Significance
Interactions between transsynaptic cell-adhesion molecules enable formation of trillions of synaptic connections that wire neurons into vast functional circuits. Neurexins are presynaptic cell-adhesion molecules that bind to diverse postsynaptic ligands, and are genetically linked to neuropsychiatric disorders. Here we show that CA10, a secreted, catalytically inactive carbonic-anhydrase homolog, binds in a cis configuration to all neurexins, including Nrxn1γ. In cultured neurons, CA10 enhances surface transport of neurexins, indicating that the neurexin–CA10 complex assembles in the secretory pathway. Our results suggest that neurexins bind to a larger repertoire of trans and cis ligands than previously envisioned to mediate multifarious synaptic interactions with diverse functions.
Keywords: Car10, Car11, synapse, cell adhesion
Abstract
Establishment, specification, and validation of synaptic connections are thought to be mediated by interactions between pre- and postsynaptic cell-adhesion molecules. Arguably, the best-characterized transsynaptic interactions are formed by presynaptic neurexins, which bind to diverse postsynaptic ligands. In a proteomic screen of neurexin-1 (Nrxn1) complexes immunoisolated from mouse brain, we identified carbonic anhydrase-related proteins CA10 and CA11, two homologous, secreted glycoproteins of unknown function that are predominantly expressed in brain. We found that CA10 directly binds in a cis configuration to a conserved membrane-proximal, extracellular sequence of α- and β-neurexins. The CA10–neurexin complex is stable and stoichiometric, and results in formation of intermolecular disulfide bonds between conserved cysteine residues in neurexins and CA10. CA10 promotes surface expression of α- and β-neurexins, suggesting that CA10 may form a complex with neurexins in the secretory pathway that facilitates surface transport of neurexins. Moreover, we observed that the Nrxn1 gene expresses from an internal 3′ promoter a third isoform, Nrxn1γ, that lacks all Nrxn1 extracellular domains except for the membrane-proximal sequences and that also tightly binds to CA10. Our data expand the understanding of neurexin-based transsynaptic interaction networks by providing further insight into the interactions nucleated by neurexins at the synapse.
A plethora of protein–protein interactions orchestrate the robust yet plastic wiring of the central nervous system. A well-known example at the level of synapses is that of presynaptic neurexins, which mediate transsynaptic interactions with a number of structurally diverse postsynaptic ligands (1, 2). Neurexins [NRXN (human) and Nrxn (mouse)] are essential for normal synaptic function (3), and genetic variation in NRXN genes predisposes to autism and schizophrenia (4). Although the exact synaptic function of neurexins is incompletely understood, their cis- and trans-synaptic interactions are believed to shape the molecular architecture and properties of specific synapses. Each of the three mammalian neurexin genes (Nrxn1 to Nrxn3 in mice) encodes two principal isoforms, longer α-neurexins and shorter β-neurexins, from distinct promoters (5–7). The extracellular domains of α-neurexins contain six LNS (laminin-neurexin-sex hormone-binding globulin) and three EGF (epidermal growth factor-like) domains, whereas the extracellular sequence of β-neurexins only includes the last LNS6 domain of α-neurexins (5–7). Neurexin transcripts are subject to extensive alternative splicing at six conserved sites [splice site (SS)1 to SS6] to generate >1,000 unique transcripts (8–10), creating a molecular diversity that may contribute to synaptic diversity. For example, alternative splicing at SS4 regulates the affinity of LNS6 for neuroligins (11), leucine-rich repeat transmembrane neuronal proteins (LRRTMs) (12), and cerebellin-1/glutamate receptor delta-2 (GluRδ2) (13), and influences synaptic properties in vivo (14).
Carbonic anhydrases (EC 4.2.1.1) are ubiquitous enzymes that play a key role in acid–base homeostasis by catalyzing the interconversion of carbon dioxide and water to bicarbonate. Carbonic anhydrase-related proteins (CARPs) belong to the family of α-carbonic anhydrases but are catalytically inactive due to the loss of one or more of the histidine residues that coordinate the zinc atom in the catalytic core (15, 16). CARPs are evolutionarily conserved in metazoan genomes—interestingly, even more so than the catalytic CAs (17)—suggesting they have alternative cellular functions that are yet to be established. Indeed, a subset of receptor tyrosine phosphatases (PTPRG and PTPRZ) contains extracellular catalytically inactive carbonic-anhydrase domains (18) that mediate the interactions of PTPRG and PTPRZ with contactins, a class of Ig-domain cell-adhesion molecules (19, 20). In mammals, all CARPs that contain no other major domains (CA8, CA10, and CA11) are predominantly expressed in the CNS. Of these, CA8—the most extensively studied gene—encodes an intracellular protein that is almost exclusively expressed in cerebellar Purkinje cells and that is essential for normal Purkinje cell development in mice and humans (21–23). Less is known about CA10 and CA11, both of which carry predicted signal peptides, suggesting a role in the secretory pathway and/or extracellular space.
Here we report the identification of CA10 and CA11 as extracellular binding partners for neurexins. We show that CA10 directly interacts in a cis configuration with conserved residues present in all neurexin isoforms, and assembles into a stable complex with neurexins that leads to the formation of a specific intermolecular disulfide bond. CA10 promotes the surface levels of both α- and β-neurexins when overexpressed in neurons. We also show that the Nrxn1 gene expresses an isoform from an internal third promoter, Nrxn1γ, and that CA10 tightly binds to this isoform as well. Our data expand the known diversity of neurexin ligands, and identify a high-affinity stoichiometric interaction partner for a conserved family of carbonic anhydrase-related proteins in brain.
Results
Identification of CA10 as a Neurexin Ligand.
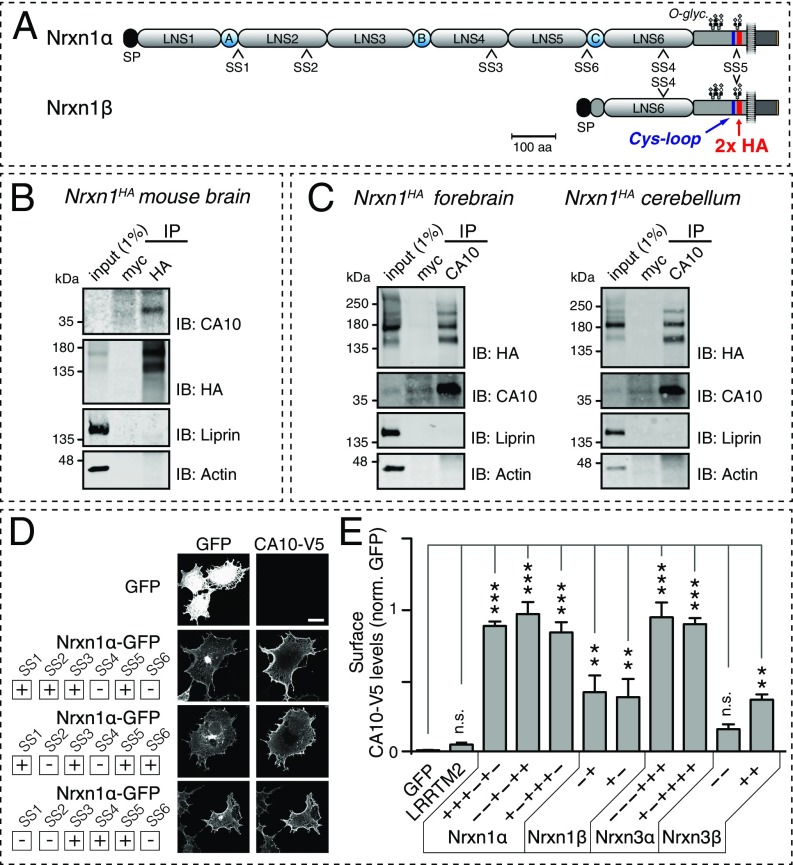
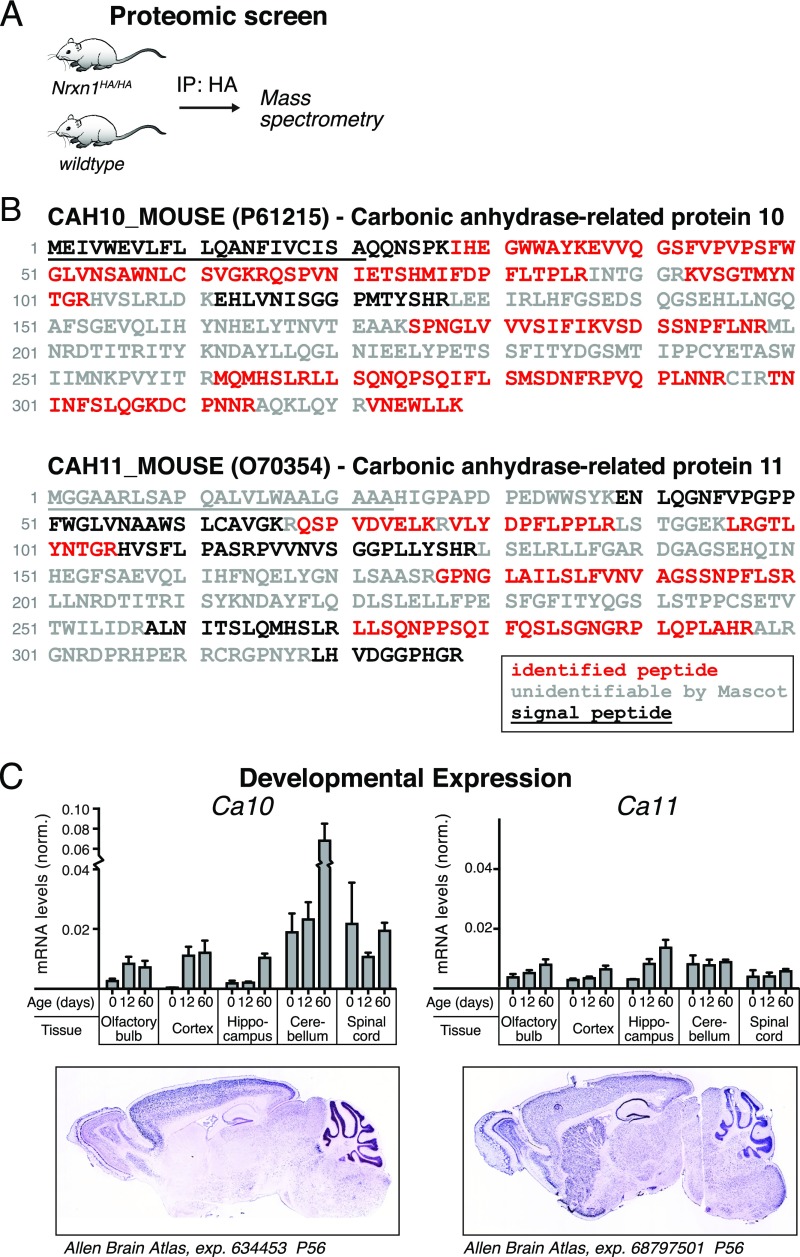
To learn more about the molecular architecture and regulation of neurexin-based synaptic complexes, we screened for proteins that form complexes with neurexins in brain. We prepared brain lysates from knockin mice in which a double-hemagglutinin (2× HA) tag was inserted in Nrxn1 between the transmembrane region and the O-glycosylated “stalk” sequence (Nrxn1HA mice; Fig. 1A; note that Nrxn1HA mice will be described in detail in a future publication). We immunoprecipitated Nrxn1 using HA antibodies (Fig. S1A), and consistently identified peptides corresponding to CA10 and CA11 by mass spectrometry of the immunoprecipitates (Fig. S1B). Using quantitative (q)RT-PCR (Fig. S1C), we found that CA10 is most highly expressed in cerebellum, followed by spinal cord and cerebral cortex. Expression of CA11 was generally lower but more widespread than that of CA10, and included the basal ganglia. Due to its higher expression and better availability of reagents, we focused our studies on CA10.
Fig. 1.
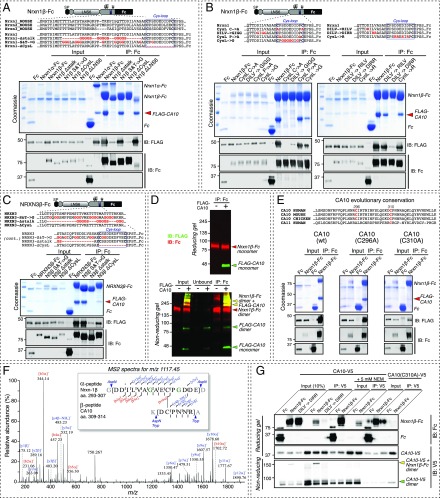
CA10 and Nrxn1 form a complex in murine neurons. (A) Schematic of the Nrxn1α and Nrxn1β domain organizations (gray, LNS1 to LNS6 domains; blue, EGF-like domains). The positions of the tandem HA tag in the Nrxn1HA knockin mice and of the conserved cysteine-loop sequences in neurexins are indicated. SS1 to SS6 identify sites of alternative splicing. O-glyc., O-linked glycosylation sequence; SP, signal peptide. (B) Immunoprecipitation of Nrxn1 from mouse brain lysates from Nrxn1HA mice, using an antibody against the HA epitope or to the myc epitope (as a negative control), identifies an endogenous Nrxn1–CA10 complex. Samples were analyzed by immunoblotting (IB) for CA10, HA (to detect HA-tagged endogenous Nrxn1), α-liprins, and β-actin (the latter two as specificity controls). (C) IPs of endogenous CA10 from forebrain (Left) and cerebellar (Right) lysates of Nrxn1HA mice confirm the Nrxn1–CA10 complex. Controls and immunoblots were performed as in B. (D) Representative confocal images to illustrate the analysis of CA10 surface binding to various neurexins in COS cells that were cotransfected with V5-tagged CA10 and the indicated GFP-tagged neurexins, and then surface-labeled with an antibody against the V5 tag (for additional representative images, Fig. S3A). (Scale bar, 20 µm.) (E) Summary graph of CA10 binding to various neurexins on the surface of COS cells as described in A. Surface-V5 signal was quantified and normalized to that of GFP. Data are means ± SEM (n = 3 independent experiments, total ≥15 cells per condition; **P < 0.01, ***P < 0.001 by one-way ANOVA and Holm–Sidak’s post hoc test for multiple comparisons to that of the GFP control); n.s., not significant. GFP-tagged LRRTM2 was included as an additional control.
Fig. S1.
Identification of the carbonic anhydrase-related proteins CA10 and CA11 as neurexin ligands in a proteomic screen for proteins coimmunoprecipitating with Nrxn1. (A) Outline of the proteomic approach to identify novel neurexin-binding proteins. Adult homozygous mice carrying a knocked-in HA tag in the neurexin-1 gene (Nrxn1HA/HA; mice will be described in detail in a future publication; available from The Jackson Laboratory) were dissected, HA-tagged neurexin-1 was immunoprecipitated, and coimmunoprecipitated proteins were analyzed by mass spectrometry, identifying among others CA10 and CA11. (B) Amino acid sequence of murine CA10 and CA11 and peptide coverage in the mass spectrometry measurements of proteins that coimmunoprecipitated with HA-tagged neurexin-1. Residues of identified peptides are shown in red, nonidentified peptides in black, and those unidentifiable by the Mascot algorithm in gray. Signal peptides as predicted by SignalP 4.1 (www.cbs.dtu.dk/services/SignalP/) are underlined. (C) Developmental expression of murine Ca10 and Ca11 during mouse development as measured by quantitative RT-PCR. Data shown are mean ± SEM (n = 3); levels are normalized to the mean expression of β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as internal controls (SI Materials and Methods). (C, Lower) In situ hybridizations of sagittal brain sections of 56-d-old mice from the Allen Brain Atlas (www.brain-map.org). © 2004 Allen Institute for Brain Science, Allen Mouse Brain Atlas.
We first validated our proteomic findings by coimmunoprecipitations (co-IPs). We used brains from Nrxn1HA mice and an antibody against the HA epitope that was introduced into Nrxn1 in these mice to pull down endogenous, HA-tagged Nrxn1 (Fig. 1B). IP of Nrxn1 caused co-IP of endogenous CA10 [note that as described previously (24), Nrxn1 is primarily expressed as Nrxn1α (>90%) in multiple alternatively spliced variants, with Nrxn1β accounting for a much smaller proportion (<5%)]. In a reciprocal experiment, we performed IPs of CA10 from mouse forebrain and cerebellum, and observed high enrichment of Nrxn1 with CA10 but with an apparently more restricted set of specific Nrxn1α isoforms (Fig. 1C).
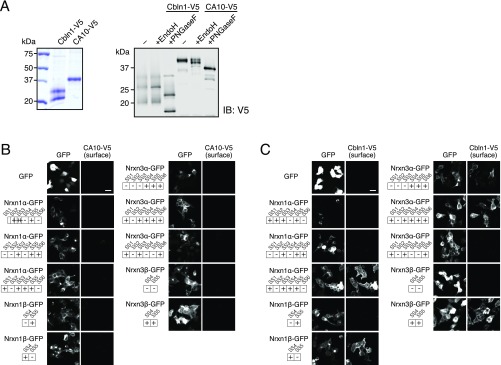
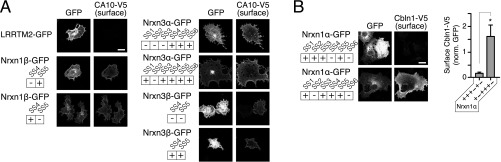
To test the binding of CA10 to neurexins by an independent approach and examine whether neurexins other than Nrxn1α also bind, we purified recombinant CA10 with C-terminal V5 and hexahistidine tags (CA10-V5) from the medium of HEK293 cells. This confirmed that CA10 is a secreted glycoprotein (Fig. S2A). Using purified CA10-V5, we tested whether CA10 binds directly to neurexins by applying recombinant proteins to cells expressing various neurexin isoforms. In contrast to Cbln1, which showed robust surface binding to cells expressing neurexin isoforms that contain the SS4 insert (13), CA10 surprisingly did not exhibit significant binding to neurexins in this experiment (Fig. S2 B and C). However, when we coexpressed CA10 together with a series of splice variants of Nrxn1α, -1β, -3α, and -3β all neurexins were able to tether the normally secreted CA10 to the surface of COS cells, as assessed by surface labeling of the V5 tag (Fig. 1 D and E and Fig. S3A). This interaction was specific to neurexins, as CA10 did not bind to other cell-surface receptors such as LRRTM2. Moreover, coexpression of V5-tagged Cbln1 with neurexin isoforms in COS cells confirmed the specificity of this assay because only SS4-containing isoforms tethered Cbln1 to the cell surface (Fig. S3B). Viewed together, these results suggest that CA10 is a neurexin ligand that binds to both α- and β-neurexins in a cis configuration in the secretory pathway.
Fig. S2.
CA10 is a secreted glycoprotein (A), and cell-surface binding of recombinant CA10 and cerebellin to neurexin-expressing cells (B and C). (A) Coomassie-stained gel of recombinant cerebellin-1 (Cbln1; used as a positive control) and CA10 purified from the media of transfected HEK293 cells. CA10 produced in HEK293 cells is only partially sensitive to EndoH, but quantitatively shifts to a lower size upon treatment with PNGaseF. (B) Representative confocal images of HEK293 cells transfected to express the indicated GFP-tagged neurexin constructs. Cells were incubated with recombinant, V5-tagged CA10 described in A and subsequently surface-labeled for the V5 tag. (Scale bar, 20 µm.) (C) Same as in B, using recombinant V5-tagged Cbln1 (as a positive control). (Scale bar, 20 µm.)
Fig. S3.
Measurements of the sequence dependence of CA10 binding to neurexins: representative images and controls complementing Fig. 1 D and E. (A) Representative confocal images of COS cells that had been cotransfected with expression vectors for V5-tagged CA10 and the indicated GFP-tagged neurexins or GFP-tagged LRRTM2 (used as a control). COS cells were surface-labeled for the V5 tag of CA10, and the GFP and V5 signals were imaged. Data show representative images for those quantifications in Fig. 1E for which no representative images are shown in Fig. 1D. (Scale bar, 20 µm.) (B) Same as A and Fig. 1 but for V5-tagged cerebellin-1 (Cbln1-V5) instead of CA10 to demonstrate the specificity of the assay. Neurexin-1α containing an insert in splice site 4 binds cerebellin-1, whereas neurexin-1α lacking an insert does not. Data are means ± SEM (n = 3 independent experiments, total ≥15 cells per condition; *P < 0.05 by one-way ANOVA. (Scale bar, 20 µm.)
CA10 Localizes to Synapses.
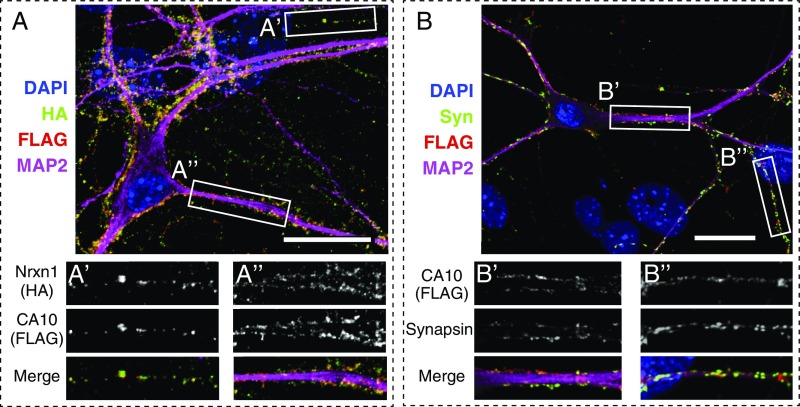
To assess the subcellular localization of CA10, we expressed FLAG-tagged CA10 in cultured cortical neurons via lentiviral transduction, as none of the tested antibodies against endogenous CA10 was suitable for immunocytochemistry. Surface labeling of tagged CA10 showed near-complete colocalization of CA10 with endogenous Nrxn1 in mouse Nrxn1HA neurons (Fig. 2A). Moreover, surface-labeled CA10 colocalized with synaptic puncta labeled by the synaptic marker synapsin (Fig. 2B). Thus, at least some CA10 is a synaptic protein in neurons that colocalizes with Nrxn1.
Fig. 2.
CA10–Nrxn1 complex is synaptic. (A) Confocal image of cortical neurons cultured from Nrxn1HA mice and infected with a lentivirus expressing FLAG-tagged CA10. Cells were surface-labeled using antibodies to the HA epitope for detection of endogenous HA-tagged Nrxn1 (green), and to the FLAG epitope for detection of CA10 (red). Cells were counterstained for the somatodendritic marker MAP2 (magenta) and nuclear DAPI (blue). (Scale bar, 20 µm.) (A′ and A″) Magnifications of areas outlined in A. (B) Confocal image of cultured cortical neurons from wild-type mice infected with a lentivirus expressing FLAG-tagged CA10. Cells were surface-labeled using an antibody against FLAG (red) and subsequently permeabilized for counterstaining for synapsin (green), MAP2 (magenta), and DAPI (blue). (B′ and B″) Magnifications of areas outlined in B. (Scale bar, 20 µm.)
CA10 Forms a Stable Stoichiometric Complex with Nrxn1α.
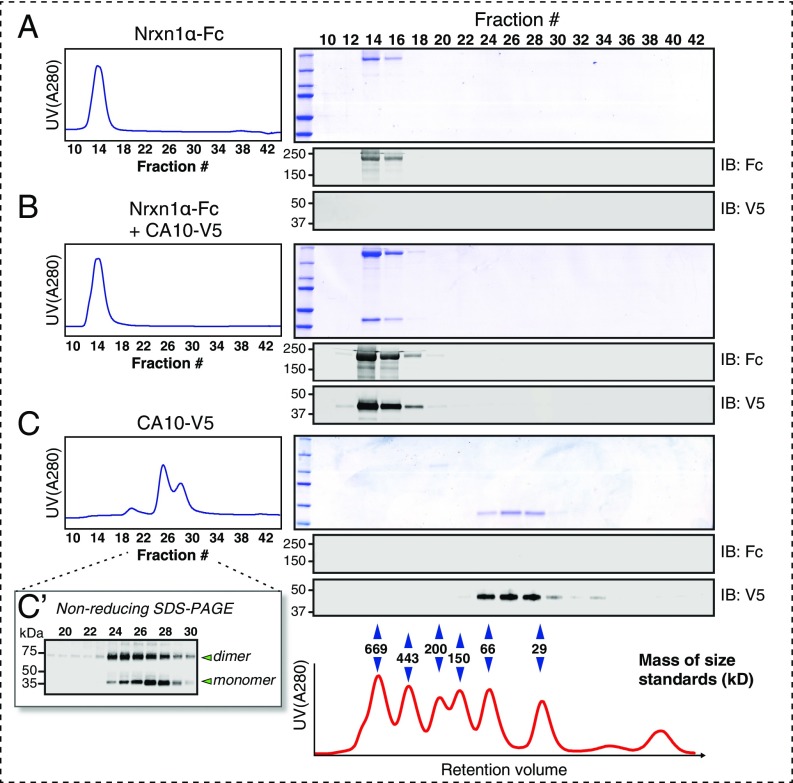
The finding that CA10 bound to neurexins in the secretory pathway but not when added to neurexins displayed on the cell surface raised the question of whether the neurexin–CA10 complex is stable and stoichiometric. To test this question, we constructed vectors encoding the recombinant Nrxn1α ectodomains fused to IgG-Fc domains (Nrxn1α-Fc) or CA10 with C-terminal V5 and hexahistidine tags (CA10-V5). Nrxn1α-Fc and CA10-V5 were purified from the medium of HEK293 cells that had been transfected with these vectors either alone or in combination either by protein A–Sepharose or cobalt metal-affinity chromatography and analyzed by size-exclusion chromatography (Fig. 3). Strikingly, coexpression of Nrxn1α-Fc with CA10 produced a secreted stoichiometric complex that was fully stable during gel filtration and migrated with a molecular weight that differed from that of its components (Fig. 3). His-tagged CA10-V5 purified and separated alone showed a broad, bimodal absorbance peak with apparent sizes that roughly corresponded to monomers and dimers. Indeed, analysis of the CA10-eluting fractions by nonreducing immunoblotting confirmed the presence of both monomeric and dimeric species (Fig. 3C′).
Fig. 3.
Gel-filtration analyses reveal a stable stoichiometric CA10–Nrxn1 complex. The Nrxn1α ectodomain fused to the IgG Fc domain and V5-tagged CA10 was expressed in HEK293 cells alone or together, and purified by protein A–Sepharose chromatography (Nrxn1α-Fc or CA10 together with Nrxn1α-Fc) or by cobalt metal-affinity chromatography (CA10-V5 also contains a hexahistidine sequence). Purified Nrxn1α-Fc (A), Nrxn1α-Fc purified with CA10-V5 (B), or CA10-V5 alone (C) were then subjected to gel filtration (Left, normalized UV-absorbance profiles), and samples were analyzed by reducing SDS/PAGE followed by Coomassie staining or immunoblotting as indicated. Migration of protein standards on the same column is shown (Bottom). Note that the Nrxn1α ectodomain migrates near the void volume of the column because its Fc tag renders it dimeric, explaining the lack of a retention-volume shift upon CA10 binding. (C′) CA10 fractions separated under nonreducing conditions, resolving bands that correspond in size to CA10 monomers and dimers (see also Fig. S4).
Fig. S4.
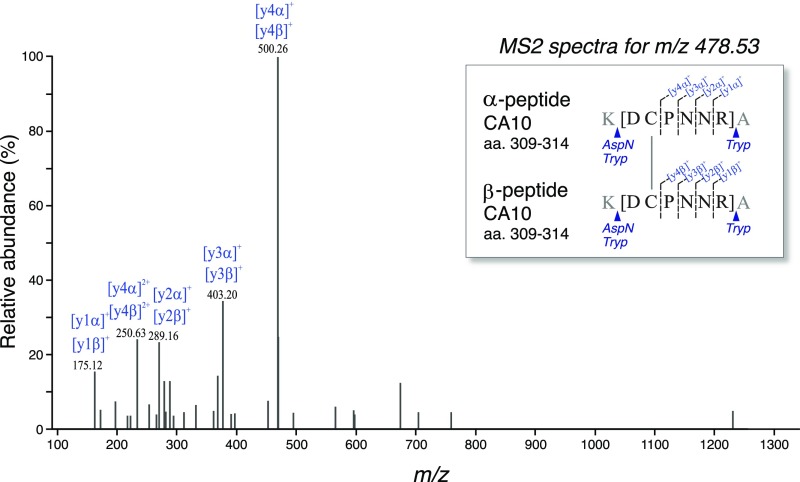
Characterization of the covalent CA10 homodimer. Fragmentation spectrum and sequence (Inset) of disulfide-bridged peptides from CA10 dimers. The spectrum was obtained by digests of recombinant CA10-V5 protein with trypsin and AspN under native conditions (Materials and Methods). The m/z of the fragment was 478.53, z = 3; calculated [M + H]+ 1433.56, observed [M + H]+ 1433.57.
A Juxtamembranous Stalk Sequence of Neurexins Binds to CA10.
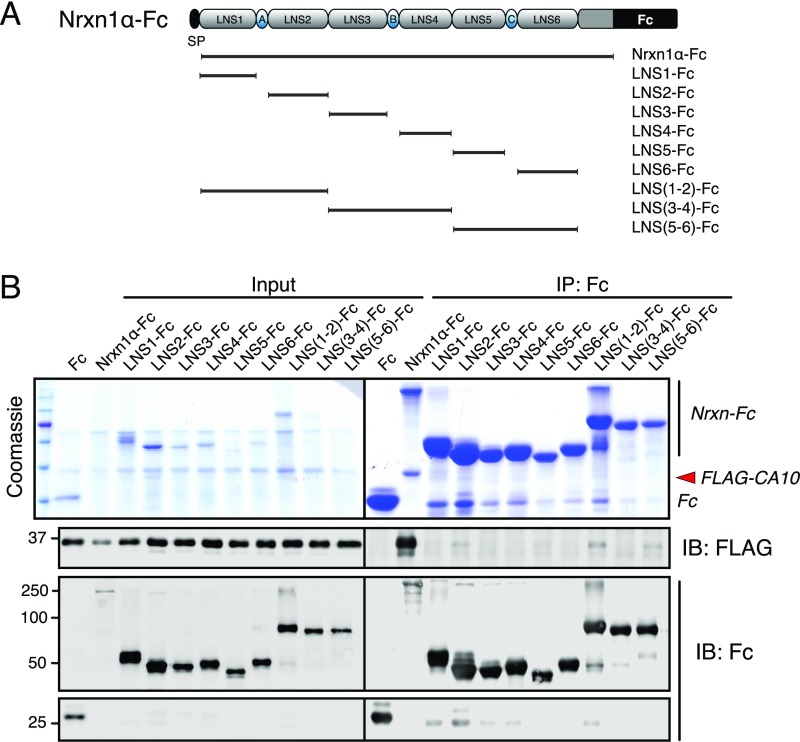
Which neurexin sequences mediate CA10 binding? To address this question, we coexpressed FLAG-tagged CA10 and various soluble Fc-tagged neurexins in HEK293 cells, purified the neurexins from the medium using protein A beads, and probed for copurification of CA10. We first tested truncation mutants of the Nrxn1α ectodomain, divided into either each individual LNS domain or each EGF-repeat domain with its flanking LNS domains (Fig. S5). In contrast to the full-length Nrxn1α ectodomain (Fig. 1), none of the individual LNS or EGF-repeat domains supported CA10 binding. These results, in combination with the binding of CA10 to both α- and β-neurexins in the cell-surface experiment (Fig. 1 D and E and Fig. S3A), suggested that the CA10-binding site of neurexins could be situated in the stalk region between the LNS6 domain and the transmembrane region. This conserved region contains a threonine-rich sequence of ∼40 residues with several putative O-linked glycosylated sites as well as a predicted short cysteine-loop sequence composed of two conserved cysteines flanking an eight-residue acidic sequence (25) (Fig. 1A).
Fig. S5.
LNS domains of Nrxn1α are not involved in CA10 binding. (A) Diagram of recombinant protein constructs. Recombinant proteins were produced as fusion proteins with the Fc domain of IgG. Nrxn1α constructs included either the entire extracellular sequences (Nrxn1α-Fc), or the indicated single or double LNS domains of Nrxn1α. FLAG-tagged CA10 was coexpressed in transfected HEK293 cells with the various Nrxn1α proteins, and CA10 was coimmunoprecipitated from the media using protein A-coated beads. (B) Analysis of proteins in the media from transfected HEK293 cells was obtained as described in A, and of proteins immunoprecipitated from the media using protein A-coated beads. Proteins were examined by SDS/PAGE under reducing conditions, followed by Coomassie blue staining (Top) or immunoblotting with antibodies to Fc or FLAG (Bottom).
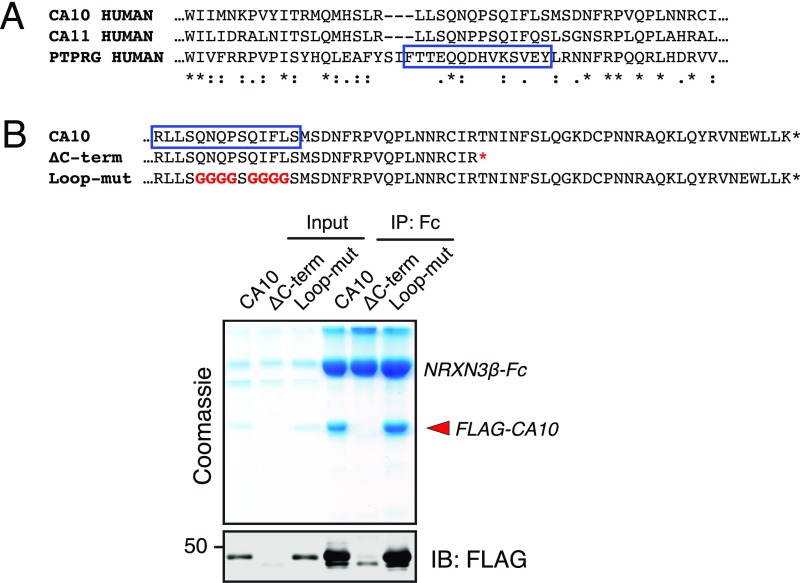
To test binding of the stalk region of neurexins to CA10, we produced Fc-tagged Nrxn1β with mutations in this region and examined their ability to copurify CA10 after coexpression in HEK293 cells (Fig. 4). We found that deletion of either the threonine-rich sequence or the cysteine-loop sequence nearly blocked CA10 binding, whereas replacement of all serine and threonine residues in this region did not appear to alter CA10 binding (Fig. 4A). Further mutations of the stalk region revealed that specific sequence elements are selectively essential for CA10 binding. We found that mutation in the conserved DILV motif of the stalk region in Nrxn1 to either GIGG or DIRR blocked CA10 binding, as did substitution of the two cysteines in the conserved cysteine loop to alanines (Fig. 4B). Moreover, neutralizing the negative charges in the cysteine loop severely impaired binding, whereas mutation of the proline residues flanking the cysteines did not (Fig. 4B). To test whether this interaction is conserved across isoforms and species, we examined a corresponding set of mutants of human NRXN3β and obtained similar results (Fig. 4C). Thus, neurexins capture CA10 via a conserved stalk region whose two components—a threonine-rich sequence and a cysteine loop—are both essential for CA10 binding.
Fig. 4.
Mapping of the CA10–neurexin interaction by coimmunoprecipitation and mass spectrometry. (A and B) Analysis of the Nrxn1 sequence determinants for binding CA10 by copurification of FLAG-tagged CA10 with Fc-tagged neurexins in HEK293 cells, using the Fc tag only as a negative control. Harvested media (input) and Fc-enriched fractions obtained by incubation with protein A-beads (IP: Fc) were analyzed by reducing SDS/PAGE, followed by Coomassie staining and immunoblotting using antibodies to the FLAG and Fc tags as indicated. The sequences of the various mutants of Nrxn1β are shown with mutated residues in red (Top). The ∼70-kDa band seen in input fractions of Coomassie-stained gels represents bovine albumin. (C) Same as A and B, but for human NRXN3β. (D) The Nrxn1β–CA10 complex involves disulfide bonds. Nrxn1β-Fc expressed alone or coexpressed with FLAG-CA10 was purified (IP: Fc) from the media (input) of transfected HEK293 cells, and the indicated fractions were analyzed by SDS/PAGE under reducing (Top) and nonreducing conditions (Bottom). Two-color detection was used for simultaneous IB with antibodies to the FLAG epitope (to detect CA10; green) and to the Fc domain fused to Nrxn1β (red). Under nonreducing conditions, most FLAG-tagged CA10 comigrates with Fc-tagged Nrxn1β (overlap; yellow). (E) CA10 cysteine mutations impair Nrxn1β binding. Alignment of the C-terminal sequences of CA10 and CA11 with cysteines marked in red (Top). Copurification as described in A of FLAG-tagged wild-type, C296A-mutant, and C310A-mutant CA10 with Nrxn1β-Fc, followed by SDS/PAGE and Coomassie staining or IB using antibodies against the FLAG and Fc tags, shows that the conserved CA10 cysteine residue C310 is essential for Nrxn1β binding. (F) Analysis of the covalent CA10–neurexin disulfide bond by mass spectrometry. Fragmentation spectrum with masses of b (red) and y (blue) ion series, and the deduced sequence (Inset) of the disulfide-bridged CA10–neurexin peptide resulting from sequential digests by trypsin and AspN. The m/z was 1117.45, z = 2, and the calculated [M + H]+ in agreement with the observed (2233.90). To avoid interference of possible O-linked glycosylation sites, the S&T>G construct described in A was used that replaces two serine residues with glycines (green). (G) Purification of wild-type but not C310A-mutant V5-tagged CA10 copurifies Fc-tagged Nrxn1β. Serum-free media from cells producing Nrxn1β-Fc or the “DIRR” mutant described in B were mixed with recombinant V5-tagged wild-type or C310A-mutant CA10. Samples were immunoprecipitated for the V5 tag (IP: V5) and analyzed by SDS/PAGE under reducing (Top) and nonreducing conditions (Bottom), followed by IB using antibodies against the V5 and Fc tags. NEM, N-ethyl-maleimide (added before mixing the proteins).
The Neurexin–CA10 Complex Includes a Disulfide Bond.
Comparison of SDS/PAGE and immunoblotting analyses performed on the Nrxn1β-Fc–FLAG-CA10 complex under reducing vs. nonreducing conditions revealed that its constituents migrated as monomers under reducing but as a heterodimer under nonreducing conditions (Fig. 4D). This result suggests that CA10 and neurexins may form intermolecular disulfide bonds.
Mature CA10 contains four cysteines, of which the conserved Cys60 and Cys244 are predicted to form an intramolecular disulfide bridge (26). We thus tested the role of the two other, more C-terminal cysteines, Cys296 and Cys310, in neurexin binding. Cys296 is located near the end of the carbonic-anhydrase domain and is not found in CA11, whereas the Cys310 is located in the hydrophilic C-terminal stretch of CA10 and CA11 and is conserved between the two proteins. We performed the same copurification assay as described above to test the role of these cysteines in Nrxn1β binding (Fig. 4E). Mutation of the conserved Cys310, but not of Cys296, dramatically reduced Nrxn1β binding, suggesting that it is involved in forming a disulfide bond with neurexins.
To further map the neurexin–CA10 disulfide bond, we copurified the complex formed by CA10 with Nrxn1β(S&T>G)-Fc (Fig. 4A) and subjected it to in-solution proteolytic digests followed by LC-MS/MS analysis. We identified a single pair of disulfide-linked peptides that contained a disulfide bond formed between Cys310 of CA10 and the N-terminal of the two cysteines in the neurexin cysteine loop of Nrxn1β-Fc (Fig. 4F). Pull-downs of wild-type and mutant Nrxn1β-Fc with wild-type and C310A-mutant V5-CA10 confirmed the requirement for C310 in CA10 for binding Nrxn1β (Fig. 4G).
Nrxn1γ Represents a Third Principal Nrxn1 Isoform That Specifically Binds to CA10.
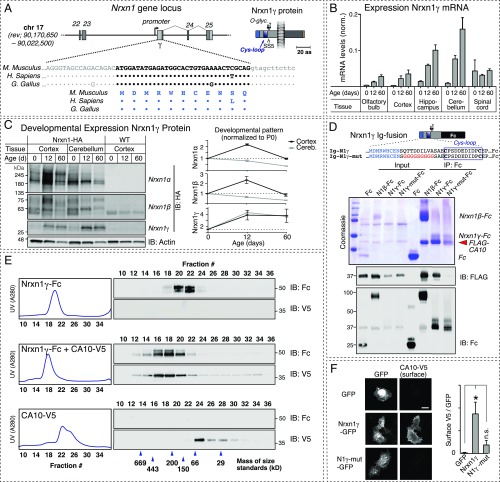
Deep sequencing of RNA from mouse brain recently identified a novel, very short Nrxn1 transcript that lacks all extracellular Nrxn1 sequences except for the stalk region (27). Thus, this isoform—referred to as Nrxn1γ—is predicted to bind to CA10 but not to other known neurexin ligands. Nrxn1γ transcripts originate from an internal promoter in the Nrxn1 gene, and include a unique 5′ sequence that encodes a short Nrxn1γ-specific N-terminal sequence (Fig. 5A). Nrxn1γ-specific genomic sequences are highly conserved in mammals, birds, and reptiles but not in fish (Fig. 5A), and are absent from Nrxn2 or Nrxn3 genes.
Fig. 5.
Identification of Nrxn1γ protein as a Nrxn1 gene-derived CA10 receptor. (A) Organization of the Nrxn1γ-specific promoter and exon in the Nrxn1 gene (Top; exon numbering is according to ref. 9), and sequences of the 5′ end and N terminus of the Nrxn1γ mRNA and protein, respectively, from mouse, human, and chicken (Bottom). Identity of mouse, human, and chicken Nrxn1γ sequences is indicated by dots, with nonconserved residues shown. A schematic of the Nrxn1γ domain organization is shown (Right). (B) Levels of Nrxn1γ transcript assessed by quantitative RT-PCR targeting the Nrxn1γ-specific 5′ sequence in mouse brain regions of the indicated ages. Data are means ± SEM (n = 3), normalized to the mean expression of the internal controls β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by the ΔΔCt method (SI Materials and Methods). (C) Levels of Nrxn1α, Nrxn1β, and Nrxn1γ protein assessed by IB of cortical and cerebellar lysates from Nrxn1HA mice at the indicated ages. Equal amounts of total protein were loaded, and Nrxn1 levels detected with antibodies to HA were quantified using fluorescent secondary antibodies and normalized to the levels at birth. Data are means ± SEM (n = 5 per time point and brain region). See also Fig. S6. (D) Coimmunoprecipitation of FLAG-tagged CA10 with soluble, Fc-tagged Nrxn1γ. FLAG-CA10 was coexpressed with wild-type or mutant Nrxn1γ-Fc fusion proteins (see sequences; Top). Harvested media (input) and Fc-enriched fractions (IP: Fc) were separated under reducing conditions and analyzed by Coomassie staining and IB using antibodies to the FLAG and Fc tags as indicated. Note that Nrxn1γ-Fc and FLAG-CA10 overlap in size on the Coomassie-stained gel. (E) Analysis of the stable stoichiometric Nrxn1γ–CA10 complex by gel filtration. Experiments were carried out as described for Fig. 3. (F) Confocal micrographs of COS cells cotransfected with V5-tagged CA10 plus GFP-tagged Nrxn1γ and surface-labeled with V5 antibodies [(Left) representative images; (Right) summary graph of surface-V5 signal normalized to that of GFP, shown as means ± SEM (n = 3 independent experiments, total ≥15 cells per condition); *P < 0.05 by one-way ANOVA and Holm–Sidak’s post hoc test for comparison with the control condition]. (Scale bar, 20 µm.) Nrxn1γ-mutant sequence is as in D. n.s., not significant.
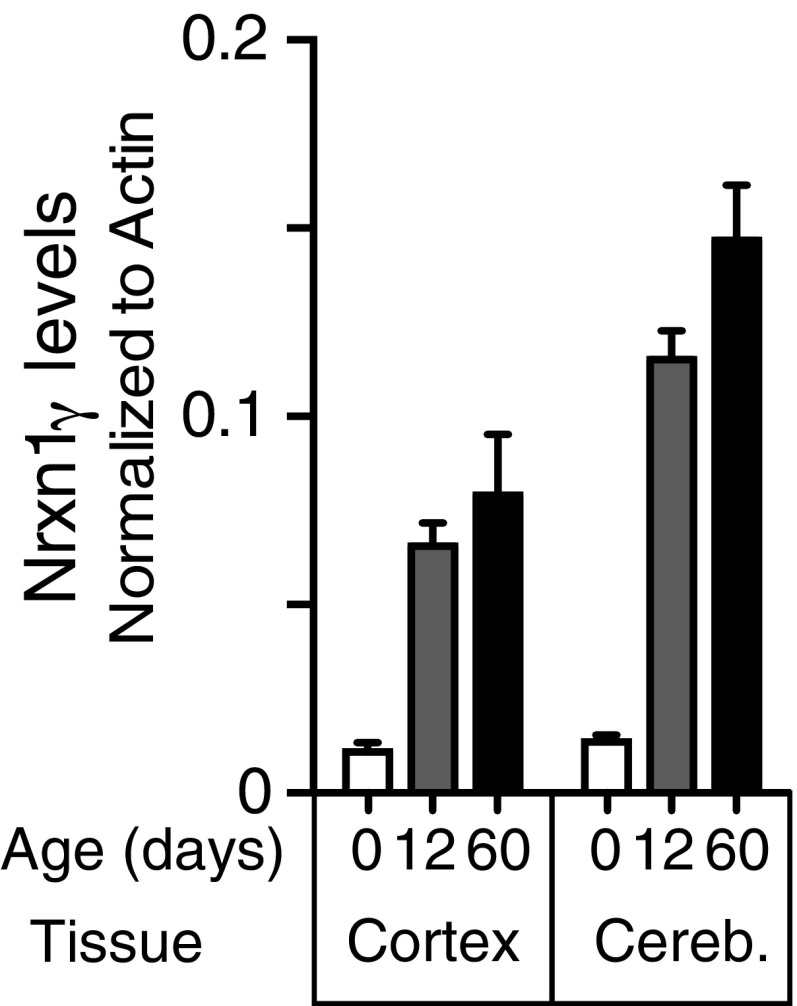
We found that the Nrxn1γ transcript, as assayed by qRT-PCR, is most highly expressed in cerebellum and increases with postnatal age (Fig. 5B). Fortunately, the HA epitope in our Nrxn1HA mice was inserted into the stalk region of Nrxn1, allowing us to probe for the presence of Nrxn1γ protein using these mice. We identified a Nrxn1γ band of the predicted size in brain lysates of cortex and cerebellum; Nrxn1γ protein levels were higher in cerebellum than in cortex, consistent with the qRT-PCR results, and increased postnatally in a pattern that differed from that of Nrxn1α and Nrxn1β (Fig. 5C and Fig. S6).
Fig. S6.
Steep postnatal increase in neurexin-1γ protein levels as determined by quantitative immunoblotting of homogenates from cortex and cerebellum of mice as a function of age. Nrxn1γ levels are shown as means ± SEM (n = 5 per time point and brain region), normalized for actin as an internal control. Data complement those shown in Fig. 4C.
Next, we tested whether CA10 could act as an extracellular ligand for Nrxn1γ. We coexpressed a recombinant Nrxn1γ-Fc fusion protein with FLAG-CA10 in HEK293 cells, and found that Nrxn1γ-Fc efficiently pulled down FLAG-CA10, suggesting that they form a complex (Fig. 5D). Mutation of the DILV stalk sequence in Nrxn1γ blocked the interaction. To confirm the complex, we performed gel-filtration analyses analogous to those shown in Fig. 3 with Nrxn1γ, and again observed a stable stoichiometric complex (Fig. 5E). Finally, we found that coexpression of CA10 and Nrxn1γ, but not of the nonbinding mutant of Nrxn1γ, was sufficient to tether CA10 to the cell surface of transfected COS cells (Fig. 5F). This result demonstrates not only binding but also that the short extracellular domain of Nrxn1γ is able to reach the cell surface despite lack of a predicted signaling peptide. Thus, Nrxn1γ forms an endogenous receptor for CA10.
CA10 Increases the Surface Levels and Changes the Posttranslational Processing of Neurexins.
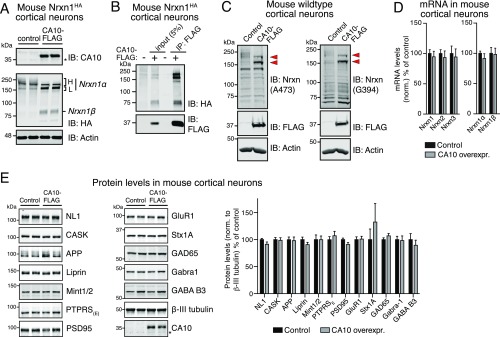
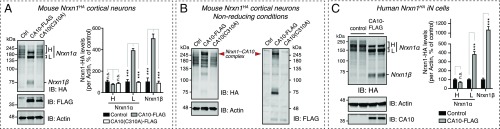
To explore potential functional effects of CA10 on neurexins, we overexpressed CA10 via lentiviral transduction in cultured cortical mouse neurons that normally synthesize only low levels of endogenous CA10 (Fig. S7A). CA10 overexpression dramatically increased the levels and altered the migration of endogenous Nrxn1 in these neurons (Fig. 6A). Specifically, CA10 overexpression at least doubled Nrxn1α and Nrxn1β levels, and transformed the many Nrxn1α bands that are normally observed in neurons —ranging in size from 160 to 260 kDa—to two major bands of ∼160 and ∼190 kDa (Fig. 6A). All of these Nrxn1 isoforms were enriched by IP of CA10 (Fig. S7B), indicating that the increased levels are associated with complex formation. Accordingly, expression of the CA10 Cys310Ala mutant, which does not bind neurexins (Fig. 4D), did not affect endogenous neurexin levels (Fig. 6A). Moreover, overexpressed CA10 comigrated with Nrxn1α in a large complex on SDS/PAGE under nonreducing conditions (Fig. 6B). To exclude the possibility that insertion of the HA tag near the binding site of CA10 (Fig. 1A) affected our results, we repeated these experiments in wild-type neurons using immunoblotting with two different pan-neurexin antisera and obtained the same results (Fig. S7C). Overexpression of CA10 did not change the levels of neurexin transcripts, as measured by qRT-PCR (Fig. S7D). Furthermore, we used human embryonic stem cells that had been genome-edited to carry an HA-tagged NRXN1 allele (28). We differentiated those cells into neuronal cells (iN cells) by a direct lineage-conversion protocol (29) and found that CA10 had a similar effect on neurexin protein levels and isoform distribution in human neurons (Fig. 6C). Importantly, the effect of CA10 was specific to neurexins, as we saw no change in the levels of other pre- and postsynaptic proteins (Fig. S7E), including APP and PTPRS.
Fig. S7.
Effect of CA10 overexpression on neurexin mRNA and protein levels in cultured cortical neurons (A–D) and on the levels of various other neuronal proteins (E). (A) CA10 overexpression increases neurexin-1 levels in cultured cortical neurons from Nrxn1HA mice infected with CA10-expressing or control lentiviruses, as analyzed by immunoblotting to CA10, the neurexin-1 HA tag, or endogenous actin. Note the additional apparent molecular weight shift in neurexin-1 bands. In the CA10 immunoblot, the asterisk marks the band corresponding to endogenous, untagged CA10. (B) Immunoprecipitation of FLAG-tagged CA10 expressed in cortical neurons coimmunoprecipitates endogenous neurexin-1α and -1β as revealed by immunoblotting of the endogenous HA-tagged proteins. (C) Immunoblotting with two independent antibodies to nontagged neurexins in cultured cortical neurons demonstrates that the CA10 overexpression increases the levels of endogenous neurexins and shifts their apparent molecular weight distribution. Cortical neurons cultured from wild-type mice were infected with control or CA10-expressing lentiviruses, and analyzed by immunoblotting with antibodies raised against the C terminus of neurexin-2 (A473) or neurexin-3 (G394). Red arrowheads emphasize the appearance of the ∼160 kDa and ∼190 kDa Nrxn1α bands upon expression of CA10. As a control, neurons were also examined by immunoblotting for the expressed FLAG-tagged CA10 and for actin. (D) CA10 overexpression does not alter neurexin mRNA levels. mRNA levels of the indicated neurexins in mouse cortical neurons infected with control or CA10-expressing lentiviruses as described for C, and assessed by quantitative RT-PCR (means ± SEM; n = 6). (E) CA10 overexpression does not alter expression of other neuronal proteins besides neurexins. Synaptic proteins were analyzed by quantitative immunoblotting in cortical neurons infected with control or CA10-expressing lentiviruses, and analyzed by immunoblotting to the indicated proteins [Left, representative immunoblots; Right, summary graph of protein levels (means ± SEM; n = 5–12), determined using fluorescence-labeled secondary antibodies and LI-COR detection and normalized to β-III tubulin as measured with Tuj-1 antibodies]. Asterisk as in A. All lentiviral infections were performed at DIV4 to 5 and analyzed at DIV14 to 15.
Fig. 6.
Overexpression of CA10 increases Nrxn1 protein levels and alters the migration pattern of Nrxn1α on SDS/PAGE, suggesting a change in posttranslational processing. (A) Nrxn1 levels in cortical neurons cultured from Nrxn1HA mice as a function of CA10 expression. Neurons were infected with control lentiviruses (empty vector) or lentiviruses expressing FLAG-tagged wild-type or C310A-mutant CA10, and analyzed by IB (Left) with antibodies to HA (to detect tagged endogenous Nrxn1), FLAG (to detect CA10), and actin (as a loading control). Nrxn1α isoforms migrating at higher (H) or lower (L) apparent molecular weights and Nrxn1β were quantified (Right) by normalizing to levels of actin (n = 3). (B) Experimental samples described in A were analyzed by SDS/PAGE and immunoblotting under nonreducing conditions. Fluorescent two-color immunoblotting using antibodies to the HA and FLAG epitopes was used to detect tagged Nrxn1 and CA10, respectively, on the same membrane. Actin was used as loading control. (C) Nrxn1 levels in human-induced neurons infected with lentivirus to express control (empty vector) or FLAG-tagged CA10. The iN cells are genome-edited to carry an HA tag in the NRXN1 locus (28). Samples were immunoblotted (Left) using antibodies against HA (to detect tagged NRXN1), CA10 (to detect overexpressed CA10; endogenous levels are low in these cultures), and actin (as loading control). Neurexin levels were quantified (Right) as described in A. Bar graphs show means ± SEM; ***P < 0.001 by two-way ANOVA and Holm–Sidak’s test for multiple comparisons. n.s., not significant.
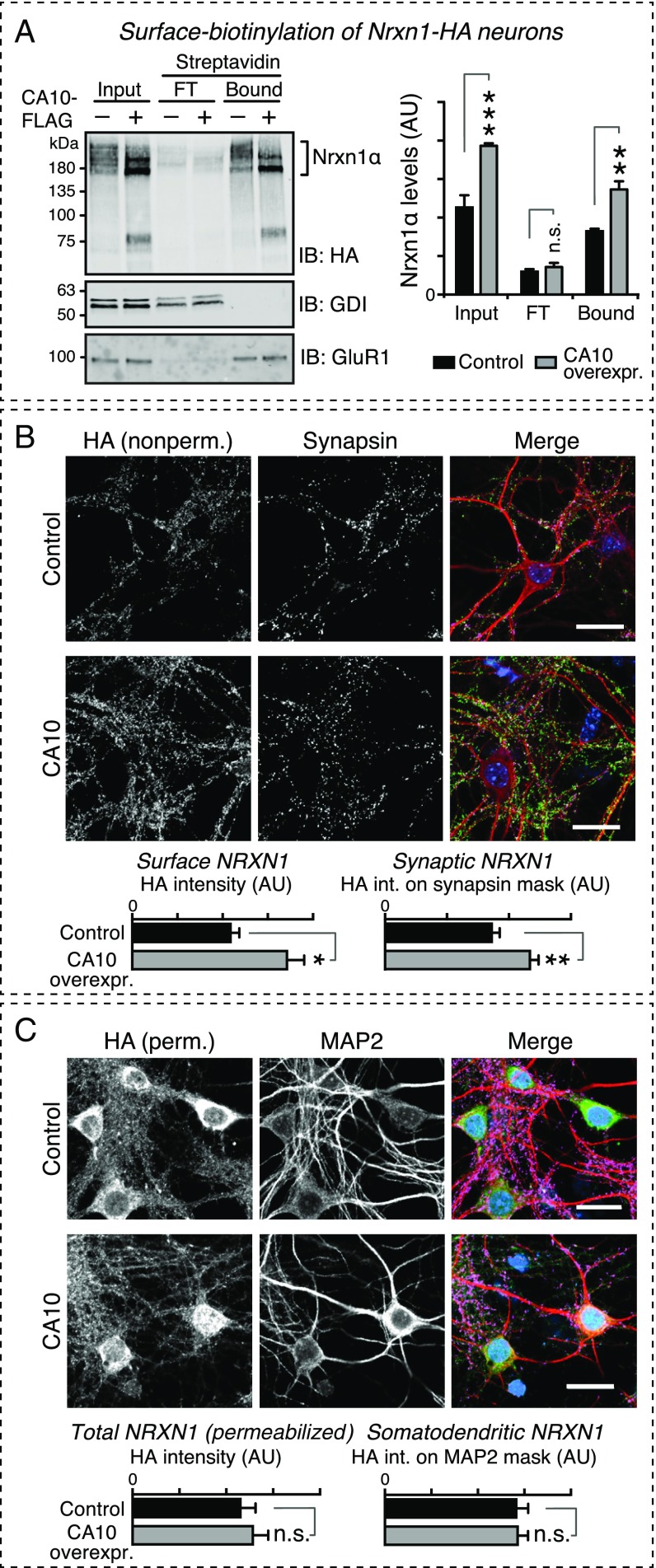
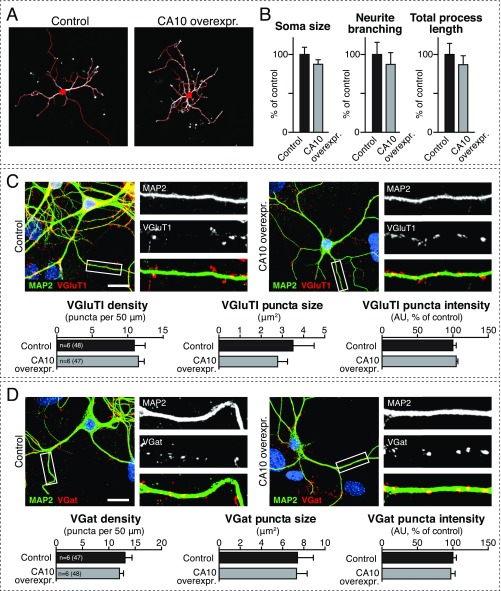
The change in apparent size and increase in abundance of Nrxn1 induced by CA10 overexpression are consistent with the association of Nrxn1 with CA10 in the secretory pathway. Does this association increase the surface levels of Nrxn1? After cell-surface biotinylation of Nrxn1HA mouse neurons with and without CA10 overexpression, streptavidin-mediated pull-down of biotinylated proteins demonstrated increased levels of Nrxn1 in CA10-expressing neurons, whereas the levels of the AMPA-receptor subunit GluR1 did not change (Fig. 7A). Moreover, surface labeling of Nrxn1HA mouse neurons without and with CA10 overexpression with an antibody to the Nrxn1-HA epitope, followed by counterstaining for the synaptic marker synapsin, demonstrated that CA10 expression increases the surface and synaptic levels of Nrxn1 (Fig. 7B). At the same time, the levels of total HA intensity as stained in permeabilized neurons did not change (Fig. 7C). These experiments suggest that CA10 actively promotes surface expression of Nrxn1. However, CA10 overexpression did not alter any morphological parameter of neurons thus far analyzed. Specifically, upon imaging neurons sparsely transfected with GFP, we observed no CA10-induced change in neuronal soma size, neurite length, or neurite branching (Fig. S8 A and B). Moreover, we found no change in excitatory or inhibitory synapse numbers or size, as analyzed by immunocytochemistry for the excitatory and inhibitory markers VGluT1 and VGAT, respectively (Fig. S8 C and D). Thus, CA10 does not cause a major transformation of neuronal shape or differentiation or synapse formation and/or elimination.
Fig. 7.
CA10 overexpression increases surface levels of endogenous Nrxn1 in cortical neurons. (A) Nrxn1 surface levels assessed by surface biotinylation as a function of CA10 expression. Nrxn1HA cortical neurons were infected with control lentiviruses (empty vector) or lentiviruses expressing FLAG-tagged CA10, and surface proteins were biotinylated. Input, unbound flow-through (FT) or streptavidin-bound proteins were analyzed by IB (Left) using antibodies against HA (to detect tagged Nrxn1α), the AMPA-receptor subunit GluR1/GluA1, and the cytosolic protein GDI, the latter two as controls for enrichment of surface and intracellular compartments, respectively. Summary graphs of absolute quantifications (Right) show means ± SEM (n = 3). Statistical comparison by two-way ANOVA with Sidak’s multiple comparison test (**P < 0.01, ***P < 0.001). AU, arbitrary units. (B) Immunocytochemical assessment of Nrxn1 surface levels in Nrxn1HA cortical neurons infected with lentivirus to express CA10-FLAG or control (empty vector). Nonpermeabilized cells were stained with an HA antibody to label endogenous Nrxn1 (green) and subsequently counterstained with antibodies against the synaptic marker synapsin (magenta) and somatodendritic MAP2 (red); nuclear DAPI is in blue. The HA signal intensity and the intensity of signal overlapping with synapsin were quantified. Bar graphs show means ± SEM (n = 6 cultures, with three fields imaged and averaged per culture; *P < 0.05, **P < 0.01 by Student's t test). (C) Immunocytochemical assessment of total neurexin levels. Cells described in B were stained using an HA antibody (to detect endogenous, tagged Nrxn1) under permeabilizing conditions (green). MAP2 and DAPI as in B. Intensity of the total HA signal and that overlapping with the somatodendritic marker MAP2 were quantified. Bar graphs show means ± SEM (n = 6 cultures, with three fields imaged and averaged per culture; n.s., not significant by Student’s t test). Lookup tables of the representative images in B and C have been adjusted for presentation. (Scale bars, 20 μm.)
Fig. S8.
CA10 overexpression does not alter neuronal morphology or synapse numbers. (A) Example images of cortical neurons that overexpress CA10 via lentiviral delivery and were subsequently sparsely transfected with a GFP-encoding plasmid for visualization (GFP shown in white). Soma and neurites from nonoverlapping neurons were automatically outlined (red) and measured using the MetaMorph Neurite Outgrowth module. (B) Measurements of soma size, number of neurite branch points, and total neurite length of neurons described in A. Data presented are means ± SEM (n = 6 independent cultures, total of 64 to 67 cells analyzed). (C) Representative images of cortical neurons infected with control or CA10-expressing lentivirus stained for the excitatory synaptic marker VGluT1 (Slc17a7; red), MAP2 (green), and nuclear DAPI (blue). (Scale bar, 20 µm.) (C, Bottom) Summary graphs of the density, size, and intensity of VGluT1-positive synaptic puncta along secondary dendrites, represented as means ± SEM; total numbers of cells and independent cultures analyzed are listed in the bars. (D) Representative images of cortical neurons described in A stained for the inhibitory synaptic marker VGAT (Slc32a1; red), MAP2 (green), and nuclear DAPI (blue). (D, Bottom) Summary graphs of the density, size, and intensity of VGAT-positive synaptic puncta along secondary dendrites, represented as means ± SEM; total numbers of cells and independent cultures analyzed are listed in the bars. Lookup tables of the representative images in C and D have been adjusted for presentation.
Discussion
The properties of CA10 and CA11, described two decades ago as catalytically inactive extracellular carbonic anhydrase-related proteins (30, 31), suggested they were extracellular binding partners for unknown receptors in brain, but their targets remained unknown. Here we show that CA10—and, by extension, CA11—is an endogenous ligand for neurexins that binds to a conserved juxtamembranous sequence present in all neurexins. This sequence consists of a short threonine-rich, presumably O-glycosylated sequence followed by an acidic cysteine loop comprising eight residues (25). In addition, we report an isoform of Nrxn1, called Nrxn1γ, that is surprisingly small because it lacks all extracellular Nrxn1 sequences except for the short threonine-rich sequence and cysteine loop to which CA10 binds but that includes the regular transmembrane region and cytoplasmic tail of neurexins. Of all extracellular neurexin domains, Nrxn1γ therefore contains only the CA10-binding region, and appears to be specialized for this neurexin ligand and possibly other neurexin ligands binding to the threonine-rich sequence and/or cysteine loop of neurexin. Thus, our results suggest the following five conclusions.
First, CA10 and, by extension, CA11 are stoichiometric neurexin ligands that bind to neurexins in vitro and in vivo (Figs. 1–5). We demonstrated that immunoprecipitations of endogenous proteins coisolate CA10 and CA11 with Nrxn1 (Fig. 1), and that this interaction could be replicated with recombinant proteins (Figs. 3–5). Most importantly, we showed that secreted truncated forms of Nrxn1α or Nrxn1γ formed a stable 1:1 complex with CA10 that could be purified from the medium of the expressing cells on a gel-filtration column, implying a high-affinity interaction (Figs. 3 and 5). In this complex, Nrxn1 and CA10 proteins were covalently linked by a disulfide bond formed by conserved cysteine residues in both proteins (Fig. 4). Although most of our experiments were performed with Nrxn1, the CA10-binding sequence in Nrxn1 that we identified (see below) is highly conserved in all neurexins, suggesting that all neurexins bind.
Second, CA10 localizes to synapses when overexpressed as a tagged protein in neurons. We demonstrated this localization using both endogenous Nrxn1 (Fig. 2A) and synapsin (Fig. 2B) as markers, suggesting that CA10 forms a complex with Nrxn1 at the synapse.
Third, CA10 binds to a juxtamembranous sequence in Nrxn1 for which no previous ligand was known (Fig. 5). This sequence is composed of two elements, a threonine-rich more N-terminal sequence and a short “cysteine loop” in which two cysteine residues flank a conserved acidic eight-residue sequence (25). The threonine-rich N-terminal sequence appears to be essential for CA10 binding, whereas the cysteine loop also contributes (Fig. 4). We hypothesize that the cysteine loop normally forms an intramolecular disulfide bridge in neurons that, owing to the short loop sequence, is strained, and that upon CA10 binding to neurexin the intramolecular disulfide bond is converted into an intermolecular disulfide bond because Nrxn1 is bound to CA10 by a disulfide bond formed by the N-terminal cysteine of the cysteine-loop sequence (Fig. 4F).
Fourth, the Nrxn1 gene expresses from a third promoter, Nrxn1γ, a developmentally and spatially regulated short isoform whose gene product was previously predicted computationally (27) (Fig. 5). The extracellular sequence of Nrxn1γ contains only the juxtamembranous threonine-rich and cysteine-loop sequences of Nrxn1α and Nrxn1β but none of the other canonical extracellular neurexin sequences. Nrxn1γ is evolutionarily conserved in tetrapods, but no equivalent γ-neurexin isoform appears to be expressed from the Nrxn2 or Nrxn3 genes. Strikingly, Nrxn1γ avidly binds to CA10 similar to Nrxn1α (Fig. 5), confirming the CA10-binding site of neurexins and assigning to this short Nrxn1 variant a specific biological activity.
Fifth and finally, overexpression of CA10 in neurons creates a shift in neurexin isoforms in mouse and human neurons and increases surface trafficking of neurexins (Figs. 6 and 7), suggesting that CA10 associates with neurexins in the secretory pathway and serves, at least in part, as a neurexin chaperone during surface transport. Alternatively, it is possible that CA10 attenuates the turnover of surface neurexins by blocking their proteolytic turnover by metalloprotease and γ-secretase (32, 33).
Very little is known at present about CA10 and/or CA11, apart from studies in zebrafish using morpholino- and CRISPR-mediated disruption of the zebrafish orthologs ca10a and ca10b, which resulted in developmental abnormalities and mortality of unknown mechanism (26). However, these results do not reveal the actual functions of CA10 and CA11. Our data provide insights on the molecular role of CA10 and CA11, and expand the panoply of extracellular neurexin ligands. Up to now, all neurexin ligands were interactors of their LNS domains, but now with CA10 we have identified a ligand for their juxtamembranous sequences. Moreover, our data define a Nrxn1 isoform (Nrxn1γ) whose short extracellular region is composed only of the CA10-binding sequence and that also binds tightly to CA10 and presumably its close homolog, CA11.
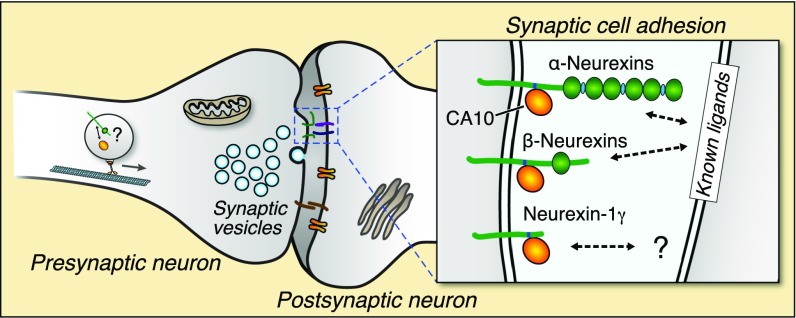
Based on the results of our studies and on analogies to related proteins, at least two overall functions that are not mutually exclusive can be envisioned (Fig. 8). First, CA10 and CA11 may act primarily as neurexin chaperones, as suggested by our findings that CA10 overexpression increases the total levels of neurexin proteins, changes their pattern of posttranslational modifications, and enhances their surface levels. These data suggest that CA10 associates with neurexins in the secretory pathway and may facilitate their folding. Second, CA10 and CA11 may function as adaptors that enable indirect interactions of neurexins with unknown postsynaptic target molecules, thus mediating formation of novel transsynaptic complexes. This hypothesis is suggested not only by the analogous role of cerebellins that enable transsynaptic complexes of presynaptic neurexins with postsynaptic GluRδ2 proteins (13) but also by the similar activities of catalytically inactive carbonic-anhydrase domains in other proteins. Specifically, the extracellular carbonic-anhydrase domains of PTPRG and PTPRZ mediate their interaction with Ig domains 2 and 3 of contactins (19, 20). The crystal structures of the PTPRG–CNTN4 and PTPRZ–CNTN1 complexes revealed a β-hairpin loop that protrudes from the CARP fold and accounts for the majority of the interacting surface (20, 34). It is possible that the carbonic-anhydrase domain of CA10 and CA11 mediates similar interactions, even though the amino acids involved in the PTPRG–contactin and PTPRZ–contactin complexes are not well-conserved to CA10/11 and mutation of the corresponding stretch did not impair binding between CA10 and NRXN3β (Fig. S9). Addressing the overall functions of CA10 and CA11 by defining their molecular interactions and probing their biological roles using specific genetic manipulations will be a major goal of future experiments.
Fig. 8.
Schematic illustration of a synaptic cleft with the various neurexin isoforms in complex with CA10. A synaptic vesicle protein transport vesicle (STV) containing neurexin and CA10 illustrates the possibility that the complex forms in a cis configuration already in the secretory pathway.
Fig. S9.
CA10 residues corresponding to the β-hairpin of PTPRG are not required for binding to Nrxns. (A) Sequence alignment between CA10, CA11, and the CA-like domain of PTPRG, with conservation indicated by ClustalW symbols (below). Marked in blue are residues of the CA-like domain of PTPRG that, based on crystal structure data, form a β-hairpin structure that makes up the major binding interface with CNTN4. (B) Binding of the indicated CA10 mutants to secreted NRXN3β. Residues of CA10 that align to the β-hairpin loop in the CA-like domain of PTPRG are marked in blue, mutated residues shown in red, and stop codons as asterisks (Top). Proteins were analyzed from media of cotransfected HEK293 cells and immunoprecipitated using protein A-coated beads. Proteins were examined by SDS/PAGE under reducing conditions, followed by Coomassie blue staining (Middle) or immunoblotting with antibodies against FLAG (Bottom). Albumin is seen as a prominent ∼70-kDa band in input fractions. Note that the C-terminal truncation mutant does not express.
Materials and Methods
Constructs.
A cDNA clone of human CA10 (GenBank accession no. BC020577.1) from Harvard PlasmID was used to clone expression constructs (human CA10 protein shares 100% amino acid identity with mouse Ca10). Tagged constructs were subcloned using PCR (PrimeSTAR; Takara) and isothermal assembly (Gibson Assembly Mix; New England Biolabs). CA10-V5 consists of full-length CA10 cloned into the pEBMulti vector (Wako Chemicals) with C-terminal addition of V5 and hexahistidine tags separated by GGQ linkers. For expression in neurons, full-length CA10 with a C-terminal FLAG tag was cloned after the human synapsin promoter in a lentiviral vector. FLAG-CA10 was generated by replacing the predicted signal peptide (N-terminal 21 amino acids) with that of IgK followed by a hexahistidine tag, 2× FLAG tags, and a TEV protease site separated by GGQ linkers. Cysteine mutants were generated by site-directed mutagenesis using KOD polymerase (Roche).
Plasmids encoding Fc-tagged ectodomains of mouse Nrxn1α (splice isoform SS1−, SS2− SS3+ SS4−, SS5+, SS6+) or Nrxn1β (SS4−, SS5−) were generated by PCR and isothermal assembly into a pEBMulti vector containing the Fc fragment of human IgG (35). Further mutations and truncations of Nrxn1β—as illustrated in the figures—were generated by isothermal assembly of overlapping PCR fragments or (for cysteine mutants) by site-directed mutagenesis. The Nrxn1β ΔLNS6 construct lacks all but the 21 most C-terminal residues of the LNS domain. Human NRXN3 constructs (SS4−) were based on previously described plasmids (25). Plasmids encoding separate LNS domains of Nrxn1α with C-terminal Fc tags have been previously described (36). PCR products from various neurexin cDNA clones and isothermal assembly into the pEGFP vector (Clontech) were used to generate plasmids encoding full-length Nrxn1 and Nrxn3 isoforms with C-terminal (intracellular) GFP fusion. The sequences of all plasmids were verified by Sanger sequencing.
Recombinant Proteins.
Recombinant V5- and His-tagged CA10 and Cbln1 were produced in FreeStyle 293 cells (Life Technologies). Suspension cultures (100 to 150 mL) were transfected using FreeStyle MAX reagent (Invitrogen), and the medium volume was expanded on the following day to ∼2.5 to 3 times the original volume. Media were harvested 5 to 7 d after transfection and concentrated to 10 to 20% of the harvested volume using Kvick Start cassettes (GE Healthcare). His-tagged proteins were bound in batch to washed TALON cobalt resin (Clontech) overnight at 4 °C. Resins were packed on gravity-flow columns and washed twice with Hanks’ balanced salt solution (HBSS; Gibco) supplemented with 1 mM MgCl2. Bound protein was eluted using HBSS + 500 mM imidazole and subsequently dialyzed against HBSS for at least 16 h with a minimum of three buffer exchanges. For Fig. S2A, enzymatic deglycosylation of recombinant CA10-V5 and Cbln1-V5 with PNGaseF or EndoH (both from New England Biolabs) was performed according to instructions.
Fc-tagged proteins used for size-exclusion chromatography were produced in monolayer HEK293 cells grown in DMEM (Gibco) + 10% (vol/vol) FBS (Atlanta Biologicals). Cells (150 to 450 cm2) were cotransfected using calcium phosphate with plasmids encoding the Fc-tagged protein and either CA10 or control (GFP). Four to 6 h after transfection, the media were replaced by serum-free FreeStyle 293 media (Gibco) that were harvested 3 d after transfection. Fc-tagged proteins and protein complexes were bound in batch to washed protein A resin (Sepharose 4B; Invitrogen) overnight at 4 °C. Beads were packed on gravity-flow columns and washed twice with 5 mL HBSS followed by elution of the protein using 500 to 1,000 μL of 0.1 M glycine (pH 2.5). Eluates were immediately neutralized in Tris⋅HCl (pH 8.0) followed by dialysis into HBSS as described above.
Size-Exclusion Chromatography.
Proteins purified in isolation or in complex were separated over a Superdex 200 10/300 GL column (GE Healthcare) in HBSS (Gibco) with a flow rate of 0.75 mL/min controlled by an NGC Quest 10 Chromatography System (Bio-Rad). Gel-filtration size standards (Sigma; MWGF1000) were used to calibrate the relationship between retention time and apparent size. Fractions were collected every 0.5 mL and analyzed by SDS/PAGE.
Coimmunoprecipitations from Mouse Brain.
Forebrains and cerebella were dissected from two 8-wk-old homozygous Nrxn1HA/HA mice and homogenized in 10× (vol/wt) of IP buffer (100 mM NaCl, 4 mM KCl, 2 mM CaCl2, MgCl2, 20 mM Tris, pH 7.4) with protease inhibitors (Roche cOmplete EDTA-free and 1 mM PMSF) using a Potter–Elvehjem tissue grinder. Triton X-100 was added to a final concentration of 1% and incubated for 1 h, followed by centrifugation at 20,000 × g for 30 min. Half a milliliter of cleared lysate was diluted with 1 mL IP buffer and incubated with 5 µg antibody overnight. Protein A–Sepharose (4B; Invitrogen) washed three times in IP buffer was added to each sample and incubated for 1 h. Beads were washed four times with IP buffer containing 0.1% Triton X-100 and eluted with 50 µL Laemmli sample buffer containing 200 mM DTT. All steps were carried out on ice or at 4 °C.
The following antibodies were used for IPs: HA (mouse clone 16B12; Covance), myc (mouse clone 9E10; DSHB), and CA10 (mouse clone 862319; R&D Systems).
Copurification and Binding Assays.
Monolayer HEK293 cells were cotransfected using calcium phosphate with constructs encoding FLAG-tagged CA10 and various Fc-tagged neurexin constructs. Four to 10 h after transfection, the cells were washed once with HBSS and the media were replaced with serum-free FreeStyle 293 media (Gibco). Two days later, the media were harvested and kept on ice. The media were cleared by centrifugation (1,500 × g, 10 min), and Hepes (pH 7.5) was added to a final concentration of 10 mM. For 3 mL of media, 0.1 mL was saved as the input fraction and the remaining volume was incubated with protein A beads (Invitrogen; protein A–Sepharose 4B; 40 μL of a 1:1 beads:HBSS slurry) under rotation at 4 °C overnight. The beads were collected by centrifugation at 1,000 × g for 1 min at 4 °C and washed four times with 0.5 mL HBSS, followed by elution in 40 μL Laemmli buffer [2% (wt/vol) SDS, 60 mM Tris, pH 6.8, 10 mM EDTA, 10% (vol/vol) glycerol, 0.1 M DTT, 0.1 μg/mL bromophenol blue] at 65 °C for 10 min.
For incubations with separately expressed proteins (Fig. 4G), conditioned serum-free media from HEK293 cells expressing Fc or Nrxn-Fc were filtered (0.2 μm), and Tris (pH 7.4) was added to a final concentration of 10 mM. To 600-μL aliquots, roughly equimolar amounts (final concentration 30 nM) of CA10-V5 or C310A-mutant CA10-V5 was added and the samples were incubated overnight at 4 °C under orbital rotation. After collecting 50-μL input fractions, 30 μL of a 1:1 slurry of washed (in HBSS) anti-V5 beads (Sigma) was added. After a 1-h incubation under orbital rotation at 4 °C, the beads were washed three times with 0.5 mL HBSS. Seventy percent of the bead volume was eluted with nonreducing Laemmli buffer, and the remaining 30% was collected for in-solution digests for mass spectrometry.
Mass Spectrometric Analyses of Disulfides.
Native CA10-V5–Nrxn1β complexes captured on V5-beads or recombinant CA10-V5 in solution were subjected to in-solution digestion with trypsin (1:100; Promega) in 25 mM NH4CO3 buffer (pH 8.0) for 12 h at 37 °C. PMSF (1 mM) was used to terminate the trypsin activity, followed by digestion with AspN (1:50; Promega) for 12 h at 37 °C. Removal of salts and buffer was performed on in-house stage tips with C18 resin (C18 Empore; 3M), and the peptides were dissolved in 0.2% formic acid. LC-MS analysis was performed as described (37). Cross-linked products were identified with the StavroX (v. 3.5.1) software tool (38) using the following settings: (i) mass tolerance of the precursor ion of 2 ppm, (ii) tolerance for fragment ions of 20 ppm, (iii) cleavage C-terminal of Arg and Lys and N-terminal of Asp with up to two missed cleavages allowed, (iv) oxidation of Met as a variable modification with one modification allowed, and (v) loss of two hydrogens between cross-linked cysteines. Candidate fragmentation spectra were further analyzed manually. In a separate experiment, the proteins were denatured by boiling for 10 min in nonreducing Laemmli buffer before separation by SDS/PAGE and in-gel digestion. In this experiment, all possible combinations of CA10–CA10 and CA10–neurexin disulfide bonds could be observed by LC-MS/MS. Although this likely represents disulfide reshuffling due to sample handling (39), it demonstrated that all possible dipeptide combinations could be detected.
Immunoblotting and Antibodies.
Cells were lysed directly in Laemmli sample buffer [1×; 10% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.1% bromophenol blue, 10 mM EDTA, 60 mM Tris, pH 6.8], unless otherwise stated containing 100 mM DTT. Samples were boiled for 5 min, separated by SDS/PAGE on 4 to 20% TGX gradient gels (Bio-Rad), and transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked and incubated in Tris-buffered saline with 0.1% Tween with 5% (wt/vol) nonfat milk. Primary antibodies were incubated overnight at 4 °C. Near-infrared secondary antibodies (LI-COR) were used for detection. Membranes were scanned using an Odyssey CLx System (LI-COR). Quantification of band densities were performed using LI-COR Image Studio software.
The following cultured antibodies were used for immunoblotting: HA (mouse clone 16B12; Covance; 1:2,000), V5 (mouse clone R960-25; Invitrogen; 1:5,000), actin (mouse clone AC-74; Sigma; 1:5,000), liprin (rabbit antisera 4396; 1:5,000), neuroligin-1 (mouse clone 4C12; Synaptic Systems; 1:1,000), CASK (mouse; BD Biosciences; 1:1,000), APP (mouse clone 22C11; Millipore; 1:1,000), Mint1 (rabbit antisera U1693; 1:1,000), GluR1 (rabbit polyclonal AB1504; Millipore; 1:1,000), GAD65 (mouse clone GAD-6; DSHB; 1:500), PSD-95 (mouse clone 73-028; NeuroMab; 1:5,000), PTPRS (rabbit antisera PAC9986; 1:2,000), CA10 (rabbit polyclonal HPA054825; Sigma-Aldrich; 1:800), syntaxin-1A (rabbit antisera 438B; 1:5,000), β-tubulin III (mouse clone TUJ1; Covance; 1:2,000), Gabra-1 (mouse clone N95/35; NeuroMab; 1:500), and GABA B3 (mouse clone N87/25; NeuroMab; 1:500).
Surface Biotinylation.
Primary cortical neurons grown in 12-well plates were washed twice with ice-cold Dulbecco’s PBS supplemented with 1 mM MgCl2 and 0.5 mM CaCl2 (PBS/MC) and incubated with Sulfo-NHS-Biotin (Thermo Fisher; EZ-Link) at 1 µg/mL in PBS/MC on ice for 12 min under gentle agitation. The cells were subsequently washed three times with ice-cold 50 mM glycine and 0.5% BSA in PBS/MC and twice with PBS/MC, followed by lysis for 15 min on ice in RIPA buffer (120 µL per well) containing protease inhibitors (Roche cOmplete EDTA-free and 1 mM PMSF). Lysates were cleared by centrifugation at 20,000 × g for 15 min, diluted 1:1 in RIPA, and, following removal of the input fraction, incubated with magnetic streptavidin beads (Pierce) under orbital rotation for 3 h at 4 °C. Unbound proteins were collected and the beads were washed once in RIPA buffer, after which bound proteins were eluted in Laemmli sample buffer. Unbound and bound fractions were normalized to the same relative amount of starting material.
Immunocytochemistry.
For surface labeling of neurons, cells were washed twice in equilibrated PBS containing 1 mM MgCl2, 0.5 mM CaCl2, and 4% sucrose (PBS/MCS) and incubated with cultured antibody in PBS/MCS at 37 °C for 20 min followed by three washes in PBS/MCS. Cells were subsequently fixed in 4% (wt/vol) paraformaldehyde (PFA) with 4% (wt/vol) sucrose for 10 min at room temperature (RT), rinsed three times in PBS, and blocked using 5% (vol/vol) goat serum with 1% BSA and 0.01% sodium azide for 30 min at RT. Alexa 488- or 546-conjugated secondary antibodies (Life Technologies; 1:1,000) were applied for 1 h at RT and washed (3 × 10 min in PBS) before permeabilization. Subsequently, cells were permeabilized in blocking buffer containing 0.2% Triton X and incubated with cultured antibodies for intracellular epitopes overnight at 4 °C. Following three 10-min washes, Alexa-conjugated secondary antibodies were applied in 1:600 to 1:1,000 dilution for 1 h at RT, followed by three 10-min washes and mounting of coverslips in Fluoromount (SouthernBiotech) containing DAPI. Surface labeling of COS cells was performed in HBSS with 10 mM Hepes, 2 mM CaCl2, 1 mM MgCl2, and 2% BSA for 30 min at 37 °C followed by two 3-min washes and fixation in 4% PFA for 10 min, and then stained as above. The following cultured antibodies were used for immunocytochemistry: HA (mouse clone 16B12; affinity-purified; Covance; 1:500), V5 (mouse clone R960-25; Invitrogen; 1:5,000), FLAG (rabbit polyclonal F7425; Sigma-Aldrich; 1:500), synapsin (rabbit antisera E028; 1:500), VGluT1 (rabbit polyclonal 135002; Synaptic Systems; 1:500), VGAT (rabbit polyclonal 131003; Synaptic Systems; 1:1,000), and MAP2 (chicken polyclonal; Aves Labs; 1:5,000).
Confocal Microscopy and Image Analysis.
Images were acquired using an A1Rsi confocal microscope (Nikon) equipped with a 63×/1.4 N.A. objective. Sequential scanning was used to acquire 12-bit images with the detector gain adjusted for each piece of experiment to maximize dynamic range and avoid pixel saturation. Pixel dimensions were set according to Nyqvist criteria. Maximum z-stack projections were analyzed using High Content Image Analysis software (Nikon). Images used for quantifications were subtracted from the background signal measured in empty image regions. The General Analysis module (Nikon) was used to generate binary masks overlaying MAP2 or synaptic markers. The binary masks were defined by a threshold intensity (kept constant throughout each experiment) manually set to best define the structure. For analysis of total and surface HA labeling, the HA-derived signal was quantified in either entire images or restricted to the binary masks. For analysis of synaptic puncta, rectangular regions of interest (ROIs) (one to three per cell) along nonoverlapping secondary dendrites were defined manually with the experimenter blinded to the synaptic staining. Next, the General Analysis module (Nikon) was used to automatically generate a binary mask for the synaptic marker. A signal threshold corresponding to the lowest intensity that (based on a pilot experiment) reliably defined synaptic puncta was used. Within each ROI, the objects (synaptic puncta) defined by the binary mask were automatically counted and their sizes and signal intensities were measured. The lengths of the analyzed dendritic segments (i.e., the lengths of the ROIs) were measured and used to normalize synaptic counts. To determine soma size, neurite outgrowth, and neurite branching (Fig. S8 A and B), neurons sparsely transfected with GFP were imaged using a 10×/0.45 N.A. objective and an A1Rsi confocal microscope (Nikon). Soma intensities were allowed to saturate to allow visualization of thinner neurites. Images were converted to 8-bit grayscale images, imported into MetaMorph software (Molecular Devices), and analyzed using the Neurite Outgrowth application module. The algorithm automatically identifies soma, based on intensity (>40,000 gray-levels over background), size (maximum width 37 μm), and protruding neurites (>3,000 gray-levels over background) and calculates soma size, number of neurite branch points, and the total neurite length for each neuron. Correctness of fit was judged manually. All morphological experiments were performed with the experimenter blinded to the experimental condition.
SI Materials and Methods.
For quantitative RT-PCR, primary neuronal culture, transfection of cultured neurons, lentivirus production and transduction, and iN culture, please SI Materials and Methods. All experiments involving the use of live mice were performed with approval of the Stanford Institutional Animal Care and Use Committee.
SI Materials and Methods
Quantitative RT-PCR.
Total RNA was prepared from dissected mouse brain regions or cultured neurons using the PrepEase RNA Spin Kit (Affymetrix). RNA was normalized to 50 ng (cells) or 8 ng (tissues) per reaction. Reverse transcription and amplification were carried out using VeriQuest Probe One-Step qRT-PCR Master Mix (Affymetrix) in a 7900HT Real-Time PCR System (Applied Biosystems). Primers and FAM-labeled, ZEN/IBFQ-quenched hydrolysis probes for Ca10 (primer 1, primer 2, probe: 5′-GCCCTCCTGAAATATTGACCA-3′, 5′-TTCTGACACCTCTTCGCATC-3′, 5′-TCCCACTGACCTTCCTGCCC-3′), Ca11 (5′-TGCAGGTAGGAAGGAGACA-3′, 5′-CGTGGATGTGGAGCTGAAG-3′, 5′-CGGCCAGTGTTGTACAAGGTTCC-3′), Nrxn1 (5′-CGATGTCATCTGTCCCAACA-3′, 5′-GCCATCGGATTTAGCACTGTC-3′, 5′-TGGAGCTGCACATACACCAAGGAA-3′), Nrxn2 (5′-TGATATTTGCCGTCGCTCAC-3′, 5′-GGTGCGAGTGGACAGTG-3′, 5′-TCAATTGTAATGTCGTCCGTGCCCA-3′), Nrxn3 (5′-CACTGATAATGAACGCCTCCA-3′, 5′-CCTTTGTCCTTTCCTCCGATG-3′, 5′-CCTTTTTCCTGCAGCCACTCCTCT-3′), Nrxn1α (5′-TTCAAGTCCACAGATGCCAG-3′, 5′-CAACACAAATCACTGCGGG-3′, 5′-TGCCAAAACTGGTCCATGCCAAAG-3′), Nrxn1β (5′-CCTGTCTGCTCGTGTACTG-3′, 5′-TTGCAATCTACAGGTCACCAG-3′, 5′-AGATATATGTTGTCCCAGCGTGTCCG-3′), and Nrxn1γ (5′-GCCAGACAGACATGGATATGAG-3′, 5′-GTCAATGTCCTCATCGTCACT-3′, 5′-ACAGATGACATCCTTGTGGCCTCG-3′) were obtained from IDT. Efficacy of the qPCR assays was tested by serial dilutions and found to be 0.96 to 1.00.
Data were normalized to the expression of GAPDH (Applied Biosystems; 4352932) using the ΔΔCt method. For developmental expression profiles, both GAPDH and ACTB (Applied Biosystems; 4352933) were used for normalization of the data, to reduce the contribution of changes in housekeeping gene expression during development. The Ct values of ACTB and GAPDH for each sample were averaged and subsequently used for normalization by the ΔΔCt method.
Primary Neuronal Culture.
Mouse cultures were prepared from postnatal day (P)0 to P0.5 pups of CD1 or Nrxn1HA/HA breeding pairs. Cortices were freed from meninges, olfactory bulbs, and hippocampi in cold HBSS (Sigma-Aldrich) containing 1 mM Hepes and enzymatically digested [2% papain suspension (Worthington), 2 U/mL DNaseI, 0.5 mM EDTA in HBSS] for 20 min at 37 °C. Digested tissue was washed with warm plating medium [MEM (Gibco) supplemented with 10% FBS (HyClone), 2 mM l-glutamine, 0.5% glucose, 0.2 mg/mL NaHCO3, 0.1 mg/mL transferrin (Calbiochem), 0.25 μg/mL insulin (Sigma)] and gently triturated using a 1-mL micropipette. The dissociated cells were strained through a 70-μm filter and plated on Matrigel (BD)-coated glass coverslips or plastic wells. The media were replaced the following morning by neuronal growth media [MEM with 5% FBS, 2% B27 supplement (Gibco), 0.5 mM l-glutamine, 0.5% glucose, 0.2 mg/mL NaHCO3, 0.1 mg/mL transferrin, 0.25 μg/mL insulin]. Thirty-six hours after plating, 50% of the medium volume was replaced and supplemented with arabinose C (Sigma-Aldrich; 2 mM final concentration).
Transfection of Cultured Neurons.
To outline neurites, neuronal cultures transduced with lentivirus on day in vitro (DIV)4 were transfected using calcium phosphate with a GFP-expressing plasmid (pEGFP-N1; Clontech) on DIV6. Conditioned media were collected from cultures and, following two washes with equilibrated DMEM (Gibco), replaced with DMEM with 10 mM MgCl2. Per coverslip, 30 μL DNA crystals (1 μg of DNA in 16.7 μL 250 mM CaCl2 mixed with an equal volume of 2× HBSS; see below) was added dropwise. After 40 min, crystals were washed with equilibrated HBSS (2 × 5 min). After a brief wash with neuronal growth media, cultures were left in the saved conditioned media.
Lentivirus Production and Transduction.
Recombinant lentiviral particles were produced in HEK293 cells cotransfected with lentiviral vectors encoding CA10-FLAG or control (empty vector) with helper plasmids pRSV-REV, pMDLg/gRRE, and pVSVG. Per 75 cm2 of cells, 0.5 mL 250 mM CaCl2 containing molar equivalents of DNA (12 μg of CA10-FLAG or control, 3.9 μg pRSV-REV, 8.1 μg pMDLg/gRRE, and 6.0 μg pVSVG) was added dropwise to an equal volume of 2× HBSS (0.4 M NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 0.2% glucose, 38.4 mM Hepes, pH 7.05) under vigorous mixing and immediately added dropwise to the cells. Approximately 6 h after transfection, cells were washed with equilibrated HBSS and media were replaced by neuronal growth media. Viral supernatant was harvested 48 h after transfection, cleared by a centrifugation (1,500 × g, 10 min), aliquoted, and snap-frozen. Neuronal cultures were infected with lentivirus on DIV4 or 5 by adding 50 or 100 μL of viral supernatant per well of a 24- or 12-well plate, respectively. Neurons were analyzed on DIV14 to 16.
iN Culture.
Human ES cells genome-edited to carry an HA insertion in the NRXN1 locus (cTr allele) (28) were differentiated into iN cells as described (29). Lentivirus to express the Cre recombinase [to convert the truncated allele to the NRXN1HA allele (28)] was added with lentivirus expressing either control (mock) or CA10 on DIV6. Cells were analyzed on DIV23.
Acknowledgments
We thank Iryna Huryeva for technical support, Anna J. Khalaj and Erica M. Seigneur for sharing developmental mouse RNA, Chang-Hui Pak for help with iN cultures, and Markus Missler for sharing plasmids encoding the Fc-tagged Nrxn1α ectodomain truncations. F.H.S. is funded by the Swedish Research Council (Grant 350-2012-6543) and the Sweden-America Foundation. This study was supported by grants from the NIMH (MH052804 and MH104172; to T.C.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621321114/-/DCSupplemental.
References
- 1.Reissner C, Runkel F, Missler M. Neurexins. Genome Biol. 2013;14(9):213. doi: 10.1186/gb-2013-14-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol. 2012;22(3):412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423(6943):939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 4.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: Synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257(5066):50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 6.Ushkaryov YA, Südhof TC. Neurexin III alpha: Extensive alternative splicing generates membrane-bound and soluble forms. Proc Natl Acad Sci USA. 1993;90(14):6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ushkaryov YA, et al. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;269(16):11987–11992. [PubMed] [Google Scholar]
- 8.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14(3):497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 9.Treutlein B, Gokce O, Quake SR, Südhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA. 2014;111(13):E1291–E1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiner D, et al. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84(2):386–398. doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Ichtchenko K, et al. Neuroligin 1: A splice site-specific ligand for beta-neurexins. Cell. 1995;81(3):435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 12.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64(6):791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141(6):1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154(1):75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sjöblom B, Elleby B, Wallgren K, Jonsson BH, Lindskog S. Two point mutations convert a catalytically inactive carbonic anhydrase-related protein (CARP) to an active enzyme. FEBS Lett. 1996;398(2–3):322–325. doi: 10.1016/s0014-5793(96)01263-x. [DOI] [PubMed] [Google Scholar]
- 16.Nishimori I, et al. Restoring catalytic activity to the human carbonic anhydrase (CA) related proteins VIII, X and XI affords isoforms with high catalytic efficiency and susceptibility to anion inhibition. Bioorg Med Chem Lett. 2013;23(1):256–260. doi: 10.1016/j.bmcl.2012.10.103. [DOI] [PubMed] [Google Scholar]
- 17.Aspatwar A, Tolvanen ME, Parkkila S. Phylogeny and expression of carbonic anhydrase-related proteins. BMC Mol Biol. 2010;11:25. doi: 10.1186/1471-2199-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnea G, et al. Identification of a carbonic anhydrase-like domain in the extracellular region of RPTP gamma defines a new subfamily of receptor tyrosine phosphatases. Mol Cell Biol. 1993;13(3):1497–1506. doi: 10.1128/mcb.13.3.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peles E, et al. The carbonic anhydrase domain of receptor tyrosine phosphatase beta is a functional ligand for the axonal cell recognition molecule contactin. Cell. 1995;82(2):251–260. doi: 10.1016/0092-8674(95)90312-7. [DOI] [PubMed] [Google Scholar]
- 20.Bouyain S, Watkins DJ. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc Natl Acad Sci USA. 2010;107(6):2443–2448. doi: 10.1073/pnas.0911235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, et al. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics. 2005;171(3):1239–1246. doi: 10.1534/genetics.105.044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Türkmen S, et al. CA8 mutations cause a novel syndrome characterized by ataxia and mild mental retardation with predisposition to quadrupedal gait. PLoS Genet. 2009;5(5):e1000487. doi: 10.1371/journal.pgen.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaya N, et al. Phenotypical spectrum of cerebellar ataxia associated with a novel mutation in the CA8 gene, encoding carbonic anhydrase (CA) VIII. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(7):826–834. doi: 10.1002/ajmg.b.31227. [DOI] [PubMed] [Google Scholar]
- 24.Anderson GR, et al. β-Neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell. 2015;162(3):593–606. doi: 10.1016/j.cell.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gokce O, Südhof TC. Membrane-tethered monomeric neurexin LNS-domain triggers synapse formation. J Neurosci. 2013;33(36):14617–14628. doi: 10.1523/JNEUROSCI.1232-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aspatwar A, et al. Inactivation of ca10a and ca10b genes leads to abnormal embryonic development and alters movement pattern in zebrafish. PLoS One. 2015;10(7):e0134263. doi: 10.1371/journal.pone.0134263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Q, et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci USA. 2015;112(11):3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pak C, et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell. 2015;17(3):316–328. doi: 10.1016/j.stem.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78(5):785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewett-Emmett D, Tashian RE. Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Mol Phylogenet Evol. 1996;5(1):50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 31.Lovejoy DA, et al. Evolutionarily conserved, “acatalytic” carbonic anhydrase-related protein XI contains a sequence motif present in the neuropeptide sauvagine: The human CA-RP XI gene (CA11) is embedded between the secretor gene cluster and the DBP gene at 19q13.3. Genomics. 1998;54(3):484–493. doi: 10.1006/geno.1998.5585. [DOI] [PubMed] [Google Scholar]
- 32.Bot N, Schweizer C, Ben Halima S, Fraering PC. Processing of the synaptic cell adhesion molecule neurexin-3beta by Alzheimer disease alpha- and gamma-secretases. J Biol Chem. 2011;286(4):2762–2773. doi: 10.1074/jbc.M110.142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saura CA, Servián-Morilla E, Scholl FG. Presenilin/γ-secretase regulates neurexin processing at synapses. PLoS One. 2011;6(4):e19430. doi: 10.1371/journal.pone.0019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamprianou S, Chatzopoulou E, Thomas J-L, Bouyain S, Harroch S. A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells. Proc Natl Acad Sci USA. 2011;108(42):17498–17503. doi: 10.1073/pnas.1108774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SJ, Uemura T, Yoshida T, Mishina M. GluRδ2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J Neurosci. 2012;32(13):4688–4701. doi: 10.1523/JNEUROSCI.5584-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reissner C, et al. Dystroglycan binding to α-neurexin competes with neurexophilin-1 and neuroligin in the brain. J Biol Chem. 2014;289(40):27585–27603. doi: 10.1074/jbc.M114.595413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Recktenwald CV, Hansson GC. The reduction-insensitive bonds of the MUC2 mucin are isopeptide bonds. J Biol Chem. 2016;291(26):13580–13590. doi: 10.1074/jbc.M116.726406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Götze M, et al. StavroX—A software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23(1):76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 39.Rombouts I, et al. Formation and reshuffling of disulfide bonds in bovine serum albumin demonstrated using tandem mass spectrometry with collision-induced and electron-transfer dissociation. Sci Rep. 2015;5:12210. doi: 10.1038/srep12210. [DOI] [PMC free article] [PubMed] [Google Scholar]