Significance
Despite its widespread application to the species delimitation problem, our study demonstrates that what the multispecies coalescent actually delimits is structure. The current implementations of species delimitation under the multispecies coalescent do not provide any way for distinguishing between structure due to population-level processes and that due to species boundaries. The overinflation of species due to the misidentification of general genetic structure for species boundaries has profound implications for our understanding of the generation and dynamics of biodiversity, because any ecological or evolutionary studies that rely on species as their fundamental units will be impacted, as well as the very existence of this biodiversity, because conservation planning is undermined due to isolated populations incorrectly being treated as distinct species.
Keywords: multispecies coalescent, species delimitation, coalescent theory
Abstract
The multispecies coalescent model underlies many approaches used for species delimitation. In previous work assessing the performance of species delimitation under this model, speciation was treated as an instantaneous event rather than as an extended process involving distinct phases of speciation initiation (structuring) and completion. Here, we use data under simulations that explicitly model speciation as an extended process rather than an instantaneous event and carry out species delimitation inference on these data under the multispecies coalescent. We show that the multispecies coalescent diagnoses genetic structure, not species, and that it does not statistically distinguish structure associated with population isolation vs. species boundaries. Because of the misidentification of population structure as putative species, our work raises questions about the practice of genome-based species discovery, with cascading consequences in other fields. Specifically, all fields that rely on species as units of analysis, from conservation biology to studies of macroevolutionary dynamics, will be impacted by inflated estimates of the number of species, especially as genomic resources provide unprecedented power for detecting increasingly finer-scaled genetic structure under the multispecies coalescent. As such, our work also represents a general call for systematic study to reconsider a reliance on genomic data alone. Until new methods are developed that can discriminate between structure due to population-level processes and that due to species boundaries, genomic-based results should only be considered a hypothesis that requires validation of delimited species with multiple data types, such as phenotypic and ecological information.
Major advances in understanding the patterns of biodiversity and the processes that generate those patterns are being made with increasing rapidity, scope, and scale, as we study species across the tree of life (e.g., refs. 1–3). These advances take the boundaries of the species that constitute their fundamental units as known. Identification of these boundaries—that is, species delimitation—currently is making increasing use of genomic data under statistical model-based approaches, typically using the multispecies coalescent model (4), as opposed to traditional taxonomic work based on phenotypic data. Coupled with technological advances for collecting genomic datasets across many individuals per putative species, inference under the multispecies coalescent model provides us with considerable power in identifying recently diverged taxon boundaries. For example, simulations show that we can delimit lineages with extremely short divergence times, as with the popular program BPP (Bayesian Phylogenetics and Phylogeography) (5). However, these remarkable gains have also paradoxically given rise to a new challenge—determining whether what we delimited genetically represents lineages isolated due to speciation or simply within-species population structure. Population genetic structure is ubiquitous among all taxa in which gene flow is reduced by geographic and/or environmental barriers (6). However, the extent of population structure varies as the level of gene flow and time of separation results in more or less evolutionary independence among populations (7). With more loci, there is the possibility of detecting ever-more-fine population structure and potentially confounding population structure with species boundaries. As a consequence, the increased resolution of genomic data makes it possible to not only detect divergent species lineages, but also local population structure within them—that is, a fractal hierarchy of divergences.
Misidentification of population structure as putative species is therefore emerging as a key issue (8) that has received insufficient attention, especially with respect to methodologies for delimiting taxa based on genetic data alone. Because species delimitation is inextricably linked to patterns of species diversity, the models used to delimit species are not just limited to issues about species boundaries, but are also paramount to understanding the generation and dynamics of biodiversity (9–12). Consequently, when the lines become blurred—delimiting species in some cases, but populations in other cases—species delimitation becomes a critical issue with ramifications across multiple fields of study, such as, for example, the estimation of diversity in macroecological studies (13–15), analyses of food webs (16), or conservation, where oversplitting of small, isolated populations based on genetic data could be detrimental (17). More fundamentally, distinguishing between species- and population-level lineages is central to understanding the speciation process itself (18, 19).
Not all populations become species. Instead, speciation theory points to a continuum for the probability that a population lineage will evolve into a new species (20). Depending on the extent and duration of isolation and the form and strength of selection, speciation becomes more or less a protracted process, with new lineages only gradually and stochastically evolving from the initially isolated lineages into true species over time (18, 19, 21). This process is in contrast to commonly applied birth–death models, in which speciation is abstracted as an instantaneous event, such that all divergent lineages are treated as immediately forming true species (22). Here, we investigate the robustness of species delimitation under the multispecies coalescent model when speciation is considered as an extended process rather than an instantaneous event. Specifically, we simulate genetic data under a protracted speciation model (21), such that the degree of genetic structure accumulating among lineages differs (7), depending on the duration of speciation (21), and investigate the accuracy of inferences from the popular model-based species delimitation program BPP (4, 23, 24). Unlike birth–death models, which treat all lineages equivalently, under the protracted speciation model, population lineages are distinguished from species lineages (Fig. 1). In other words, a lineage-splitting event does not correspond to an instantaneous speciation event, but, rather, an incipient speciation representing, for example, the initial isolation of a new population. This incipient species can go extinct or merge back into the parent population at a particular rate, or, alternatively, if remaining isolated for a long enough time, converts or develops into a true species. The phylogeny generated by the protracted speciation process, therefore, consists of a mosaic of population-level as well as species-level lineages (Fig. 1), representing the dynamic and chimeric nature of real-world speciation (7, 20, 25). For example, the opportunities for geographic isolation and/or rates of reproductive isolation differ among taxa (18, 19, 21). A large number of species might be generated under a short period either due to high rates of completion of speciation (e.g., the evolution of reproductive isolation; ref. 25) or many opportunities for initiation of species (e.g., many relatively isolated populations across a landscape that contribute to the initiation of speciation; ref. 26). However, depending on which of these two parameters predominate, the potential for inflating estimates of species diversity through the delimitation of populations may differ substantially among clades.
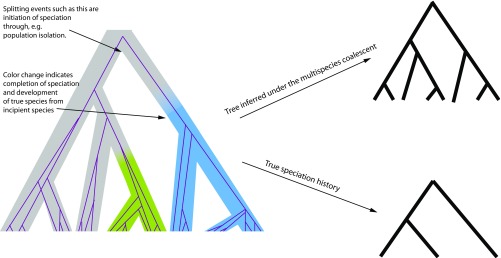
Fig. 1.
(Left) The tree represents the true history upon which the gene genealogies (shown by thin purple lines) are conditioned, with the colors representing species. Note that here we show a single representative gene genealogy, with four individuals sampled per lineage, whereas in practice multiple gene genealogies are sampled for delimiting species. This tree shows speciation as a process rather than speciation as an event. That is, the internal nodes of the containing tree do not represent instantaneous complete speciation events, but, rather, initiation of the speciation process due to microevolutionary processes that result in population isolation. Not all of the lineages that arise due to population isolation develop into true species. For example, some may merge back into the other lineages of the same species in the future if whatever barriers led to their isolation were to be removed (i.e., the evolutionary independence will be ephemeral because two populations belonging to the same species do not have an impediment to reproduction). Some of these isolated incipient species lineages, however, do stochastically develop into true species (indicated by shift in color), so that they will remain distinct lineages with independent evolutionary trajectories that do not merge back into their parental species, even if the isolation barriers were to disappear. (Upper Right) The tree shows the results of inference under the multispecies coalescent using BPP, which includes the structuring both due to species boundaries as well as due to lineage splitting as a result of population isolation. As such, the inference corresponds to the full structural history, but not the true speciation history, which is shown by the tree in Lower Right. That is, the multispecies coalescent does not distinguish between structuring due to population isolation vs. structuring due to speciation: It only identifies genetic structure.
The objective here is to assess this potential degree of bias for inferred patterns of species diversity with the multispecies coalescent model to diagnose species given that (i) the speciation process is not instantaneous (18–21, 25, 26), and (ii) genetic structure is a chimeric pattern resulting from isolation due to both population divergence as well as species boundaries because of the extended nature of the speciation process (18, 19, 21, 26). Note that our study is not restricted to any particular species concept. Instead, it is based simply on the basic principle that there is a distinction between populations and species, which is recognized both empirically and theoretically (7). Different researchers need not agree on how the distinction between populations vs. species might be operationalized or how divergence might proceed (i.e., we model divergence in the absence of gene flow, but other demographic models might be applied; for a model of divergence with gene flow, see ref. 27). Unless the investigator explicitly assumes that all and any genetic structure of any kind is attributable to species boundaries, then distinguishing between these two sources of structure is fundamental to the species-delimitation analysis. Addressing this knowledge gap—can the multispecies coalescent distinguish population-level structure and species boundaries—is critical, given the advocacy for delimiting taxa under the multispecies coalescent (28).
Results
Across all of the species conversion rates, BPP tends to overestimate the number of true species (Figs. 2 and 3). Moreover, at the lowest species conversion rates (0.001 and 0.1), the species delimited by BPP are essentially random with respect to the actual number and specific boundaries of species, averaging 5–13 times more estimated species than actually present in the data (Table 1). It is also worth noting that the errors are all positive: BPP never underestimates the number of true species and only overestimates them (i.e., there is a systematic bias in the inferred number of species). Only at the highest species conversion rates (10 or 1,000 times the rate of species initiation) do the error rates become more moderate, but the inferred number of species is still almost double the actual number of species used in simulations (i.e., three to six more species inferred than the actual number of species, five; Table 1).
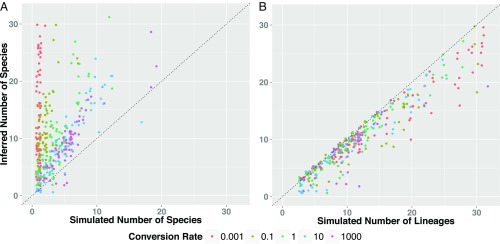
Fig. 2.
The performance of species delimitation under the multispecies coalescent when the data are generated under the protracted speciation model, from simulations run for a fixed duration of time (5 units) under different species conversion rates. Speciation initiation rate was fixed at 0.5, while extinction rates were either 0.0 and 0.2 (the plot does not distinguish between these different extinction rates, because these had no meaningful effect on the main results or our argument). (A) Shown is the number of species per replicate inferred at a 0.95 probability vs. the number of true species on the input tree. (B) Shown is the number of species per replicate vs. the number of lineages (i.e., both true species as well as lineages representing incipient species or population structure). Generally, across all conversion rates, BPP tends to overestimate the number of species. However, what is striking is that BPP does not track species, as seen in A, but, rather, tracks structure of any sort, whether incipient species or true species, as seen in B.
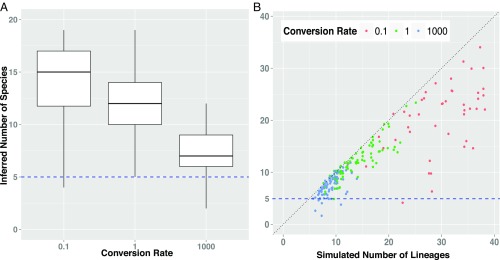
Fig. 3.
The performance of species delimitation under the multispecies coalescent when the data are generated under the protracted speciation model, from simulations run until five true species (indicated with dashed line) were produced under different species conversion rates. Speciation initiation rate was fixed at 0.5, while extinction rates were either 0.0 and 0.2. (A) The number of inferred species at different speciation conversion rates is shown in the box plots. The multispecies coalescent tends to overestimate the number of species when conversion rates are small and there is a long lag between initiation and speciation, because it is diagnosing structure rather than species per se, and does not distinguish between structure due to incipient species or population processes and structure due to actual species. This is clearly shown in B, where the number of lineages (which include incipient or population-level lineages as well as true species lineages) is shown by the diagonal. The inference tends to track the number of lineages, rather than the number of true species (five; shown by the dashed horizontal line), and thus not distinguishing between population or species.
Table 1.
Sizes of species trees with different number of species generated under the protracted speciation process under different species conversion rates, c, the rate at which isolated lineages develop into true species
| Lineages | Species | |||||
| c | Max | Min | Mean | Max | Min | Mean |
| 0.001 | 47 | 3 | 17.35 | 2 | 1 | 1.01 |
| 0.1 | 38 | 4 | 13.29 | 7 | 1 | 1.83 |
| 1 | 36 | 3 | 11.43 | 12 | 1 | 3.75 |
| 10 | 39 | 3 | 10.43 | 17 | 1 | 4.87 |
| 1,000 | 39 | 3 | 10.67 | 19 | 1 | 5.46 |
At the lowest conversion rate (0.001, or 500 times slower than the speciation initiation rate), on average only one actual species was generated in each species phylogeny, even though the number of lineages ranged between 3 and 47. At the highest conversion rate (1,000 or 2,000 times faster than the speciation initiation rate), on average approximately five species were expected on phylogenies that ranged in size from 3 to 39 tips. Results here are based on a birth rate of 0.5 and extinction rates of 0.0 and 0.2 (see Materials and Methods for details).
In contrast, if we focus our assessment on how much structure is recovered by BPP (i.e., the delimitation of lineages, regardless of whether they are true species or incipient divergences reflecting population structure), we see that BPP does very well (Fig. 2). Although BPP tends to generally underestimate the number of lineages, the error values are relatively low: a factor of 0.15 to 0.20, and rms errors (rmses) range from 1.76 to 3.17, across all speciation conversion rates.
Our results show that, given the realities of speciation as an extended process, the phylogeny that conditions the coalescent process is a mosaic of population-level as well as species-level structure (Fig. 1; e.g., ref. 29 vs. ref. 30). The number of true species associated with a phylogeny will be much lower than the number of lineages, and the degree of discrepancy between the number of lineages and true species will vary as a function of the conversion rate—that is, the rate at which isolated lineages develop into true species (Table 2).
Table 2.
Performance of the multispecies coalescent model as implemented in BPP to delimit species under different species conversion rates, c
| Absolute Error | Normalized | ||||
| c | Max | Min | Mean | Error | RMSE |
| 0.001 | 31.00 | 2.00 | 13.16 | 12.96 | 13.16 |
| 0.1 | 29.00 | 1.00 | 9.25 | 5.65 | 9.25 |
| 1 | 20.00 | −1.00 | 6.01 | 1.83 | 6.04 |
| 10 | 14.00 | −4.00 | 3.70 | 0.92 | 3.88 |
| 1000 | 11.00 | −3.00 | 3.46 | 0.72 | 3.68 |
“Absolute error” is the difference between the number of inferred species and the number of true species in the simulation. “Normalized error” is the absolute error divided by the number of true species simulated. rmse is the square root of the square of the normalized error.
Discussion
The multispecies coalescent model diagnoses genetic structure and not species. Specifically, we show that the number of “species” identified by the multispecies coalescent actually reflects the genetic structure of the data, which includes both population structure within species as well as structure between species. Note that the focus of our argument is on the validity of species delimitation under the multispecies coalescent model in general, and not on the particular implementation of this model as exemplified by BPP. There are other programs that implement species delimitation under the multispecies coalescent, but they all use the same underlying model, and differences in results in terms of the accuracy and level of resolution of the genetic structure will be attributable to particularities of the implementation. Regardless of which implementation is used, as long as the program or implementation uses the multispecies coalescent, our central thesis holds: what is diagnosed is genetic structure, with no distinction between structure due to populations or due to species. What we present here are the conceptual, rather than statistical or computational, implications of using the multispecies coalescent model for species delimitation, and these implications are invariant with respect the particular program or approach that implements inference under this model.
Likewise, we use the protracted speciation model (21) because it allows for the generation of data that include genetic structure due to both population isolation and species. Obviously, data might have been simulated under alternative demographies (e.g., divergence with gene flow; ref. 27). Irrespective of the divergence process used to generate the genetic data, the conclusions of the manuscript will not change radically—the multispecies coalescent does not distinguish between genetic structure due to population vs. species. It is possible that, under certain specific conditions, the erroneous inferences under the multispecies coalescent may be lower than those we document here. For example, the rate of gene flow between populations may be high enough so that the structure due to populations will not be detected under the multispecies coalescent, yet, conversely, gene flow between species may be low enough such that the structure due to species boundaries is strongly evident. However, in cases like this, the multispecies coalescent has not suddenly gained the ability to distinguish between genetic structure due to populations vs. structure due to species. Rather, this apparent discrimination is an artefactual one due to contriving the relative gene flow rates between populations so as to erode the signal due to population structure below the detection threshold of the multispecies coalescent, while at the same time maintaining the signal due to species structure above this threshold.
Consequently, if all the multispecies coalescent can identify is genetic structure, it is not a justifiable model for delimiting species, irrespective of which species concept an investigator might apply. Our results indicate that the only way that BPP in its current form can be validly used as a species delimitation approach is if external information is used to make the determination. This external information can be an a priori hypothesis or knowledge that all of the structure in the genetic data identified by BPP do indeed correspond to species rather than populations, or, equivalently, but perhaps much more unrealistically, the rate of speciation completion is so rapid compared with the rate of population isolation that species instantaneously form. Alternatively, morphological, ecological, ethological, or other classes of data must be used to correctly attribute the elements of structure delimited by BPP to either species-level (31) or population-level processes.
Dense spatial sampling of individuals and genomes, which increasingly characterize many current studies, is leading more and more to datasets where the primary structuring is a mix between lineages that have undergone speciation as well as others that reflect spatial structuring of populations (32). We expect this to be particularly a problem in cases where species delimitation analysis is most needed—recent, rapid radiations, where the processes promoting divergence will result in both species and population structure. Even if it is recognized by researchers using programs that implement the multispecies coalescent for species delimitation that what is being delimited is not so much species but structure—which the researchers are assuming to be coincident with species boundaries—this distinction may be lost when their results are used by other studies or efforts (33). These efforts include not only research in other fields, such as ecology or evolutionary biology, but also in more applied contexts, such as conservation reporting, management, and policy development, either at local or global levels.
When the results of species delimitation under the multispecies coalescent are used in secondary studies, the fact that the species identities follow from a postanalytical assumption of the original researchers that the genetic structure corresponds only to species boundaries and not populations, rather than a fundamental result of the analysis itself, is lost. Not only will the possible inflation of species numbers have consequences that may go beyond the immediate findings of these secondary studies or reports, but an important source of error will be overlooked by reviewers, readers, or other consumers of these secondary studies. As such, until we develop genomic-based species delimitation approaches that are able to discriminate between population- and species-level structuring, it is important not just to recognize, but for researchers to treat and report the units delimited under the multispecies coalescent at best as tentative hypotheses of species, to be confirmed or rejected through subsequent analysis or application of other data or information, rather than as true species as such.
Materials and Methods
Our approach consisted of the following steps:
-
1.
Generation of trees with population-level (incipient species) and species-level (true species) lineages, under a model that explicitly models the processes of speciation initiation and speciation completion.
-
2.
Generation of coalescent gene genealogies conditioned by the structure of the above trees under the multispecies coalescent, with multiple individuals per lineage of the structuring tree for multiple independent loci.
-
3.
Generation of sequence data alignments on gene trees for each locus.
-
4.
Inference of species trees based on sequence data using a multispecies coalescent inference program.
-
5.
Comparison of the inferred vs. actual number of species in the original species tree.
The simulation of the species trees was carried out by using the “ProtractedSpeciationProcess” class of DendroPy (34), which provides for sampling trees from the protracted speciation model. The protracted speciation model is described in detail in refs. 21, 35, and 36, and we refer the reader to those works for more information, because the focus of the current work is not on the protracted speciation model as such. Here, we use the protracted speciation model as a generative model that allows us to simulate speciation as an extended process rather than an event, with a lag between initial population isolation or divergence of a lineage from an ancestral species and its development into true species. In the original protracted speciation model terminology, a lineage on an isolated evolutionary track that has not yet developed into a true species is known as an incipient species, whereas lineages that have developed into true species are known as “full” or “good species” (here we use the term “true” species for this concept). In contrast to the simple birth–death model, which simulates speciation as instantaneous events and has only two parameters, a birth and death rate, the protracted speciation model has five parameters: the incipient species birth rate (the rate at which incipient species produce new incipient species), the true species birth rate (the rate at which true species produce new incipient species), the incipient species extinction rate (the rate at which incipient species lineages become extinct or merge back into their parent species), the true species extinction rate (the rate at which true species lineages go extinct), and the species conversion rate (the rate at which incipient species develop into true species).
In our study, we set both the incipient species birth rate as well as the true species birth rate to be equal (i.e., a common “species initiation rate”). Similarly, we set both the incipient species extinction rate as well as the true species extinction to be equal (i.e., a common “extinction” rate). Thus, our use of the protracted speciation model can be considered to produce trees under the conventional birth–death process, with the birth rate corresponding to the species initiation rate and the death rate corresponding to the extinction rate, with lineages on the resulting tree transitioning to true species at the given conversion rate.
Two classes of simulation regimes were used: a “fixed duration” regime, where the simulations were run for a total of 5.0 time units (resulting in varying numbers of total lineages and true species), and two “fixed species number” regimes, where the simulations were run until five true species were generated on each phylogeny (with varying numbers of total lineages per phylogeny).
For the fixed-duration regime, 20 phylogenies were simulated under each combination of the following parameters for 5.0 time units:
-
•
Species initiation rate: 0.5
-
•
Species extinction rate: 0.0, 0.2
-
•
Species conversion rate: 0.001, 0.1, 1.0, 10.0, and 1,000.0.
For the fixed-species-number regime, 20 phylogenies consisting of five true species were simulated under each combination of the following parameters:
-
•
Species initiation rate: 0.5
-
•
Species extinction rate: 0.0, 0.2
-
•
Species conversion rate: 0.1, 1.0, and 1,000.0.
For all subsequent stages of analysis, we use the phylogeny so produced (i.e., a mosaic consisting of both true species lineages, as well as the incipient species lineages) as the species tree.
Gene trees were simulated under the multispecies coalescent model by using the “ContainingTree” class of DendroPy (34). The phylogenies sampled from the protracted speciation process in the previous step were used as the containing tree, with haploid population sizes of each lineage fixed to 100,000, and 10 individual genes sampled from each lineage. Sequence alignments were simulated on the gene trees produced in the previous step by using Seq-Gen. A total of 10 loci, consisting of 1,000 characters in each locus, were simulated for each individual on each gene tree under the Jukes–Cantor model using two different mutation rates: and mutations per site per unit of time.
The set of alignments generated for each species tree were passed to BPP along with the corresponding species tree as source data. BPP was set to use the (true) species tree as a guide tree with searches under algorithm 0 (i.e., mode “10”); consequently, any errors in the delimitation of species was not due to upstream analyses (37). Uniform rooted trees were used as the species model prior. Priors for the and parameters were set such that the mean corresponded to the true (simulated) values of each input dataset. A total of 10,000 samples from the posterior were used, with samples taken every 100 generations after automatic fine-tuning of proposal parameters and a burn-in of 4,000 samples.
The BPP results were processed such that only clades with a 0.95 or greater posterior probability were interpreted as true species: The summary tree produced by BPP was traversed in preorder, and any internal node in the original species tree that had a posterior probability of 0.95 or greater was retained, whereas all other internal nodes were collapsed. The number of tips in the postprocessed tree was taken to the be the number of species inferred by BPP with a posterior probability of 0.95 or greater.
All plots were made by using the R package ggplot2.
Acknowledgments
We thank Dr. Mark T. Holder at the University of Kansas for his generosity with the high-performance computing resources of his lab, without which this work would not have been possible. We also thank four anonymous reviewers for thoughtful comments and suggestions, all of which considerably improved and strengthened this paper. This work was supported by National Science Foundation Grant DEB 11-45989 (to L.L.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:1–5. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinchliff CE, et al. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci USA. 2015;112(41):12764–12769. doi: 10.1073/pnas.1423041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci USA. 2010;107(20):9264–9269. doi: 10.1073/pnas.0913022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Zhang DX, Zhu T, Yang Z. Evaluation of a Bayesian coalescent method of species delimitation. Syst Biol. 2011;60:747–761. doi: 10.1093/sysbio/syr071. [DOI] [PubMed] [Google Scholar]
- 6.Avise JC. 2000. Phylogeography: The History and Formation of Species (Harvard Univ Press, Cambridge, MA)
- 7.Hey J, Pinho C. Population genetics and objectivity in species diagnosis. Evolution. 2012;66(5):1413–1429. doi: 10.1111/j.1558-5646.2011.01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens B, Pelletier T, Reid N, Satler J. How to fail at species delimitation. Mol Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 9.Moritz C, Schneider C, Wake B. Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Syst Biol. 1992;41:273–291. [Google Scholar]
- 10.Agapow P, et al. The impact of species concept on biodiversity studies. Q Rev Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- 11.de Quieroz K. Different species problems and their solutions. Bioessays. 2005;26:67–70. doi: 10.1002/bies.20325. [DOI] [PubMed] [Google Scholar]
- 12.de Quieroz K. Ernst Mayr and the modern concept of species. Proc Natl Acad Sci USA. 2005;102:6600–6607. doi: 10.1073/pnas.0502030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaac NJ, Mallet J, Mace GM. Taxonomic inflation: Its influence on macroecology and conservation. Trends Ecol Evol. 2004;19(9):464–469. doi: 10.1016/j.tree.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Payo DA, et al. Extensive cryptic species diversity and fine-scale endemism in the marine red alga Portieria in the Philippines. Proc Biol Sci. 2013;280(1753):20122660. doi: 10.1098/rspb.2012.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vodă R, Dapporto L, Dincă V, Vila R. Cryptic matters: Overlooked species generate most butterfly beta-diversity. Ecography. 2015;38(4):405–409. [Google Scholar]
- 16.Hambäck PA, et al. Bayesian species delimitation reveals generalist and specialist parasitic wasps on Galerucella beetles (Chrysomelidae): Sorting by herbivore or plant host. BMC Evol Biol. 2013;13(1):92. doi: 10.1186/1471-2148-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankham R, et al. Implications of different species concepts for conserving biodiversity. Biol Conserv. 2012;153:25–31. [Google Scholar]
- 18.Rosenblum EB, et al. Goldilocks meets Santa Rosalia: An ephemeral speciation model explains patterns of diversification across time scales. Evol Biol. 2012;39(2):255–261. doi: 10.1007/s11692-012-9171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dynesius M, Jansson R. Persistence of within-species lineages: A neglected control of speciation rates. Evolution. 2014;68(4):923–934. doi: 10.1111/evo.12316. [DOI] [PubMed] [Google Scholar]
- 20.Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24(3):145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Etienne RS, Morlon H, Lambert A. Estimating the duration of speciation from phylogenies. Evolution. 2014;68(8):2430–2440. doi: 10.1111/evo.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nee S. Birth-death models in macroevolution. Annu Rev Ecol Evol Syst. 2006;37:1–17. [Google Scholar]
- 23.Yang Z, Rannala B. Unguided species delimitation using DNA sequence data from multiple loci. Mol Biol Evol. 2014;31:3125–3135. doi: 10.1093/molbev/msu279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z. The BPP program for species tree estimation and species delimitation. Curr Zool. 2015;61(5):854–865. [Google Scholar]
- 25.Weir JT, Wheatcroft DJ, Price TD. The role of ecological constraint in driving the evolution of avian song frequency across a latitudinal gradient. Evolution. 2012;66(9):2773–2783. doi: 10.1111/j.1558-5646.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 26.Botero CA, Dor R, McCain CM, Safran RJ. Environmental harshness is positively correlated with intraspecific divergence in mammals and birds. Mol Ecol. 2014;23(2):259–268. doi: 10.1111/mec.12572. [DOI] [PubMed] [Google Scholar]
- 27.Heled J, Bryant D, Drummond AJ. Simulating gene trees under the multispecies coalescent and time-dependent migration. BMC Evol Biol. 2013;13(1):44. doi: 10.1186/1471-2148-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol. 2012;27(9):480–488. doi: 10.1016/j.tree.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Knowles LL, Carstens BC. Estimating a geographically explicit model of population divergence. Evolution. 2007;61(3):477–493. doi: 10.1111/j.1558-5646.2007.00043.x. [DOI] [PubMed] [Google Scholar]
- 30.Carstens BC, Knowles LL. Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: An example from Melanoplus grasshoppers. Syst Biol. 2007;56(3):400–411. doi: 10.1080/10635150701405560. [DOI] [PubMed] [Google Scholar]
- 31.Solís-Lemus C, Knowles LL, Ané C. Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution. 2015;69(2):492–507. doi: 10.1111/evo.12582. [DOI] [PubMed] [Google Scholar]
- 32.Moritz C, et al. Multilocus phylogeography reveals nested endemism in a gecko across the monsoonal tropics of Australia. Mol Ecol. 2016;25(6):1354–1366. doi: 10.1111/mec.13511. [DOI] [PubMed] [Google Scholar]
- 33.Huang JP, Knowles LL. The species versus subspecies conundrum: Quantitative delimitation from integrating multiple data types within a single Bayesian approach in Hercules beetles. Syst Biol. 2015;65(4):685–99. doi: 10.1093/sysbio/syv119. [DOI] [PubMed] [Google Scholar]
- 34.Sukumaran J, Holder MT. DendroPy: A Python library for phylogenetic computing. Bioinformatics. 2010;26(12):1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 35.Etienne RS, Rosindell J. Prolonging the past counteracts the pull of the present: Protracted speciation can explain observed slowdowns in diversification. Syst Biol. 2012;61(2):204–213. doi: 10.1093/sysbio/syr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert A, Morlon H, Etienne RS. The reconstructed tree in the lineage-based model of protracted speciation. J Math Biol. 2015;70(1-2):367–397. doi: 10.1007/s00285-014-0767-x. [DOI] [PubMed] [Google Scholar]
- 37.Olave M, Solà E, Knowles LL. Upstream analyses create problems with DNA-based species delimitation. Syst Biol. 2014;63(2):263–271. doi: 10.1093/sysbio/syt106. [DOI] [PubMed] [Google Scholar]