig. 5.
Position of Glu325 in C-terminal six-helix bundle of WT LacY (Protein Data Bank ID code 2V8N). LacY is presented as rainbow-colored backbone (from blue to red for helices 1–12) with hydrophilic cavity open to cytoplasmic side. Side chain of Glu325 located in helix X is shown as spheres. The area around Glu325 is enlarged with hydrophobic environment displayed as a space-filled cartoon (cyan).
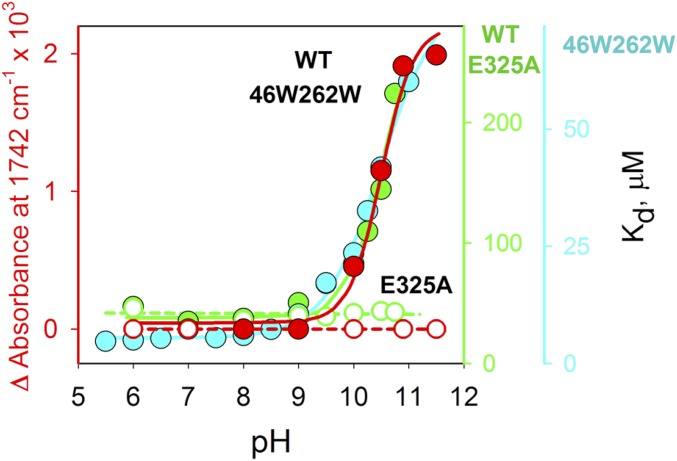
The pKa of Glu325 was determined by plotting the Δ in signal intensity at 1,742 cm−1 versus pH (Fig. 6). From the ΔIR fit observed (red), Glu325 has a pKa of 10.5 ± 0.1, a value that agrees remarkably well with the pKapps obtained previously for either WT LacY (green) (12, 16) or mutant LacYww (cyan) (15). Although the E325A mutant binds galactoside with high affinity over the entire pH range measured, essentially no value for Δ is observed (open circles).
Fig. 6.
pH dependence of Δ-IR intensity change at 1,742 cm−1 measured with LacYww in the absence or presence of NPG (filled red circles) or E325A LacY (open red circles), and Kd values for NPG binding to WT LacY (filled green circles), E325A LacY (open green circles), or LacYww (filled cyan circles). Kd values were calculated as the ratio of rate constants (koff/kon) measured by stopped flow (12, 35).
Effect of NPG.
Notably, the IR signal at 1,742 cm−1 exhibits no significant difference in the absence or presence of NPG at pH 10.5, 10.9, or 11.5 with either WT LacY or the LacYww mutant. Moreover, the pH titration is unaffected by the absence or presence of the galactoside (Fig. 6).
Discussion
These studies provide convincing evidence that Glu325 in LacY, which plays a central role in galactoside/H+ symport, has a pKa of 10.5, as suggested by studies on the effect of pH on galactopyranoside affinity. Glu325 is an essential part of the coupling mechanism in LacY, because its protonation or deprotonation determines whether or not galactoside binds effectively. Clearly, this represents an important aspect of coupling, which initiates the transport mechanism. Importantly, other symporters with a carboxyl group that behaves like E325 with respect to tranwport have also been described recently for FucP (19), and XylE (20) and GlcPSe (21).
Stabilization of the protonated form of Glu325 is likely due to the hydrophobic microenvironment within transmembrane helix X (Fig. 5). But, deprotonation is also necessary for turnover, and with an apparent pK of 10.5, how does deprotonation occur? Possibly, the pKa of Glu325 may decrease by becoming more accessible to water. In contrast, however, Arg302 may be important in this capacity (24, 36). Like neutral replacements for Glu325, certain neutral-replacement mutants for Arg302 are also defective in lactose/H+ symport, but catalyze equilibrium exchange (24). Perhaps the positively charged guanidinium group at position 302 facilitates deprotonation of Glu325 after the galactoside dissociates. Although Arg302 and Glu325 are relatively far apart with the hydroxyl group of Tyr236 in between in the current structure, the double-mutant R302C/E325C exhibits excimer fluorescence when labeled with pyrene maleimide (37) and the double-mutant R302H/E325H binds Mn(II) with micromolar affinity (38). Therefore, Arg302 and Glu325 may be in closer proximity in another conformation of LacY.
Examples of residues with perturbed pKas in important mechanistic positions have been reported for several membrane proteins, as reviewed by Harris and Turner (39) for example. It was concluded that the pKa is often modulated by a combination of several types of interactions such as long-range and local electrostatic effects. A prominent example is residue D96 in bacteriorhodopsin. A high pKa is also observed for cytochrome c oxidase, where residue E278 (Paracoccus denitrificans numbering), localized in the membrane, was found to have a pKa higher than 11 (40). Electrochemically induced FTIR difference spectra revealed a signal at 1,748 cm−1, at a position close to that observed here for E325 in LacY. A similar hydrogen-bonding environment is thus expected.
One important observation in the current work is that the titration of Glu325 is not altered by binding of NPG. This is unexpected because protonation of Glu325 is coupled to increased affinity of the galactoside. In a straightforward thermodynamic model, the H+ and sugar affinities should demonstrate reciprocity, (i.e., if H+ binding enhances sugar affinity by 100-fold, then sugar binding should also enhance the affinity of the H+ by 100-fold). However, in addition to the IR findings, binding of sugar to LacY does not cause any change in ambient pH, as discussed previously (15).
In conclusion, these finding provide direct experimental evidence that Glu325 has a pKa of 10.5, a value that coincides precisely with the variation of the affinity of LacY for galactoside as a function of pH. The conclusion provides strong confirmation for the critical role of this residue in the reaction mechanism postulated for LacY (2).
Materials and Methods
Materials.
Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. Restriction enzymes were purchased from New England Biolabs. The QuikChange II kit was purchased from Stratagene. NPG was from Sigma. Talon superflow resin was purchased from BD Clontech. Dodecyl-β-d-maltopyranoside (DDM) and octyl-β-d-glucoside (OG) were from Affimetrix. Synthetic phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) were from Avanti Polar Lipids, Inc. All other materials were of reagent grade obtained from commercial sources.
Construction of Mutants, Purification of LacY, and Reconstitution into Proteoliposomes.
Construction of mutants, expression in E. coli, and purification of LacY were performed as described (41). All constructs contained a C-terminal 6His-tag that was used for affinity purification with Talon resin. Purified proteins (10–15 mg/mL) in 50 mM sodium phosphate (NaPi/0.02% DDM; pH 7.5) were frozen in liquid nitrogen and stored at −80 °C until use. Reconstitution into proteoliposomes was carried out with synthetic phospholipids (POPE/POPG ratio 3:1) by using the dilution method (42). Briefly, purified LacY in 0.02% DDM (wt/vol) was mixed with phospholipids dissolved in 1.2% OG maintaining a lipid-to-protein ratio of 5 (wt/wt). The mixture was kept on ice for 20 min and then quickly diluted 50 times in 50 mM NaPi buffer (pH 7.5). The proteoliposomes (PLs) were harvested by centrifugation for 1 h at 100,000 g, suspended in the same buffer, and subjected to two cycles of freeze/thaw/sonication. Flash-frozen PLs were stored at −80 °C. Before use, the PLs were subjected to an additional two cycles of freeze/thaw/sonication.
Surface Modification of the Silicon Crystal and Protein Immobilization.
First, a gold layer was cast on the surface of a Si ATR crystal by etching the Si with hydrofluoric acid (HF) and reduction of AuCl4 as described previously (30). Before the deposition of the gold film, the ATR crystal was polished with 0.3-µm alumina, rinsed with copious amounts of Millipore water, acetone, and water again. The crystal was then dried under an argon stream and immersed in 40% NH4F (wt/vol) for 1 min, rinsed, and dried again. It was then heated at 65 °C for 10 min together with the plating solution. This solution was a 1:1:1 mix (vol/vol/vol) of (i) 15 mM NaAuCl4, (ii) 150 mM Na2SO3, 50 mM Na2S2O3, and 50 mM NH4Cl, and (iii) HF 2% (wt/vol; total volume: 1 mL). Once the plating temperature was reached, the prism was covered with the solution for 40 s, and the reaction was stopped by washing the plating solution off with water, followed by drying with a stream of argon. The resulting gold film was then tested for electrical conductance with a multimeter (the typical electric resistance of the layer as measured from one corner to another of the crystal should be ∼15 Ω for a thickness of 50 nm).
The experimental procedure for the nickel nitrilotriacetic acid self-assembled monolayer (Ni-NTA SAM) was adapted from refs. 30, 43. First, the gold modified silicon ATR crystal was covered with 1 mg/mL of 3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester) (DTSP) in dry dimethyl sulfoxide and the monolayer was allowed to self-assemble for 1 h. The excess DTSP was then washed away with dry DMSO and the crystal was dried under an argon stream. Afterward, it was covered with 100 mM Nα′,Nα′′-bis(carboxymethyl)-l-lysine in 0.5 M K2CO3 at pH 9.8 for 3 h and then rinsed with water. Finally, the surface was incubated in 50 mM Ni(ClO4)2 for 1 h before being washed one last time with water. For immobilization of the protein, 5 µL of 7.2 mg/mL LacY was deposited on the modified gold surface for 1h.
Infrared Spectroscopy.
A configuration allowing the simultaneous acquisition of FTIR spectra in the ATR mode with perfusion of solutions with given composition was used. As a multireflection ATR unit, we used silicon crystal with 3-mm surface diameter. All experiments were carried out with a Bruker Vertex 70 FTIR spectrometer (Globar source, KBr Beamsplitter, mercury cadmium telluride detector) at 8-mm aperture and 40-kHz scanner velocity. The measurements were carried out at ∼7 °C. Solutions was kept on ice before use. For the data presented here, the pump speed was kept constant at a flow rate of 0.2 mL/min. Before each perfusion step, the input tube was carefully washed with water and buffer.
Difference Spectra.
To monitor pH-induced difference spectra, we used one perfusion buffer with constant pH value 7.0 (25 mM KPi/100 mM KCl/0.01% DDM) and a second perfusion solution with the same composition but at different pH values ranging from 8.0 to 11.5. At the beginning, the system was equilibrated with the KPi (pH 7.0) for 30 min. Thereafter, the spectrum was recorded as background and the perfusion solution was changed to the second solution (pH range 8.0–11.5). After 20 min (pH 8.0–11.5) minus pH 7.0 difference spectra were recorded. The new state of the protein was recorded as background, and the solution was changed to pH 7.0. Again after 20 min, the pH 7.0 minus (pH 8.0–11.5) difference spectra were obtained. The same procedure was repeated five times and the difference spectra were averaged and smoothed. Baseline correction was done, where necessary. The data were normalized on the basis of the absorbance spectra obtained at the beginning of each experiment, when the data from different samples had to be compared.
Difference Spectra of LacY in the Presence of NPG.
NPG was dissolved in 25 mM KPi/100 mM KCl/0.01% DDM (pH 7.0) at a final concentration of 80 µM. A second perfusion solution of the same composition at pH values ranging from 9.0 to 11.5 was also prepared. The experiments have then been performed in analogy to the data obtained in the absence of NPG.
Acknowledgments
We are deeply indebted to Prof. Robert B. Gennis for arranging the marriage between our two laboratories. We also thank Irina Smirnova for many insightful discussions, critically reading the manuscript, and helping with preparation of some of the figures. John Lim provided excellent technical support. P.H. acknowledges the icFRC, the CNRS, and the University of Strasbourg for financial support. The work was also supported by NIH Grants DK51131, GM120043, and National Science Foundation Eager Grant MCB1547801 to H.R.K., who also gratefully acknowledges the University of California, Los Angeles and a gift from Ruth and Bucky Stein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621431114/-/DCSupplemental.
References
- 1.Saier MH, Jr, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1(2):257–279. [PubMed] [Google Scholar]
- 2.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112(5):1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 4.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25(6):1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104(39):15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaptal V, et al. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc Natl Acad Sci USA. 2011;108(23):9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar H, et al. Structure of sugar-bound LacY. Proc Natl Acad Sci USA. 2014;111(5):1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar H, Finer-Moore JS, Kaback HR, Stroud RM. Structure of LacY with an α-substituted galactoside: Connecting the binding site to the protonation site. Proc Natl Acad Sci USA. 2015;112(29):9004–9009. doi: 10.1073/pnas.1509854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, et al. Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation. Proc Natl Acad Sci USA. 2016;113(44):12420–12425. doi: 10.1073/pnas.1615414113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: A novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12(13):1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 12.Smirnova I, Kasho V, Sugihara J, Choe JY, Kaback HR. Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry. 2009;48(37):8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrasco N, Antes LM, Poonian MS, Kaback HR. lac permease of Escherichia coli: Histidine-322 and glutamic acid-325 may be components of a charge-relay system. Biochemistry. 1986;25(16):4486–4488. doi: 10.1021/bi00364a004. [DOI] [PubMed] [Google Scholar]
- 14.Carrasco N, et al. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989;28(6):2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- 15.Smirnova I, Kasho V, Sugihara J, Vázquez-Ibar JL, Kaback HR. Role of protons in sugar binding to LacY. Proc Natl Acad Sci USA. 2012;109(42):16835–16840. doi: 10.1073/pnas.1214890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci USA. 2008;105(26):8896–8901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madej MG, Kaback HR. 2014. The life and times of Lac permease: Crystals ain't enough, but they certainly do help. Membrane Transporter Function: To Structure and Beyond, Springer Series in Biophysics: Transporters, Vol 17, eds Ziegler C, Kraemer R (Springer, Berlin), pp 121–158.
- 19.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467(7316):734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490(7420):361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 21.Iancu CV, Zamoon J, Woo SB, Aleshin A, Choe JY. Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc Natl Acad Sci USA. 2013;110(44):17862–17867. doi: 10.1073/pnas.1311485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan L, Jakkula SV, Hodkoff AA, Su Y. Role of Gly117 in the cation/melibiose symport of MelB of Salmonella typhimurium. Biochemistry. 2012;51(13):2950–2957. doi: 10.1021/bi300230h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ethayathulla AS, et al. Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahin-Tóth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichiacoli. Proc Natl Acad Sci USA. 2001;98(11):6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwaki M, et al. Redox-induced transitions in bovine cytochrome bc1 complex studied by perfusion-induced ATR-FTIR spectroscopy. Biochemistry. 2003;42(38):11109–11119. doi: 10.1021/bi0343020. [DOI] [PubMed] [Google Scholar]
- 26.Siebert F, Mäntele W, Gerwert K. Fourier-transform infrared spectroscopy applied to rhodopsin. The problem of the protonation state of the retinylidene Schiff base re-investigated. Eur J Biochem. 1983;136(1):119–127. doi: 10.1111/j.1432-1033.1983.tb07714.x. [DOI] [PubMed] [Google Scholar]
- 27.Wolf SFE, Cui Q, Gerwert K. Infrared spectral marker bands characterizing a transient water wire inside a hydrophobic membrane protein. J Chem Phys. 2014;141(22):22D524. doi: 10.1063/1.4902237. [DOI] [PubMed] [Google Scholar]
- 28.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1999;96(10):5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellwig P, et al. Involvement of glutamic acid 278 in the redox reaction of the cytochrome c oxidase from Paracoccus denitrificans investigated by FTIR spectroscopy. Biochemistry. 1998;37(20):7390–7399. doi: 10.1021/bi9725576. [DOI] [PubMed] [Google Scholar]
- 30.Kriegel S, Uchida T, Osawa M, Friedrich T, Hellwig P. Biomimetic environment to study E. coli complex I through surface-enhanced IR absorption spectroscopy. Biochemistry. 2014;53(40):6340–6347. doi: 10.1021/bi500955a. [DOI] [PubMed] [Google Scholar]
- 31.Barth A. Infrared spectroscopy of proteins. Biochim Biophys Acta. 2007;1767(9):1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Yang K, et al. Glutamate 107 in subunit I of the cytochrome bd quinol oxidase from Escherichia coli is protonated and near the heme d/heme b595 binuclear center. Biochemistry. 2007;46(11):3270–3278. doi: 10.1021/bi061946+. [DOI] [PubMed] [Google Scholar]
- 33.Barth A. The infrared absorption of amino acid side chains. Prog Biophys Mol Biol. 2000;74(3-5):141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 34.Wolpert M, Hellwig P. Infrared spectra and molar absorption coefficients of the 20 alpha amino acids in aqueous solutions in the spectral range from 1800 to 500 cm(-1) Spectrochim Acta A Mol Biomol Spectrosc. 2006;64(4):987–1001. doi: 10.1016/j.saa.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 35.Smirnova I, Kasho V, Sugihara J, Kaback HR. Trp replacements for tightly interacting Gly-Gly pairs in LacY stabilize an outward-facing conformation. Proc Natl Acad Sci USA. 2013;110(22):8876–8881. doi: 10.1073/pnas.1306849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson M, et al. Proton-coupled dynamics in lactose permease. Structure. 2012;20(11):1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung K, Jung H, Wu J, Privé GG, Kaback HR. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32(46):12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 38.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal-binding sites to study helix proximity in the lactose permease of Escherichia coli. 2. Proximity of helix IX (Arg302) with helix X (His322 and Glu325) Biochemistry. 1995;34(48):15667–15670. doi: 10.1021/bi00048a010. [DOI] [PubMed] [Google Scholar]
- 39.Harris TKTG, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53(2):85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 40.Brändén G, Pawate AS, Gennis RB, Brzezinski P. Controlled uncoupling and recoupling of proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103(2):317–322. doi: 10.1073/pnas.0507734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104(42):16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viitanen P, Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification, reconstitution, and characterization of the lac permease of Escherichia coli. Methods Enzymol. 1986;125:429–452. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- 43.Ataka K, Heberle J. Use of surface enhanced infrared absorption spectroscopy (SEIRA) to probe the functionality of a protein monolayer. Biopolymers. 2006;82(4):415–419. doi: 10.1002/bip.20501. [DOI] [PubMed] [Google Scholar]
- 44.Smirnova IN, Kasho VN, Kaback HR. Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry. 2006;45(51):15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]