Significance
The DNA damage response (DDR) is a well-orchestrated and tightly regulated process. The DDR pathway does not act in isolation; indeed, evidence of cross-talk between the DDR and numerous signaling pathways affecting cytoskeletal integrity, nutrient sensing, and autophagy has been demonstrated. In this paper, we report that the DDR induces a distinct pathway of autophagy: genotoxin-induced targeted autophagy (GTA). GTA requires the action of the checkpoint kinases Mec1/ATR, Tel1/ATM, and Rad53/CHEK2. Rad53 mediates the transcriptional up-regulation of autophagy genes via negative regulation of the repressor Rph1/KDM4. GTA requires components of the selective autophagy machinery and is distinct from canonical autophagy pathways. A genome-wide screen for GTA modulators identifies genes required for genotoxin-induced autophagy and starvation-induced autophagy.
Keywords: DNA damage, autophagy, ATM kinase, ATR kinase, budding yeast
Abstract
Autophagy plays a central role in the DNA damage response (DDR) by controlling the levels of various DNA repair and checkpoint proteins; however, how the DDR communicates with the autophagy pathway remains unknown. Using budding yeast, we demonstrate that global genotoxic damage or even a single unrepaired double-strand break (DSB) initiates a previously undescribed and selective pathway of autophagy that we term genotoxin-induced targeted autophagy (GTA). GTA requires the action primarily of Mec1/ATR and Rad53/CHEK2 checkpoint kinases, in part via transcriptional up-regulation of central autophagy proteins. GTA is distinct from starvation-induced autophagy. GTA requires Atg11, a central component of the selective autophagy machinery, but is different from previously described autophagy pathways. By screening a collection of ∼6,000 yeast mutants, we identified genes that control GTA but do not significantly affect rapamycin-induced autophagy. Overall, our findings establish a pathway of autophagy specific to the DNA damage response.
Exposure of budding yeast cells to DNA damage initiates a signaling cascade that is mediated by a pair of central PI3-kinase–like kinases (PIKKs): Mec1/ATR and Tel1/ATM (1).The activation of these kinases results in the phosphorylation and activation of various downstream effector proteins: notably Rad53/CHEK2, Rad9/TP53BP1, and Chk1/CHEK1 (2, 3). Rad53 and Chk1 are kinases that phosphorylate additional effector proteins, which serve to enforce cell cycle arrest or modify enzymes required for the repair of damaged DNA. Cell cycle arrest after DNA damage occurs in large part by stabilizing the securin Pds1/PTTG1/Securin, the inhibitor and chaperone of the Esp1/ESPL1 cohesin protease, separase. Phosphorylation of Pds1 prevents its ubiquitination by the anaphase-promoting complex (APC) and its degradation by the proteasome (4).
The best described targets of Mec1/ATR and Tel1/ATM are proteins that are directly involved in the processes of cell cycle progression and DNA repair (5); however, work from several laboratories has demonstrated that these kinases target many other proteins. For example, in human cells, Mec1/ATR-Tel1/ATM signaling results in at least 900 unique phosphorylation events on 700 proteins (6). A high proportion of these targets are enriched for the DNA damage response (DDR); however, a significantly large fraction are enriched for other diverse cellular processes that include protein metabolism, cytoskeletal functions, and developmental processes (6). Of note, the transcription factor TP53/p53, which is itself activated by ATM after DNA damage, is required for the transcriptional up-regulation of genes involved in the cytoplasmic process of autophagy (7). Similarly, in DNA-damaged budding yeast, Mec1 and Tel1 have been implicated in the phosphorylation of hundreds of targets, again, both those directly implicated in cell cycle regulation and DNA repair as well as others in diverse cellular functions (8, 9).
Autophagy is a catabolic process that occurs in response to a variety of cellular stresses and aids cellular survival in adverse conditions. For example, in response to nutrient starvation, cells recycle internal reserves of basic metabolites by the degradation of proteins, or even whole organelles, by their digestion in the vacuole/lysosome. This process is initiated by the formation of double-membraned cytosolic sequestering vesicles, termed phagophores, which can engulf either random portions of the cytoplasm, organelles, or specific proteins (10). Phagophores mature into autophagosomes and, along with their respective cargo, are targeted to the vacuole wherein their contents are degraded by resident vacuolar hydrolases, and the resulting macromolecules are released back into the cytosol for reuse (10).
In budding yeast, autophagy pathways can generally be categorized into selective or nonselective (hereafter referred to as macroautophagy) pathways. This distinction is based on the cargo captured within the autophagosomes. Engulfment of random portions of bulk cytosol after nutrient limitation is referred to as macroautophagy whereas targeted encapsulation of specific proteins or organelles is termed selective autophagy (11). Macroautophagy in yeast is principally controlled by the nutrient-sensing kinase target of rapamycin complex 1 (TORC1). Under normal growing conditions, when nutrients are plentiful, TORC1 inhibits autophagy by maintaining a key autophagy-regulating protein, Atg13, in a hyperphosphorylated state (12). The interaction of Atg13, and a stable ternary complex consisting of Atg17–Atg31–Atg29, with Atg1 is required for the induction of autophagy as it promotes the kinase activity of Atg1 (12, 13). When nutrients are scarce, TORC1 activity is down-regulated, resulting in the accumulation of hypophosphorylated Atg13 and the formation of the Atg13–Atg1–Atg17–Atg31–Atg29 complex.
In contrast to macroautophagy, selective autophagy can occur even in nutrient-replete conditions where TORC1 activity is high. This form of autophagy results in the degradation of specific proteins or organelles by the autophagic machinery and is largely dependent on the central scaffold-like protein Atg11 (14). Selectivity in this process is achieved by the binding of a receptor protein to its cognate target (e.g., Atg32, the receptor for mitophagy). These receptor–cargo complexes then bind to Atg11 (11, 15), which links it to core autophagy machinery. The interaction between Atg11 and receptors stimulates Atg1’s kinase activity, thus promoting phagophore formation to encapsulate the selected cargo (16).
Our laboratory and others have demonstrated a role for autophagy in the DDR. Specifically, in budding yeast, hyperactive autophagy after DNA damage causes the vacuolar localization and degradation of Pds1 by autophagy, resulting in the inability of cells to resume cell cycle progression due to the nuclear exclusion of Esp1 (17). Furthermore, subunits of the ribonucleotide reductase enzyme are also targets of DNA damaged-induced autophagy (18). Together, these results led us to hypothesize that the DNA damage checkpoint induces autophagy to control the levels of mitosis-promoting proteins and DNA repair enzymes and thus restrain cell cycle progression (17, 19).
Recently, it was shown that mammalian CHEK1 itself is a target of chaperone-mediated autophagy, a selective, autophagy-like pathway that exists in higher eukaryotes (20). In metazoans, DNA damage caused by genotoxin agents [such as ionizing radiation, etoposide, doxorubicin, cisplatin, and reactive oxygen species (ROS)] induces autophagy; however, the signaling mechanisms connecting the DDR to autophagy are unclear (7, 21–23). Autophagy induction after DNA damage seems to require TP53 (7, 22), but whether this role is a truly DNA damage-specific role is not known because TP53’s control over autophagy is complex—it may act either to stimulate or repress autophagy depending on its subcellular localization or cellular context (24). Moreover, a definitive role for the core components of the DDR signaling pathway in the induction of autophagy have not been investigated. Finally, it is unclear what components of the autophagic machinery are required for DNA damage-induced autophagy and whether this phenomenon represents a pathway of autophagy distinct from canonical autophagy pathways. In this paper, we answer these questions by demonstrating that DNA damage induces a pathway of autophagy in budding yeast that we term genotoxin-induced targeted autophagy (GTA), which is distinct from previously described autophagy pathways. GTA requires the core components of the DDR machinery (notably Mec1/ATR, Tel1/ATM, and Rad53/CHEK2), but these proteins do not have a significant role in starvation-induced autophagy, indicating that the signals mediating GTA are distinct from those involved in starvation-induced autophagy. Analysis of known ATG genes revealed that GTA requires components of the selective autophagy machinery and requires a distinct subset of autophagy genes. Using screening approaches, we identified genes that are specifically involved in GTA and not in starvation-induced autophagy pathways.
Results
DNA Damage Induces Autophagy in a Mec1-Rad53–Dependent Manner.
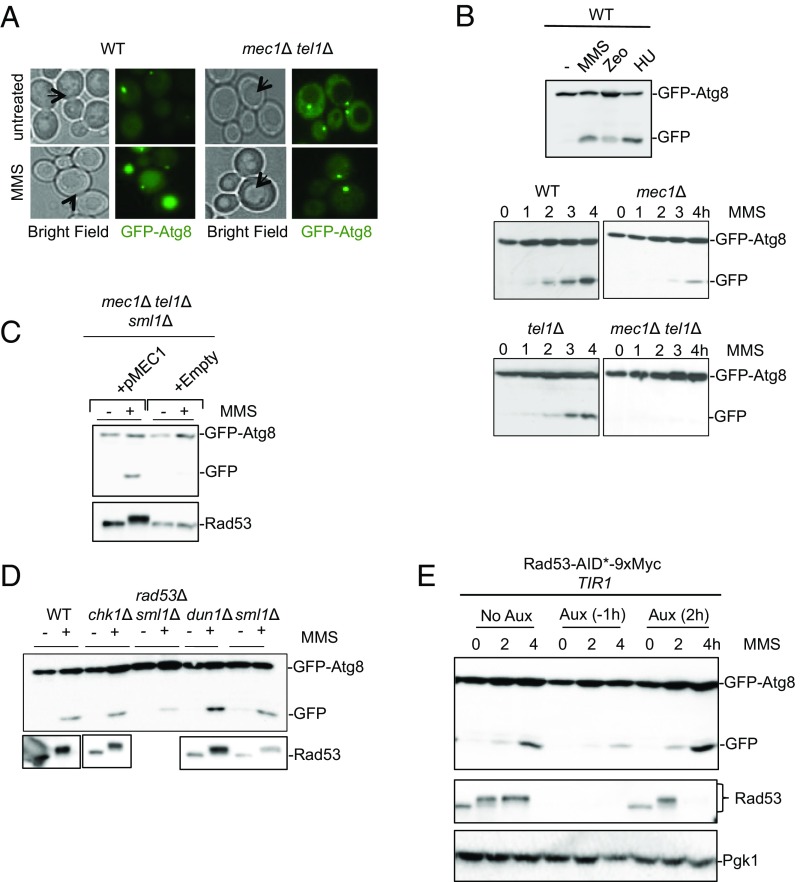
To monitor autophagy after DNA damage, we used the well-established GFP-Atg8 processing assay (25, 26). Autophagosomes are formed near the vacuole, at the phagophore assembly site (PAS). Atg8 is a ubiquitin-like protein critical for the induction of autophagy and the formation of the autophagosome at the PAS; its delivery into the vacuole is an indication of autophagy. When Atg8 tagged with GFP at its N terminus enters the vacuole, it is degraded, but, because the GFP moiety is relatively resistant to degradation by vacuolar proteases, it is possible to monitor autophagy by the appearance of a free GFP intermediate by immunoblot (14, 27) as well as by its vacuolar localization. We transformed our WT strain with a plasmid expressing GFP-Atg8 and induced DNA damage by the addition of the alkylating agent methyl methanesulfonate (MMS). As seen in Fig. 1A, MMS treatment resulted in an increase in the proportion of cells displaying both punctate (PAS) and vacuolar localization of GFP-Atg8 (100% of cells after 4 h of exposure displayed vacuolar GFP-Atg8 localization) as well as the appearance of a free GFP-band, indicative of autophagy (Fig. 1B). To determine whether this autophagic induction was a consequence of DNA damage, we treated cells lacking both MEC1 and TEL1, which are required for the detection of DNA damage, with MMS. As seen in Fig. 1A, GFP-Atg8 did not localize to the vacuole in mec1Δ tel1Δ cells, indicating a complete block in autophagy.
Fig. 1.
DNA damage induces autophagy in a Mec1-, Tel1-, and Rad53-dependent manner. (A) WT or mec1Δ tel1Δ cells carrying GFP-Atg8 were left untreated or treated with 0.04% MMS for 4 h and then visualized using fluorescence microscopy. Representative cells undergoing autophagy display a vacuolar signal of GFP-Atg8 (arrows denote position of vacuoles). (B, Top) WT cells were left untreated (−) or treated with 0.04% MMS, 300 mg/mL zeocin (Zeo), or 0.2 M hydroxyurea (HU) for 4 h, after which cells were collected, lysates were extracted, and immunoblots were performed using an α-GFP antibody. (Middle and Bottom) Cells of the indicated genotypes were treated with MMS as in A and were collected at various time points indicated to monitor autophagy by GFP-Atg8 processing. Lethality of the mec1Δ mutant is suppressed by deletion of SML1. (C) The mec1Δ tel1Δ sml1Δ cells were transformed with either empty vector or pMEC1 and examined for autophagy as in B and for Rad53 hyperphosphorylation, indicating DNA damage checkpoint activation. (D) Cells were treated with MMS as in B and monitored for GFP-Atg8 cleavage. The lethality of rad53Δ is suppressed by deletion of SML1. (E) A strain carrying the Rad53-AID (auxin inducible degron) with the GFP-Atg8 plasmid was (i) left untreated (No Aux), (ii) pretreated with auxin 1 h before addition of MMS [Aux (−1h)], or (iii) treated with auxin 2 h after the addition of MMS [(Aux 2h)]. Autophagy was determined by GFP immunoblot, and Rad53 levels were monitored by Rad53 immunoblot.
To verify that the effect of MMS reflected its creation of DNA damage, we treated cells with two other DNA-damaging agents: zeocin, which creates primarily DNA double-strand breaks (DSBs), or hydroxyurea, which inhibits DNA replication (28). As seen in Fig. 1B, both agents were able to induce autophagy. Additionally, we examined autophagy in strains that suffered site-specific DNA damage. A single irreparable DNA DSB caused by expressing HO endonuclease (29) is sufficient to induce autophagy (SI Appendix, Fig. S1A) albeit with slower kinetics and not to the same extent as seen with genotoxic agents. Importantly, an isogenic strain in which the HO endonuclease cleavage site had been mutated and was grown in identical culture conditions did not induce autophagy, indicating that the autophagy induction in this system was a specific response to the formation of DSBs (SI Appendix, Fig. S1A). We hypothesized that the more robust response to agents such as MMS could be due to the fact that genotoxins created more DNA damage than a single HO break. Consistent with this hypothesis, autophagy is induced to a higher extent in strains suffering eight HO-induced irreparable DSBs (SI Appendix, Fig. S1A), suggesting that autophagy induction is proportional to the amount of DNA damage.
To investigate which components of the DDR signaling pathway are required, we examined immunoblots of extracts from cells expressing GFP-Atg8. WT and mutant cells were harvested at 1-h intervals for 4 h after inducing DNA damage by MMS. An increase of autophagic flux, as judged by the relative amount of the free GFP band to total signal, occurred by 2 h and steadily increased until at least 4 h in the WT strain (Fig. 1B, Lower panels). The absence of MEC1 slowed down the initial appearance of the GFP cleavage product and reduced the level of autophagy (Fig. 1B). In contrast, the deletion of TEL1 had only a minor effect on autophagy induction after DNA damage; however, the absence of both MEC1 and TEL1 completely eliminated the appearance of the free GFP band (Fig. 1B), consistent with the blocked vacuolar localization of GFP-Atg8 (Fig. 1A). To verify that these effects were primarily due to MEC1, we complemented the mec1Δ tel1Δ strain with a centromeric plasmid containing MEC1 under its endogenous promoter, which restored autophagy in response to DNA damage (Fig. 1C). To test whether Mec1 and Tel1 are also implicated in macroautophagy, we used the GFP-Atg8 processing assay and treated cells with rapamycin. Rapamycin was able to induce autophagy in mec1Δ cells to the same extent as WT and to about 50% of WT in mec1Δ tel1Δ cells (SI Appendix, Fig. S1B) showing that the DNA damage checkpoint kinases are largely dispensable for macroautophagy. Together, these results demonstrate that DNA damage induces autophagy in a pathway we call genotoxin-induced targeted autophagy (GTA) that is largely Mec1-dependent, with a smaller, redundant role played by Tel1.
We next directed our focus to the downstream effectors of Mec1-Tel1 signaling, notably, Chk1 and Rad53. We found that deletion of RAD9 or CHK1 did not significantly affect GTA (Fig. 1D and SI Appendix, Fig. S1C). However, deletion of RAD53 severely reduced GTA but had little effect on rapamycin-induced macroautophagy (Fig. 1D and SI Appendix, Fig. S1B). These results suggest that Mec1 signals through Rad53 to mediate autophagy induction after DNA damage. Surprisingly, analysis of the rad53Δ chk1Δ double mutant revealed that GTA was restored to WT levels (SI Appendix, Fig. S1D), suggesting that Chk1 may negatively regulate GTA. One checkpoint response carried out by Rad53 is the activation of the serine/threonine kinase Dun1, which promotes the transcriptional up-regulation of a subset of damage-responsive genes (30); however, deletion of DUN1 had no effect on GTA (Fig. 1D).
We next became interested in whether continuous checkpoint activity is required for GTA. To address this question, we fused an auxin-inducible degron (AID) (31) onto Rad53 in a strain expressing GFP-Atg8. A 1-h treatment with auxin was sufficient to markedly reduce Rad53 abundance (Fig. 1E). When auxin was added 1 h before DNA damage was induced, GTA was reduced, similar to the effect observed in rad53Δ cells (Fig. 1E). However, GTA was not affected when Rad53 was degraded 2 h after the initiation of DNA damage and when autophagy was measured 2 h later (Fig. 1E). These results suggest that continuous checkpoint activity is not required for the maintenance of GTA.
GTA Leads to Rad53-Mediated Transcriptional Up-Regulation of Autophagy Genes via Negative Regulation of Rph1.
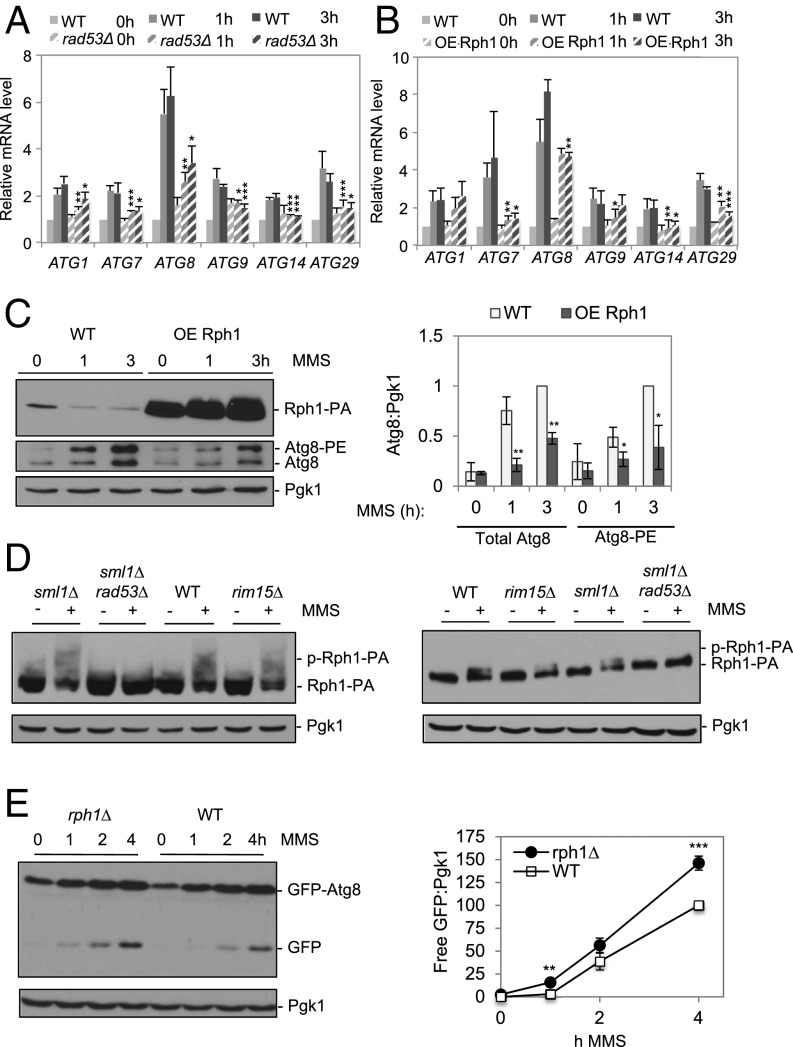
An important aspect of the control of autophagy occurs at the transcriptional level, as a number of ATG genes are transcriptionally up-regulated during autophagy (32). ATG gene transcription controls both the magnitude and the kinetics of the autophagy response (33). Indeed, fusion of single autophagy genes, such as ATG8 or ATG9, to promoters of varying strengths reveals a correlation between ATG gene expression and autophagy activity after nitrogen starvation (34, 35). The transcription of ATG genes is repressed in nutrient-rich conditions through the action of various transcriptional repressors, notably the Rpd3–Sin3–Ume6 histone deaceyltase (HDAC) complex and Rph1/KDM4 (33, 36). We monitored ATG gene transcription by quantitative (q)RT-PCR in cells suffering DNA damage. A representative set of ATG genes (ATG1, ATG7, ATG8, ATG9, ATG14, and ATG29) that play various roles in the autophagy process were selected for analysis. We observed a two- to eightfold increase in all genes tested after DNA damage, the largest increase being in ATG8 transcription (Fig. 2A). This transcriptional up-regulation was significantly reduced in the absence of RAD53 (Fig. 2A), suggesting that transcriptional up-regulation of ATG genes after DNA damage is mediated by the Rad53 checkpoint kinase.
Fig. 2.
DNA damage leads to transcriptional up-regulation of autophagy-related genes in a Rad53-dependent manner. (A) Transcript abundance of autophagy genes in sml1Δ and rad53Δ sml1Δ mutants were assayed 0, 1, and 3 h after MMS treatment. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test. (B) Overexpression (OE) of Rph1 prevents transcriptional up-regulation of autophagy genes. (C) Overexpression of Rph1 inhibits Atg8 induction and lipidation after MMS-induced DNA damage. Quantification of the blot is presented to the Right. *P < 0.05, **P < 0.01. (D) Rph1 is phosphorylated by Rad53 after MMS. Strains of the indicated genotype were treated with 0.04% MMS and analyzed for Rph1 phosphorylation by Western blot. (Left) A Phos-tag gel. (Right) SDS/PAGE gel without phos-tag. (E) The rph1Δ cells have higher autophagy after DNA damage. Quantification of the blot is presented to the Right. **P < 0.05, ***P < 0.01. Error bars reflect SD from 3 independent experiments.
Rph1, the only known demethylase targeting histone H3-K36 trimethylation in yeast, is involved in DDR. Rph1 negatively regulates the transcription of the photolyase gene PHR1 that mediates resistance to UV-induced DNA damage (37). Rph1 is also phosphorylated in a Rad53-dependent manner after DNA damage, which causes its dissociation from chromatin, thereby relieving repression of target genes (38). Given that Rph1 acts as an autophagy repressor (36) and is also linked to the DDR, we investigated whether Rph1 plays any role in ATG gene transcription after DNA damage. Consistent with its role as a negative regulator, overexpression of Rph1 after DNA damage prevented the up-regulation of ATG1, ATG7, ATG8, and ATG14, but not ATG1; these results largely phenocopied the effect seen in rad53∆ cells (Fig. 2B). Furthermore, we observed that Rph1 overexpression also limited Atg8 protein up-regulation and lipidation as well as GTA (Fig. 2C and SI Appendix, Fig. S2A). We confirmed that Rph1 is phosphorylated after DNA damage in a Rad53-dependent manner (Fig. 2D) and that deletion of RPH1 resulted in the induction of autophagy with faster kinetics and to a higher extent after DNA damage (Fig. 2E).
If Rad53 is acting to stimulate GTA solely via inactivation of Rph1, then deletion of RPH1 in rad53Δ cells would restore GTA. The rad53Δ rph1Δ double mutants partially restored autophagy, but only to about ∼20% of WT (SI Appendix, Fig. S2B). These results suggest that Rad53 acts in both an Rph1-dependent and -independent fashion to induce GTA. Taken together, our results demonstrate that transcriptional up-regulation of ATG genes after DNA damage is facilitated by Rad53-mediated inactivation of Rph1, which is required for the maximum induction of GTA. A further indication that the GTA response is distinct from the canonical nutrient starvation-mediated control comes from examining the requirement for the Rim15 kinase, which is required under nitrogen starvation conditions for the phosphorylation of Rph1, thus allowing for the full induction of starvation-induced autophagy (36). We found that, under DNA damage conditions, Rim15 is not required for Rph1 phosphorylation (Fig. 2D), further supporting the idea that GTA-induced autophagy is distinct from starvation-induced degradation.
Genotoxin-Induced Autophagy Requires a Distinct Subset of Autophagy Genes.
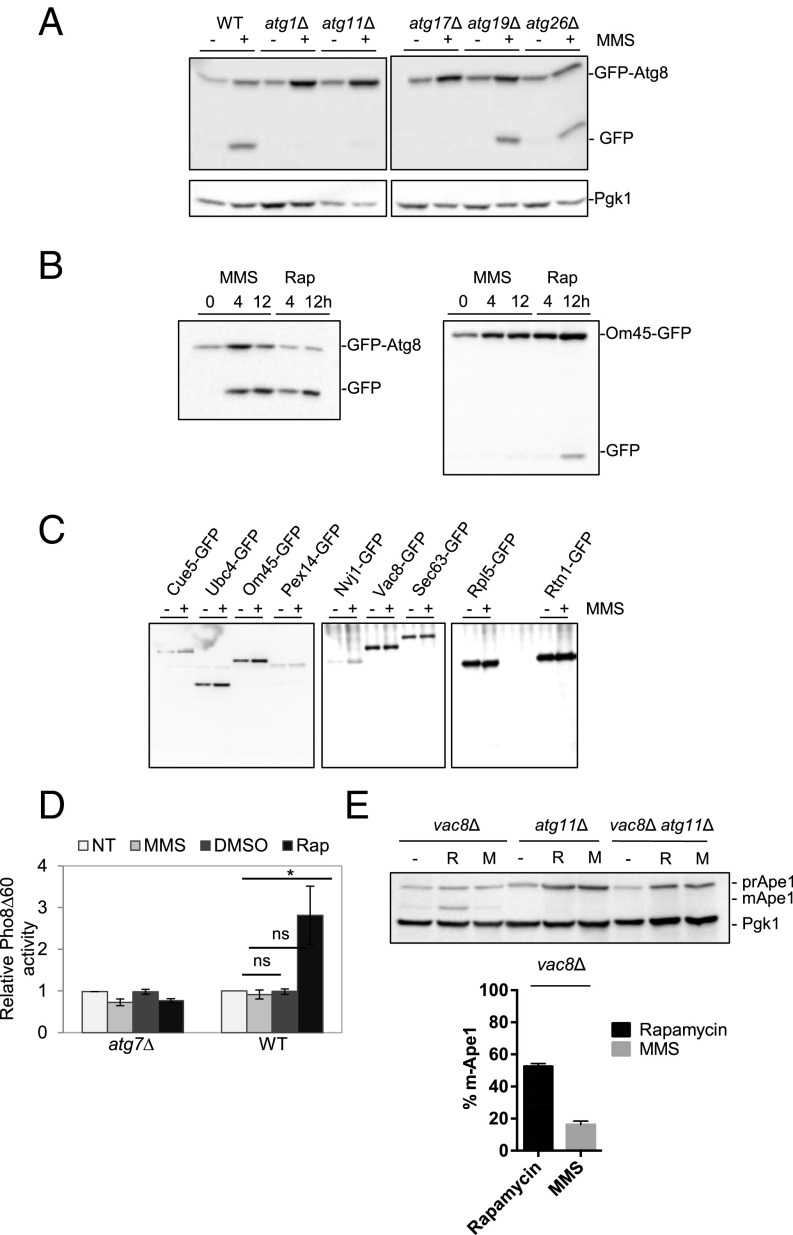
Having established that GTA requires the core components of the DDR machinery, we turned our attention to the specific pathway of autophagy that DNA damage might induce. Analysis of the genetic requirements for autophagy has led to the identification of ∼35 ATG genes in budding yeast (10). These genes can be categorized into two groups: core factors that are required for all pathways of autophagy and selective autophagy genes, defining several different pathways. We undertook a systematic analysis of ATG gene requirements after DNA damage, the results of which are summarized in SI Appendix, Table S1. Core ATG genes, such as ATG1, ATG13, ATG14, ATG16, and ATG18, are required for GTA (Fig. 3 and SI Appendix, Fig. S3A and Table S1); however, genes required for selective autophagy displayed different requirements. Most forms of selective autophagy in budding yeast require the central scaffold/adaptor protein Atg11, which binds to different receptor proteins with their cognate cargo to activate Atg1 (16). We found that GTA is completely dependent on ATG11 (SI Appendix, Table S1 and Fig. 3A).
Fig. 3.
ATG gene requirements for GTA. (A) ATG deletion strains expressing GFP-Atg8 were treated with 0.04% MMS for 4 h, and GFP immunoblots were performed. Representative blots are shown in A, and all other blots are shown in SI Appendix, Fig. S3A. Results of all ATG mutants tested are summarized in SI Appendix, Table S1. (B) GTA does not induce canonical selective autophagy pathways. WT cells expressing either GFP-Atg8 or Om45-GFP were treated with MMS (0.04%) or rapamycin (200 ng/mL) for 4 and 12 h, respectively. GFP immunoblot was performed to monitor GTA and mitophagy. (C) Strains expressing various GFP fusions as indicated were treated with MMS to monitor different forms of selective autophagy. (D) GTA does not induce macroautophagy. The Pho8∆60 assay was performed on MMS-treated WT and atg7Δ cells expressing pho8Δ60. (E) Analysis of prApe1 processing after rapamycin or MMS. Strains of the indicated genotypes were treated with either rapamycin or MMS, and prApe1 processing was judged by immunoblot. Error bars reflect SD from three independent experiments.
Because ATG11 is also required for several selective autophagy pathways, including mitophagy, pexophagy, nucleophagy, and reticulophagy/ER-phagy (11), it is possible that GTA could induce one or more of these pathways; however, GTA did not require the associated receptor proteins for any of these processes (Fig. 3A and SI Appendix, Fig. S3A and Table S1). To monitor these various forms of selective autophagy, we used well-established reporter strains harboring a single GFP fusion to a specific organellar or autophagy target protein, each of which is degraded on the same principle as GFP-Atg8. For example, a GFP fusion of Om45, a mitochondrial matrix protein, is delivered into the vacuole under starvation conditions and can be used to specifically monitor mitophagy, both by vacuolar localization of the GFP chimera or by the appearance of the free GFP band by Western blot (39). As seen in Fig. 3B, treatment of cells with rapamycin induced mitophagy, as previously reported, whereas treatment of cells with MMS did not, indicating that GTA does not induce mitophagy. In the same fashion, we did not observe cleavage of reporters for pexophagy, reticulophagy, nucleophagy, ribophagy, or ubiquitin-mediated aggrephagy (Fig. 3C): Pex11-GFP (40), Sec63-/Rtn1-GFP (41), Rpl5-GFP (42),Nvj1-/Vac8-GFP (43), and Cue5-GFP (44). Thus, GTA does not induce these selective autophagy pathways.
Whereas Atg11 is required for most selective autophagy variants, it has recently been shown to play a role in autophagosome–vacuole fusion during macroautophagy as well (45). We could directly monitor macroautophagy after DNA damage by using the Pho8Δ60 assay (46). The PHO8 gene in budding yeast encodes an alkaline phosphatase enzyme that normally resides in the vacuole. The enzyme is activated after transport into the vacuolar lumen by resident proteases. Removal of the amino-terminal 60 amino acids from Pho8 renders the protein unable to enter into the vacuole and can only do so if phagophores nonselectively engulf the protein and transport it during autophagy-inducing conditions. Therefore, activation of the truncated form of Pho8 is an indication of macroautophagy (46). As shown in Fig. 3D, we did not observe significant induction of alkaline phosphatase activity after DNA damage, in contrast to the increase seen with cells treated with rapamycin for the same duration (Fig. 3D).
As an alternative method to assess macroautophagy, we monitored the delivery and processing of aminopeptidase I (Ape1), a vacuolar resident peptidase. Ape1 is synthesized in the cytosol as a precursor (prApe1) that forms a higher order oligomer and, under nutrient-rich conditions, is delivered to the vacuole by the cytoplasm-to-vacuole targeting (Cvt) pathway, wherein it is processed to its faster migrating and active form (47). When the Cvt pathway is blocked (e.g., by deletion of VAC8), prApe1 is delivered to the vacuole by macroautophagy. We monitored the processing of prApe1 in vac8Δ cells after rapamycin and MMS treatment. MMS induced prApe1 processing, although not to the same extent seen in rapamycin-treated cells (52% vs. 16%) (Fig. 3E), indicating that some amount of prApe1 is apparently delivered by macroautophagy under DNA damage conditions. We found that, under these conditions, the delivery of Ape1 is completely dependent on Atg11 in response to either rapamycin or MMS (Fig. 3E), suggesting that Atg11 could have roles outside of the selective autophagy pathways (45). These results suggest that macroautophagy could be partially induced during the DNA damage response, but, when the Cvt pathway is functional (in VAC8 cells), GTA is entirely dependent on Atg11, but not Atg19. Overall, our results strongly suggest that GTA is an ATG11-dependent process that is distinct from the canonical pathways of selective autophagy and macroautophagy.
DNA Damage Activates the Atg1 Kinase in an Atg11-Dependent Fashion.
Both selective autophagy and macroautophagy require the activity of the Atg1 kinase. Nutrient deprivation stimulates the autoactivation of Atg1 by negative regulation of the TOR and PKA kinases (12, 13, 48, 49). Therefore, this mode of control links the nutrient status of a cell to the activation of autophagy. In contrast, selective autophagy can occur in conditions of plentiful nutrients (47). Recently, it has been demonstrated that selective receptor-bound cargo stimulates Atg1 activity in an Atg11-dependent fashion, thereby driving selective autophagy even in nutrient-replete conditions (16). We wished to assess whether the regulation of GTA occurs in a similar way because GTA shares some of the hallmarks of this process—most notably, its dependence on Atg11 and occurrence in nutrient-rich conditions. To address this question, we monitored Atg1 kinase activity after DNA damage by purifying epitope-tagged Atg1 (FLAG-Atg1), from WT and atg11Δ cells with or without DNA damage, and incubating it with myelin basic protein (MBP) and ATP-γ32P. DNA damage robustly stimulated the kinase activity of Atg1 in a manner that was completely dependent on Atg11 (Fig. 4A).
Fig. 4.
GTA activates the Atg1 kinase in an Atg11-dependent manner. (A) FLAG-Atg1 was immunoprecipitated from atg18Δ atg19Δ (referred to as WT) or atg11Δ atg18Δ atg19Δ cells with or without 0.04% MMS. Kinase activity was determined by incubation with MBP and ATP-γ32P. To improve the efficiency of Atg1 complex immunoprecipitation from cell lysates, ATG18 was deleted to block autophagosome degradation. Because a high proportion of Atg1 kinase activity in cells arises from Atg19-bound cargo, ATG19 was also deleted to accurately determine GTA-specific activation of Atg1. Note that ATG19 is dispensable for GTA (Fig. 5B). (B) GTA partially requires Atg11’s receptor binding domain. WT, atg11Δ, and cells expressing Atg11 lacking the receptor-binding domain (atg11ΔRBD) were transformed with GFP-Atg8 and analyzed for GTA. A representative blot and quantification are shown. Error bars reflect SEM from three independent experiments.
During selective autophagy, Atg11 also acts as a scaffold, aiding the recruitment of autophagy factors to the phagophore assembly site (14). This process is mediated by one or more of four coiled-coil (CC) domains present in the protein (14). Of these, the CC4 domain directly binds autophagy receptors, such as Atg19, Atg34, and Atg32, and thus is also called the receptor-binding domain (RBD). Atg11 lacking the RBD (atg11ΔRBD) phenocopies atg11Δ for the Cvt and mitophagy pathways and fails to activate Atg1 in these selective autophagy pathways (16). We found that atg11ΔRBD is partially defective for GTA, about 50% of WT (Fig. 4B). This finding suggests that, in contrast to other selective autophagy pathways, Atg11 may engage additional domains along with the RBD to drive GTA. Overall, our findings demonstrate that Atg11 can promote Atg1 activity to stimulate GTA and enforce the idea that GTA requires components of the selective autophagy machinery and that regulation of GTA is similar to that of selective autophagy pathways.
GTA Occurs in a Pathway Parallel to That of TORC1 Signaling.
In nutrient-rich conditions, autophagy is inhibited by the action of TORC1 via the hyperphosphorylation of Atg13, which limits its ability to activate the Atg1 kinase (12, 13). It is possible that Mec1-Rad53 might act upstream of TORC1 signaling to induce autophagy: i.e., Mec1-Rad53 might act either directly or indirectly to inhibit TORC1 signaling. In higher eukaryotes, cytosolic pools of ATM can inhibit MTORC1 in response to reactive oxygen species, but, interestingly, this inhibition does not occur in response to the DNA-damaging agent etoposide (23); therefore, the role of TOR signaling in GTA remains enigmatic. We monitored TORC1 activity by examining the phosphorylation of Atg13, a direct target of TORC1. Atg13 is hyperphosphorylated by TORC1 under nutrient-rich conditions and can be observed as slower migrating bands on SDS/PAGE gels (12, 13). As shown previously (12), treatment of cells with rapamycin resulted in dephosphorylation of Atg13, resulting in faster migrating bands of Atg13 (Fig. 5A). DNA damage caused by MMS also resulted in Atg13 dephosphorylation, which occurred even in the absence of MEC1 and/or TEL1 (Fig. 5A). Interestingly, we observed two phenotypes in tel1∆ and mec1Δ tel1∆ cells, even in the absence of DNA damage (Fig. 5A). First, Atg13 migrated faster in nondamaging conditions in the double knockout, suggesting that Tel1 is involved in maintaining Atg13 phosphorylation in nutrient-rich conditions. Second, there was a reduction of Atg13 protein levels in tel1Δ and mec1Δ tel1Δ cells in nondamage conditions, suggesting that Tel1 and, possibly, Mec1 contribute to Atg13 protein stability (Fig. 5A). This result may help explain the 50% reduction in rapamycin-induced autophagy of mec1Δ tel1Δ cells (SI Appendix, Fig. S1C). Reduced protein abundance is not a peculiarity of Atg13 because we observed a similar reduction with Sch9, another TORC1 effector. Sch9, an AGC family kinase, is a direct target and a central effector of TORC1 signaling (50). Sch9 is also dephosphorylated after DNA damage, and this dephosphorylation occurred independently of Mec1 (Fig. 5B). Furthermore, Sch9 protein levels were also reduced in mec1Δ tel1Δ cells, reminiscent of the condition observed with Atg13 (Fig. 5B). In contrast, we observed no effect on TORC2 signaling as judged by phosphorylation on the TORC2 effector Ypk1 (Fig. 5B). Treatment of cells with the combination of MMS and rapamycin caused no further change in the migration of Atg13 (Fig. 5C). Together, our results suggest that GTA occurs in a pathway parallel to that of TORC1 signaling.
Fig. 5.
GTA occurs parallel to TORC1 signaling. (A) Strains were transformed with a 2m plasmid expressing Atg13 and treated with either rapamycin or MMS. Atg13 phosphorylation was evaluated by separating extracts by 7% SDS/PAGE followed by Atg13 immunoblot. (B) Analysis of TORC1 and TORC2 signaling. sml1Δ and mec1Δ tel1Δ sml1Δ strains were treated with rapamycin, cycloheximide, or MMS, and the levels of phosphorylated Sch9 and Ypk1 were determined. The relative levels of phosphorylated (pSch9, pYpk1) and total (Sch9, Ypk1) are indicated. (C) Cells were treated as in A and B with MMS, rapamycin, or the combination of both. Endogenous Atg13 phosphorylation was evaluated by separating extracts on SDS/PAGE gels followed by Atg13 immunoblot.
Screen for Regulators of DNA Damage-Induced Autophagy.
Our observations indicate that GTA is a DNA damage-specific phenomenon that does not require any known autophagy receptors and is distinct from any known selective autophagy pathway. These findings indicate that there might be genes that specifically control GTA but not other forms of autophagy. To identify such genes, we used synthetic genetic array (SGA) technology to screen a library of yeast mutants (either knockouts or DAmP alleles with decreased protein abundance) (51–54). Using SGA, we introduced GFP-Atg8 and the vacuolar marker Vph1-mCherry to create a collection of ∼6,000 unique yeast strains, each one harboring GFP-Atg8 and Vph1-mCherry in the background of a single gene mutation (SI Appendix, Fig. S6). Using an automated culture and image acquisition platform (55), we treated cells with MMS and imaged them for GFP-Atg8 and Vph1-mCherry to assess autophagy induction and vacuolar morphology. Each mutant was imaged in triplicate before and after MMS treatment, and the subsequent images were manually inspected to identify two categories: those that had lower than WT GFP-Atg8 vacuolar signal or cells that had higher than WT vacuolar GFP-Atg8 signal (SI Appendix, Fig. S6). This approach enabled us to generate a list of candidate mutations that appeared to either block or hyperactivate GTA.
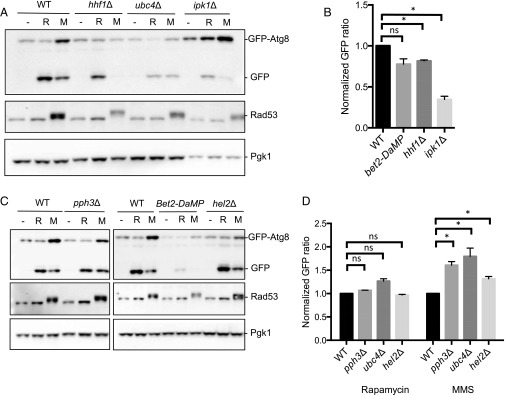
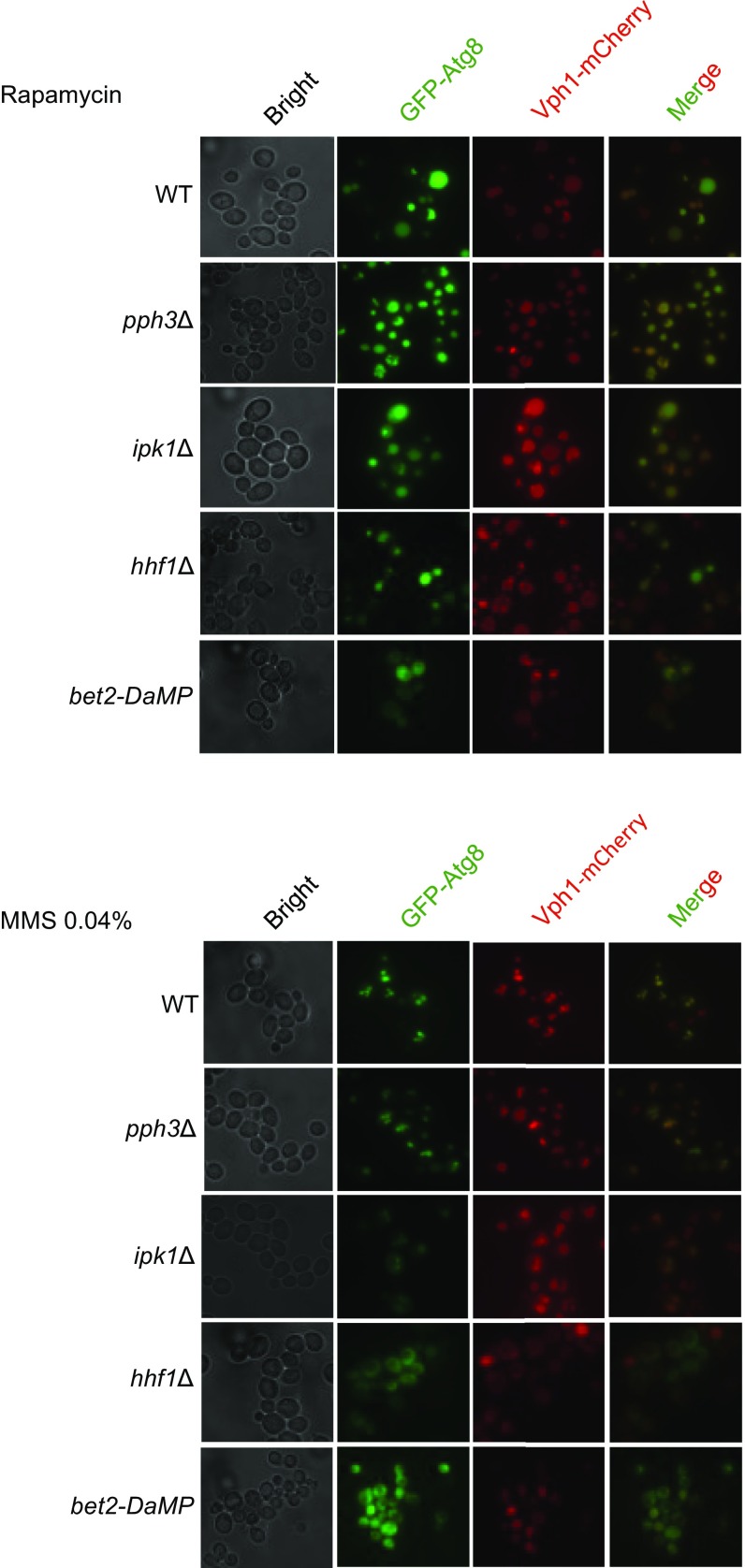
As expected, some of the genes we identified were known autophagy genes (e.g., ATG1, ATG5, ATG10, ATG13) or had previously defined roles in the autophagy process (e.g., YPT1, MON1) (56). Because we were interested in mutations that specifically affected GTA, but not starvation-induced autophagy, we rescreened these hits with rapamycin and eliminated those that also showed the same phenotype as with MMS. This process led to the identification of 28 candidate genes that were affected for GTA but not general autophagy (SI Appendix, Table S2). These candidates were subsequently verified for their autophagy induction after MMS and rapamycin treatment both by Western blot and fluorescence microscopy, the results of which are summarized in SI Appendix, Table S2 and Figs. 6 and 7. In this way, we narrowed down the list of genes to five with the most prominent phenotypes. Null mutations or DAmP alleles of the genes HHF1 and BET2 completely blocked GTA in response to MMS whereas null mutants in PPH3, HEL2, and UBC4 displayed significantly elevated GTA. The HHF1 mutant displayed a slight but statistically significant reduction in autophagy in response to rapamycin; nevertheless, this mutant completely blocked GTA (Fig. 6). Thus, we have identified genes largely specific to the process of genotoxin-induced autophagy. We also identified one mutant, IPK1, that displayed significantly reduced autophagy in response to either MMS or rapamycin and therefore would be considered a general autophagy gene (Fig. 6B).
Fig. 6.
Analysis of screen hits from SGA screen by GFP-Atg8 processing assay. Cells were treated with either 0.04% MMS or 200 ng/mL rapamycin. (A) HHF1 and IPK1 are required for GTA, whereas UBC4 represses GTA. (B) PPH3 and HEL2 repress GTA, while BET2 is required. Representative blots are shown. (C) Quantification of rapamycin-induced autophagy for all mutants is shown as normalized GFP ratios. Error bars reflect SEM from three independent experiments. *P < 0.0125 (Bonferroni correction of unpaired t test value). (D) Quantification of rapamycin- and MMS-induced autophagy for all mutants is shown as normalized GFP ratios. Error bars reflect SEM from three independent experiments. *P < 0.0125 (Bonferroni correction of unpaired t test value). ns, not significant.
Fig. 7.
Analysis of screen hits from SGA screen by fluorescence microscopy. Strains of the indicated genotypes were treated with either 0.04% MMS or 200 ng/mL rapamycin for 4 h and imaged for GFP-Atg8 localization to assess autophagy. Vph1-mCherry decorates vacuoles. Representative images for each genotype are shown.
Because GTA correlates with the strength of the DNA damage response and requires the DNA damage kinases Mec1, Tel1, and Rad53 (Fig. 1 and SI Appendix, Fig. S1A), it is possible that the genes we identified in our screen could affect GTA by modulating the strength of the DDR. Accordingly, we monitored the phosphorylation of Rad53 after DNA damage but observed no significant changes in Rad53 hyperphosphorylation in any of our candidate mutations (Fig. 6 A and B), suggesting that the DDR functions normally in these mutants. In summary, we identified five genes that are specifically involved in controlling GTA: HHF1 and BET2 are positive regulators of GTA whereas PPH3, HEL2, and UBC4 negatively regulate GTA.
Discussion
The cellular response to DNA damage involves the coordinated action of numerous, diverse pathways to ensure survival in the face of genotoxic stress. We have previously shown that the activation of autophagy by the DDR serves to enforce robust cell cycle arrest through the degradation of Pds1/PTTG1/Securin (17, 19). Additionally, DDR-induced autophagy may control the repair of DNA lesions through the degradation of ribonucleotide reductase or other DNA repair enzymes (18). Although it has been demonstrated that DNA damage can stimulate autophagy, the molecular mechanisms connecting these pathways have remained largely unexplored. Here, we report that several agents causing different types of DNA damage, or even a single DNA double-strand break, activate autophagy in a process we term genotoxin-induced targeted autophagy (GTA). GTA requires the action of the central DDR kinases Mec1/ATR, Tel1/ATM, and Rad53/CHEK2. The roles of these proteins in autophagy activation are specific to DNA damage because these kinases are largely dispensable for rapamycin-induced autophagy. GTA requires the transcriptional up-regulation of numerous autophagy genes, which are controlled by Rad53-mediated inactivation of the transcriptional repressor Rph1/KDM4. This transcriptional regulation is reminiscent of the damage-induced autophagy pathway in higher eukaryotes, which occurs by TP53-mediated up-regulation of numerous autophagy genes (7, 22). However, it has also been demonstrated that cytoplasmic TP53 can inhibit autophagy under basal growth conditions (57), suggesting a dual role for TP53 in autophagy control (24). We have not observed elevated autophagy under basal growth conditions in the checkpoint kinase mutants tested, suggesting that these proteins do not normally inhibit autophagy in budding yeast. Rad53 does not act to induce GTA solely via inactivation of Rph1 because the rad53Δ rph1Δ double mutant restored autophagy to only 20% of WT levels. Therefore, we suggest that Rad53 acts in both an Rph1-dependent and -independent manner to induce GTA. Additionally, our data indicate that Chk1 may act as a negative regulator of GTA because the double rad53Δ chk1Δ restored GTA, raising the possibility of dual regulation by the DDR on autophagy. We note that one candidate identified in our screen as a regulator of GTA, Vhs2, has been recently identified to have potential Chk1 phosphorylation sites; also Atg13 was identified to have Chk1 phosphorylation sites (9).
To find additional regulators of GTA, we undertook a genome-wide high content screen, which led to the identification of five genes that we consider being GTA-specific: that is, their mutations displayed either significantly reduced or elevated autophagy in response to DNA damage but were otherwise similar to WT for rapamycin-induced autophagy. We identified two genes that are specifically required to induce GTA: BET2 and HHF1. BET2 is an essential gene that encodes the β subunit of the type II geranyl-geranyl transferase (GGTase) enzyme and is responsible for the attachment of prenyl groups onto the C termini of proteins (58). This modification is thought to aid in the recruitment of proteins to various subcellular membranes, which could explain its role in the autophagic process. Along these lines, however, we note that Bet2 is involved in a range of trafficking pathways and as such may have an indirect role in GTA. Interestingly, the human homolog of BET2, RABGGTB, can rescue the inviability of the yeast mutant, suggesting a strong conservation of function (59). It will be interesting to know whether other GGTase proteins are also involved in GTA. HHF1 encodes one of two copies of the histone H4 gene. Histones play central roles in transcription; we note that the up-regulation of GFP-Atg8 in the hhf1Δ strain appears to be diminished in response to damage (Fig. 6). An alteration in histone subunit balance may affect the expression of other genes as well.
We also discovered three genes that negatively regulate GTA: namely, PPH3, UBC4, and HEL2. PPH3 encodes the catalytic subunit of the PP4 phosphatase and is involved in Rad53 dephosphorylation (60). Ubc4 and Hel2 are E2 and E3 ubiquitin ligases that are involved in the degradation of excess histones. Interestingly, we noted above that reduction of histone H4 levels (hhf1Δ) blocks GTA, suggesting that there may be a network of regulation around histone dosage that may be critical for GTA. Further work will help clarify the exact roles of these genes in GTA.
One mutant, ipk1Δ, was found to significantly reduce autophagy in response to both MMS and rapamycin, indicating that IPK1 is involved in general autophagy. Ipk1 is part of the inositol polyphosphate synthesis pathway, which is responsible for the generation of various inositol polyphosphates (IP3–IP8), which have multiple functions in cells. We note that others have found no effect of IPK1 deletion on general autophagy in response to nitrogen starvation (61). The reason for this discrepancy could be due either to differences in experimental conditions used (rapamycin treatment vs. nitrogen starvation) or to differences in strain background. Nevertheless, both results suggest an involvement of inositol polyphosphate synthesis in the regulation of autophagy.
In higher eukaryotes, DNA damage stimulates chaperone-mediated autophagy, a selective autophagy-like pathway, which does not use core autophagy proteins (20, 62). However, it is not entirely clear which other pathways of autophagy DNA damage might induce, and which components of the autophagic machinery are required. We find that GTA in budding yeast requires the core components of the autophagic machinery. Selective autophagy requires the binding of receptor protein-bound cargo complexes to the Atg11 complex (11). We find that GTA is completely dependent on ATG11 but does not use any known autophagy receptor nor does it induce any previously described selective autophagy pathway. Moreover, the activation of the Atg1 kinase during GTA requires the presence of Atg11 (Fig. 4), a mode of regulation that is similar to other forms of selective autophagy. However, GTA is also accompanied by the down-regulation of TORC1 and indeed may partially induce nonselective autophagy as well (Fig. 3E). Therefore, we conclude that GTA is a largely selective autophagy process but may occur under conditions wherein nonselective autophagy is active. In conclusion, the ensemble of necessary ATG genes and the identification of GTA-specific genes from our screen indicate that GTA is distinct from other selective “phagy” pathways. We suggest that GTA may engage unknown receptor(s) to direct the degradation of target proteins, which implies that the regulation of GTA is distinct from that of canonical autophagy pathways.
Materials and Methods
Yeast Media, Strains, and Plasmids.
All yeast strains used are described in SI Appendix, Table S3 and are derivatives of either JKM179, BY4741, or YMS721 (16, 63, 64). Most mutant strains were constructed using single-step PCR-mediated transformation of yeast colonies or by genetic crosses. The Rad53-AID strain was construct as described previously (31). Primer sequences used in this study are listed in Table S4. The strains encoding C-terminal fusions of selective autophagy targets were obtained from the yeast GFP collection (Invitrogen) (65). The GFP-Atg8 and Atg13 plasmids were a kind gift from Yoshiaki Kamada, National Institute for Basic Biology, Okazaki, Japan (13). Most experiments were performed in either YEPD [1% yeast extract, 2% (wt/vol) peptone, 2% (wt/vol) dextrose] or YEP-lactate [3% (wt/vol) lactic acid] media.
GFP-Atg8 Processing Assay.
Strains containing GFP-Atg8 were grown either in YEP-lactate or YEPD media overnight to a cell concentration of ∼5 ×106 cells per milliliter, which corresponds to an OD600 of 0.2. To assess GTA levels, we treated cells with methyl-methane sulphonate (MMS) (final concentration 0.04%), hydroxyurea (0.2 M), or zeocin (300 mg/mL) and collected cells at the time points indicated for fluorescence microscopy or Western blotting. To monitor GTA in response to DSBs, cells were pregrown in YEP-lactate, and the HO endonuclease was induced by the addition of 2% galactose to the media. To assess rapamycin-induced autophagy, we treated cells with rapamycin at a final concentration of 200 ng/mL for 4 h.
Western Blotting.
Western blotting was carried out using the trichloroacetic acid (TCA) protocol as previously described (66). Frozen cell pellets from time courses were thawed on ice and resuspended in 200 μL of 20% (wt/vol) TCA. An equal volume of acid-washed glass beads was added to the tubes, and the cells were lysed by vortexing for 2 min. Cell lysates were collected by centrifugation, and additional washes of the beads were performed twice with 200 μL of 5% TCA. The precipitated proteins were collected by centrifugation and solubilized in 2× Laemmli buffer (60 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, 0.2% bromophenol blue) and boiled at 100 °C for 5 min. The insoluble material was removed by centrifugation, and the supernatant was separated on SDS/PAGE gels of the appropriate percentage. After transfer of separated proteins onto membranes, immunoblots were performed using α-GFP (ab290 or ab6556; Abcam), α-Rad53 (ab166859), α-Pgk1 (Abcam), α-Atg13, α-Sch9, α-p-SCH9S758 (67), α-Ypk1 (Santa Cruz Biotechnology), and α-pYpk1T662 (68) antibodies.
SGA.
The synthetic genetic array was performed as described previously (63).
Transcription Assay.
Transcript abundance after MMS treatment was performed as previously described (36).
Atg1 Kinase Assay.
Atg1 kinase assay was performed as previously described (16).
Statistical Analysis.
To calculate statistical significance of data, an unpaired Student’s t test was performed using GraphPad Prism software (Version 6). Multiple comparisons of data were corrected using the Bonferroni correction metric to define significance.
Supplementary Material
Acknowledgments
We thank Dr. Yoshiaki Kamada (National Institute for Basic Biology, Okazaki, Japan) for the kind gift of plasmids. V.V.E. was supported by a Howard Hughes Medical Institute international predoctoral fellowship from 2013 to 2015. D.P.W. and B.L. have been supported by National Institutes of Health (NIH) Genetics Training Grant TM32 GM007122. This work was supported by NIH Grants GM61766 and GM20056 (to J.E.H.) and GM053396 (to D.J.K.) and by a Bronfman Brandeis–Israel collaborative research grant (to J.E.H. and M.S.). Funding from Harvard University supported V.D. The M.S. laboratory is supported by BSF Grant 2013101. E.S. and R.J.L. acknowledge funding from the Canton of Geneva and Singal X of the Swiss National Science Foundation’s Systems X program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614364114/-/DCSupplemental.
References
- 1.Harrison JC, Haber JE. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez Y, et al. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286(5442):1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 3.Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281(5374):272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal R, Tang Z, Yu H, Cohen-Fix O. Two distinct pathways for inhibiting pds1 ubiquitination in response to DNA damage. J Biol Chem. 2003;278(45):45027–45033. doi: 10.1074/jbc.M306783200. [DOI] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 7.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27(9):1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104(25):10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou C, et al. Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks. Proc Natl Acad Sci USA. 2016;113(26):E3667–E3675. doi: 10.1073/pnas.1602827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reggiori F, Klionsky DJ. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20(1):43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015;59(3):372–381. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dotiwala F, et al. DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase. Proc Natl Acad Sci USA. 2013;110(1):E41–E49. doi: 10.1073/pnas.1218065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyavaiah M, Rooney JP, Chittur SV, Lin Q, Begley TJ. Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1. Mol Cancer Res. 2011;9(4):462–475. doi: 10.1158/1541-7786.MCR-10-0473. [DOI] [PubMed] [Google Scholar]
- 19.Eapen VV, Haber JE. DNA damage signaling triggers the cytoplasm-to-vacuole pathway of autophagy to regulate cell cycle progression. Autophagy. 2013;9(3):440–441. doi: 10.4161/auto.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823. doi: 10.1038/ncomms7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 22.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 23.Alexander A, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10(6):637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3(3):181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279(29):29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6(9):a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94(3):399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75(6):1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 31.Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30(9):341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin M, Klionsky DJ. Regulation of autophagy: Modulation of the size and number of autophagosomes. FEBS Lett. 2014;588(15):2457–2463. doi: 10.1016/j.febslet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartholomew CR, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA. 2012;109(28):11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin M, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014;24(12):1314–1322. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard A, et al. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol. 2015;25(5):546–555. doi: 10.1016/j.cub.2014.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang YK, Wang L, Sancar GB. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol Cell Biol. 1999;19(11):7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EM, Jang YK, Park SD. Phosphorylation of Rph1, a damage-responsive repressor of PHR1 in Saccharomyces cerevisiae, is dependent upon Rad53 kinase. Nucleic Acids Res. 2002;30(3):643–648. doi: 10.1093/nar/30.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283(47):32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31(13):2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522(7556):359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 42.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 43.Pan X, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11(7):2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, et al. The Atg17-Atg31-Atg29 complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr Biol. 2016;26(2):150–160. doi: 10.1016/j.cub.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 47.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584(7):1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephan JS, Yeh Y-Y, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106(40):17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279(20):20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26(5):663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 51.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5(8):711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 54.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 55.Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200(6):839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107(17):7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi G, Yu JA, Newman AP, Ferro-Novick S. Dependence of Ypt1 and Sec4 membrane attachment on Bet2. Nature. 1991;351(6322):158–161. doi: 10.1038/351158a0. [DOI] [PubMed] [Google Scholar]
- 59.Kachroo AH, et al. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348(6237):921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Neill BM, et al. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci USA. 2007;104(22):9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor R, Jr, Chen PH, Chou CC, Patel J, Jin SV. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy. 2012;8(9):1300–1311. doi: 10.4161/auto.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cuervo AM, Wong E. Chaperone-mediated autophagy: Roles in disease and aging. Cell Res. 2014;24(1):92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–159. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- 64.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(5):2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 66.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7(2):293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 67.Gaubitz C, et al. Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol Cell. 2015;58(6):977–988. doi: 10.1016/j.molcel.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 68.Berchtold D, et al. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol. 2012;14(5):542–547. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.