Significance
Wnt signaling, named after the secreted proteins that bind to cell surface receptors to activate the pathway, is crucial for normal cell functions, and its deregulation can culminate in cancer. Several surveillance mechanisms have evolved as a precautionary measure to ensure proper development and the avoidance of cancer. Here we reveal that the E3 ubiquitin ligase Mule can target β-catenin for degradation to stop the Wnt signal during constitutive activation. Significantly, our data indicate that a combined loss of Mule and Apc accelerates the conversion of normal intestinal stem cells into cancer stem cells, paving the way to colorectal cancer development. It may be possible to use this knowledge to manipulate Mule, β-catenin, or Wnt pathway functions to reduce cancer initiation.
Keywords: β-catenin, Mule, Wnt signaling, stem cells, colorectal cancer
Abstract
Wnt signaling, named after the secreted proteins that bind to cell surface receptors to activate the pathway, plays critical roles both in embryonic development and the maintenance of homeostasis in many adult tissues. Two particularly important cellular programs orchestrated by Wnt signaling are proliferation and stem cell self-renewal. Constitutive activation of the Wnt pathway resulting from mutation or improper modulation of pathway components contributes to cancer development in various tissues. Colon cancers frequently bear inactivating mutations of the adenomatous polyposis coli (APC) gene, whose product is an important component of the destruction complex that regulates β-catenin levels. Stabilization and nuclear localization of β-catenin result in the expression of a panel of Wnt target genes. We previously showed that Mule/Huwe1/Arf-BP1 (Mule) controls murine intestinal stem and progenitor cell proliferation by modulating the Wnt pathway via c-Myc. Here we extend our investigation of Mule’s influence on oncogenesis by showing that Mule interacts directly with β-catenin and targets it for degradation under conditions of hyperactive Wnt signaling. Our findings suggest that Mule uses various mechanisms to fine-tune the Wnt pathway and provides multiple safeguards against tumorigenesis.
Activation of the Wnt pathway, named after the secreted proteins that bind to cell surface receptors to activate the pathway, plays a critical role in stem cell self-renewal, and thus the homeostasis of normal mammalian tissues. However, constitutive activation of Wnt signaling has been implicated in a broad range of cancers, including melanoma, hepatocellular carcinoma, and cancers of the prostate, thyroid, ovary, and breast (1–3). Perhaps the best-studied malignancy arising from the loss of regulation of the Wnt signaling pathway is colorectal cancer, which develops through a multistage process driven by the progressive accumulation of genetic mutations (4). The key initial activating mutation occurs in components of the Wnt pathway, but most commonly in APC.
Canonical Wnt signaling involves a relay of protein interactions that serve to transmit a signal from the extracellular space to the plasma membrane. This signal is amplified in the cytoplasm and then directed to the nucleus to culminate in the activation of the Wnt target gene program. In molecular terms, when the glycoprotein Wnt is secreted into the extracellular space, it binds to a cell’s Frizzled receptors in conjunction with the low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6). The engagement of these receptors recruits the scaffolding protein Dishevelled (Dvl), leading to LRP5/6 phosphorylation and consequent recruitment of the scaffolding protein, Axin2 and glycogen synthase kinase 3 beta (GSK-3β). This recruitment results in disruption of the “destruction complex” that contains APC and normally binds to the signal transduction protein β-catenin to trigger its degradation. On Wnt signaling, however, β-catenin is released from the destruction complex, accumulates in the cytoplasm, and then translocates into the nucleus, where it associates with the T-cell factor/lymphoid enhancing factor transcription factors to activate the transcription of the battery of genes composing the Wnt program (5, 6). These genes influence cellular processes such as proliferation, differentiation, migration, and adhesion (7). Conversely, in the absence of Wnt, the destruction complex binding to β-catenin phosphorylates it and marks it for ubiquitination by the E3 ubiquitin (Ub) ligase β-TRCP, encoded by the beta-transducin repeat-containing gene. The ubiquitinated β-catenin is then targeted to the proteasome for degradation (5).
Numerous mechanisms of positive and negative regulation have evolved to fine-tune the Wnt pathway. In a previous study (8), we showed that the E3 Ub ligase Mule (Huwe1/Arf-BP1) is critical for regulating the Wnt pathway in the intestine. By studying the effects of Mule deficiency in mice of an Apc competent (Mule conditional knockout, herein referred to as cKO) or Apcmin background (Mule cKO Apcmin), we demonstrated that a lack of Mule alone could promote tumorigenesis in the intestine, and that Mule deficiency accelerated adenoma development caused by the Apcmin mutation. Our data established that Mule is a bona fide tumor suppressor in the gut.
Our previous work also established that, in the normal intestine, Mule regulates the protein levels of the receptor tyrosine kinase EphB3 by targeting it for proteasomal and lysosomal degradation. EphB/ephrinB interactions position cells along the intestinal crypt/villus axis and can compartmentalize incipient colorectal tumors. We further demonstrated that Mule controls murine intestinal stem and progenitor cell proliferation via its effects on c-Myc, which is a Mule substrate and a Wnt target. We found that c-Myc was up-regulated in Mule cKO mice, not only because of the absence of Mule-mediated c-Myc degradation but also because of hyperactivated Wnt signaling.
Previous work has shown that Mule regulates the Wnt pathway in a negative feedback loop by ubiquitinating Dvl in a Wnt ligand-dependent manner (9). However, our previous examination of Mule cKO Apcmin mice suggested that Mule targets other components of the Wnt pathway. Here we demonstrate that Mule can bind directly to β-catenin, the Wnt signal transducer. Mule-mediated β-catenin degradation occurs only under conditions of cellular hyperproliferation, as would arise when APC mutations permanently inactivate the destruction complex and allow β-catenin to stabilize and promote constitutive Wnt signaling. Our findings indicate that β-catenin degradation is an important mechanism used by Mule under conditions of Wnt pathway hyperactivation to execute its function as a tumor suppressor to prevent colon cancer.
Results
Loss of Mule Accelerates Morphological Alterations in Apcmin Organoids.
Previous in vitro work has shown that intestinal organoids established from tissue lacking a functional APC lose their crypt villus architecture and adopt an abnormal spheroid cyst-like morphology (10), and that this altered morphology is a result of up-regulated Wnt signaling instructing the cells to adopt a proliferative progenitor phenotype (11). Our earlier study of intestinal adenoma development in mice showed that Mule ablation on the Apcmin background further increased Wnt signaling over that induced by Apc mutation alone (8). Moreover, single cells isolated from adenomas that developed in Mule cKO Apcmin mice formed spheroid cysts more efficiently than cells isolated from Apcmin adenomas (8). These cystic organoids are reminiscent of the organoids grown from Lgr5-GFP-ires-CreERT2 X Apc fl/fl mice. The Lgr5-EGFP-IRES cre ERT2 “knock-in” allele ablates Lgr5 (leucine-rich repeat-containing G-coupled receptor 5) gene function and expresses GFP and the Cre ERT2 fusion protein. APCfl/fl are mice having an APC-floxed allele. Tamoxifen-mediated cre recombinase activation results in the deletion of Apc specifically in the Lgr5 stem cells (11). Loss of Apc in Lgr5 stem cells has been reported to be the cell of origin of intestinal cancer (12). These findings suggested that loss of Mule under conditions of Wnt hyperactivity promotes stem cell proliferation and expansion. This prompted us to generate and further study intestinal organoids from Apcmin and Mule cKO Apcmin mice.
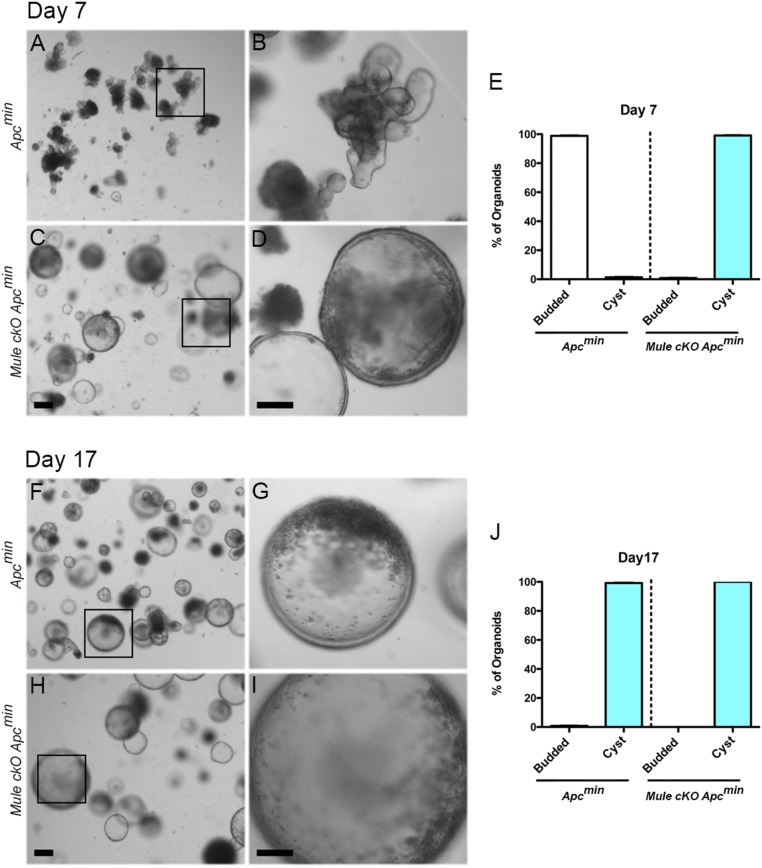
Organoids were cultured from crypts isolated from Apcmin and Mule cKO Apcmin mice. After 7 d in culture and 1 d after passaging, Apcmin organoids showed essentially normal morphology (Fig. 1 A and B). At this point, only 1% of the organoids displayed the cystic morphology (Fig. 1E). It was not until after 2 wk in culture that 99% of the organoids developed the cyst-like spheroid morphology (Fig. 1 F, G, and J). In sharp contrast, after 7 d in culture, all the Mule cKO Apcmin organoids already displayed the cyst-like morphology (Fig. 1 C–E). This altered morphology was retained by Mule cKO Apcmin organoids for the full 2 wk of culture (Fig. 1 H–J). Interestingly, there were some Mule cKO Apcmin organoids that were significantly larger than average cystic organoids, demonstrating that the loss of Mule further enhances proliferation (Fig. 1 H and I). The fact that the concurrent loss of Mule caused Apcmin organoids to display the morphology defect so quickly after establishment suggests that under conditions of Wnt hyperactivation, Mule has a direct regulatory effect on Wnt signaling. Further, the fact that these observations were made on an Apcmin background suggests Mule’s regulatory role is downstream of the destruction complex.
Fig. 1.
Mule-deficient Apcmin organoids instantly take on a cystic morphology. Bright-field microscopy of intestinal organoids that were derived from Apcmin (A, B, F, and G) or Mule cKO Apcmin (C, D, H, and I) mice and cultured for 7 d (A–D) or 17 d (F–I). Low-magnification images (A, C, F, and H) and high-magnification images of the boxed areas (B, D, G, and I) are shown. (Scale bars, 100 µm.) (E) Quantification for Apcmin and Mule cKO Apcmin organoids displaying a budded or cystic morphology on day 7 was obtained by counting four fields in three wells per sample (n = 2/genotype). (J) Quantification for Apcmin and Mule cKO Apcmin organoids displaying a budded or cystic morphology on day 17. Data are representative of two independent experiments in 3-mo-old mice. The data were also reproduced in a pair of 2-mo-old mice, albeit at different kinetics.
Loss of Mule in Addition to APC Results in Elevated β-Catenin Protein.
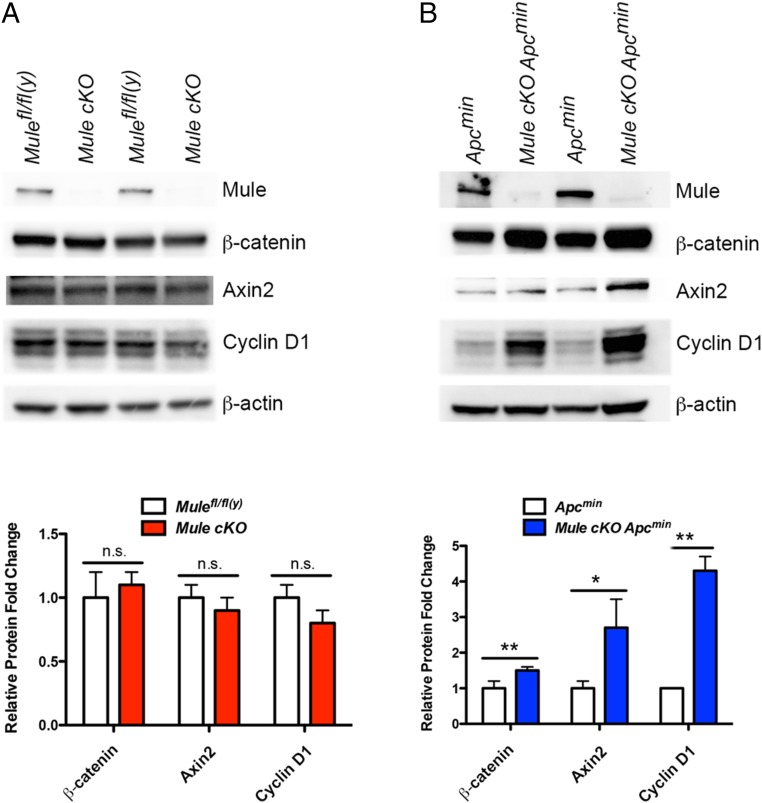
Our previous immunohistochemical examinations of WT and Mule cKO intestines revealed no differences in β-catenin staining (8). However, we suspected that Mule might regulate a Wnt pathway element upstream of β-catenin because Wnt target genes were up-regulated in Mule cKO organoids (8). Indeed, de Groot et al. reported that Mule-mediated K63-linked ubiquitination of Dvl normally inhibits Wnt pathway activation (9). Consistent with this observation, we previously identified accumulation of nuclear β-catenin in our Mule cKO adenomas (8), which could account for deregulation of the Wnt pathway in Mule-deficient cells at steady-state. Because the Apcmin background precludes any study of Mule-mediated regulation of Dvl, we focused on the only element downstream of the destruction complex: β-catenin. We first carried out immunoblotting analyses of WT (Mulefl/fl(y)) and Mule cKO organoids, but found no differences in levels of β-catenin protein or Wnt target genes Axin2 and Cyclin D1 at the protein level (Fig. 2A, quantification shown in the bar graph). Mule cKO Apcmin organoids displayed an increased level of β-catenin, Axin2, and Cyclin D1 protein compared with Apcmin organoids (Fig. 2B, quantification shown in the bar graph). Thus, loss of Mule leads to excessive β-catenin accumulation in the absence of a functional APC.
Fig. 2.
Loss of Mule increases the β-catenin protein. (A and B) Immunoblots to detect the indicated proteins in lysates of intestinal organoids generated from the indicated mouse strains (n = 3 mice/group). β-actin, loading control. Relative protein fold change is shown in the bar graphs below each immunoblot. Values were determined by quantitation of bands by Image J, followed by normalization first to the loading control and then to Apcmin. n.s., not significant. *P < 0.05; **P < 0.01.
The Change in β-Catenin Protein Levels Translates into an Elevated Wnt Target Gene Expression.
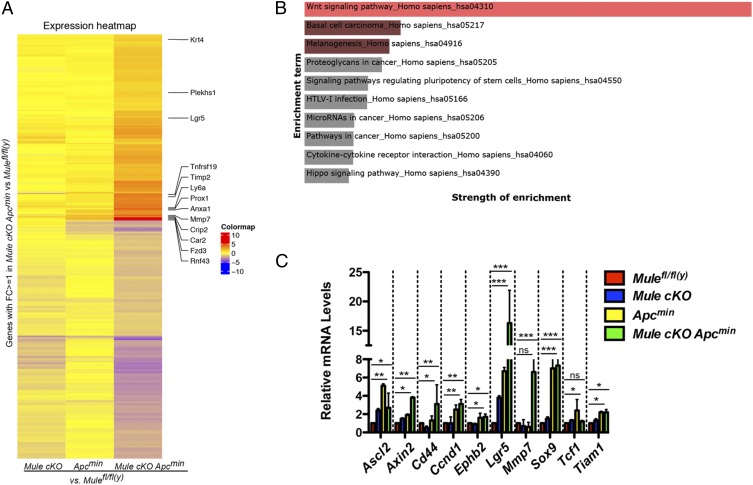
To determine the influence the increased levels of nuclear β-catenin have on global gene expression, we subjected RNA isolated from WT, Mule cKO, Apcmin, and Mule cKO Apcmin organoids to comparative gene expression profiling. The heat map generated in Fig. 3A shows that Wnt target genes Lgr5, Tnfrsf19, Mmp7 (metalloproteinase 7), and Rnf43 are among the genes most highly enriched in the Mule cKO Apcmin condition when normalized to WT. Pathway analysis of the most highly expressed genes reported by our profiling was used to enlist the top 10 enriched pathways from the KEGG (Kyoto Encyclopedia of Genes and Genomes) Database (Fig. 3B). The width of the bar and brightness of color correspond to the strength of the enrichment for the corresponding pathway. The Wnt pathway is at the top of the list (Fig. 3B). Although these results are intuitive, given that we are looking at the global transcriptional effect on an Apcmin background where Wnt signaling is elevated, the significance is that the concomitant loss of Mule serves to further increase the expression of some Wnt target genes (Fig. 3A). These results were confirmed by quantitative RT-PCR. Here, we show that Wnt target genes such as Axin2, Cd44, Ccnd1 (CyclinD1), Lgr5, and Mmp7 were up-regulated much more in the double mutants (Fig. 3C). These data further support our contention that ablation of Mule on an Apcmin background results in an increase in β-catenin protein that consequently up-regulates Wnt target genes. Particularly interesting in this context was the 16-fold increase in mRNA encoding Lgr5, which is a Wnt target and stem cell marker (Fig. 3 A and C). This observation suggests the theory that in a hyperproliferative setting, such as that resulting from APC mutation, loss of Mule may deregulate the Wnt pathway such that the cancer stem cell population can proliferate and expand.
Fig. 3.
The change in β-catenin protein levels translates into an elevated Wnt target gene expression. (A) Heat map of a microarray experiments comparing gene enrichment in Mule cKO, Apcmin, and Mule cKO Apcmin normalized to WT (n = 2/genotype). Some of the genes whose expression is elevated in the Mule cKO Apcmin are listed. (B) Pathway analysis of the most highly expressed genes reported by our profiling was used to enlist the top 10 enriched pathways from the KEGG Database. (C) Quantitative RT-PCR analysis of relative transcript levels of the indicated genes in cells isolated from intestinal organoids derived from the indicated mouse strains. Data are the mean ± SEM and are from four independent experiments. Results are expressed as fold change relative to β-actin. ***P < 0.0001; **P < 0.01; *P < 0.05. ns, not significant.
Mule Interacts Directly with β-Catenin and Targets It for Ubiquitination.
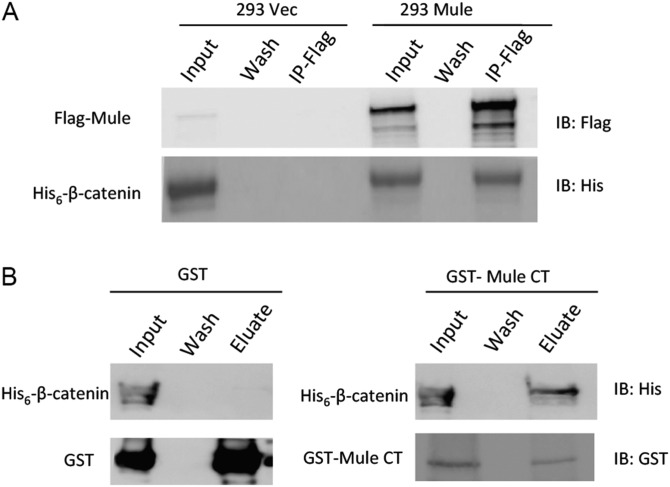
To determine the mechanism by which loss of Mule leads to an increase in β-catenin protein, we first tested whether Mule and β-catenin can physically interact. We expressed Flag-tagged human WT Mule (Mule-WT) in HEK293T and carried out coimmunoprecipitation experiments. We found that anti-Flag Ab was able to coimmunoprecipitate β-catenin just as well as anti-β-catenin (Abc) was able to coimmunoprecipitate with Flag (Mule) (Fig. 4A). A similar reciprocal experiment conducted in HCT116 cells to detect the interaction between endogenous β-catenin and Mule yielded the same results (Fig. 4B). We next examine whether the interaction of Mule and β-catenin is binary. We incubated M2 Agarose-immobilized Flag-tagged Mule purified from Mule-WT cells with recombinant His6 tagged β-catenin from Escherichia coli. Flag-tagged Mule was able to pull down recombinant His6 tagged β-catenin, suggesting Mule directly interacts with nonmodified β-catenin, and thus the interaction is not phosphorylation-dependent (Fig. S1A). An in vitro GST pull down assay using an N-terminal truncated GST-Mule protein and recombinant His6 tagged β-catenin further confirms the binary interaction between Mule and β-catenin (Fig. S1B).
Fig. 4.
Mule interacts with β-catenin in vitro and targets it for ubiquitination. (A, Left) Immunoblot to detect the indicated proteins in TCL of HEK293T cells that were stably transfected with empty vector or vector-expressing Flag-tagged WT Mule (Mule-WT). Cells were treated with the proteasome inhibitor MG132 to prevent β-catenin degradation and immunoprecipitated with anti-Flag Ab or Abc Ab. GAPDH, loading control. (Middle and Right) Immunoblot to detect β-catenin or Flag (Mule), respectively, in the immunoprecipitate from the TCL. (B) HCT116 cells were stably transfected with empty vector or vector-expressing Flag-tagged WT Mule (Mule-WT), treated with MG132 and subjected to immunoprecipitations as in A. (C) Immunoblot to detect endogenous ubiquitinated β-catenin in Mule overexpressing HEK293T cells that were transfected with HA-Ub. (D) Immunoblot to detect ubiquitinated β-catenin after an in vitro ubiquitination assay (Materials and Methods), using recombinant His-tagged β-catenin as the substrate and N-terminal truncated GST-tagged Mule (GST-Mule CT) as the E3 ligase. Experiments were performed twice with similar results.
Fig. S1.
Interaction between Mule and β-catenin. (A) Two hundred ninety-three cells overexpressing vector (293 Vec) or Flag-tagged Mule (293 Mule) were subjected to immunoprecipitation using Flag Ab conjugated agarose (IP-Flag). The resulted precipitants were incubated with recombinant His6-tagged β-catenin. His6-tagged β-catenin immobilized with Flag-Mule was detected by immunoblot using anti-His Ab. (B) His6-tagged β-catenin and GST-tagged Mule CT (3760-4374) (Input) were incubated with glutathione Sepharose. After washing (“Wash”), protein were eluted with reduced glutathione (“Eluate”). Samples were subjected to immunoblotting analysis, using anti-His or anti-GST Abs. The experiments were performed twice with similar results.
Because Mule cKO Apcmin organoids showed such a large accumulation of β-catenin protein, we hypothesized that Mule might serve as an E3 ligase regulating its stability. To test this hypothesis, we performed an intracellular ubiquitination assay in which Mule-overexpressing HEK293T cells (or HEK293T cells expressing control empty vector) were transfected with plasmids expressing HA-tagged Ub. We then subjected lysates of these cells to immunoprecipitation with Abc and showed that the endogenous β-catenin protein can be polyubiquitinated in the presence of Mule (Fig. 4C). To determine whether Mule does indeed directly ubiquitinate β-catenin, we performed in vitro ubiquitination assays. The results confirmed that recombinant full-length β-catenin protein was readily polyubiquitinated by the E3 ligase activity of an N-terminal truncated GST-Mule protein (Fig. 4D), suggesting β-catenin is a bona fide Mule substrate.
Mule Destabilizes the β-Catenin Protein and Down-Regulates Wnt Target Gene Expression.
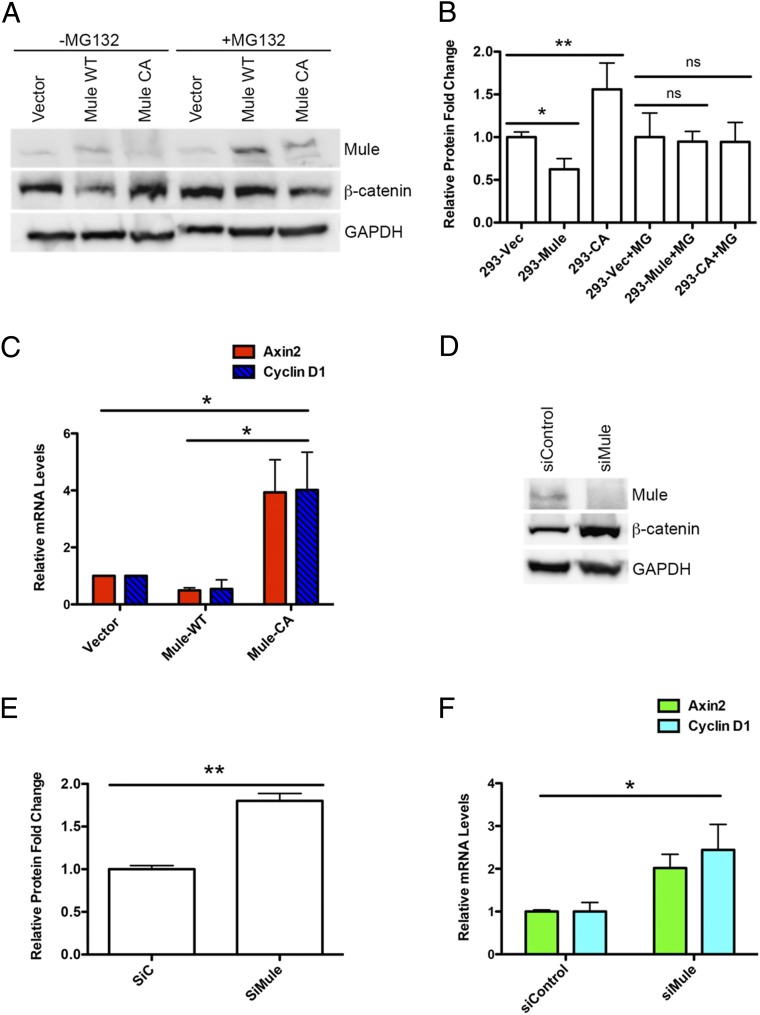
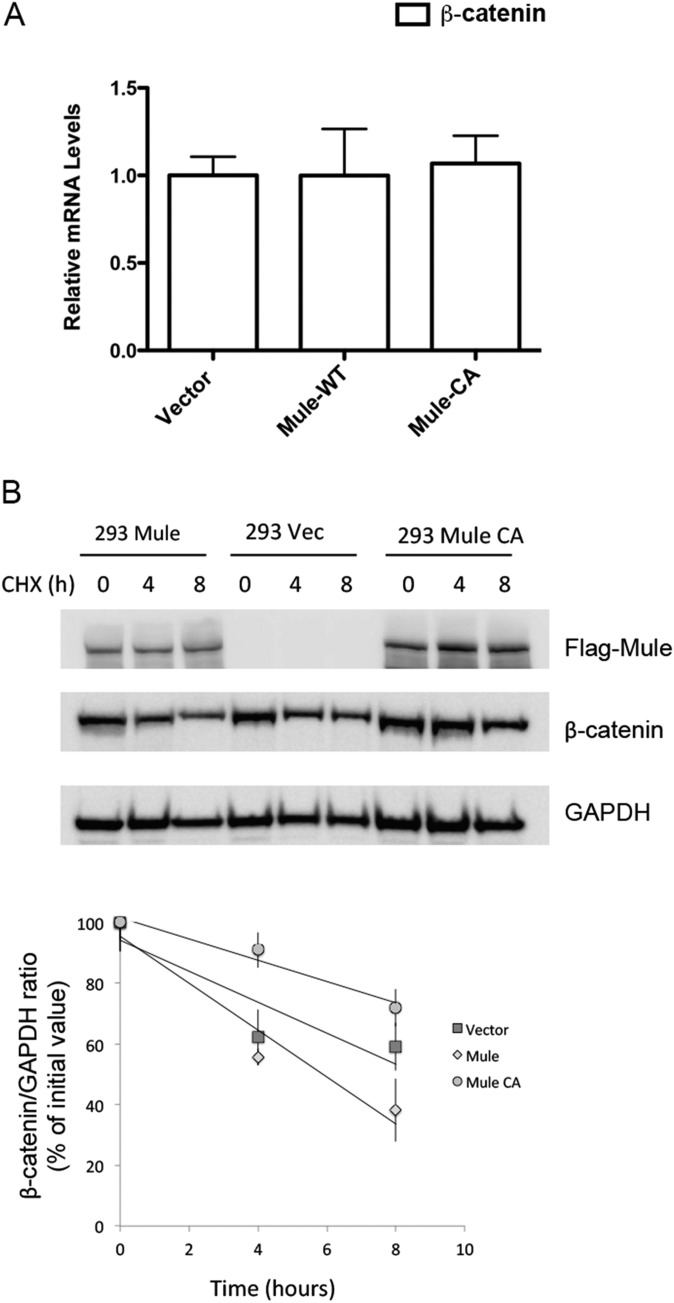
To test whether Mule-mediated ubiquitination of β-catenin actually destabilized it, we determined steady-state levels of β-catenin in HEK293T cells that overexpressed Mule-WT or Mule-CA (the catalytically inactive mutant in which cysteine 4341 is replaced by an alanine) and were treated (or not) with the proteasome inhibitor MG132. In line with our hypothesis, β-catenin levels were decreased in untreated cells expressing Mule-WT but increased in untreated cells expressing the inactive Mule-CA protein (Fig. 5 A and B). Conversely, MG132 treatment prevented Mule-mediated β-catenin destabilization, suggesting Mule can indeed target β-catenin for proteasomal degradation. Consistent with our hypothesis, Mule-WT-overexpressing cells showed a twofold down-regulation of the Wnt target genes AXIN2 and CCND1 (Cyclin D1) compared with empty vector-expressing controls, whereas these genes were up-regulated fourfold in Mule-CA-overexpressing cells (Fig. 5C). The gene expression levels of β-catenin in Mule-WT or Mule-CA-overexpressing cells were similar to empty vector-expressing controls (Fig. S2A). We next measured β-catenin protein stability by a cycloheximide chase assay in HEK293T cells when overexpressing Mule-WT or the mutant. β-catenin was degraded more quickly in Mule-WT-overexpressing cells and became stabilized in Mule-CA-overexpressing cells compared with in the empty vector-expressing controls (Fig. S2B).
Fig. 5.
Mule destabilizes β-catenin and down-regulates Wnt target gene expression. (A) Immunoblot to detect the indicated proteins in TCL of HEK293T cells that were stably transfected with empty vector or vector-expressing Flag-tagged WT Mule (Mule-WT) or inactive Mule (Mule-CA). Cells were treated (or not) with MG132 for 6 h before harvest. Values shown below the blot are the fold change in β-catenin protein, as determined by quantitation of bands by Image J, followed by normalization first to the loading control and then the vector control. Immunoblot is representative of three independent experiments. (B) Quantitative bar graph showing relative protein fold change of the immunoblot in A. Values were determined by quantitation of bands by Image J, followed by normalization first to the loading control and then to 293-Vec. (C) quantitative (q)RT-PCR analysis of relative transcript levels of the indicated genes in HEK293T cells that were stably transfected with empty vector or vector-expressing Flag-tagged WT Mule (Mule-WT) or inactive Mule (Mule-CA). Results are the mean ± SD (n = 4). (D) Immunoblot to detect the indicated proteins in HCT116 cells that were transfected with siRNA specific for Mule or control siRNA. Immunoblot is representative of three independent experiments. (E) Quantitative bar graph showing relative protein fold change of the immunoblot in D. Values were determined by quantitation of bands by Image J, followed by normalization first to the loading control and then to si-Control. (F) qRT-PCR analysis of relative transcript levels of the indicated genes in HCT116 cells that were transfected with siRNA specific for Mule or control siRNA. Results are the mean ± SD (n = 3). ns, not significant. *P < 0.05; **P < 0.01.
Fig. S2.
CHX treated 293 cells with overexpressed Flag Mule or Flag CA. (A) qRT-PCR analysis of relative transcript levels of β-catenin in HEK293T cells that were stably transfected with empty vector or vector-expressing Flag-tagged WT Mule (Mule-WT) or inactive Mule (Mule-CA). Results are the mean ± SD (n = 3). (B) Two hundred ninety-three cells transfected with Mule (293 Mule), Mule CA (293 Mule CA), and vector (293 Vec) were treated with cycloheximide 100 μg/mL and harvested at the indicated points. Cell lysates were immunoblotted for Flag-Mule and β-catenin, using Abs against Flag and β-catenin, respectively. Immunoblot is representative of three independent experiments.
We further demonstrated the ability of Mule to promote the degradation of β-catenin by examining the effect of siRNA-mediated Mule knockdown in HCT116 cells. HCT116 cells carry a β-catenin allele with a mutation in exon 3 that deletes the GSK3β phosphorylation site, resulting in constitutive activation of the β-catenin protein. We found that steady-state levels of β-catenin were higher in HCT116 cells with Mule knockdown compared with HCT116 cells treated with control siRNA, but not the gene expression levels of β-catenin (Fig. 5 D and E and Fig. S3 A and B). Consistent with this result, AXIN2 and CCND1 mRNAs were up-regulated twofold in Mule knockdown HCT116 cells compared with Mule-expressing controls (Fig. 5F).
Fig. S3.
Mule targets β-catenin for degradation. (A) Immunoblot to detect the indicated proteins in HCT116 cells transfected with two different siRNAs specific for Mule and two control siRNA. Control and Mule-specific knockdown was performed three times with similar results. (B) qRT-PCR analysis of relative transcript levels of β-catenin in HCT116 cells that were transfected with siRNA specific for Mule or control siRNA. Results are the mean ± SD (n = 3).
Last, to further understand the mechanism behind Mule’s regulation of β-catenin, we costained Apcmin and Mule cKO Apcmin organoids with Mule and β-catenin antibody (Ab) to visualize their colocalization. We were able to capture their colocalization in the nucleus (Fig. S4). The merged image for Apcmin appears purple, which is clearly absent in the Mule cKO Apcmin representative organoid shown. Collectively, these results establish that Mule can regulate the stability of β-catenin via ubiquitination, leading to proteasomal degradation.
Fig. S4.
Mule and β-catenin colocalize in the nucleus. Confocal microscopy of representative images for Apcmin and Mule cKO Apcmin organoids stained with β-catenin (green) and Mule (red). Presence of colocalization appears purple in the merged image. (Scale bar, 10 μm.) Three independent experiments were performed.
Discussion
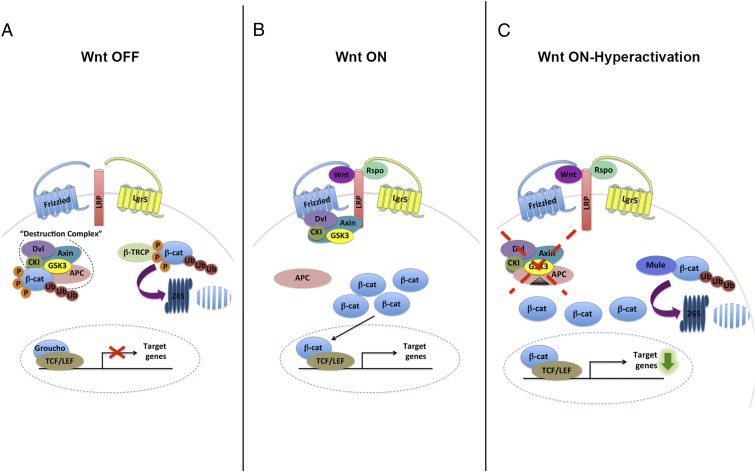
Mule, similar to many other E3 Ub ligases, regulates numerous substrates. Most Mule substrates have been discovered in vitro, and some have been found to have opposing roles in cell proliferation and tumorigenesis, such as p53 and c-Myc (13, 14). For these reasons, the role of Mule in various tissues, whether at steady state or in disease, had been elusive. During the last year, we and others have exploited different Mule knockout mouse models to demonstrate that Mule is involved in the regulation of intestinal, hematopoietic, and neural stem cells (8, 15, 16). Significantly, the developmental programs of all these tissues are driven by the Wnt pathway (17–19). In the absence of Wnt, the protein stability of β-catenin is strictly controlled by the destruction complex, resulting in its phosphorylation and subsequent ubiquitination by β-TRCP (Fig. 6A). In the presence of Wnt, β-catenin can accumulate and translocate into the nucleus to activate the Wnt transcriptional program (Fig. 6B). Rspondin/Lgr5 is thought to further enhance the Wnt signal (20). In our study, we show that, at least in a hyperproliferative setting, Mule can serve as a “back up” E3 Ub ligase to β-catenin to target it for degradation, and thereby help to quench Wnt signaling in the intestine (Fig. 6C). Our data suggest the association between Mule and β-catenin occurs under conditions when both proteins are highly expressed (Fig. 4D). We have also demonstrated that the interaction is not phosphorylation-dependent (Fig. S1A). It remains to be determined whether this regulation is tissue-specific or extends to other Wnt-driven tissues.
Fig. 6.
Model of the Wnt signaling pathway and the role of Mule during its constitutive activation. (A) In the absence of Wnt ligand, β-catenin is sequestered by the destruction complex. On phosphorylation, β-catenin is ubiquitinated by β-TRCP and degraded by the proteasome. No Wnt target genes are transcribed. (B) Wnt associates with Frizzled and LRP receptors, resulting in disruption of the destruction complex. R-spondin binds to Lgr5, which is believed to also associate with LRP receptors to enhance Wnt signaling. β-catenin is free to accumulate in the cytoplasm, translocate into the nucleus, and associate with the T-cell factor/lymphoid enhancing factor transcription factor to activate the transcription of Wnt target genes. (C) During constitutive activation of the pathway, as when APC is mutated (black triangle) rendering the destruction complex inactive, Mule targets β-catenin for degradation by the proteasome.
In our studies, we used 293T cells because they respond better to Mule overexpression than many other cell lines, allowing us to study the biology behind Mule as an E3 ligase. Although we claim that Mule and β-catenin interact in instances of hyperactive Wnt signaling, we are uncertain about the status of Wnt signaling in 293T cells. What we have gathered from generating the data for this manuscript is that the expression level of Mule matters a great deal to the biological outcome. In mice, we use conditional knockouts and compare them with WT littermates, where the expression of Mule as gauged by Western blot is significantly lower. We believe that in such a scenario, Mule is more selective of its substrates and only targets proteins for which it has the highest affinity for degradation. When Mule is expressed at a high level (that which is achieved in 293 cells), it is more promiscuous. Thus, hyperactive Wnt signaling is equivalent to elevated levels of Mule.
Our data have shown that ablation of Mule in mice of the Apcmin background heightens Wnt signaling to the point that intestinal crypts, which are isolated from these animals and placed in culture, quickly adopt the undifferentiated spheroid cyst-like morphology of cancer stem cells (10, 11). We have shown previously that Mule cKO organoids alone also adopt the same undifferentiated spheroid cyst-like morphology after 66 d in culture (8). We believe that the difference in latency is the increased Wnt feedback that is present in the Mule cKO Apcmin organoids. Although it would be interesting to speculate that the loss of Mule contributes to the acquisition of mutations in Apc or other Wnt components to drive this phenotype, our previous RNA sequencing of Mule cKO adenomas suggests this is not the case but, rather, the loss of Mule may function as the Wnt initiating event. Should this hold to be true, our Mule cKO organoid should grow independent of exogenous R-spondin. This is a key experiment that cannot be carried out in organoids derived from mice on an Apcmin background. We have never conducted this experiment in our Mule cKO organoids, but expect to do so in the future. For now, we acknowledge that this phenotype may not be entirely a result of loss of Mule-mediated regulation of β-catenin, but we can conclude that Mule plays an important part in fine-tuning Wnt signaling in both the normal and diseased intestine. These findings are significant because Wnt signaling is the main proliferative pathway in the gut that controls cell fate along the crypt/villus axis (21). Thus, in the absence of Mule, constitutive Wnt signaling can drive stem cell expansion and the conversion of a normal stem cell into a cancer stem cell.
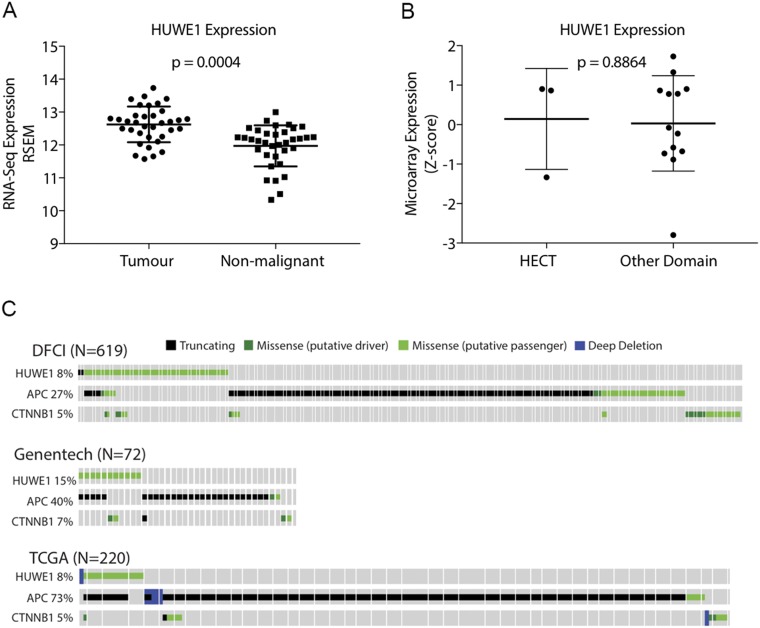
Mule has been shown to be overexpressed in cancers of the breast, lung, colon, liver, pancreas, thyroid, prostate, and larynx (14, 22). It is also subject to somatic mutations, homozygous deletion, or amplification (www.cbioportal.org). In respect to colon cancer, Mule is overexpressed in 50% of all cases; moreover, its expression is higher in tumor samples compared with nonmalignant tissue, and increases with tumor grade (Fig. S5A) (14) (cancergenome.nih.gov/publications/publicationguidelines). Mutations within Mule’s HECT (homologous to the E6-AP carboxyl terminus) domain have been identified in patients with colon and rectal adenocarcinomas (sanger.ac.uk). We searched available databases to specifically determine whether mutations within Mule, whether in its functional (HECT) domain or elsewhere, would correlate differentially with expression. Although we found no significant difference in expression with mutational site, no conclusion can be made, given the small number of tumors reported (Fig. S5B). At this point, we favor the hypothesis that a mutation within Mule, regardless of location, would alter its structure in such a way that it either will not effectively bind its substrates or will render it inactive.
Fig. S5.
Mutation and expression profiling in patient samples. (A) HUWE1 (Mule) gene expression (based on RNA-seq) for matched tumor and nonmalignant pairs (n = 35) was obtained from the University of California, Santa Cruz’s Cancer Genomics Browser. Expression between tumor and nonmalignant colorectal tissues was compared using a Wilcoxon matched-pairs signed rank test. TCGA data include mutations and putative copy number alterations. Genentech and DFCI cohorts include mutation data only. (B) HUWE1 mutation and microarray gene expression data for the TCGA’s colorectal adenocarcinoma cohort were obtained from cBioPortal. Tumors with missense mutations in HUWE1 were divided into those harboring mutations within and outside of the HECT domain. (C) Oncoprints for HUWE1, APC, and CTNNB1(β-catenin) in colorectal adenocarcinoma datasets. Data and plots were obtained from cBioPortal (www.cbioportal.org/). Percentages indicate the fraction of tumors harboring an alteration. Only tumors with alterations in the genes indicated are shown.
Further mutational analyses of available databases show that the mutations that arise within Mule are classified as putative passenger mutations. Although the label undermines the potential significance of Mule in colon cancer, only about a dozen alterations are considered functionally important and positively selected for during colorectal cancer (“putative driver”) (23). The fact that most tumor cohorts reported show that mutations within Mule occur in conjunction with mutations in APC would classify it as a redundant “not significant” hit on the Wnt pathway (Fig. S5C). Our data show, however, that loss of function of both proteins leads to rapid conversion of stem cells into cancer stem cells and consequent stem cell expansion (Fig. 1 C and D) (8). Interestingly, the oncoprints generated from cBioPortal show that some tumors only have mutations within Mule, not APC or β-catenin; we speculate that in these cases, Mule serves as the activating mutation within the Wnt pathway (Fig. S5C; www.cbioportal.org).
Our genetic mouse knockout data thus far strongly support the role of Mule being a tumor suppressor in the gut. We do acknowledge, however, that there are conflicting data published (14, 24), and further acknowledge the possibility that Mule may function differently if it is overexpressed. Thus, we are attempting to understand whether the role of Mule in an established adenoma or tumor would differ from its role in adenoma initiation, and whether Mule overexpression in colon cancer results because of its signaling feedback that may occur if mutations render it inactive, as is classically the case for p53 in cancer (25).
In conclusion, our in vitro and in vivo efforts have shown that Mule’s regulation of the Wnt pathway extends to include its main signal transducer, β-catenin. We have demonstrated that Mule plays a vital role in fine-tuning the Wnt pathway in the normal intestine, as well as in helping extinguish hyperactive Wnt signaling in the diseased intestine. Thus, Mule has an indirect but important influence on intestinal stem cell self-renewal and cell fate.
Materials and Methods
Mice.
Mulefl/fl(y) mice were as described (26) and were crossed to Villin-Cre (27) and Apcmin (28) mice to generate Mulefl/fl(y) Villin-Cre (Mule cKO) mice and Mulefl/fl(y) Villin-Cre Apcmin (Mule cKO Apcmin) mice, respectively. All animal experiments were approved by the University Health Network Animal Care Committee (AUP985).
Organoids.
Mouse intestinal organoids were established from crypts isolated from the proximal small intestine of 2–4-mo-old WT and Mule cKO littermate mice, or Apcmin and Mule cKO Apcmin littermate mice, as described (29). Organoids were maintained in Advanced Dulbecco’s Modified Eagle’s medium/F12 supplemented with 100 U/mL penicillin and streptomycin, 10 mM Hepes, 2 mM Glutamax, 1 × B27, and 1 × N2 (all from Life Technologies). This base medium was further supplemented with 1 mM N-acetylcysteine (Sigma), 50 ng/mL murine recombinant EGF (Invitrogen), 100 ng/mL Noggin (Peprotech), and 5% (vol/vol) R-spondin-1 conditioned medium (C. Kuo, Stanford University).
Cell Lines.
HEK293T cells (ATCC) were cultured in DMEM H21 (Invitrogen). HCT116 cells (ATCC) were cultured in McCoy’s medium. Media were supplemented with 10% (vol/vol) FBS (Sigma) and penicillin/streptomycin.
Immunoblotting.
Organoids were prepared for immunoblotting according to previously reported protocols (30). Pelleted organoids were lysed in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris at pH 8.0) on ice for 30 min and centrifuged at 13,000 rpm (Eppendorf centrifuge 5417R, rotor 45-30-11) for 10 min. The protein content of the supernatants was determined by BCA assay (Pierce). Protein samples (50 µg) were fractionated by 3–8% or 4–12% (vol/vol) reducing SDS/PAGE and transferred to PVDF membranes. Blots were incubated overnight with the following primary Abs: anti-Huwe1 (Bethyl Laboratories), anti-Abc (Cell Signaling, #9562), anti-Cyclin D1 (Cell Signaling, #2978), anti-Axin2 (abcam, ab32197), and anti-β-actin (Abcam). Primary Abs were detected with HRP-conjugated secondary Ab against rabbit IgG (GE Healthcare). Proteins were visualized using enhanced luminol-based chemiluminescence (GE Healthcare). As appropriate, bands on immunoblots were quantified with Image J densitometric software.
Microarray: Analysis Procedure.
The microarray gene-expression data from the Affymetrix scanner were read using the Oligo R package and then processed downstream using the Limma R package from the Bioconductor suite of tools. The background-corrected and RMA-normalized intensity values were fitted into linear models, after which fold changes of differential gene expression between selected contrasts were computed [Mule cKO Apcmin, Apcmin, and Mule cKO, all vs. Mulefl/fl(y) (WT)].
Heat Map.
The gene expression fold change values from the reduced list of 383 genes were also plotted as a heat map, with three columns showing the different comparisons (Fig. 3A). The rows of the heat map are hierarchically clustered, using Euclidean distance and complete linkage. The most interesting genes, which are clearly up-regulated in Mule cKO Apcmin compared with Mulefl/fl(y), are labeled.
Pathway Analysis.
The gene symbols of the most interesting genes that are marked in the heat map were fed into the pathway analysis tool Enrichr (nar.oxfordjournals.org/content/early/2016/05/03/nar.gkw377.full.pdf). This tool compares the input gene list against preannotated gene sets in its database and outputs ranked lists of enrichment terms (pathways, cell lines, or diseases), one per gene-set library. The most highly ranked enrichment terms for the input gene list provide knowledge about the list. In our case, the tool was used to enlist the top 10 enriched pathways in the KEGG Database.
qRT-PCR.
RNA was isolated from organoids and cells with TRIzol (Invitrogen), according to the manufacturer’s instructions. RNA was processed by RT-PCR as described (8). Primer sequences are available on request.
Immunoprecipitation.
Cellular interaction of Mule and β-catenin was carried out in HEK293T stable cell lines overexpressing Flag-tagged Mule or vector. Total cell lysates (TCL) were prepared in lysis buffer [50 mM Tri at pH 8.0, 150 mM NaCl, 0.6% Nonidet P-40, 1× protease inhibitor mixture (Roche)]. For immunoprecipitation studies, TCL were incubated at 4 °C overnight with anti-Flag M2 Ab (Sigma #F3165) to immunoprecipitate Flag-tagged Mule or incubated with an Abc Ab (CST #8480) to immunoprecipitate endogenous β-catenin. Cellular interaction of endogenous Mule and β-catenin was carried out in HCT116 cell lines. TCL were incubated at 4 °C overnight with anti-Mule Ab (monoclone Ab 7BF12) to immunoprecipitate endogenous Mule or incubated with an Abc Ab (CST #8480) to immunoprecipitate endogenous β-catenin. IgG was used as a negative control. In both cases, the immune complexes bound to Protein A/G resin were washed 5× in lysis buffer, detached from the agarose using SDS/PAGE loading buffer, and detected by immunoblotting using anti-Flag (or anti-Mule) and Abc Abs.
In Vitro and GST Pull-Down Assays.
Cell lysates of HEK293T overexpressing Mule-WT or empty vector (3 mg) were mixed with 20 μL Anti-Flag M2 agarose (Sigma #M8823) precleared with 5% (wt/vol) BSA at 4 °C in 1 × PBS buffer for 2 h. After incubation, the Anti-Flag M2 agarose resin was washed extensively with a buffer of 50 mM Tris at pH 7.6, 500 mM NaCl, 10% (vol/vol) glycerol, 0.1% Triton X-100, and protease inhibitor mixture (Roche) and mixed with recombinant His6 tagged β-catenin (20 μg) in 500 μL binding buffer containing 50 mM Tris at pH 7.6, 150 mM NaCl, 10% (vol/vol) glycerol, 0.1% Triton X-100, and protease inhibitor mixture at 4 °C for 2 h. After second incubation, Anti-Flag M2 agarose was washed extensively with the same buffer, and the resulting immobilized proteins were detected using immunoblotting. GST pull-down assay was carried out by mixing an equal amount of recombinant GST-tagged N-terminal truncated Mule (3760–4374) and His6 tagged β-catenin (20 μg) in the binding buffer at 4 °C for 2 h. After extensive wash with the binding buffer, the immobilized proteins were eluted using reduced glutathione and detected by immunoblotting using anti-His (Novagen #71841) or GST Abs (Thermo #8–326). Recombinant GST tagged N-terminal truncated Mule (3760-4374) and His6 tagged β-catenin were expressed in E. coli BL21 cells and purified using standard GST- and Ni-affinity chromatography methods.
Protein Stability: Cycloheximide Chase Assay.
Cells were cultured with cycloheximide at a concentration of 100 μg/mL and incubated for various hours. Cells were harvested at the indicated points. Lysates were prepared and subjected to immunoblotting analysis. The protein stability was determined by percentage of β-catenin remaining at an indicated point compared with the initial point. Data were plotted from three independent trials.
In vivo ubiquitination.
HEK293T stable cell lines overexpressing FLAG-tagged WT Mule or inactive Mule C4341A were cultured as described earlier. Near-confluent cells were transfected with pCDNA 3.1-HA-Ub, using PolyJet transfection reagent (FraggaBio), recovered after 6 h, and grown for a total of 48 h. Cells were treated with 25 μM MG132 for 4 h before harvesting, and TCL were prepared in lysis buffer of 50 mM Tris at pH 8.0, 150 mM NaCl, 0.6% Nonidet P-40, 1 × protease inhibitor mixture (Roche) with 10 mM freshly prepared N-ethylmaleimide. β-catenin was immunoprecipitated from TCL by incubating with Abc Ab (CST #8480) and Protein A/G resin. Ubiquitinated β-catenin was detected on immunoblots using anti-Ub Ab (P4G7, Covance MMS-258R) and Abc Ab (CST #8480).
In vitro ubiquitination.
Recombinant His-tagged β-catenin (full-length), GST-Mule (3760-4374) were expressed in E. coli BL-21 DE3 strain and purified using standard affinity purification methods. Ubiquitination was performed as previously described (22496338) by mixing 10 µM each E1, E2, and Ub and various amounts of GST-Mule as the E3 ligase and His-tagged β-catenin as the substrate in reaction buffer (50 mM Tris at pH 7.6, 2 mM ATP, 5 mM MgCl2, 2.5 mM DTT). Reactions were carried out for 90 min at 30 °C and terminated by adding SDS/PAGE loading buffer. Ubiquinated β-catenin was detected by immunoblotting using anti-Ub (P4G7, Covance MMS-258R) and Abc (CST #8480) Abs.
siRNA knockdown.
HCT116 cells were transfected with siRNA against Mule (AAUUGCUAUGUCU CUGGGACA) (13) or negative control siRNA (both from Genepharma). siRNAs were transfected into cells using Lipojet (SignaGen) according to the manufacturer’s protocol, and cells were harvested at 48 h posttransfection. Lysates were resolved by 7.5% (vol/vol) SDS/PAGE, followed by immunoblotting, as described earlier.
Immunostaining.
Organoids were collected in cold medium, pelleted and resuspended in cold Cell Recovery Solution (BD). After incubation on ice for 20 min with gentle mixing, organoids were washed in PBS and were allowed to settle by gravity to a pellet in the bottom of the tubes. The pellet was resuspended in 4% (vol/vol) paraformaldehyde and incubated for 20 min with gentle mixing. Organoids were washed twice with 1 × PBS, resuspended in Histogel (Thermo Scientific) according to manufacturer’s instructions, processed, and embedded in paraffin. Sections (4.5 μm) were rehydrated and incubated with blocking solution for 30 min, then with β-catenin (CST #8480) and Mule (homemade mouse, clone 7B15) Abs overnight at 4 °C. Slides were washed with PBS and incubated with secondary Abs (Jackson Donkeyxmouse-Cy5 #715–175-150 and goatxrabbit-Cy3 #111–165-144) for 35 min at room temperature. After washing with PBS, they were treated with DAPI and mounted in Entellan mounting medium. Fluorescent images were made from an Olympus FV1000 confocal microscope and Zeiss AxioImager wide-field fluorescence microscope.
Clinical Data Profiling.
Normalized RNA-seq derived gene expression data were downloaded from the University of California, Santa Cruz's Cancer Genomics Browser (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/) (31). Genomic alterations affecting APC, CTNNB1, and HUWE1 were queried in publicly available TCGA (The Cancer Genome Atlas), DFCI (Dana-Farber Cancer Institute), and Genentech colorectal adenocarcinoma datasets using cBioPortal (cbioportal.org) (32–36). HUWE1 (Mule) mutation and microarray Z-score normalized expression data were downloaded from cBioPortal. Statistical analyses were performed using GraphPad Prism software.
Statistical Analyses.
P values for intergroup comparisons were determined using the unpaired Student t test. P < 0.05 was considered significant. Unless otherwise indicated, all experiments contained 3 replicates per condition, and all experiments were repeated at least 3 times.
Acknowledgments
We are grateful to K. Gill of the Genotyping Facility, Dr. Chao Lu of the Center for Applied Genomics at the Hospital for Sick Children, and the staff of the Animal Resource Centre at the Princess Margaret Hospital (Toronto, Ontario, Canada). We thank M. Saunders for scientific editing of the manuscript. We thank the TCGA Research Network for sharing their comprehensive genomic data for colorectal adenocarcinoma tumors (cancergenome.nih.gov/publications/publicationguidelines). This research was supported by grants from the Canadian Institutes of Health Research (to T.W.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621355114/-/DCSupplemental.
References
- 1.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 2.Brown AM. Wnt signaling in breast cancer: Have we come full circle? Breast Cancer Res. 2001;3(6):351–355. doi: 10.1186/bcr321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinfeld B, et al. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275(5307):1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10(7):468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 7.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int J Mol Sci. 2013;14(8):16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Brauer C, et al. Mule regulates the intestinal stem cell niche via the Wnt pathway and targets EphB3 for proteasomal and lysosomal degradation. Cell Stem Cell. 2016;19(2):205–216. doi: 10.1016/j.stem.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Groot RE, et al. Huwe1-mediated ubiquitylation of dishevelled defines a negative feedback loop in the Wnt signaling pathway. Sci Signal. 2014;7(317):ra26. doi: 10.1126/scisignal.2004985. [DOI] [PubMed] [Google Scholar]
- 10.Fatehullah A, Appleton PL, Näthke IS. Cell and tissue polarity in the intestinal tract during tumourigenesis: Cells still know the right way up, but tissue organization is lost. Philos Trans R Soc Lond B Biol Sci. 2013;368(1629):20130014. doi: 10.1098/rstb.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121(7):1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 14.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 15.King B, et al. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat Immunol. 2016;17(11):1312–1321. doi: 10.1038/ni.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbán N, et al. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353(6296):292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes Dev. 2005;19(8):877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 18.Lento W, Congdon K, Voermans C, Kritzik M, Reya T. Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol. 2013;5(2):a008011. doi: 10.1101/cshperspect.a008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lie DC, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437(7063):1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 20.Schuijers J, Clevers H. Adult mammalian stem cells: The role of Wnt, Lgr5 and R-spondins. EMBO J. 2012;31(12):2685–2696. doi: 10.1038/emboj.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radtke F, Clevers H. Self-renewal and cancer of the gut: Two sides of a coin. Science. 2005;307(5717):1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 22.Confalonieri S, et al. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene. 2009;28(33):2959–2968. doi: 10.1038/onc.2009.156. [DOI] [PubMed] [Google Scholar]
- 23.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 24.Peter S, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6(12):1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues NR, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci USA. 1990;87(19):7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Z, et al. The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med. 2012;209(1):173–186. doi: 10.1084/jem.20111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277(36):33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 28.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256(5057):668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 29.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 30.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21(3):256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 31.Cline MS, et al. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannakis M, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;17(4):1206. doi: 10.1016/j.celrep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]